SUMMARY
Phenotypic robustness requires a process of developmental buffering that is largely not understood, but which can be disrupted by mutations. Here we show that in mef2cab1086 loss of function mutant embryos and early larvae, development of craniofacial hyoid bones, the opercle (Op) and branchiostegal ray (BR), becomes remarkably unstable; the large magnitude of the instability serves as a positive attribute to learn about features of this developmental buffering. The OpBR mutant phenotype variably includes bone expansion and fusion, Op duplication, and BR homeosis. Formation of a novel bone strut, or a bone bridge connecting the Op and BR together occurs frequently. We find no evidence that the phenotypic stability in the wild type is provided by redundancy between mef2ca and its co-ortholog mef2cb, or that it is related to the selector (homeotic) gene function of mef2ca. Changes in dorsal-ventral patterning of the hyoid arch also might not contribute to phenotypic instability in mutants. However, subsequent development of the bone lineage itself, including osteoblast differentiation and morphogenetic outgrowth, shows marked variation. Hence, steps along the developmental trajectory appear differentially sensitive to the loss of buffering, providing focus for future study.
Keywords: canalization, developmental instability, osteoblast, skeleton, pharyngeal arch, mef2c
INTRODUCTION
How is an intricate, complex and integrated morphological pattern, such as we see for bones of the vertebrate skull, reliably achieved during development? Leading studies and ideas about phenotypic stability and its regulation derive from Schmalhausen and particularly from Waddington, who proposed that development is “canalized”, or buffered against the perturbing effects of genetic mutation and environmental disturbances (Dworkin, 2005; Flatt, 2005; Schmalhausen, 1949; Waddington, 1957). Waddington depicted canalization graphically in an icon that has become well known – canalization is a sloping landscape of hills and valleys. Development is a ball rolling down the landscape, and genes regulating canalization sculpt the contours guiding the pathway of the ball – elevating the hills, deepening the valleys, and hence stabilizing the developmental trajectory. Waddington also considered that buffering might not effect just the influences of genetic mutation and environmental irregularity but also the “inherent noisiness of a developmental pathway” (Waddington, 1957). We understand developmental noise to be the product of nondeterministic fluctuations in molecular mechanisms that underlie development, for example stochastic variation in the collisions between small numbers of regulatory macromolecules that need to bind together in order to serve as effectors of progression along developmental pathways (Dongen, 2006; Polak, 2003). Because of noise, development might fail to reach a specific ‘target’ phenotype even in the absence of genetic and environmental variation (Polak, 2003). Such failure, a change in morphology due to random noise, is termed developmental instability (Polak, 2003). Later authors have debated whether buffering the effects of mutation and environment (‘canalization’), and buffering developmental noise are the same thing (Breuker et al., 2006; Debat et al., 2009; Hallgrimsson et al., 2002; Nijhout and Davidowitz, 2003), and the issue remains controversial.
Buffering can break down, resulting in increased phenotypic variation; indeed, the initial motivation for the canalization concept was to explain the increased variation commonly observed in mutants (Waddington, 1942). Apparent loss of buffering characterizes hyoid arch dermal bone development in zebrafish Endothelin1 (Edn1) pathway mutants (Kimmel et al., 2003). Edn1 provides an extracellular signal which functions along with Notch and BMP signaling in craniofacial development (Kimmel et al., 2007; Medeiros and Crump, 2012). In edn1 mutants the hyoid arch dermal bones of the early larva, the opercle (Op) and branchiostegal ray #3 (BR), show remarkably contrasting phenotypes (Kimmel et al., 2003) (for developmental anatomy see (Eames et al., 2013)). In some mutants both the BR and Op are missing, but in others the Op is enlarged. Furthermore, sometimes Op loss and Op expansion occur together on opposite sides of the same mutant (Kimmel et al., 2003), suggesting developmental instability. One interpretation of these findings is that Edn1 signaling, in a complex manner, normally regulates both activation and repression of OpBR development: Loss of one or the other downstream function – activator or repressor – variably shows up separately in the edn1 mutant.
Our craniofacial genetic screen yielded an allele of an Edn1-pathway gene that is particularly useful for understanding the OpBR phenotype (Miller et al., 2007), and is the subject of this paper. This mutation, mef2cab1086 (hereafter mef2ca−), likely causes complete loss of function of a MADS box-containing transcription factor encoding gene critical for skeletal development, Mef2c. (Arnold et al., 2007; Miller et al., 2007; Verzi et al., 2007). mef2ca functions downstream of edn1, as revealed by double-mutant and other analyses (Miller et al., 2007). As we show here, in the zebrafish strain AB genetic background in which the b1086 mutant allele was identified, the phenotype is highly variable in expressivity of the OpBR phenotype, which facilitates study and understanding of the basis of the variation. Furthermore, in extreme examples the BR resembles the Op in size and shape, suggesting the phenotype is homeotic (Miller et al., 2007). This hypothesis that mef2ca functions as a homeotic selector gene is in keeping with our current understanding of the developmental role of the gene network activated by Edn1 signaling. That is, in response to mutational loss of the Edn1 signal that is normally expressed in the ventral part of the arch (Miller et al., 2000), the more ventral BR might homeotically transform to express features of the more dorsal Op.
Here we further characterize the OpBR phenotype in mef2ca mutants, examining in particular what developmental steps appear to be associated with increased phenotypic variation. Our results show that developmental instability increases dramatically in the mutants. Phenotypic stability in the wild type is unlikely to be provided by redundancy between mef2ca and its co-ortholog mef2cb. Developmental analyses provide no evidence that disrupted early pattern specification or homeotic selector function play any direct role in the increased variation in mef2ca mutants. On the other hand we found marked variation in the location and time of appearance of ectopic osteoblasts that contribute to the expanded bone, and variation in subsequent morphogenetic bone outgrowth, including variable occurrence of a novel pattern of bone formation. We propose that loss of buffering is manifest in these relatively downstream developmental processes.
MATERIALS AND METHODS
Zebrafish lines
Zebrafish were reared according to standard protocols (Westerfield, 2007) and staged as previously described (Kimmel et al., 1995; Parichy et al., 2009). All experiments were approved by the University of Oregon Institutional Animal Care and Use Committee (IACUC). Zebrafish lines, including PCR-genotyping of mutants, were as described: mef2cab1086 (Miller et al., 2007), mef2cbfh288 (Hinits et al., 2012), furinatg419 (Walker et al., 2006), Tg(sp7:EGFP)b1212 (hereafter sp7:EGFP) (DeLaurier et al., 2010), dlx5aj107Et (hereafter dlx5a:EGFP) (Talbot et al., 2010) and trps1J1271aGt (hereafter trps1:EGFP) (Talbot et al., 2010).
Tissue labeling
Alcian Blue-Alizarin Red stains on fixed animals and vital staining with Alizarin Red were performed as previously described (Kimmel et al., 2010; Walker and Kimmel, 2007). Two-color fluorescent whole mount in situ hybridization was carried out as described (Talbot et al., 2010) Probes were as described: ihha (Avaron et al., 2006), sp7 (DeLaurier et al., 2010) to label early matrix-secreting osteoblasts (Huycke et al., 2012; Li et al., 2009), and runx2a (Flores et al., 2004) to label pre-osteoblasts (Li et al., 2009).
