Abstract
DNA methylation is a heritable epigenetic modification involved in gene silencing, imprinting, and the suppression of retrotransposons1. Global DNA demethylation occurs in the early embryo and the germline2,3 and may be mediated by Tet (ten-eleven-translocation) enzymes4–6, which convert 5-methylcytosine (mC) to 5-hydroxymethylcytosine (hmC)7. Tet enzymes have been extensively studied in mouse embryonic stem cells (ESCs)8–12, which are generally cultured in the absence of Vitamin C (VitC), a potential co-factor for Fe(II) 2-oxoglutarate dioxygenase enzymes like Tets. Here we report that addition of VitC to ESCs promotes Tet activity leading to a rapid and global increase in hmC. This is followed by DNA demethylation of numerous gene promoters and up-regulation of demethylated germline genes. Tet1 binding is enriched near the transcription start site (TSS) of genes affected by VitC treatment. Importantly, VitC, but not other antioxidants, enhances the activity of recombinant Tet1 in a biochemical assay and the VitC-induced changes in hmC and mC are entirely suppressed in Tet1/2 double knockout (Tet DKO) ESCs. VitC has the strongest effects on regions that gain methylation in cultured ESCs compared to blastocysts and in vivo are methylated only after implantation. In contrast, imprinted regions and intracisternal A-particle (IAP) retroelements, which are resistant to demethylation in the early embryo2,13, are resistant to VitC-induced DNA demethylation. Collectively, this study establishes VitC as a direct regulator of Tet activity and DNA methylation fidelity in ESCs.
Keywords: Vitamin C, DNA methylation, hydroxymethylcytosine, Tet enzymes, epigenetics, embryonic stem cells, germ cells
ESCs are derived from the inner cell mass (ICM) of the blastocyst and can be cultured in vitro to maintain a pluripotent state. Media composition has previously been shown to influence ESC heterogeneity, gene expression, and epigenetic patterns14. Our study began with the serendipitous observation that culture of mouse ESCs in Knockout Serum Replacement (KSR) strongly and reversibly induces expression of the germline gene Dazl (Supplementary Fig. 1a, b). We performed a small molecule screen and identified VitC as the KSR component responsible for Dazl induction (Supplementary Fig. 2). Dazl induction was also observed with the DNA methyltransferase (Dnmt) inhibitor 5-azacytidine, suggesting that VitC may promote DNA demethylation. As VitC enhances the activity of some Fe(II) 2-oxoglutarate dioxygenases15, we reasoned that VitC could promote the activity of Tet enzymes, leading to DNA demethylation and Dazl induction. Since mouse ESCs are commonly cultured without VitC, we set out to test the effect of VitC on the epigenetic and transcriptional state of ESCs.
VitC treatment of naïve ESCs cultured in 2i medium, which is devoid of detectable VitC (Supplementary Fig. 3), leads to a striking global increase in hmC by immunofluorescence and dot blot (Fig. 1a, b). In contrast, global levels of mC were not altered at 12 or 72 hrs of VitC treatment (Fig. 1b). To assess dynamic hmC and mC changes at specific genomic regions, hmC and mC DNA immunoprecipitation followed by deep sequencing (DIP-seq) was performed at 12 and 72 hrs following VitC treatment. We restricted our analysis to methylated regions since mC is a pre-requisite for hmC.
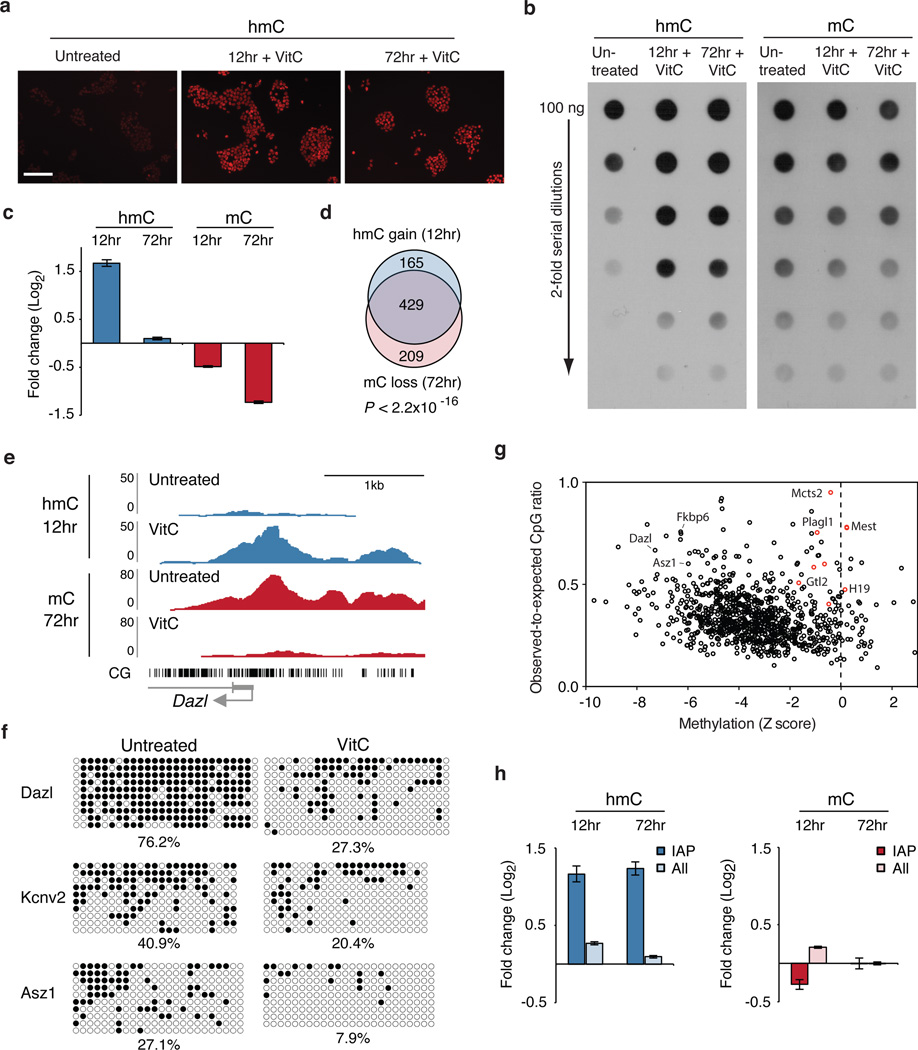
Figure 1. Vitamin C induces loss of mC at gene promoters via a transient increase in hmC.
a, Immunofluorescence for hmC. Scale bar is 200 µm. b, Global hmC and mC levels assayed by dot blot. c, Graph shows fold change in DIP-seq reads (RPKM) at methylated promoters (n = 1045) for VitC-treated cells relative to untreated cells. Values are mean ± s.e.m. d, Overlap of methylated promoters that gain hmC and those that lose mC. P value calculated using Fisher’s exact test. e, Genome browser view of Dazl.f, Bisulfite sequencing of promoters. Open circles = unmethylated, closed circles = methylated. g, Scatter plot of methylated promoters comparing change in methylation (Z score) versus CpG content. Red circles = imprinted genes. h, Graphs show fold change in RPKM at retrotransposons for VitC-treated cells relative to untreated cells. Values are mean ± s.e.m.
