Abstract
Breast cancer cells develop resistance to endocrine therapies by shifting between estrogen receptor (ER)-regulated and growth factor receptor (GFR)-regulated survival signaling pathways. To study this switch, we propose a mathematical model of crosstalk between these pathways. The model explains why MCF7 sub-clones transfected with HER2 or EGFR show three GFR-distribution patterns, and why the bimodal distribution pattern can be reversibly modulated by estrogen. The model illustrates how transient overexpression of ER activates GFR signaling and promotes estrogen-independent growth. Understanding this survival-signaling switch can help in the design of future therapies to overcome resistance in breast cancer.
Keywords: Mathematical modeling, Estrogen receptor signaling, Growth factor receptor signaling, Breast cancer, Endocrine resistance
1. Introduction
Mammalian cells can switch between different signaling pathways to achieve distinct physiological goals in response to environmental stimuli, as exemplified by immune cell differentiation [1]. This plasticity is important for normal cells to differentiate properly and to survive in stressful environments. In cancer cells, this plasticity often results in drug resistance including acquired resistance to anti-estrogenic drugs.
The estrogen receptor (ER) and growth factor receptor (GFR) pathways are major drivers of survival and proliferation in 85% of breast tumors [2,3]. In clinical practice, expression of ERa (the most prevalent of two ER genes) and HER2 (a major GFR and member of the EGFR superfamily) are validated biomarkers used to determine treatment strategies for individual patients [4]. Approximately 70% of breast cancers express ERα [5], and various endocrine therapies have been developed to interfere with ER action [5]. Antagonizing GFR pathways (e.g., using trastuzumab) in HER2+ breast cancer also improves disease-free and overall survival for breast cancer patients [6]. However, the ultimate efficacy of therapies targeting individual pathways is not satisfactory. For example, tamoxifen successfully reduces by one-third the annual death rate from breast cancer, but one-third of tamoxifen-treated women develop recurrent disease within 15 years [5]. Resistance to anti-estrogens or GFR pathway antagonists also develops in human breast cancer cell lines [7–9].
We have used mathematical modeling guided by experimental observations to explore the mechanism underlying acquired resistance to endocrine therapies as driven by the ER–GFR switch. Acquired resistance could arise by activation of a compensatory escape pathway when the normal driver pathway is inhibited [3], the so-called ‘hybrid-car’ model of breast cancer [10]. Since breast cancer cells can switch reversibly and robustly between ER and GFR pathways for proliferation and survival [3,10], blocking either the ER or GFR pathway will usually result in activation of the other, allowing some cells to survive and eventually resume proliferation. Evidence for a close regulatory relationship between ER and GFR signaling includes the reciprocal expression of ER and GFR in most breast cancers [11], and activation of GFR pathway components (HER2, EGFR, MAPK, PI3K, AKT, mTOR, NFjB etc.) as compensatory responses to anti-estrogens [5,12–14]. Interestingly, these compensatory processes are reversible after withdrawing the endocrine treatment [15]. Moreover, recent evidence indicates that ER negative (ER–) breast cancer cells may develop resistance to GFR pathway antagonists by restoring the ER pathway and hence becoming responsive to anti-estrogens [16,17].
ER and GFR are sometimes positively associated in breast cancers [18,19]. Whether ER and GFR are negatively or positively correlated depends on how ER is activated. ER can be activated either by binding to 17β-estradiol (E2, the primary estrogen present in breast tumors) to form an active E2:ER complex, or by phosphorylation (ER-P) by various kinases (e.g., ERK and AKT) at multiple sites [5,20,21]. E2:ER has an inhibitory effect on GFR. E2 withdrawal can release the inhibition of ER on GFR expression and NFjB activity [22–26], consistent with the fact that E2:ER binds the promoter region of GFR genes (e.g., HER2 and EGFR) and acts as a repressor [27,28]. However, E2-independent ER-P is positively associated with GFR, and it can up-regulate certain ligands (e.g., TGFα, EGF and amphiregulin) of the GFR signaling network, which in turn activate the kinases that phosphorylate more ER [29–31]. This auto-activation loop has been implicated in tamoxifen-resistance [31,32]. NFκB, a major integrator of the GFR signaling network, is involved with E2:ER in a mutual-inhibition feedback loop [24,33]. NFκB also controls the expression of a broad spectrum of genes regulating important cellular behaviors including cell differentiation [34,35]. In particular, NFκB activates the transcription factor TWIST and represses the expression of E-cadherin, which in turn enhances the epithelial–mesenchymal transition (EMT) in breast cancer [36]. EMT is associated with a de-differentiation process whereby epithelial-like breast cancer cells increase their ‘stemness’ and undergo a phenotypic transition from HER2– to HER2+ [37]. EMT in breast cancer cells is likely due to genome-scale epigenetic reprogramming, including the promoter activity of HER2 [38]. Epigenetic changes such as methylation or acetylation can occur during differentiation or de-differentiation and are often reversible [36–38].