Microscopy
Skeletal preparations were imaged on a Zeiss Axiophot 2. Static confocal images, either of live preparations or in situ preparations, were captured on either a Zeiss LSM 5 Pascal confocal or a Leica SD6000 spinning disk confocal with Borealis illumination technology. Images were assembled in ImageJ and Photoshop with any adjustments applied to all panels. For time-lapse recordings, animals were imaged on the spinning disk confocal as described (Huycke et al., 2012). To avoid photodamage, intervals were at least 25 min, and duration of the recordings were 24 hour or less (Jemielita et al., 2012). Movies were assembled using Metamorph (Molecular Devices) and ImageJ.
Bone size analysis
Bone size analysis used a large cross of 6 dpf (days postfertilization) larvae obtained from single pair of mef2ca heterozygotes on the strain AB background. The sizes were obtained in duplicate from digitized outlines in ImageJ, and included the Op and BR added together when two separate bones were present (as in wild types and a subset of the mutants). Sizes are reported as area1/2. Analyses of fluctuating asymmetry, quantified as the absolute difference between bone size on the left and right of single individuals, followed published guidelines (Palmer and Strobeck, 2003) with the replicates used to estimate measurement error. Statistical procedures were implemented in JMP (SAS Institute, Inc.).
RESULTS
Extraordinary variation in mutant hyoid bone phenotype
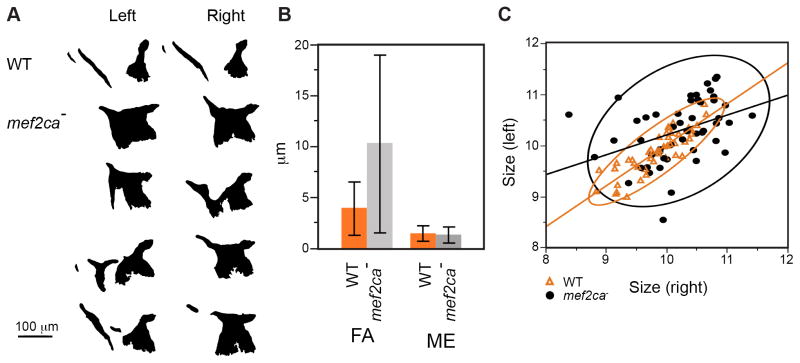
The mef2ca mutant phenotype prominently includes expansion and shape deformation of OpBR bones in the larval hyoid arch. Bone expansion in mutants signals that the wild-type gene functions as a repressor of bone development (Miller et al., 2007). Whereas the mutants can be scored reliably by their cartilage phenotypes, OpBR expressivity is variable among clutches (Supplementary Table 1). Here we focus on within-clutch variation, examined in Figs. 1 and 2 in a set of full siblings. One is immediately struck by the remarkable OpBR phenotypic variation. The ectopic bone may have the appearance of a mirror-image duplicated opercle (Fig. 1C, Op′). An ectopic bony strut (Fig. 1D, s), or a bone bridge between the Op and BR is frequently present (Fig. 1J, b). In other examples a BR may be unrecognized, likely missing (Fig. 1E). Shape variation among mutants appears dramatically greater than in the wild type.
Fig. 1. The shapes of the opercle (Op) - branchiostegal ray 3 (BR) bones are outstandingly diverse in mef2ca mutants.
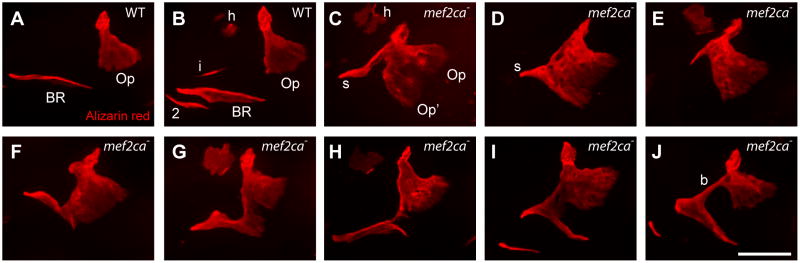
Confocal projections (dorsal up, anterior to the left), bones vitally stained with Alizarin Red and live-imaged at 6 dpf. (A, B) wild type (WT), with (B) evidently the more advanced (h: hyomandibula, i: interopercle rudiment, 2: a second BR). (C–J) mef2ca mutants. The expanded mutant bone in (C) appears to a mirror-image duplicated Op (Op′), and a strut (s) of bone may, or may not, include a BR rudiment. Examples of mutants in (F–J) include the BR bridged to the OP (b, panel J). About 2/3 of the mutants in the single pair family used for this and Fig. 2 include either a strut or bridge as part of the ectopic bone formation. Scale bar 50 μm.
Fig. 2. Mutant OpBR shapes and sizes show hallmarks of developmental instability.
(A) Example left-right OpBR pairs, shown as silhouettes with right-side bones flipped horizontally to match the orientation of the left-side bones. The examples include a wild type (upper) and four individual mutants. Note that the left side of the lowermost example looks similar to wild type while the right side is very dissimilar. (B) Fluctuating asymmetry (FA) of bone sizes are higher in the mutant than in the wild type (P<0.0001, ANOVA). Measurement error (ME) is low. Error bars show standard deviations. FA showed a normal distribution among both wild types and mutants (Shapiro-Wilk test implemented in JMP). Directional (left-right) asymmetry was not detected. (C) Left-right correlations of OpBR size (WT: R2=0.74, p<0.0001*, mutant: R2=0.156, p=0.004*). Regression lines and 95% density ellipses included.
Loss of buffering of stochastic noise and genetic background influences both appear to contribute to OpBR variation in mef2ca mutants
The nature of the OpBR phenotypic variation suggests that development in mutants has become noisy – unstable due to stochastic influences. Critically examining mutants obtained from a large single pair family showed that the OpBR phenotypes on the left and right sides of the body essentially always differed from one another, sometimes very substantially (Fig. 2A, Supplementary Figure 1). Hence, loss of buffering of stochastic noise is likely a major cause of the shape variation.
Because of the extreme nature of the shape variation we were unsuccessful in attempts to use multivariate methods, including landmark analysis and Elliptical Fourier analysis, to quantify it. However, we could compare fluctuating asymmetries of bone sizes (as distinct from shapes) in wild types and mutants. Fluctuating asymmetry, a widely used measure of developmental instability, is over twice as high in mutants as in wild types (Fig. 2B). Notably, the mutant variance was more than ten times higher (the error bars in Fig. 2B show standard deviations, the square root of the variances). Hence, by fluctuating asymmetry analysis the developmental instability of the mutant OpBR phenotype is prominent.
We also used bivariate analysis to compare the left-right size correlations between wild types and mef2ca mutants (Fig. 2C). Supporting developmental instability, we observed that the correlations were considerably lower in mutants than in wild types, both as estimated from the wider shapes of the 95% ellipses shown on the plots, and from the R2 values obtained from linear regression. However, the plots show that the left and right sides are correlated (i. e., the slopes of the regression lines differ significantly from zero; mutant: P<0.004, wild type: P<0.0001). Genetic background differences and/or differences in growth rates among individual mutants might explain the correlation.