Remarkably, most methylated promoters transiently gain hmC at 12 hrs and return to baseline levels or below at 72 hrs, while mC is lost progressively at 12 and 72 hrs (Fig. 1c, Supplementary Fig. 4a, 5). Reduction of hmC at 72 hrs may be explained by loss of mC substrate. After 72 hours of VitC treatment methylation is reduced by 2-fold or more in 61% of analyzed promoters (Supplementary Table 1). Demethylation at exons, introns, and intergenic regions is also observed (Supplementary Fig. 4b). There is a highly significant overlap in the promoters that gain hmC at 12 hrs and those that lose mC at 72 hrs (P value < 2.2×10−16, Fig. 1d, Supplementary Fig. 6). Additionally, hmC gain and mC loss occur at the same genomic locations near the TSS (example in Fig. 1e). These results support a kinetic model in which oxidation of mC to hmC precedes DNA demethylation that may occur via active or passive mechanisms. We confirmed demethylation at the promoters of 3 representative genes by bisulfite sequencing (Fig. 1f). Several high-density CpG promoters show minimal demethylation and many of these were identified as imprinted genes (Fig. 1g), indicating that certain regions of the genome are resistant to VitC-induced demethylation.
While dot blot analysis indicates that the global rise in hmC is sustained at 72 hrs (Fig. 1b), DIP-seq indicates that promoters return to baseline hmC levels at this time point (Fig. 1c). This apparent discrepancy may be explained by prolonged retention of hmC on repetitive elements, which cover a large portion of the genome. Indeed, we find that IAP endogenous retroviruses (ERVs), gain hmC at 12 hrs and maintain elevated levels after 72 hrs of VitC treatment (Fig. 1h). IAP retroelements are also resistant to VitC-induced demethylation, as are other repetitive elements, which may explain the maintenance of mC observed in the dot blot (Fig. 1h, Supplementary Fig. 7a,b, Supplementary Table 2 ). The increase in hmC at IAP retroelements does not correspond to a loss in mC, which could occur if only a small fraction of methylated CpGs within these ERVs gain hmC, resulting in no detectable loss of overall methylation by DIP. Indeed, bisulfite sequencing reveals that IAP retroelements are not demethylated with VitC treatment at 72 hrs (Supplementary Fig. 7c).
The effects of VitC are specific, as several other antioxidants tested did not increase global hmC (Supplementary Fig. 8). The effects of VitC are also reversible. The global increase in hmC is lost rapidly after 3 days of VitC withdrawal, while promoter mC increases gradually following VitC removal (Supplementary Fig. 9).
To determine how VitC affects gene expression in ESCs, microarray experiments were performed. Only ~200 genes are changed by more than 2-fold, and most are up-regulated (Fig. 2a, Supplementary Table 3), consistent with loss of a silencing mark like mC. Up-regulated genes are enriched on the X chromosome (32.7% observed vs. 3.8% expected) and for germline Gene Ontology terms16 (Fig. 2b, c). Pluripotency gene expression is not affected (Supplementary Table 3) and VitC treatment does not impair differentiation (Supplementary Fig. 10). Importantly, the expression of Tet and Dnmt genes is not affected by VitC treatment (Supplementary Fig. 11).
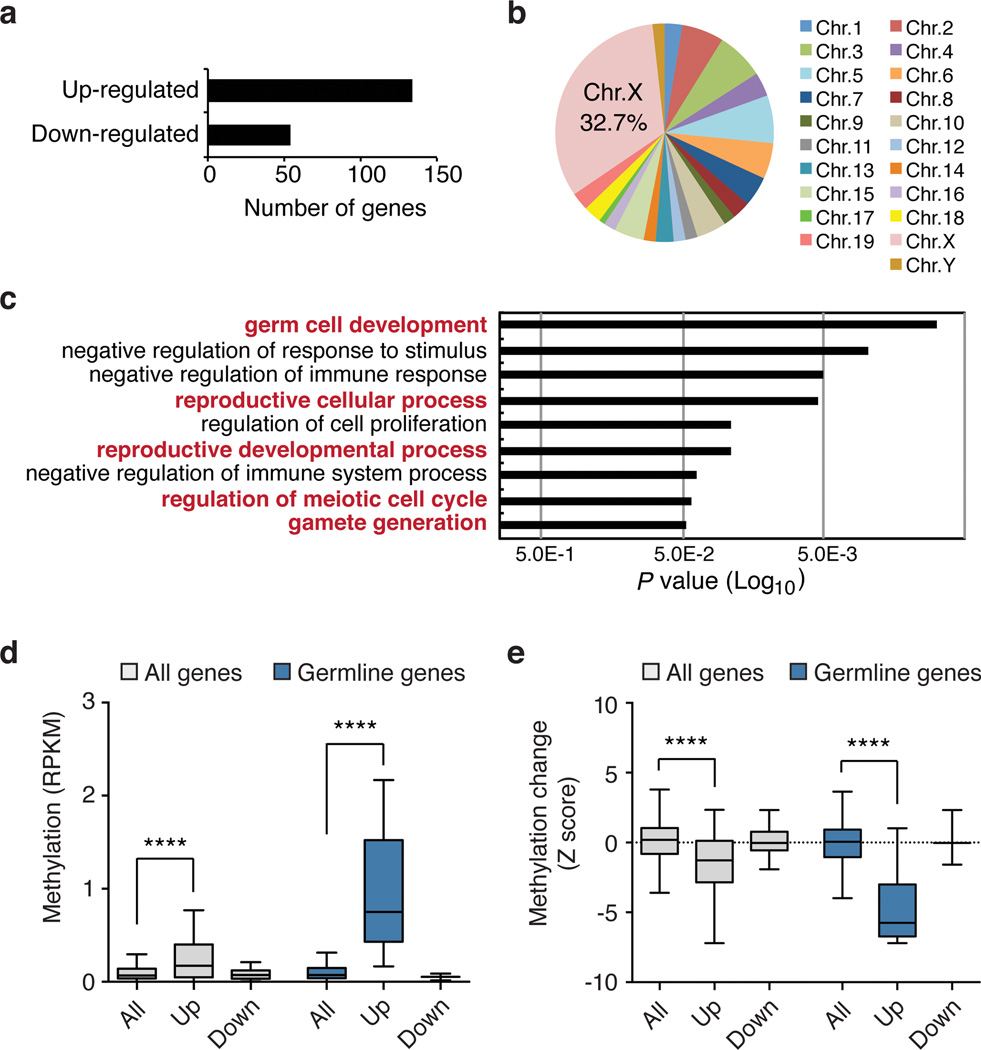
Figure 2. Vitamin C-induced DNA demethylation leads to expression of germline genes.