While the crosstalk between ER and GFR pathways in breast cancer, especially in MCF7 cells, has been widely studied [5,13,20,22,31,39,40], a comprehensive, dynamic view of ER–GFR crosstalk is still lacking. Previously, we proposed a simplified model that could account for the effects of E2 withdrawal on the bimodal distribution of GFR (HER2 or EGFR) in MCF7 cells [41]. However, this model combined all components of the GFR pathway into one variable and required an unreasonably slow rate constant to fit the experimental data. A more realistic model would allow the GFR pathway to exhibit both rapid (e.g., post-translational modifications of GFR proteins) and slow modifications (e.g., epigenetic modifications of GFR promoters). Moreover, a recent report indicates that transient ER overexpression can robustly activate E2-independent growth of MCF7 cells [42], suggesting further modifications to achieve a more realistic model.
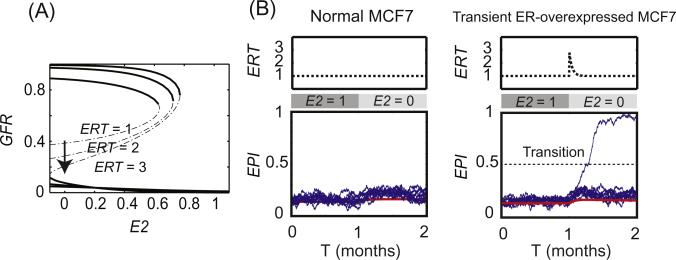
Here we present a new model to explore the mathematical characteristics of the ER–GFR switch that is a central determinant of breast cancer cell fate in response to endocrine therapies. The model explains many aspects of the available experimental data (Supplementary document, Fig. S1–S4), for example: (1) in sub-clones of MCF7 cells transfected with GFR (HER2 or EGFR), there are three different distribution patterns of GFR [43,44] (Fig. S1), (2) for sub-clones with a bimodal distribution of GFR, the distribution can be reversibly manipulated by varying E2 levels [43,44] (Fig. S2), (3) whereas E2 withdrawal in GFR-transfected MCF7 cells switches on GFR expression within weeks, E2 addition takes months to switch off expression [43,44] (Fig. S2), (4) E2 withdrawal can up-regulate GFR expression within 5 weeks in GFR-transfected MCF7 cells, but fails to do so in wild type MCF7 cells [43,44] (Fig. S3), and (5) transient ER overexpression in MCF7 cells can switch on the GFR pathway and promote E2-independent growth [42] (Fig. S4). The model provides a new tool to understand and evaluate these intriguing experimental observations, and it may help in finding new strategies to overcome anti-estrogen resistance in breast cancer.
2. Materials and methods
We postulate a highly condensed model of the interaction between ER and GFR (Fig. 1A and Supplementary documents). The protein level of GFR is down-regulated by E2:ER complex [27,28]. After E2 withdrawal, GFR is released from inhibition and its downstream kinases phosphorylate ER to an E2-independent form, ER-P [5,20,21]. ER-P can activate and stabilize the GFR pathway, creating a positive feedback loop [29–31]. In addition, GFR further activates transcription factors such as NFκB, promoting a series of epigenetic changes contributing to increased GFR expression and establishing another positive feedback loop [34,35]. For simplicity, we combine the epigenetic factors contributing to GFR expression into the quantity ‘EPI’. ‘E2ER’ and ‘ERP’ are used to represent [E2:ER] and [ER-P]. The wiring diagram in Fig. 1A was translated into ordinary differential equations (ODEs) by a formalism that allows us to capture complex dependencies in a simple manner [45] for simulation and analysis. We used the program XPP-AUT, available freely at http://www.math.pitt.edu/~bard/xpp/xpp.html, to simulate the model and to draw bifurcation diagrams. The ensemble stochastic simulations were performed with Matlab Version 7.9.0. A detailed version of materials and methods is provided in the Supplementary document.