These data also support that the mutant bones are enlarged, as compared with wild type (ANOVA, F1,93=15.5; P=0.0002). Development of enlarged bones in mutants is in keeping with our understanding that mef2ca in the wild type functions as a genetic repressor (Miller et al., 2007). Whether the requirement of mef2ca for repression and for developmental stability are one and the same is not understood.
The mef2ca co-ortholog mef2cb may not function in OpBR development
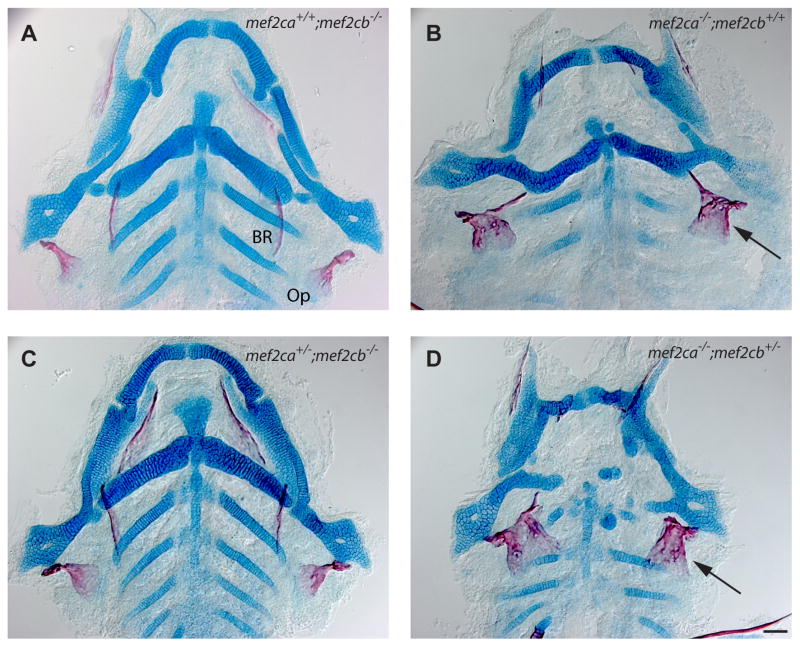
Reducing genetic redundancy can result in an increase in phenotypic variation (Cook et al., 1998; Klingenberg, 2003; Wagner, 1999; Wilkins, 1997). We used double mutant analyses to investigate whether mef2ca might function redundantly with its co-ortholog mef2cb in OpBR development, and hence could contribute to phenotypic stability. By the redundancy hypothesis, the mef2cb single mutant OpBR phenotype might resemble that of the mef2ca single mutant. Furthermore, partial redundancy predicts that subtracting wild-type alleles of mef2cb in the mef2ca mutant background might enhance the OpBR phenotype, converting bone expansion to bone loss, thus matching the change in phenotypic enhancement with increased loss of function of edn1 (Kimmel et al., 2003). However, for the OpBR phenotypes with mef2cb mutants, we observe neither result. The craniofacial phenotypes of mef2cb and mef2ca single mutants are dissimilar, with respect to both cartilage and bone (Fig. 3A, B). In fact the mef2cb mutant phenotype is indistinguishable from wild type (not shown in Fig. 3, compare with the wild type in Supplementary Figure 2A). We cannot meaningfully interpret the homozygous double mutant craniofacial phenotype; these mutants show edema and nonspecific defects due to disruptions during embryogenesis (including heart malformation; (Hinits et al., 2012)). However, removing a single wild-type mef2cb allele in the compound mutant (genotype mef2ca−/−;mef2cb+/−) avoids such defects, and for the OpBR phenotype of these compound mutants we do not see the enhancement predicted by the redundancy hypothesis. Rather, the mef2ca−/−;mef2cb+/− bone phenotype matches the mef2ca−/−;mef2cb+/+ single mutant (Fig, 3B, D, in D 36 out of 41 mutants (88%) show the expanded OpBR phenotype). In contrast, the cartilage phenotype is strongly enhanced (compare B and D). Compound mutants with the reciprocal genotype (mef2ca+/−;mef2cb−/−) resemble the phenotypically normal mef2cb single mutant (Fig. 2A, C, in C 32 out of 36 mutants (90%) show a wild-type like OpBR phenotype).
Fig. 3. mef2cb may not function in OpBR development, but functions partially redundantly with mef2ca in cartilage development.
Ventral views of dissected flat mounts at 6 dpf, stained with Alcian Blue and Alizarin Red. (A, C) The skeletons (bone, red; cartilage, blue) appear phenotypically normal. (B, C) The OpBR bones show the expansion phenotype (arrows), the cartilages are disrupted, substantially more so in (D) than (B). Scale bar 50 μm.
These data suggest that mef2ca and mef2cb function partially redundantly in cartilage development, but provide no evidence for mef2cb participation in OpBR development. Hence we find no indication for partial redundancy between the two genes providing OpBR phenotypic stability in the wild type. We also examined furina;mef2ca double mutants (using furina− to partially lower edn1 function, see (Walker et al., 2006)), looking for evidence of genetic interaction with respect to the expanded OpBR phenotype. However the furina;mef2ca double mutants show OpBR reduction, not expansion, and hence were not useful for this analysis (Supplementary Figure 2).
Variation in mef2ca homeotic selector gene function
Ectopic bone derived from the BR region in the mef2ca mutant sometimes resembles a duplicated Op, suggesting homeosis (Op′ in Fig. 1C, Supplementary Fig. 3). To study whether the ectopic bone has other features of Op identity, and characterize its variation, we examined markers that are normally expressed differentially between the Op and BR.
One such marker is the trps1J1271aGt transgene (abbreviated here as trps1:EGFP). In wild type this transgene is highly expressed in the joint cells of the Op, where it articulates with the hyosymplectic cartilage (Fig. 4A, arrow; Talbot et al., 2012). Expression at lower levels more generally characterizes skeletal tissue, both cartilage and bone (Huycke et al., 2012; Nichols et al., 2013). The wild-type BR expresses trps1:EGFP only at low levels. However, in mef2ca mutants in which the BR is expanded there is a new patch of bright trps1:EGFP expression resembling the Op-hyosymplectic cartilage articulation, just where the bone attaches to its supporting ceratohyal cartilage, (Fig 4B, arrow; 5 out of 5 cases where the BR is included in the ectopic bone). In another case, where the BR appears to not contribute to the ectopic bone, and there is no cartilage attachment, there is also a region of bright expression suggesting a duplicated Op joint (Fig. 4C, arrow). Notably, in this example we also see a small element that we identify by its position as a poorly developed BR – here not transformed into an Op, and there is no associated bright trps1:EGFP expression (Fig. 4C, asterisk). Finally, in cases where expanded bone is present, but to a lesser extent than the others in this set, expression resembles wild type (Fig. 4D; n=3 out of 9 total).
Fig. 4. Loss of mef2ca results in the ectopic differentiation of cells with Op joint identity.