a, Number of genes differentially expressed following VitC treatment (2-fold and P < 0.05 by t-test). b, Chromosomal distribution of up-regulated genes. c, Gene ontology analysis of up-regulated genes. d, Box plot showing basal promoter methylation levels (RPKM) in untreated ESCs for all genes on the microarray (n = 18,023), up-regulated genes (n = 102), down-regulated genes (n = 48), all germline genes (n = 865), up-regulated germline genes (n = 8), and down-regulated germline genes (n = 3). e, Box plot showing the extent of VitC-induced demethylation (Z score) at gene promoters categorized as in panel d. The box plots have Tukey whiskers, a line for the median, and edges for the 25th/75th percentile. ****P < 0.0001 by ANOVA throughout the figure.
We and others have previously reported that germline genes are induced in ESCs lacking Dnmts17,18. Of the 134 VitC-induced genes, 48 (36%) are also up-regulated in Dnmt1−/−;Dnmt3a−/−;Dnmt3b−/− (Dnmt TKO) ESCs, which are devoid of DNA methylation (Supplementary Fig. 12a, b). Interestingly, VitC further increases expression of a subset of these genes in Dnmt TKO ESCs (Supplementary Fig. 12b), suggesting that VitC may regulate gene expression by additional mechanisms. For example, VitC may also stimulate histone demethylases, as has been shown in induced pluripotent stem (iPS) cell generation19.
Genes up-regulated by VitC have higher basal levels of promoter methylation in untreated cells (Fig. 2d). When analysis is restricted to germline genes (associated with Gene Ontology term “reproduction”), genes up-regulated by VitC show even higher basal levels of promoter methylation (Fig. 2d). Furthermore, up-regulated genes, especially up-regulated germline genes, show significant loss of methylation (Fig. 2e). Taken together, these results indicate that widespread promoter demethylation induced by VitC promotes the up-regulation of predominantly germline-associated genes. However, promoter demethylation is not sufficient to induce expression of most methylated genes, possibly due to redundant epigenetic silencing mechanisms or a lack of activating transcription factors.
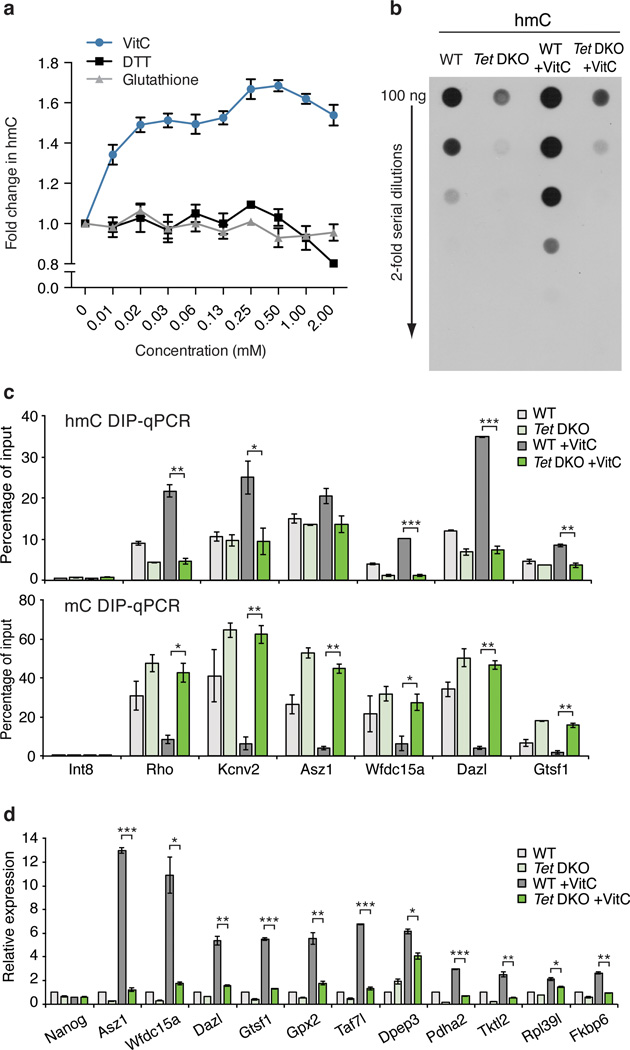
As Tet1–3 are the only known enzymes that oxidize mC to hmC, we hypothesized that the effects of VitC would be mediated by Tet enzymes. Indeed, VitC, but not other antioxidants like glutathione or DTT, dose-dependently increases recombinant Tet1 activity in a biochemical assay (Fig. 3a). ESCs express two Tet family members, Tet1 and Tet2, and these enzymes appear to be highly redundant20,21. Tet1 binding10 is enriched near the TSS of promoters that gain hmC or lose mC with VitC treatment (Supplementary Fig. 13). To test if the effects of VitC are Tet-dependent, we analyzed Tet DKO ESCs21. By dot blot, Tet DKO ESCs show greatly reduced hmC signal that is not increased following VitC treatment (Fig. 3b). Residual signal in Tet DKO ESCs may be due to antibody background or low-level Tet3 expression. Importantly, DIP-qPCR reveals that in contrast to wild-type cells, VitC treatment of Tet DKO ESCs does not affect hmC or mC levels at gene promoters (Fig. 3c). Furthermore, VitC-induced gene expression is significantly attenuated in Tet DKO ESCs (Fig. 3d). The modest gene induction observed may be due to effects of VitC unrelated to DNA methylation, as already suggested by the analysis of Dnmt TKO ESCs (Supplementary Fig. 12b). Tet1−/− (Tet1 KO) ESCs20 also show an attenuated increase in global hmC, reduced promoter demethylation, and reduced gene induction in response to VitC, however, these effects are more subtle than in the Tet DKO ESCs (Supplementary Fig. 14). These data indicate that the effects of VitC are Tet-dependent and are mediated by both Tet1 and Tet2.