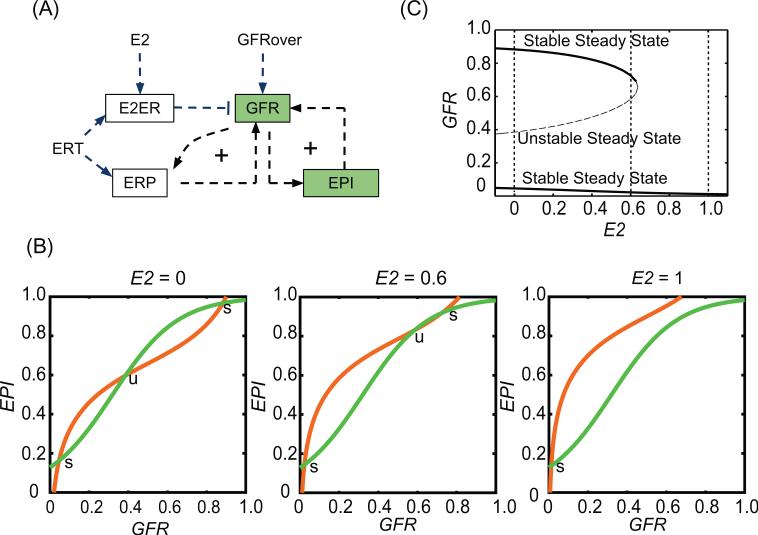
Fig. 1.
A model of the crosstalk between ER and GFR pathways exhibits bistable switching properties. (A) Influence diagram of the model. E2, estrogen level; ERT, total ERα level; E2ER, estrogen-dependent E2:ER complex; ERP, phosphorylated ER; GFR, growth factor receptor; EPI, epigenetic components in GFR pathway; GFRover, number of extra GFR gene copies. (B) Nullclines of the system at different E2 levels. s, stable steady state; u, unstable steady state. (C) Bifurcation diagram of GFR, with E2 as the bifurcation parameter. The curves trace the steady state level of GFR as a function of E2 level. For a given value of E2, a cell may express a low or high value of GFR (upper and lower solid lines; the middle dashed line indicates a branch of unstable steady states).
3. Results
3.1. Bifurcation analysis of the survival-signaling switch
The nullclines of our system of equations (Eq. S1 and S2) are plotted in Fig. 1B. The intersections of these two curves correspond to steady states of the model. The number of steady states is controlled by the value of E2 level. When E2 = 1, there is one stable steady state corresponding to low GFR and low EPI (GFR–/EPI–). When E2 is reduced below 0.65, there are three steady states, two of which are stable and a third which is unstable. The stable steady states have GFR and EPI levels that are either both low (GFR–/EPI–) or both high (GFR+/EPI+). Fig. 1C illustrates how the steady states of the system change with E2. The system has three steady states in the range of 0 < E2 < 0.65 and only one stable steady state when E2 > 0.65. However, E2 is not the only parameter that influences the system's bistability. GFRover, which represents the influence of additional GFR genes transfected into MCF7 cells, can also be used as a bifurcation parameter. Fig. 2A shows that when E2 is held constant at E2 = 1 the system is bistable only when 3.2 < GFRover < 12.8. We will show how this bistable survival-signaling switch can explain the results of several important experiments that are difficult to understand without a model.