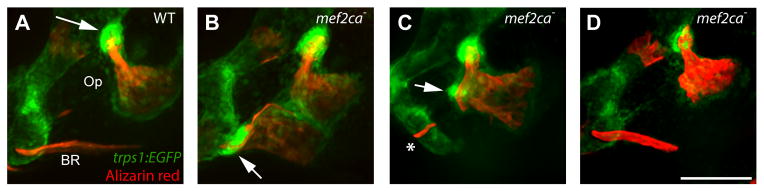
Confocal projections of larvae expressing trps1:EGFP at 6 dpf, live imaged after counterstaining vitally with Alizarin Red. (A) Wild-type cells of the Op-hyosymplectic articulation, but not BR-ceratohyal cartilage articulation, express high levels of trps1:EGFP (arrow). (B) Mutant cells of the expanded (transformed) BR-ceratohyal articulation now show high levels of trps1:EGFP (arrow). (C) A region of ectopic bone cells not involving the BR, express high levels of trps1:EGFP (arrow). A stunted bony spur at the position of the BR does not show high expression (asterisk). (D) A mutant showing less ectopic bone expansion matches the wild type (A) in trps1:EGFP expression. Scale bar 50 μm.
Finding bright trps1:EGFP expression supports the hypothesis that the ectopic bone has Op identity. Lack of trps1:EGFP expression might mean that the ectopic bone has no such identity, or alternatively, that it is an ectopic Op that does not include a duplicated joint region. We used a second method, RNA in situ analysis examining expression of ihha, to attempt to resolve between these two possibilities. ihha is differentially expressed between the Op and BR at 79 hpf, invariably showing strong expression in osteoblasts along the ventral-posterior edge of the Op in wild type at this stage, and only low expression in the early BR (Fig 5A). In mutants with prominently expanded expression of the osteoblast marker sp7 we also observe expansion of ihha expression (Fig. 5B, C). Hence, by this test the ventral-posterior growing edge of the ectopic region of bone has Op identity (n=18/18 mutants showing sp7 expansion). However, the extent of ectopic ihha expression in mutants is variable. A patch of sp7-labeled tissue in one of the mutants illustrated shows ectopic ihha expression at the location of the BR, or perhaps an ectopic spur (Fig. 5B3, arrow). However, in the second example sp7-labeled cells in apparently the same position (Fig. 5C2, asterisk) do not express ihha.
Fig. 5. sp7-expressng osteoblasts of the Op and ectopic Op′ specifically express ihha during phase 2 of Op development.
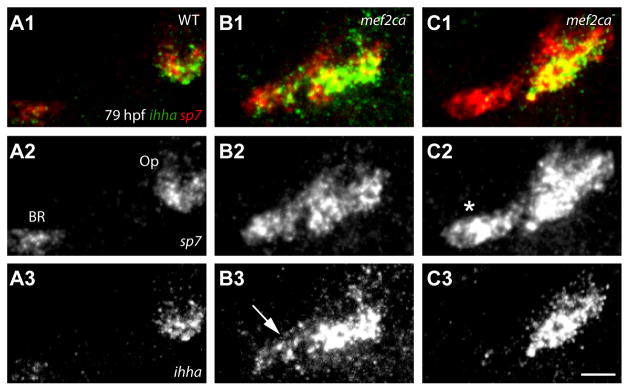
Confocal projections of larvae labeled for ihha and sp7, RNA in situ hybridization. (A) ihha is expressed at high levels in the wild-type Op, but only minimally in the BR. (B) Expanded ihha mef2ca mutant expression is present at the location of the BR (B3, arrow). (C) Op ihha expression is expanded (C3, arrow), but not detected at the location of the BR (C2, asterisk). Scale bar 50 μm.
We interpret the trps1:EGFP and ihha expression results to mean that the ectopic bone usually has the identity of an Op, likely in all cases where the expansion is prominent, and whether or not the BR is involved in the phenotype. The findings argue against an interpretation that the extreme phenotypic variation we observe in prominently expanded OpBR elements is due to their having Op identities in some instances but not others.
Mispatterning of dlx5a expression in mef2ca mutants may not explain later variation in the OpBR skeletal phenotype
Dorsal-ventral patterning of OpBR development by mef2ca, including specification of Op identity, is in part via regulation of dlx5a (Walker et al., 2006), and expression in wild type persists in the Op and BR (Verreijdt et al., 2006). We used a transgenic reporter line, dlx5a:EGFP, at two stages in the same individuals to examine dynamic aspects of dlx5a expression and its variation in live wild-type and mef2ca mutant embryos. Re-examining the same individual mutants allows us to accurately determine if early phenotypic variation in dlx5a:EGFP expression predicts the nature of variation of the subsequent skeletal phenotype.
At 3 dpf the wild type shows a striking pattern of expression of dlx5a:EGFP (Fig. 6A1). Two side-by-side largely separate expression domains, preskeletal condensations, occupy the dorsal and ventral border regions of the hyoid arch intermediate domain. These two domains meet at the center of the figure (Fig. 6A1, arrowhead). The more posterior domain marks the position of the ossifying Op rudiment (Fig. 6A1, arrow). The more anterior condensation marks the position of the nascent hyoid cartilages where the BR will subsequently develop. By 6 dpf, wild-type dlx5a:EGFP expression includes regions of active bone-secreting osteoblasts – especially the rapidly growing ventral-posterior Op edge (Fig. 6A2; vp). The mef2ca mutant pattern contrasts markedly with the wild type at both stages. At 3 dpf, the two dlx5a:EGFP expression domains are fused together because of ectopic expression in between them (Fig. 6B1-D1; the region of fusion is marked with an asterisk in 6B1). At 6 dpf, where we see striking variation in the ectopic bone shape among these same mutants; the strong labeling of the mutant OpBR includes the ventral posterior edge of the ectopic Op′ (Fig. 6B2-D2, arrow in 6B2).
Fig. 6. Variation in the expression of dlx5:EGFP in preskeletal condensations (3 dpf, upper panels) does not predict later variation in the larval mutant OpBR skeletal elements (re-imaged at 6 dpf, lower panels) of mef2ca mutants.
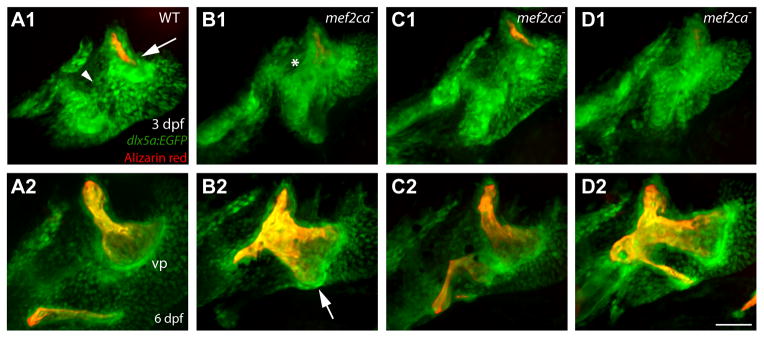
Live preparations, bone vitally counterstained with Alizarin Red. At the early stage the wild type shows two fairly well separated expression domains (A1, 6 animals imaged at 3dpf, and then again at 6 dpf). These domains are merged together in mef2ca mutants that later show different OpBR expansion phenotypes (B1-D1, 8 mutants imaged). Scale bar 50 μm.
These findings show that ectopic expression of dlx5a:EGFP at 3 dpf is generally marking out territory where we see ectopic, expanded bone later: That is, the expanded bone always occupies the region between the Op and the hyoid cartilage, and this region is just where the early fusion of the dlx5a:EGFP expression domains occurs. However, the 3 dpf expression pattern seems substantially less variable than the pattern of bone and dlx5a:EGFP expression 3 days later: The 3 dpf pattern does not clearly predict the nature of the later variation.