Figure 3. The effects of Vitamin C are Tet-dependent.
a, Dose-dependent effect of VitC on in vitro Tet activity. n = 3 technical replicates, values are mean ± s.d. b, Dot blot for hmC following a 12 hr VitC treatment. c, hmC (top) and mC (bottom) DIP-qPCR following a 12 or 72 hr VitC treatment, respectively. d, Gene expression at 72 hrs of VitC treatment. qRT-PCR data expressed relative to untreated wild-type cells. For c, d, n = 2 biological replicates, values are mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001 by t-test throughout the figure.
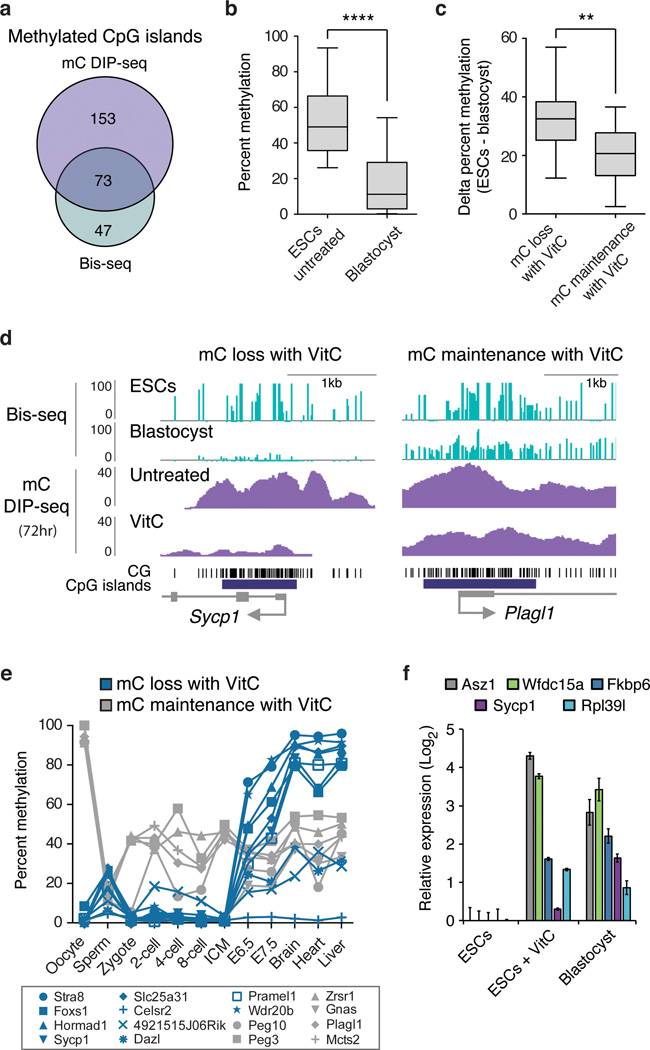
Recent studies highlight differences in the methylomes of ESCs and blastocysts, with ESCs showing higher levels of methylation13,22. Using published genome-wide bisulfite sequencing data for ESCs and blastocysts13, we investigated the relationship between VitC-induced demethylation and differences in methylation between ESCs and the blastocyst. Analysis was performed on promoter CpG islands (CGIs) methylated in both our study and ref. 13 (Fig. 4a, Supplementary Table 4), which generally show greater methylation in ESCs than blastocysts (Fig. 4b). Intriguingly, VitC induces greater demethylation at CGIs that are hypermethylated in ESCs versus blastocysts (Fig. 4c, d). Conversely, VitC has modest effects on CGIs with similar methylation levels in ESCs and blastocysts, such as imprinted regions (Fig. 4c, d). IAP ERVs, which are similarly methylated in both ESCs and the blastocyst, are also resistant to VitC-induced demethylation in ESCs (Supplementary Fig. 7d). These findings suggest that the effects of VitC are most pronounced at genes showing higher methylation in ESCs than blastocysts.
Figure 4. Vitamin C reduces DNA methylation in ESCs that is normally gained post-implantation.
a, Overlap of methylated CGIs in ESCs from this study (mC DIP-seq, RPKM >0.5) and ref. 13 (whole genome bisulfite (bis)-seq, >25% methylated CpGs). Only CGIs found to be methylated in both data sets were used for subsequent analysis in b–d. b, The box plot shows CGI methylation levels in ESCs and blastocysts (values from ref. 13, ****P < 0.0001 by t-test). c, CGIs were first categorized as having mC loss with VitC (>75% loss of mC at 72 hrs, n = 23) or mC maintenance with VitC (<25% loss of mC at 72 hrs, n = 14) in ESCs, then plotted for difference in methylation between untreated ESCs and blastocysts. CGIs demethylated upon VitC treatment show significantly greater ESC hypermethylation compared to CGIs resistant to VitC (**P < 0.01 by t-test). d, Genome browser views of a gene from each category described in panel c. e, Methylation levels during development of all genes categorized in panel c for which data exist (ref. 2). f, Gene expression in ESCs cultured with or without VitC compared to E3.5 blastocysts. Data expressed relative to untreated ESCs. n = 2 biological replicates, values are mean ± s.e.m.
Next, methylation dynamics during development were determined for these gene categories using a reduced representation bisulfite sequencing (RRBS) data set2. Gene promoters that show mC loss with VitC are generally unmethylated up to the ICM stage and then undergo extensive methylation at the epiblast stage (Fig. 4e, in blue). In contrast, gene promoters that show mC maintenance with VitC are enriched for germline differentially methylated regions (DMRs) that maintain ~50% methylation in somatic tissues throughout development (Fig. 4e, in grey). Thus, it appears that in ESCs cultured in the absence of VitC, methylation accumulates at the subset of CGIs that would normally be de novo methylated in the epiblast. Analysis of published RNA-seq data23 indicates that several VitC-induced genes are also expressed in the ICM (Supplementary Table 5). Furthermore, we find that several of these germline genes are indeed expressed in the blastocyst at levels comparable to ESCs treated with VitC (Fig. 4f). Collectively, these findings suggest that VitC remodels DNA methylation and expression patterns in ESCs, resulting in a state reminiscent of the ICM of the blastocyst.