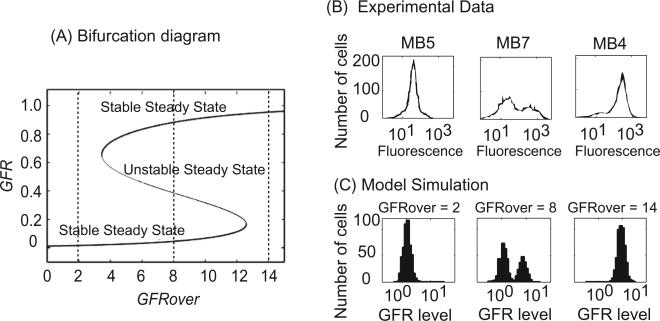
Fig. 2.
Three distribution patterns of GFR are exhibited in GFR-transfected MCF7. (A) Signal–response curve for GFR as a function of GFRover. The steady-state value of GFR is plotted as a function of GFRover (from 0 to 14). For intermediate values of GFRover, a cell may express either a low or high level of GFR (upper and lower solid lines; the middle dashed line indicates a branch of unstable steady states). (B) Different HER2 distribution patterns in HER2-overexpressed MCF7 sub-clones. Sub-clones MB5, MB7 and MB4 represent three typical distribution patterns of HER2 observed in experiment. Experimental data are adapted from [43]. (C) Distribution of GFR in 500 cells that are stochastically simulated at different values of GFRover (2, 8, and 14) for four months by starting from random initial conditions. GFR level = 10GFR in these histograms.
3.2. Three distribution patterns of GFR
Liu et al. transfected HER2 cDNA into MCF7 cells and created multiple stable sub-clones, which were further screened for HER2 protein expression levels using flow cytometry. Interestingly, three HER2 distribution patterns were observed in the sub-clones they selected [43]: (1) a single peak of cells with elevated HER2 protein (MB4 in Fig. 2B), (2) a single peak of cells with low HER2 protein, identical to control MCF7 cells (MB5 in Fig. 2B), and (3) a bimodal (two-peaked) distribution of HER2 (MB7 in Fig. 2B). Southern blotting was used to confirm that, within a sub-clone, all cells had the same number of integrated HER2 copies; hence, the bimodal distribution did not result from varying genetic conditions. Similar results in MCF7 cells transfected with EGFR have been reported [44], but a satisfactory explanation for these observations is lacking.
The major difference between these sub-clones could be that during transfection different numbers of HER2 or EGFR genes were integrated into the individual cells that formed the sub-clones. In our model, we use GFRover to denote the amount of transfected GFR and have shown that the system is bistable only when GFRover is within a specific range (Fig. 2A). When very few GFR gene copies are integrated into the cell (e.g., GFRover = 2), the system has only one stable steady state at low GFR expression, while integration of a high number of copies (e.g., GFRover = 14) results in only one stable steady state at high GFR expression. However, integration of an intermediate number of copies (e.g., GFRover = 8) creates a system with two stable steady states (either high or low GFR expression). To model distribution patterns of GFR in a population of cells, we performed 500 stochastic simulations (see Fig. 2C), starting from random initial conditions, of four month duration for each of three different cases (GFRover = 2, 8 and 14). The results of these simulations clearly replicate the experimental observations. When GFRover = 2, there is only one peak of cells at a low GFR level, corresponding to MB5 in Fig. 2B. When GFRover = 14, there is only one peak of cells at a high GFR level, corresponding to MB4 in Fig. 2B. And when GFRover = 8, two peaks of cells (bimodal distribution) coexist, corresponding to MB7 in Fig. 2B. (We note that the number of GFR gene copies actually transfected in the experiments is unknown and so the values used here for GFRover are arbitrary and can be rescaled if copy number data becomes available.)
3.3. GFR bimodal distribution manipulated by E2
By using a sub-clone of MCF7 cells with a bimodal HER2 distribution, Liu et al. further showed the sensitivity of HER2 expression to culture conditions [43]. Growing the cells for 5 weeks in charcoal-stripped fetal calf serum (CCS), which is depleted of E2, resulted in a single peak of cells expressing high levels of HER2. Similarly, Miller et al. reported that MCF7 cells transfected with EGFR and cultured in CCS create a population consisting predominantly of cells with high EGFR levels [44]. However, wild type MCF7 cells cultured in CCS for one year still show a population consisting predominantly of cells with low EGFR levels [44].