Development of early osteoblasts in mutants shows extreme phenotypic variation
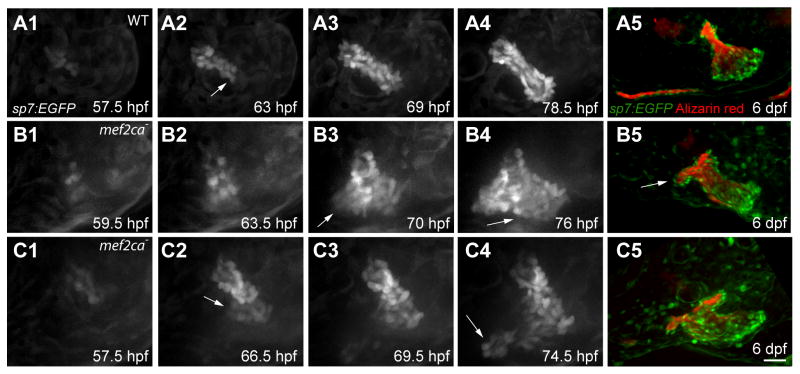
Bone lineage specification, osteoblast differentiation, and bone morphogenesis are critical developmental steps that could be vulnerable to stochastic noise in the absence of mef2ca function. We observed that, as expected from the bone phenotype, loss of mef2ca repression results in a markedly expanded region of runx2a-expressing pre-osteoblasts, as well as subsequent expansion of the early osteoblast marker sp7 associated with OpBR development (Supplementary Figure 4). Op morphogenesis itself exhibits exquisite spatiotemporal patterning in which different patterns of osteoblast recruitment occur in different phases (Kimmel et al., 2010). At first, defining phase 1, the wild-type Op grows as an elongating rod, with sp7:EGFP-expressing osteoblasts continuously recruited to the growing ventral-posterior tip (Fig. 7A; Movies in Supplementary Material) (Huycke et al., 2012; Kimmel et al., 2010). In contrast to wild types, phase 1 morphogenesis is severely disrupted in mef2ca mutants (Fig. 7B, C; Movies in Supplementary Material). Ectopic osteoblasts appear, always or nearly always during phase 1, at variable positions (compare Fig. 7B3 and 7C4) anterior to the Op principal (orthotopic) axis (n=11). The time of initial appearance of ectopic osteoblasts is also highly variable among individual mutants, occurring during a 14.5 hour interval, between 4 and 18.5 hours after the appearance of the first osteoblasts contributing to the Op principal axis itself (mean=11 hour, n=11). In the example in Fig. 7C, and in at least one other case not shown, at least two apparently independent ectopic formations arise (arrows in Fig. 7C2 and 7C4). Furthermore, there are at least two patterns of continued outgrowth of an ectopic patch once it arises. We term the two patterns ‘orthogonal’ and ‘parallel’ according to the locations of new ectopic osteoblasts incorporated into the outgrowth. Orthogonal outgrowth expands the ectopic outgrowth in a ventral-anterior direction (Fig. 7B3, arrow), orthogonal to the normal axis of Op growth, and generates the bone strut pointing out ventral-anteriorly at later stages (Fig. 7B5, arrow). In parallel outgrowth, ectopic osteoblasts add ventral-posteriorly from the site of ectopic initiation (Fig. 7B4, arrow). This pattern parallels the normal addition of osteoblasts to the growing Op during phase 1, such that the duplicated Op′ grows along side of the normal Op to which it may be fused, or not. We also used recordings at later developmental stages to follow continued outgrowth of the ectopic regions. With these later recordings we documented further outgrowth variation, including orthogonal outgrowth patterns that bridge the Op and BR, and ectopic outgrowths initially associated with the BR rather than the Op (Supplementary Fig. 5 and associated movies in Supplementary Material).
Fig. 7. Differences in position and timing of ectopic osteoblast appearance and outgrowth explains skeletal phenotypic variation.
(A1-C4) Confocal projections from time lapse recordings starting at 55 hpf. sp7:EGFP labeling. (A5-C5) The same larvae at 6 dpf, bone matrix counterstained with Alizarin Red. (A) In the wild type a small collective of sp7:EGFP-expressing osteoblasts emerge (A1), and then osteoblasts are newly recruited at the posterior-ventral apex (A2, arrow) to expand the bone linearly (A2–A4). (B) A mef2ca mutant in which ectopic osteoblasts are added to the anterior side of the Op axis (B2). The outgrowth expands with new osteoblast addition in both the orthogonal (B3, arrow) and parallel pattern (B4, arrow). At 6 dpf the Op appears duplicated; a BR is absent (B5). (C) A mef2ca mutant in which two groups of ectopic osteoblasts arise (C2, C4, arrows) and then merge. At 6 dpf the Op has a short spur, and appears duplicated. Scale bar 25 μm (A1-C4), 50 μm (A5-C5).
Our time-lapse results show that ectopic patches of osteoblasts arise and enlarge in a variety of patterns beginning during a restricted period of Op development. Ectopic outgrowth can parallel the Op phase 1 axis, expanding an Op′ domain resembling that of the Op. Orthogonal outgrowths generate bony struts or OpBR bridges.
DISCUSSION
This work explores the exceptional phenotypic variation exhibited by the OpBR in mef2ca mutants, and potential underlying genetic and developmental causes of this variation. Questions surrounding variation have been discussed for many years, and as we show here, new ways to investigate the problem provide important new insight, and new potential for future study. With loss of developmental stability and genetic repression, dlx5a:EGFP expression, and bone development are expanded ectopically, and our findings suggest that development of the bone lineage occurs in an unstable manner within this ectopic region specifically. We propose that major disturbances of developmental noise occur during initial stages of development of bone lineage itself. Future work might especially focus on the specification-morphogenesis steps of bone development particularly, and we further suggest that the remarkable extent of the variation we see in this system is a favorable attribute for such analysis.