Recently, it was reported that long-term culture of ESCs in 2i medium induces a blastocyst-like state of global hypomethylation relative to culture in serum24. As the analyses described above were carried out in 2i medium, they document effects of VitC beyond those of 2i. Nevertheless, we sought to distinguish the effects of 2i and VitC. We find that VitC induces a gain of hmC, loss of mC, and induction of germline genes in both FBS and 2i medium (Supplementary Fig. 15a–d). In contrast, culture in 2i medium alone shows little to no effect over the same 72 hr time course. The faster kinetics of action of VitC relative to 2i is likely due to their different mechanisms of action: VitC promotes Tet-mediated DNA demethylation, whereas 2i promotes passive loss of DNA methylation via up-regulation of Prdm14 and repression of Dnmt3b and Dnmt3l24 (Supplementary Fig. 15e).
In summary, this work demonstrates that VitC alters the steady-state of DNA methylation and in turn the expression of germline genes in ESCs by enhancing Tet activity. VitC reduces methylation at CGIs that normally gain methylation during the blastocyst to epiblast transition, promoting an ICM-like DNA methylation state in ESCs. Intriguingly, while human ESCs are normally cultured in medium containing VitC, they resemble mouse epiblast cells more than ICM cells. Nevertheless, VitC may also play an role in human ESCs, since it has been shown that they accumulate DNA methylation after several passages in the absence of VitC, although the underlying mechanisms were not addressed25. Notably, imprinted regions and IAP retroelements, which are resistant to DNA demethylation in the early embryo2,13, are also resistant to VitC-mediated demethylation. These regions show high levels of H3K9me318, suggesting that this mark, or readers of this mark, may play a role in protecting against Tet-mediated demethylation. VitC also improves the quality of iPS cells while preserving the fidelity of DNA methylation at imprinted regions26. Furthermore, Tets are required for methylation reprogramming during iPS cell generation and in the zygote4,27,28. VitC has also been reported to increase hmC in mouse embryonic fibroblasts29, suggesting that the mechanism characterized here may be broadly applicable to other cell types. Much work remains to be done to evaluate the ability of VitC to modulate Tet activity and DNA methylation in vivo. It will be of interest to investigate the role of VitC in contexts where Tet enzymes have been implicated30, such as in the zygote, germline, blood, and brain. Potential roles for VitC in the clinic, including in IVF culture medium or in cancers driven by aberrant DNA methylation, also deserve exploration.
Methods
Cell Culture
ESCs used in this study include: Oct4-GiP (129 × MF1), V6.5 (129/Sv × C57/BL6), Tet1−/−, Tet1−/−; Tet2−/−, J1 (129/Sv), and Dnmt TKO. All ESCs were cultured in feeder free conditions on 0.1% gelatin coated tissue culture plates. Unless indicated otherwise, ESCs were cultured in 2i medium, which is composed of a N2B27 base medium31 supplemented with the MEK inhibitor, PD0325901 (1 µM, Stemgent), the GSK3β inhibitor, CHIR99021 (3 µM, Selleck Chemicals), and with ESGRO leukemia inhibitory factor (LIF) at 1,000 U/ml (Millipore). Vitamin C (L-ascorbic acid 2-phosphate, Sigma, A8960) was added on day 1 after seeding at 100 µg/ml. Medium was replaced daily. For Knockout Serum Replacement (KSR) versus Fetal Bovine Serum (FBS) studies, a base medium composed of high glucose DMEM (Life Technologies), L-glutamine (2 mM, Life Technologies), sodium pyruvate (1 mM, Life Technologies), non-essential amino acids (1X, Life Technologies), 2-mercaptoethanol (1X, Millipore), and penicillin-streptomycin (1X, Life Technologies) was supplemented with LIF (1,000 U/ml) and either 15% KSR (KSR medium) or 15% FBS (FBS medium). For differentiation experiments, Oct4-GiP ESCs maintained in 2i medium were treated with VitC for 72 hrs. Untreated and VitC treated ESCs were then transferred to 60 mm petri dishes and cultured in suspension in FBS medium without LIF to induce embryoid body formation. Medium was replaced on day 3 and embryoid bodies were collected on day 5 of differentiation. For comparing the effects of VitC in FBS medium versus 2i medium, J1 ESCs maintained in FBS medium were switched to FBS medium plus VitC, 2i medium, or 2i medium plus VitC and harvested at 12 and 72 hrs after changing conditions. Media was replaced daily.
Small Molecule Screen
Oct4-GiP ESCs were cultured in FBS medium. Small molecules were added at the time of seeding. The small molecules used included valproic acid (2 mM, Calbiochem), trichostatin A (20 nM, Sigma), PD0325901 (1 µM, Stemgent), CHIR99021 (3 µM, Selleck Chemicals), forskolin (10 µM, Sigma), A-83–01 (1 µM, Stemgent), RG108 (250 nM, Stemgent), 5-azacytidine (1 µM, Sigma), BIX01294 (0.5 µM, Stemgent), UNC0638 (0.5 µM, Sigma), insulin (5 µg/ml, Sigma), Vitamin C (100 µg/ml, Sigma) and the lipid cocktail Albumax II (5 mg/ml, Life Technologies). The KSR components tested include VitC, Albumax II, and insulin (International Patent Application WO 98/30679).
Vitamin C quantification assay
The amount of Vitamin C in 2i medium was determined using an Ascorbic Acid Assay Kit (Abcam, ab65656). Known amounts of Vitamin C (L-ascorbic acid, Sigma, A4544) were added to the medium and tested as controls.
Antioxidant treatment
Oct4-GiP ESCs were cultured in 2i medium and treated for 24 hrs with antioxidants. The antioxidants tested included a modified glutathione, glutathione reduced ethyl ester (GMEE), at 1.5 µg/ml, sodium selenite (20 nM), vitamin B1 (9 µg/ml), vitamin E (25 µM), L-carnitine hydrochloride (15 µg/ml), and α-lipoic acid (5 µg/ml). All reagents were purchased from Sigma. Following treatment, DNA was isolated and assayed by dot blot.
Immunofluorescence staining
Cells were fixed in 4% paraformaldehyde for 15 min. After washing 3x with PBS, cells were blocked with 5% FBS in PBST (PBS + 0.5% Tween 20) for 2 hrs at room temperature. Primary antibodies were diluted in blocking solution and incubated with cells overnight at 4 °C. Cells were then washed 3x in PBS and incubated for 2 hrs at room temperature with secondary antibodies diluted in blocking solution. Cells were washed 3x in PBS prior to imaging. Primary antibodies included DAZL (1:200, Abcam) and 5-hydroxymethylcytosine (1:100, Active Motif). 594-conjugated chicken anti-rabbit (1:1,000, Life Technologies) was used for a secondary antibody.