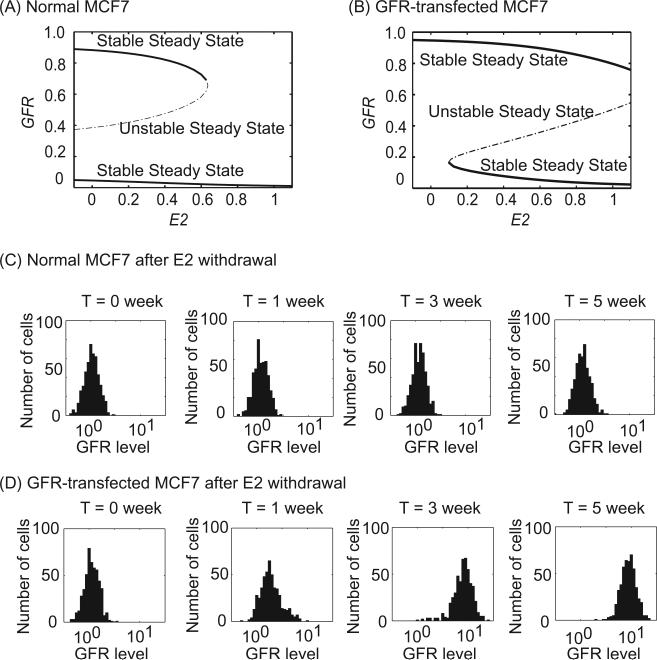
Fig. 3 provides an explanation for these results. In normal MCF7 cells cultured with E2, the cells are on the lower branch (low GFR) of Fig. 3A. When E2 is depleted, the cells will stay on this branch unless stochastic variations are strong enough to occasionally push a cell to the upper branch (high GFR, Fig. 3A). Note that there is a significant barrier separating the lower steady state from the unstable steady state (middle branch). We simulated a population of 500 MCF7 cells starting from low GFR in the E2-withdrawal condition for five weeks. No cell jumped to the high-GFR state, and there was no change in the GFR distribution pattern at the population level (Fig. 3C). However, when GFR is transfected into MCF7 cells (GFRover = 5), the barrier at E2 = 0 disappears (Fig. 3B) and the system only has one stable steady state at high GFR. Thus, when starting from the low-GFR initial condition, all cells will move to the high-GFR state. A simulation of 500 GFR-transfected MCF7 cells for five weeks produced a change in the GFR distribution pattern similar to that reported by Liu et al. and Miller et al. (Fig. 3D).
Fig. 3.
E2 withdrawal turns on GFR in GFR-transfected but not in normal MCF7 cells. (A, B) Bifurcation diagrams of GFR with E2 as bifurcation parameter in normal MCF7 cells (GFRover = 0) and in GFR-transfected MCF7 cells (GFRover = 5). (C, D) Temporal evolutions of GFR distribution under E2-withdrawal conditions for normal MCF7 cells (GFRover = 0) and for GFR-transfected MCF7 cells (GFRover = 5). In each case, 500 cells were simulated over the course of five weeks. GFR level = 10GFR in these histograms.