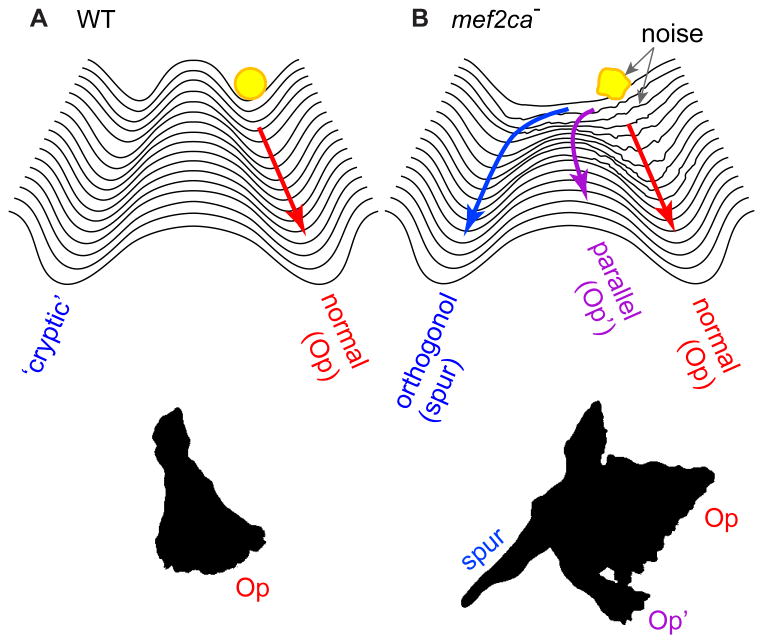
Repression by mef2ca ‘eelevates a bank’ of a phase 1 Waddington canal
Waddington’s conception of a canalized developmental trajectory is particularly apt for the first phase of Op morphogenesis (Fig. 8). Bringing to mind the metaphor of a canal, in wild-type development the bone grows out linearly. With loss of repression and increased developmental instability in the mef2ca mutant this pattern breaks down. Expression of runx2b is expanded and ectopic sp7-expressing osteoblasts are recruited to one side of the primordium. More than one seemingly independently arising ectopic initiation can occur in the same embryo. These ectopic regions of bone formation can arise either immediately along the growing primordium or at some distance away from it, but our evidence suggests they always appear to the same side of the Op primordium – toward where the BR will form, and during phase 1 of Op development, as defined as the period of linear Op outgrowth. The way in which bone development is locally misregulated in mutants strongly suggests that mef2ca function in the wild type is spatiotemporal restricted – it negatively regulates osteoblast addition within a zone adjacent to the Op primordium, and during a single temporal phase of morphogenesis. Such restriction supports the concept we proposed recently that Op development is modular, with separate morphogenetic phases being under separate genetic control (Huycke et al., 2012; Kimmel et al., 2010). In wild type, this initial phase of linear bone outgrowth, in which mef2ca function is critical, is followed by expansion of the Op ventral-posterior edge, defining phase 2. During phase 2 the Op takes on a fan shape characteristic of the early larva (e.g. Fig. 1A). In mutants with prominent bone expansion, the ectopic domain apparently enters a phase 2 of its own, as suggested by its duplicating the shape of the normal Op, and by its expanded expression of ihha (Fig. 5), a phase 2-specific regulator of growth of the Op ventral-posterior edge (Huycke et al., 2012).
Fig. 8. Devastation of a morphogenetic landscape.
(A). Using Waddington’s metaphor (Waddington, 1957), the sculpturing of a deep developmental canal keeps wild-type Op outgrowth (the yellow ball) on track. (B) Loss of repression in the mef2ca mutant lowers the left bank of the canal, such new regions of the landscape become available for bone development. Loss of buffering of developmental noise, indicated as a lumpy ball and roughened landscape, might serve to variably ‘bounce’ the trajectory into the new regions, including the ‘cryptic’ (orthogonal outgrowth) pathway.
The loss of repression normally exerted by the mef2ca gene can be seen as providing mutants with a new developmental ‘space’ to explore for bone morphogenesis, as supported by appearance of ectopic dlx5a:EGFP expression where ectopic bone will later form. The second key change in mutants – the loss of buffering of developmental noise – might facilitate entry into this space (Fig. 8). Lowering of left-right phenotypic correlation, and marked increase in fluctuating asymmetry of bone size we observe, make it clear that stochastic influences contribute in a major way to phenotypic variation in mutants. However, it is not surprising that the genetic background also seems to play a role (Waddington, 1942) Supplementary Table 1, Supplementary Figure 1), as we are currently exploring. The magnitude of the developmental instability should facilitate future efforts to understand the important question of what genetic and perhaps epigenetic networks underlie phenotypic buffering (Klingenberg, 2003; Pujadas and Feinberg, 2012).
Lack of evidence for genetic redundancy between mef2ca and mef2cb providing stability of OpBr development
We used epistasis analysis to explore the possibility that with mutational loss of mef2ca, genetic redundancy with its co-ortholog mef2cb is lost, and this loss accounts for the increase in phenotypic variation in the OpBR phenotype. Redundancy, including partial redundancy, provides backup for gene function, increases the size of developmental genetic networks, and hence increases developmental robustness (Wilkins, 1997). However, our study revealed no role of mef2cb in OpBR development. This finding for bone development is in contrast to cartilage development where our evidence clearly suggests mef2ca and mef2cb function partially redundantly. In our earlier epistasis analysis of edn1;mef2ca double mutants we concluded that mef2ca may be the single major upstream mediator of OpBR repression, whereas regulation of cartilage development (contra bone) includes an edn1 dependent but mef2ca-independent function (Miller et al., 2007). The mef2ca;mef2cb epistasis analyses reported here fully agree with this interpretation. Our data support subfunctionalization of the mef2ca, mef2cb duplicates (Force et al., 1999; Postlethwait et al., 2004), since the two genes differentially control cartilage and bone development. It is unlikely that loss redundancy of function explains variation in the OpBR phenotype in mef2ca mutants.
Is pattern specification a noisy process?
A second possible source of variation in the mef2ca mutant is instability of early, pre-skeletal pharyngeal arch patterning. Dorsal-ventral pattern is mediated in part through an Edn1 signaling-dependent network of genes in which mef2ca is prominent. Recent work makes it clear that the Edn1-signaling network is particularly important for Dlx-mediated patterning in the intermediate arch domain in which the primordia of both the Op and BR develop (Medeiros and Crump, 2012; Talbot et al., 2010). Indeed, morpholino knockdown of Dlx genes can result in the OpBR expansion phenotype studied here (Talbot et al., 2010; Walker et al., 2006). Relevant to intermediate domain patterning, our marker analyses provide support for the hypothesis that mef2ca functions as a OpBR homeotic selector gene (see Introduction), previously based only on skeletal morphologies. The ventral-to-dorsal transformation of the BR (located near the ventral border of the intermediate domain; (Talbot et al., 2010)) to take on the identity of an Op (near the dorsal border), is as predicted for an edn1-dependent selector gene. The ectopically expanded bone expresses Op identity markers whether or not the BR is involved in the expansion, thus there is no suggestion in our data that variation in selector gene function per se is particularly noisy in the mutant.
Our analysis of dlx5a:EGFP expression also suggests that instability in Dlx-mediated patterning may not be the major source of phenotypic skeletal variation. At 72 hpf the two wild-type expression domains are replaced with an enlarged, fused single domain in the mutant. We note that expanded expression at 72 hpf, while in keeping with Mef2ca functioning as a repressor, contrasts with the partial loss of dlx5a expression we previously found in RNA in situ analyses at 30 hpf (Miller et al., 2007). The difference is likely due to dynamic changes in dlx5a regulation with stage, but might also be due to perdurance in the expression GFP. A developmental time series comparing in situ and GFP expression patterns might resolve this issue. In any case, to the point of the origin of phenotypic variation, the appearance of this single dlx5a:EGFP expression domain does not seem to vary greatly among mutants, in marked contrast to the expression of the same marker in the OpBR of the same individuals three days later, where it is associated with active osteoblasts.