Genomic DNA Preparation
DNA was isolated using a Qiagen Gentra Puregene Kit or the phenol-chloroform-isoamyl alcohol method. RNase A digestion was included in the isolation procedure.
Dot blot
Isolated DNA (1 µg per sample) was denatured in 0.1M NaOH for 10 min at 95 °C. Samples were neutralized with 1 M NH4OAc on ice, and then serially diluted 2-fold. DNA samples were spotted on a nitrocellulose membrane using a Bio-Dot apparatus (Bio-Rad). The blotted membrane was washed in 2x SSC buffer, dried at 80 °C for 5 min, and UV cross-linked at 120,000 µjoules/cm2. The membrane was then blocked in Odyssey buffer (Li-Cor) diluted 1:1 in PBS (Odyssey:PBS) overnight at 4 °C. Mouse anti-5-methylcytosine monoclonal antibody (Active Motif, 1:500) or rabbit anti-5-hydroxymethylcytosine polyclonal antibody (Active Motif, 1:5,000) in Odyssey:PBS was added for 3 hrs at RT. The membrane was washed for 10 min 3x in PBS, and then incubated with either HRP-conjugated sheep anti-mouse IgG (GE, 1:10,000) or HRP-conjugated goat anti-Rabbit IgG (Abcam, 1:10,000) secondary antibodies in Odyssey:PBS for 3 hrs at RT. The membrane was then washed for 10 min 3x in PBS and visualized by chemiluminescence with GE ECL Plus.
qRT-PCR
Total RNA was isolated from cultured cells using Qiagen RNeasy with on-column DNase I treatment. cDNA was generated from 1 µg of RNA using random hexamers to prime the reaction. The cDNA was used as template for qRT-PCR. qRT-PCR was performed in combination with the KAPA SYBR Fast ABI Prism qPCR kit on an Applied BioSystems 7900HT sequence detection system. Primer sequences are listed in Supplementary Table 6. The relative amount of each gene was normalized using two housekeeping genes (L7 & Ubb), unless otherwise specified.
Blastocyst expression analysis
C57BL/6 (Simonsen) female mice were injected with 7.5 IU of pregnant mare gonadotropin (HUMC-NHPP) followed by 7.5 IU human chorionic gonadotropin (Sigma) 46 hrs later. Primed females were mated with DBA/2 (Simonsen) male mice. Detection of a vaginal plug was designated as 0.5 days post-coitum. Embryonic day 3.5 blastocysts were obtained by flushing the uterus of superovulated females with M2 medium (Sigma). Total RNA was isolated from collected blastocyst using a RNeasy Micro Kit (Qiagen) with on-column DNase I treatment. qRT-PCR was performed as described above. The relative amount of each gene was normalized to L7. All animal work was conducted in accordance with protocols approved by the Institutional Animal Care and Use Committee at the University of California, San Francisco.
Microarray analysis
Total RNA was isolated from biological triplicates of Oct4-GiP ESCs treated with or without VitC for 72 hrs and hybridized to Affymetrix mouse gene 1.0 ST GeneChip arrays (Affymetrix). Only annotated probes were considered for analysis. DChip software was used for gene expression statistical analysis.
Gene ontology analysis
Gene ontology functional annotation was performed in DAVID.
In vitro recombinant Tet1 activity assay
Recombinant human TET1 catalytic domain (1.2 ug) was incubated for 10 minutes at 37 °C with 178.2 ng of biotinylated DNA substrate in 50 mM HEPES (pH 8.0), 50 mM NaCl, 1 mM α-ketoglutarate, 3.7 uM ammonium iron (II) sulfate hexahydrate, 0.1 mg/mL BSA, 1 mM ATP and titrating concentrations of Vitamin C (Sigma, A5960), DTT (Sigma, D0632) or reduced glutathione (Sigma, G6529). The reaction was quenched by adding EDTA (11 mM). Each sample was loaded into a 384 well streptavidin coated plate in triplicates (25 µl/well) and left overnight at 4 °C on a rotating platform. The next morning the wells were washed 3x with 90 µl of 1x Tris-Buffered Saline (TBS) with 0.1 % Tween (TBST). The wells were then incubated with 50 µl per well of anti-hmC antibody (Active Motif) diluted 1:5,000 in TBST with 5% milk for 1 hr at room temperature on a rotating platform. Next the wells were washed 3x with 90 µl of TBST, followed by incubation with 50 µl of goat anti-rabbit peroxidase diluted 1:3,000 in TBST with 5% milk for 1 hr on a rotating platform. The wells were then washed 3x with TBST, and 30 µl of TMB Substrate Reagent mix (BD Biosciences) were added to each well. The reaction proceeded in the dark for 10–12 min, and was then quenched with 20 µl of 25% sulfuric acid. The absorbance of each well was measured at 450 nm. A standard curve of two fold dilutions from a fully hydroxymethylated version of the DNA substrate was used to obtain a linear regression, which was used to calculate pmols of hmC formed in the reaction from absorbance values. The data are presented as fold change in hmC relative to untreated.
Bisulfite Sanger sequencing
Oct4-GiP ESCs treated with or without VitC for 72 hrs were analyzed for promoter methylation. To analyze the methylation status of the selected genes, 0.2 µg of genomic DNA was subject to sodium bisulphite conversion using the EZ DNA Methylation-Gold kit (Zymo Research). Primers specific for the genes analyzed (Supplementary Table 6) were employed in nested or semi-nested PCR reactions. PCR products were cloned via T/A cloning using the pGEM-T easy kit (Promega) and individual inserts were sequenced using Genewiz Sequencing. Data was analyzed using Quantification Tool for Methylation Analysis32. The percent of methylated CpGs sequenced is presented for each set of samples.
mC and hmC DIP-PCR
DIP was performed using the Diagenode MagMeDIP or hMeDIP Kit with minor modifications. DNA was sonicated into short fragments (100–1,000 bp) with a Diagenode Bioruptor for 20 min with 15 sec ON / 15 sec OFF cycles at low power. Sonicated DNA was heat denatured at 95 °C for 10 min. 1 µg of sonicated DNA was immunoprecipitated with 1 µg of mouse anti-5-methylcytosine monoclonal antibody (Active Motif, 1 µg/µl) or 2.5 µg of mouse anti-5-hydroxymethylcytosine monoclonal antibody (Diagenode, 1 µg/µl). Following a 2 hr incubation at 4 °C, Magbeads (Diagenode) were added to the DNA-antibody mixture and samples were incubated at 4 °C overnight. Isolation of immunoprecipitated DNA was performed according to the kit instructions. qPCR was performed in combination with the KAPA SYBR Fast ABI Prism qPCR kit on a Applied BioSystems 7900HT Sequence Detection System. Primer sequences are listed in Supplementary Table 6.