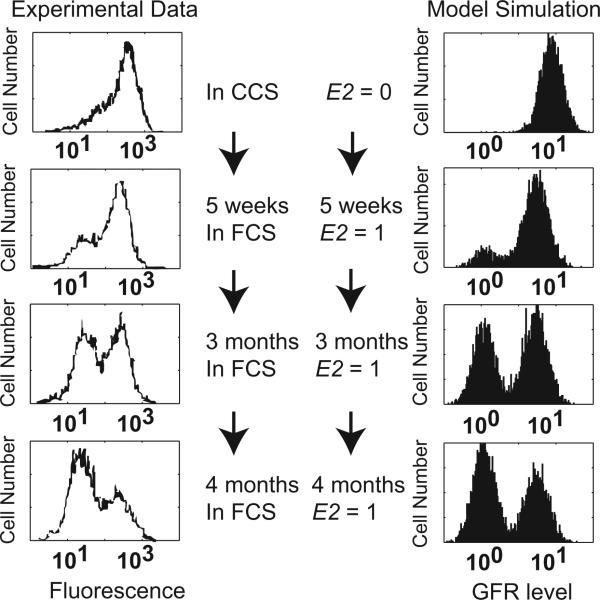
Experiments also show that the distribution pattern of GFR can be reversibly controlled by E2 [43,44]. Growing the MB8 sub-clone MCF7 cells (with bimodal HER2 distribution) in CCS for five weeks resulted in a single peak at high HER2 (Fig. 4, top left). Continuing to grow these cells in fetal calf serum (FCS), which contains E2, gradually leads to the rise of a peak at low HER2 (Fig. 4, left panel) [43]. Similar dynamics have also been reported in MCF7 cells with EGFR overexpression [44]. Southern blots confirmed that there was no variation in transfected HER2 gene copy number during E2 manipulation; Northern blots showed that HER2 mRNA expression is consistent with the protein level [43]. These data imply that genetic mutations are not the cause of HER2 heterogeneity in these MCF7 cells.
Fig. 4.
GFR bimodal distribution is reversibly controlled by different E2 levels. Left panel, experimental data adapted from [43]. The MB8 sub-clone of GFR-transfected MCF7 cells showing a bimodal HER2 distribution was treated with a series of E2 conditions: CCS, without E2, for 5 weeks; FCS, with E2, for 5 weeks, 3 months and 4 months. Right panel, model simulations, showing the distribution of GFR level in 5000 GFR-transfected MCF7 cells (GFRover = 5) under the same conditions as the experiments. GFR level = 10GFR in these histograms.
The bifurcation diagram in Fig. 3B provides an explanation for these experimental results. Consider a population of GFR-transfected MCF7 cells depleted of E2 (E2 = 0) and having high GFR expression. If E2 is now provided to the cells (E2 = 1), they will stay in the high-GFR state, but the barrier to transitioning to the low-GFR state will be diminished. Given adequate time, stochastic variations may induce some cells to transition to the low-GFR branch. We simulated a population of 5000 cells under these conditions, and the GFR distributions from the model compared well with the experimental observations (Fig. 4, right panel). Note that after E2 withdrawal, GFR-transfected MCF7 cells can switch on GFR within weeks, but cells need months to turn off GFR after E2 addition. Our simulation also replicates this asymmetry in response time (Figs. 3 and 4).
3.4. Role of ER overexpression
A recent study observed that ER overexpression in MCF7 cells activates the ER-regulated genes pS2 and PR in the absence of E2 [42]. Prolonged culturing of these cells leads to proliferation in E2-depleted conditions. However, this proliferation can still be inhibited by faslodex (a pure ER antagonist), indicating the role of E2-independent ER-P activation in maintaining cell proliferation. By contrast, long term culturing of wild type MCF7 cells in the absence of E2 failed to cause resumed cell growth. More interesting, the overexpression of ER by adenovirus gene transfection in their study was only transient, as evidenced by the complete loss of co-transfected GFP protein after culturing the cells for 12 weeks.
To understand these experimental results using our model, we first evaluated the role of ER overexpression. Fig. 5A shows bifurcation diagrams of GFR level, with E2 as the bifurcation parameter, at different ER levels (ERT = 1, 2 and 3). This figure shows that increasing ER level decreases the barrier for cells to switch from the GFR–/ERP–/EPI– state to the GFR+/ERP+/EPI + state when E2 is deprived. Consequently, a transient increase of ER will also result in a transient decrease of the barrier, opening a temporary window to an increased probability of state transitions under noise. To demonstrate this outcome, we stochastically simulated a population of 10,000 cells and evaluated how transient ER overexpression influences the frequency of transitions in E2-depleted MCF7 cells. No transition was observed during one month of E2 depletion in normal MCF7 cells (ERT = 1, Fig. 5B, left panel). However, in MCF7 cells with transiently over-expressed ER there were 80 transitions during a similar time window (Fig. 5B, right panel). Cells that transition will have high levels of GFR, EPI, and ERP, implying the capability of E2-independent cell growth. The number of transitions is influenced by the strength of the ERT pulse, which was simulated with the following equation:
| (1) |
where ER0 = 1, ERover = 1.8, T1/2 = 0.25 month, t starts from the time of ER overexpression.