Instability in bone lineage development
Whereas we find no clear evidence for variation due to loss of genetic redundancy or to instability in dorsal-ventral patterning, our results directly show marked variation in development of the bone lineage itself. Supported by RNA in situ expression studies of runx2a and sp7, the most illuminating studies are our real-time analyses. Time-lapse recordings show exactly where in the intermediate domain, and when during phase 1 ectopic domains sp7:EGFP expressing osteoblasts arise and begin to expand – in a variety of ways fitting into one or both of two generalized patterns, orthogonal and parallel (Fig. 8). But the behavior of the osteoblasts in any given embryo seems otherwise entirely unpredictable. In another recent investigation from our laboratory, in which we investigated increased craniofacial skeletal phenotypic variation in fras1 mutants, in this case, variation in the mandibular and hyoid cartilages, we also identified morphogenesis as a developmental process prone to developmental instability (Talbot et al., 2012). This coincidence makes us wonder about the extent to which our findings might be generalized. Whereas developmental instability has been investigated in a variety of systems (Polak, 2003), its actual underpinnings in terms of cellular-developmental behaviors require additional information. It is clear from our study that future work toward understanding canalization needs to be targeted at least to the processes of cell fate specification and morphogenesis.
Might regulation of developmental noise contribute to evolvability?
A feature of the mutant skeletal phenotype stands out by exhibiting a modicum of stability in the face of extreme variation. A skeletal strut, sometimes elaborated as a bone bridge completely connecting the Op and BR appears as a novelty, a result of an orthogonal pattern of outgrowth of the ectopic bone. The occurrence in about 2/3 of the mutants leads us to propose that the strut is not simply revealing developmental instability, but is providing evidence of the uncovering of a genetically based ‘cryptic’ developmental pathway. Bone can form along this pathway when, in the mutant condition, ectopic osteoblasts are generated that frequently can find it (Fig. 8). Hypothetically, increased developmental noise as well as loss of genetic repression could aid in deflecting bone development onto the cryptic pathway. Indeed, the combination of novelty and instability in the mef2ca mutant brings to mind models that biological systems inhabit a regime of critical dynamics poised between constrained regularity and random chaos (Alon, 2007; Kadanoff, 1908; Kauffman, 1993). Loss of buffering in the mef2ca mutant might plunge the system into a unstable, perhaps chaotic-like state because of its poised nature in the wild type. However, adaptive evolutionary change, which could be initially facilitated by developmental noise, cannot persist without further compensatory evolution in phenotypic buffering that allows the regaining of regularity. This scenario matches current understanding that temporary loss of canalization increases evolvability through the exposure of accumulated cryptic genetic variation, thereby leading to increased phenotypic variation (Flatt, 2005). As a possible model for such compensation, it would be interesting to learn if mef2ca modifier screens in zebrafish might yield OpBR phenotypes matching the severities of those reported here, but with increased stability.
Supplementary Material
Supplementary Table 1: mef2cab1086 Penetrance and Expressivity of the OpBR Phenotype
Highlights.
Opercle and branchiostegal ray hyoid bones are variably expanded and fused in mef2ca mutants.
Expanded bone phenotypes arise from ectopic osteoblast differentiation in mutants.
The ectopic bone exhibits opercle identity as well as novel morphological features.
Loss of mef2ca causes disruption of developmental buffering producing developmental instability.
Stability is not due to redundancy with mef2cb, and likely is not related to mef2ca homeotic function.
Acknowledgments
We thank Dayna Lamb and Bonnie Ullmann for technical assistance, and the University of Oregon fish facility for care and maintenance of animals. We enjoyed critical and useful input from Bill Cresko, Raghu Parthasarathy, and current and past members of the Kimmel Lab, particularly Jared Talbot, Gage Crump, and Johann Eberhart. Heather Jamniczky and Bill Cresko provided thoughtful comments on drafts of the paper.
Funding
The research was supported by National Institutes of Heath (NIH) [RO1DE13834, PO1HD22486], and NIH fellowships, IF32DE019345 to A.D. and F32-086027 to J.T.N. TILLING of mef2cb was supported by a NIH grant [RO1HG002995] to C.B.M.
Footnotes
Author contributions
C.B.K. conceived of, and designed the experiments. A.D., T.R.H., J.T.N., M.E.S, A.L, C.W, and J.D. performed experiments. T.R.H. and C.B. K. analyzed data. L.P. and C.B.M. performed TILLING. C.B.K. and J.T.N wrote the paper.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material available online at
References
- Alon U. Introduction to Systems Biology: And the Design Principles of Biological Networks. CRC press; 2007. [Google Scholar]
- Arnold MA, Kim Y, Czubryt MP, Phan D, McAnally J, Qi X, Shelton JM, Richardson JA, Bassel-Duby R, Olson EN. MEF2C transcription factor controls chondrocyte hypertrophy and bone development. Developmental Cell. 2007;12:377–389. doi: 10.1016/j.devcel.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Avaron F, Hoffman L, Guay D, Akimenko M. Characterization of two new zebrafish members of the hedgehog family: atypical expression of a zebrafish indian hedgehog gene in skeletal elements of both endochondral and dermal origins. Developmental Dynamics. 2006;235:478–489. doi: 10.1002/dvdy.20619. [DOI] [PubMed] [Google Scholar]
- Breuker CJ, Patterson JS, Klingenberg CP. A single basis for developmental buffering of Drosophila wing shape. PLoS One. 2006;1:e7. doi: 10.1371/journal.pone.0000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook DL, Gerber AN, Tapscott SJ. Modeling stochastic gene expression: implications for haploinsufficiency. Proceedings of the National Academy of Sciences. 1998;95:15641–15646. doi: 10.1073/pnas.95.26.15641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debat V, Debelle A, Dworkin I. Plasticity, canalization, and developmental stability of the Drosophila wing: joint effects of mutations and developmental temperature. Evolution. 2009;63:2864–76. doi: 10.1111/j.1558-5646.2009.00774.x. [DOI] [PubMed] [Google Scholar]
- DeLaurier A, Eames BF, Blanco-Sanchez B, Peng G, He X, Swartz ME, Ullmann B, Westerfield M, Kimmel CB. Zebrafish sp7:EGFP: A transgenic for studying otic vesicle formation, skeletogenesis, and bone regeneration. Genesis. 2010;48 doi: 10.1002/dvg.20639. spcone. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dongen S. Fluctuating asymmetry and developmental instability in evolutionary biology: past, present and future. Journal of Evolutionary Biology. 2006;19:1727–1743. doi: 10.1111/j.1420-9101.2006.01175.x. [DOI] [PubMed] [Google Scholar]
- Dworkin I. Canalization, cryptic variation, and developmental buffering: a critical examination and analytical perspective. Variation: A central concept in biology. 2005:131–158. [Google Scholar]
- Eames BF, DeLaurier A, Ullmann B, Huycke TR, Nichols JT, Dowd J, McFadden M, Sasaki MM, Kimmel CB. FishFace: interactive atlas of zebrafish craniofacial development at cellular resolution. BMC Developmental Biology. 2013;13:23. doi: 10.1186/1471-213X-13-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatt T. The evolutionary genetics of canalization. Quarterly Review of Biology. 2005;80:287–316. doi: 10.1086/432265. [DOI] [PubMed] [Google Scholar]