hmC and mC DIP-seq
Oct4-GiP ESCs were treated with VitC (100 µg/ml) for 12 and 72 hrs. For each sample, 12 µg of genomic DNA was isolated, split into 3 replicates of 4 µg each and sonicated to ~100–500 bp on a Covaris E210 platform (75 s, 10% duty cycle). Sheared DNA was end-repaired, A-tailed, and ligated to custom paired-end adapters as described33. Ligated genomic DNA was size selected (100–300 bp) by 8% PAGE (Nuvex, Invitrogen) to remove unligated adapters. Replicates were pooled and subjected to qPCR using truncated PE1.0/2.0 PCR primers to assess ligation efficiency and quantified by fluorometer (Qubit, Life Technologies). Adapter-ligated DNA was heat denatured at 95 °C for 10 minutes, rapidly cooled on ice, and immunoprecipitated with a mouse monoclonal anti-methylcytidine antibody (1 mg/ml, Eurogentec) or a mouse monoclonal anti-hydroxymethylcytidine (hmC) antibody (1.6 mg/ml, Diagenode). Primary antibodies were added at 1 µl per microgram of DNA and samples were incubated overnight at 4 °C with rocking agitation in 500 µl IP buffer (10 mM sodium phosphate buffer, pH 7.0, 140 mM NaCl, 0.05% Triton X-100). To recover the immunoabsorbed DNA fragments, 1 µl of rabbit anti-mouse IgG secondary antibody (2.5 mg/ml, Jackson ImmunoResearch) and 100 µl Protein A/G beads (Pierce Biotechnology) were added and incubated for 2 hrs at 4 °C with agitation. After immunoprecipitation beads were resuspended in TE with 0.25% SDS and 0.25 mg/ml proteinase K for 2 hrs at 55 °C and then allowed to cool to room temperature. Immunoprecipitated and supernatant DNA were purified using Qiagen MinElute columns and eluted in 16 µl EB (Qiagen). Sequencing libraries were generated by 10–15 cycles of PCR using custom indexed paired-end Illumina PCR primers34. The resulting reactions were purified over Qiagen MinElute columns, after which a final size selection was performed by electrophoresis in 8% PAGE. Libraries were quality controlled by spectrophotometry and Agilent DNA Bioanalyzer analysis. An aliquot of each indexed library was pooled by sample and sequenced 2 per lane on an Illumina Hiseq2000 platform (2×76nt + 7nt) following the manufacturer’s recommended protocol and V3 chemistry (Illumina Inc.).
Bioinformatics
Sequence reads (75 bp paired-end) were aligned to the mouse reference genome (mm9) using BWA v0.5.935 and default parameters. Samtools36 and Picard (http://picard.sourceforge.net/) were used to sort and mark duplicate reads respectively. Reads having identical coordinates were collapsed into a single read and reads with mapping qualities ≥ 5 passed to FindPeaks 4.0137 for generation of unthresholded and thresholded (FDR < 0.01) coverage wig files to be visualized in the UCSC genome browser38. To quantify the strength of hmC and mC DIP-seq marks, these wig files were used to calculate Reads Per Kilobase per Million mapped reads (RPKM)39,40 values in various regions of interest including RefSeq41 promoters and CGIs (downloaded from http://genome.ucsc.edu on May 15, 2012). For pair-wise sample comparisons, an empirical Z score was calculated assuming the distribution of RPKMs for each sample followed a Poisson model: Z score = (RPKMA – RPKMB)/√(RPKMA – rABRPKMB), where RPKMA and RPKMB are RPKMs in the region of interest of A and B samples respectively, and rAB = NA/NB, where Nx is the total number of aligned reads used for normalization. For promoter analysis, promoters were defined as ± 500 bp from the TSS. Methylated promoters were defined as having > 0.5 RPKM values in both 12 and 72 hr untreated mC DIP-seq samples (n = 1045). For previously published Tet1 ChIP-seq data (ref. 10), short read data were downloaded from the NCBI GEO archive (GSE24843) and remapped to mm9 (NCBI 37). ChIP-seq reads having identical coordinates were collapsed into a single read. Tet1 reads were directionally extended by 300 bp using FindPeaks and unthresholded coverage wig files were generated to create Tet heatmaps in methylated promoters using ChAsE42, an interactive analysis and exploration tool for epigenetic data. Tet1 ChIP-seq data was corrected by control data. To determine which retroelement subfamilies show global hmC and mC DIP-seq changes in VitC-treated compared to untreated ESCs, RPKM values were calculated for all subfamilies in each hmC and mC DIP-seq dataset. To generate RPKM values, we calculated the total number of reads aligned to each retroelement subfamily using both unique and multiple-aligned reads, and normalized the total number of reads by total genomic bp per subfamily and total number of reads for each dataset. For whole genome bisulfite-seq analysis of CGIs, the percentage of methylation for each individual cytosine was downloaded from ref. 13 (http://www.nodai-genome.org/mouse_en.html). CGI methylation levels were calculated as the average of the percentage of methylation for all cytosines in each CGI region. Analysis was restricted to CGIs ± 1,000 bps from a TSS.