Fig. 5.
ER overexpression increases the probability of a survival-signaling switch. (A) Bifurcation diagram of GFR with E2 as bifurcation parameter at different ER levels (ERT = 1, 2 and 3). (B) Transient ER overexpression opens a time window for transitions from low to high GFR levels. Left panel, normal MCF7 cells show no transitions in stochastic simulations of 10000 cells (20 cells are plotted for illustration). Right panel, 80 transitions are observed within a short time window during transient ER overexpression in MCF7 cells (20 cells and one example of a transition are plotted for illustration). The pulse of ER overexpression is simulated by Eq. (1) in the manuscript. Red line, trace of a deterministic simulation; blue lines, traces of 20 stochastic simulations; the horizontal dotted line shows the threshold we set to score transitions (EPI = 0.5).
To capture the experimental proliferation data in E2-depleted conditions, we created a simple population growth model (Fig. 6A) given by the equations:
| (2) |
| (3) |
Fig. 6.
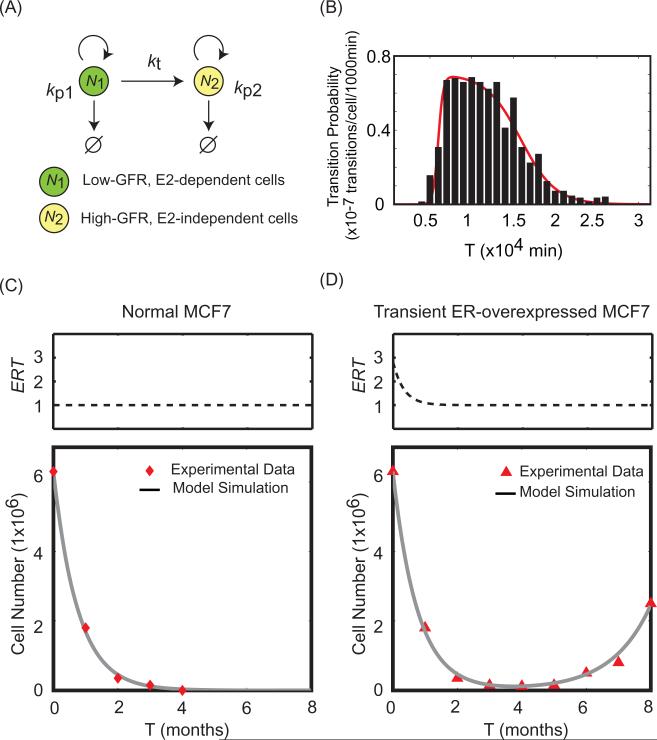
A population growth model shows the effects of transient ER overexpression. (A) Schematic representation of the population growth model. kp1, kp2 = (specific birth rate – specific death rate) for cell populations N1 and N2, respectively. kt = rate at which low-GFR cells switch to high-GFR cells. (B) Transition probability of MCF7 cells that transiently overexpress ER according to the survival-signaling switch model described in Fig. 5. Black bars, percentages of cells having transitions in given time intervals (1000 min); Red line, fitted curve showing how kt varies with time in ER-overexpressed MCF7 cells. (C) Experimental data (red diamonds) and simulation results (grey line) for the dynamics of total cell number in normal MCF7 cells after E2 depletion. (D) Experimental data (red triangles) and simulation results (grey line) for the dynamics of total cell number in transient ER-overexpressed MCF7 cells after E2 depletion. The pulse of ER overexpression is simulated by Eq. (1). Experimental data are adapted from [42].