- Flores MV, Tsang VWK, Hu W, Kalev-Zylinska M, Postlethwait J, Crosier P, Crosier K, Fisher S. Duplicate zebrafish runx2 orthologues are expressed in developing skeletal elements. Gene expression patterns. 2004;4:573–581. doi: 10.1016/j.modgep.2004.01.016. [DOI] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett F, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallgrimsson B, Willmore K, Hall BK. Canalization, developmental stability, and morphological integration in primate limbs. American journal of physical anthropology. 2002;119:131–158. doi: 10.1002/ajpa.10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinits Y, Pan L, Walker C, Dowd J, Moens CB, Hughes SM. Zebrafish Mef2ca and Mef2cb are essential for both first and second heart field cardiomyocyte differentiation. Developmental Biology. 2012 doi: 10.1016/j.ydbio.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huycke TR, Eames BF, Kimmel CB. Hedgehog-dependent proliferation drives modular growth during morphogenesis of a dermal bone. Development. 2012 doi: 10.1242/dev.079806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemielita M, Taormina MJ, Delaurier A, Kimmel CB, Parthasarathy R. Comparing phototoxicity during the development of a zebrafish craniofacial bone using confocal and light sheet fluorescence microscopy techniques. J Biophotonics. 2012 doi: 10.1002/jbio.201200144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadanoff LP. Hip bone is connected to… II. networks. 1908;1:0. [Google Scholar]
- Kauffman S. The origins of order: Self organization and selection in evolution. Oxford University Press; 1993. [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, DeLaurier A, Ullmann B, Dowd J, McFadden M. Modes of Developmental Outgrowth and Shaping of a Craniofacial Bone in Zebrafish. PLoS One. 2010;5 doi: 10.1371/journal.pone.0009475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ullmann B, Walker M, Miller CT, Crump JG. Endothelin 1-mediated regulation of pharyngeal bone development in zebrafish. Development. 2003;130:1339–51. doi: 10.1242/dev.00338. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Walker MB, Miller CT. Morphing the hyomandibular skeleton in development and evolution. J Exp Zool B Mol Dev Evol. 2007;308:609–24. doi: 10.1002/jez.b.21155. [DOI] [PubMed] [Google Scholar]
- Klingenberg CP. A developmental perspective on developmental instability: theory, models and mechanisms. Developmental instability: causes and consequences. 2003:14–34. [Google Scholar]
- Li N, Felber K, Elks P, Croucher P, Roehl HH. Tracking gene expression during zebrafish osteoblast differentiation. Developmental Dynamics. 2009;238:459–466. doi: 10.1002/dvdy.21838. [DOI] [PubMed] [Google Scholar]
- Medeiros DM, Crump JG. New perspectives on pharyngeal dorsoventral patterning in development and evolution of the vertebrate jaw. Developmental Biology. 2012;371:121–135. doi: 10.1016/j.ydbio.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CT, Schilling TF, Lee K, Parker J, Kimmel CB. sucker encodes a zebrafish Endothelin-1 required for ventral pharyngeal arch development. Development. 2000;127:3815–28. doi: 10.1242/dev.127.17.3815. [DOI] [PubMed] [Google Scholar]
- Miller CT, Swartz ME, Khuu PA, Walker MB, Eberhart JK, Kimmel CB. mef2ca is required in cranial neural crest to effect Endothelin1 signaling in zebrafish. Dev Biol. 2007;308:144–57. doi: 10.1016/j.ydbio.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols JT, Pan L, Moens CB, Kimmel CB. barx1 represses joints and promotes cartilage in the craniofacial skeleton. Development. 2013;140:2765–2775. doi: 10.1242/dev.090639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijhout H, Davidowitz G. Developmental perspectives on phenotypic variation, canalization, and fluctuating asymmetry. Developmental instability. Causes and consequences. 2003:3–13. [Google Scholar]
- Palmer AR, Strobeck C. Developmental Instability: Causes and Consequences. Oxford University Press; Oxford: 2003. CH 17. Fluctuating Asymmetry Analyses Revisited; pp. 279–319. [Google Scholar]
- Parichy DM, Elizondo MR, Mills MG, Gordon TN, Engeszer RE. Normal table of postembryonic zebrafish development: staging by externally visible anatomy of the living fish. Dev Dyn. 2009;238:2975–3015. doi: 10.1002/dvdy.22113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak M. Developmental instability: causes and consequences. Oxford University Press; 2003. [Google Scholar]
- Postlethwait J, Amores A, Cresko W, Singer A, Yan YL. Subfunction partitioning, the teleost radiation and the annotation of the human genome. Trends in Genetics. 2004;20:481–490. doi: 10.1016/j.tig.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Pujadas E, Feinberg AP. Regulated Noise in the Epigenetic Landscape of Development and Disease. Cell. 2012;148:1123–1131. doi: 10.1016/j.cell.2012.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki MM, Nichols JT, Kimmel CB. edn1 and hand2 Interact in Early Regulation of Pharyngeal Arch Outgrowth during Zebrafish Development. PLoS One. 2013;8:e67522. doi: 10.1371/journal.pone.0067522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmalhausen Factors of evolution: the theory of stabilizing selection 1949 [Google Scholar]
- Talbot JC, Johnson SL, Kimmel CB. hand2 and Dlx genes specify dorsal, intermediate and ventral domains within zebrafish pharyngeal arches. Development. 2010;137:2506–2516. doi: 10.1242/dev.049700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot JC, Walker MB, Carney TJ, Huycke TR, Yan YL, Bremiller RA, Gai L, Delaurier A, Postlethwait JH, Hammerschmidt M, et al. fras1 shapes endodermal pouch 1 and stabilizes zebrafish pharyngeal skeletal development. Development. 2012;139:2804–13. doi: 10.1242/dev.074906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verreijdt L, Debiais-Thibaud M, Borday-Birraux V, Van der Heyden C, Sire JY, Huysseune A. Expression of the dlx gene family during formation of the cranial bones in the zebrafish (Danio rerio): differential involvement in the visceral skeleton and braincase. Dev Dyn. 2006;235:1371–89. doi: 10.1002/dvdy.20734. [DOI] [PubMed] [Google Scholar]
- Verzi MP, Agarwal P, Brown C, McCulley DJ, Schwarz JJ, Black BL. The transcription factor MEF2C is required for craniofacial development. Developmental Cell. 2007;12:645–652. doi: 10.1016/j.devcel.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH. Canalization of development and the inheritance of acquired characters. Nature. 1942;150:563–565. doi: 10.1038/1831654a0. [DOI] [PubMed] [Google Scholar]
- Waddington CH. The strategy of genes 1957 [Google Scholar]
- Wagner A. Redundant gene functions and natural selection. Journal of Evolutionary Biology. 1999;12:1–16. [Google Scholar]
- Walker MB, Kimmel CB. A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotech Histochem. 2007;82:23–8. doi: 10.1080/10520290701333558. [DOI] [PubMed] [Google Scholar]
- Walker MB, Miller CT, Coffin Talbot J, Stock DW, Kimmel CB. Zebrafish furin mutants reveal intricacies in regulating Endothelin1 signaling in craniofacial patterning. Dev Biol. 2006;295:194–205. doi: 10.1016/j.ydbio.2006.03.028. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book; A Guide for the Laboratory Use of Zebrafish (Danio rerio) Eugene, OR: University of Oregon Press; 2007. [Google Scholar]
- Wilkins AS. Canalization: a molecular genetic perspective. Bioessays. 1997;19:257–262. doi: 10.1002/bies.950190312. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: mef2cab1086 Penetrance and Expressivity of the OpBR Phenotype