Supplementary Material
Acknowledgements
The authors wish to acknowledge the epigenomics and sequencing groups at Canada’s Michael Smith Genome Sciences Centre, Vancouver, Canada for performing mC DIP and hmC DIP Illumina sequencing, as well as for technical support. We thank Chih-Jen Lin for technical advice on isolation of blastocysts. We especially thank Meelad M. Dawlaty and Rudolf Jaenisch for the Tet1−/− and Tet1−/−;Tet2−/− ESCs. We also wish to thank Masaki Okano for the Dnmt1−/−;Dnmt3a−/−;Dnmt3b−/− ESCs. We thank Jun S. Song for statistical advice, as well as Marco Conti, Stavros Lomvardas, Joseph Costello and members of the Ramalho-Santos lab for critical reading of the manuscript. K.B. is a recipient of an NSF pre-doctoral fellowship and K.T.E. is supported by a California Institute of Regenerative Medicine postdoctoral training grant (TG2-01153). M.M.K. is supported by a postdoctoral fellowship from the Michael Smith Foundation for Health Research. M.C.L. is supported by a CIHR New Investigator Award. This work was funded by a CIHR grant (92093) to M.C.L. and M.H., NIH grants HD065812 and CA151535 and a grant from the California Institute of Regenerative Medicine to A.R., and an NIH New Innovator Award (DP2OD004698) and R01 (OD012204) to M.R.-S.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions M.R.-S. directed the study. K.B. and K.T.E. designed and performed experiments. K.B., K.T.E., M.M.K., M.C.L. and M.R.-S. analyzed and interpreted the data. M.M.K. and M.C.L. performed bioinformatics analyses. P.G. performed bisulfite sequencing. S.M. generated the human recombinant Tet1-catalytic domain, and J.A.Z.-M. performed the in vitro Tet activity assay under the direction of A.R., who provided expertise on Tet activity. A.T. and M.H. developed and performed DIP-seq and sequencing data processing. D.J.L. provided to K.T.E. financial support, advice, and lab space. K.B., K.T.E., M.C.L. and M.R.-S. wrote the manuscript.
Author Information Microarray and DIP-seq data have been deposited in the Gene Expression Omnibus under the accession number GSE46403. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
References
- 1.Bird A. DNA methylation patterns and epigenetic memory. Genes & Development. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 2.Smith ZD, et al. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484:339–344. doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seisenberger S, et al. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol. Cell. 2012;48:849–862. doi: 10.1016/j.molcel.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gu T-P, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- 5.Yamaguchi S, et al. Tet1 controls meiosis by regulating meiotic gene expression. Nature. 2012;492:443–447. doi: 10.1038/nature11709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hackett JA, et al. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science. 2013;339:448–452. doi: 10.1126/science.1229277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pastor WA, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473:394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu H, et al. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011;473:389–393. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams K, et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ficz G, et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 12.Koh KP, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobayashi H, et al. Contribution of intragenic DNA methylation in mouse gametic DNA methylomes to establish oocyte-specific heritable marks. PLoS Genet. 2012;8:e1002440. doi: 10.1371/journal.pgen.1002440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marks H, et al. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loenarz C, Schofield CJ. Expanding chemical biology of 2-oxoglutarate oxygenases. Nat. Chem. Biol. 2008;4:152–156. doi: 10.1038/nchembio0308-152. [DOI] [PubMed] [Google Scholar]
- 16.Wang PJ, McCarrey JR, Yang F, Page DC. An abundance of X-linked genes expressed in spermatogonia. Nat. Genet. 2001;27:422–426. doi: 10.1038/86927. [DOI] [PubMed] [Google Scholar]
- 17.Fouse SD, et al. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell. 2008;2:160–169. doi: 10.1016/j.stem.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karimi MM, et al. DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell. 2011;8:676–687. doi: 10.1016/j.stem.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang T, et al. The histone demethylases Jhdm1a/1b enhance somatic cell reprogramming in a vitamin-C-dependent manner. Cell Stem Cell. 2011;9:575–587. doi: 10.1016/j.stem.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 20.Dawlaty MM, et al. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell. 2011;9:166–175. doi: 10.1016/j.stem.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dawlaty MM, et al. Combined deficiency of tet1 and tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev. Cell. 2013;24:310–323. doi: 10.1016/j.devcel.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borgel J, et al. Targets and dynamics of promoter DNA methylation during early mouse development. Nat. Genet. 2010;42:1093–1100. doi: 10.1038/ng.708. [DOI] [PubMed] [Google Scholar]
- 23.Tang F, et al. Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-Seq analysis. Cell Stem Cell. 2010;6:468–478. doi: 10.1016/j.stem.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leitch HG, et al. Naive pluripotency is associated with global DNA hypomethylation. Nat. Struct. Mol. Biol. 2013;20:311–316. doi: 10.1038/nsmb.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chung T-L, et al. Vitamin C promotes widespread yet specific DNA demethylation of the epigenome in human embryonic stem cells. Stem Cells. 2010;28:1848–1855. doi: 10.1002/stem.493. [DOI] [PubMed] [Google Scholar]
- 26.Stadtfeld M, et al. Ascorbic acid prevents loss of Dlk1-Dio3 imprinting and facilitates generation of all-iPS cell mice from terminally differentiated B cells. Nat. Genet. 2012;44:398–405. doi: 10.1038/ng.1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doege CA, et al. Early-stage epigenetic modification during somatic cell reprogramming by Parp1 and Tet2. Nature. 2012;488:652–655. doi: 10.1038/nature11333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao Y, et al. Replacement of Oct4 by Tet1 during iPSC induction reveals an important role of DNA methylation and hydroxymethylation in reprogramming. Cell Stem Cell. 2013;12:453–469. doi: 10.1016/j.stem.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 29.Minor EA, Court BL, Young JI, Wang G. Ascorbate induces Ten-eleven translocation (Tet) methylcytosine dioxygenase-mediated generation of 5-hydroxymethylcytosine. J. Biol. Chem. 2013;288:13669–13674. doi: 10.1074/jbc.C113.464800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan L, Shi YG. Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development. 2012;139:1895–1902. doi: 10.1242/dev.070771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ying Q-L, et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumaki Y, Oda M, Okano M. QUMA: quantification tool for methylation analysis. Nucl. Acids Res. 2008;36:W170–5. doi: 10.1093/nar/gkn294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morin RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wiegand KC, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N. Engl J.Med. 2010;363:1532–1543. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fejes AP, et al. FindPeaks 3.1: a tool for identifying areas of enrichment from massively parallel short-read sequencing technology. Bioinformatics. 2008;24:1729–1730. doi: 10.1093/bioinformatics/btn305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kent WJ, Zweig AS, Barber G, Hinrichs AS, Karolchik D. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics. 2010;26:2204–2207. doi: 10.1093/bioinformatics/btq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Meth. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 40.Pepke S, Wold B, Mortazavi A. Computation for ChIP-seq and RNA-seq studies. Nat. Meth. 2009;6:S22–32. doi: 10.1038/nmeth.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pruitt KD, Maglott DR. RefSeq, LocusLink: NCBI gene-centered resources. Nucl. Acids Res. 2001;29:137–140. doi: 10.1093/nar/29.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Younesy H, et al. An Interactive Analysis and Exploration Tool for Epigenomic Data. Computer Graphics Forum (Proceedings of EuroVis 2013) 2013;32 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.