In these equations, N1 denotes the number of low-GFR, E2-dependent MCF7 cells and N2 denotes the number of high-GFR, E2-independent MCF7 cells. The two cell types have different proliferation rates (kp1, kp2), and there is a transition rate, kt, which describes how fast a low-GFR cell can switch to a high-GFR cell under E2-depleted conditions. Notice that kt = 0 in normal MCF7 cells, since no transition is observed in Fig. 5B. Furthermore, kt is not a constant in MCF7 cells with transient ER overexpression. To determine the dynamics of kt we stochastically simulated a population of 6 × 106 cells with transiently overexpressed ER. The percentage of cells having transitions within independent time intervals is plotted in Fig. 6B (black bars), and the histogram was fit by a function of the form:
| (4) |
where fitting parameters a, b, c, d and h are all positive values. The red curve in Fig. 6B is the result of the fit and describes how kt varies with time in MCF7 cells with the transient ER overexpression we consider. Using a standard curve fitting method (Matlab Version 7.9.0, Curve Fitting Toolbox), we determined the best fitting parameter values to be a = 3.322 × 10–3, b = 6 × 103, c = 4.802 × 10–4, d = 1.587 × 104 and h = 7.029 × 10–8.
Choosing the parameters ERover = 1.8, kp1 = –3 × 10–5, and kp2 = 2 × 10–5, our simulation results for total cell number (N1 + N2) in Fig. 6 C,D match the experimental results (red diamonds and triangles) for both the normal and ER-overexpressed cells. Thus, by combining models at two different scales (molecular and population), we provide a plausible explanation for how transient ER overexpression can promote E2-independent growth in MCF7 breast cancer cells.
4. Discussion
Breast cancer is the most common invasive cancer in women. Endocrine therapy, as the most successful targeted cancer therapy, has been very effective in reducing breast cancer mortality. However, resistance often develops and the recurrence rate of breast cancer after targeted therapies remains unacceptably high. The molecular mechanisms of acquired endocrine resistance have been intensively studied both in vivo and in breast cancer cell lines, such as ER+ MCF7 cells. Many genes have been postulated as key players in acquired endocrine resistance [5,20,22,39], but despite knowledge of their roles in cell survival and growth, little is known about how the ‘escape’ paths are mechanistically activated, dynamically regulated, and epigenetically maintained during the development of endocrine resistance. Understanding the molecular mechanisms used by breast cancer cells to acquire resistance to endocrine treatment is critical for designing new therapies for breast cancer.
Emergence of resistance is closely associated with cellular heterogeneity. It has been widely recognized that breast cancer cells, including the well-studied MCF7 cells, are inherently heterogeneous. Distinct cell phenotypes in the same MCF7 cell population can be observed. Upon treatment, individual MCF7 cells can have diverse responses: while some cells die through apoptosis, others remain alive [23]. MCF7 cells also exhibit heterogeneous expression of a few key proteins. For example, although the MCF7 cell line is generally classified as ER+, a minority of these cells express low levels of ER [46]. The bimodal experimental data considered in the present work also supports the heterogeneity of cancer cells [43,44].
What is the source of this heterogeneity? A population comprised of cancer cells with distinct phenotypes is usually attributed to mutations resulting from the genomic instability of cancer cells. However, this emphasis on genetic causes for heterogeneity has been challenged, as summarized by Huang et al. [47]. Recent studies indicate that viability within a population of cancer cells can also result from noisy gene expression and the fact that gene networks have multiple stable states, providing a non-genetic source of heterogeneity [47]. The presence of cancer stem cells (CSCs) in breast cancer strongly supports a non-genetic basis for tumor heterogeneity. There should be no genetic difference between CSCs and differentiated ‘bulk’ cells unless they acquire new mutations. CSCs, unlike bulk cells, are usually ER– and dependent on the GFR pathway to survive [48,49]. For example, evidence shows that NFκB inhibitors can preferentially inhibit CSCs in MCF7 cells, but not MCF7 bulk cells [50]. Moreover, CSCs have the potential to develop into bulk cells, and vice versa. These reversible transitions between stem cells and bulk cells are implicated in the well-documented cellular mesenchymal–epithelial transition and epithelial–mesenchymal transition. During either of these transitions, cells are reported to have an increased or decreased ‘stemness’ associated with changes in expression of specific stem cell surface markers [37].
In this work, a mathematical model based on a bistable switch with an epigenetic component successfully explains five intriguing experimental observations in MCF7 cells [42–44] (Figs. S1–S4). (1) The bifurcation analysis in Fig. 2A shows that GFR-transfected MCF-7 cells have three distinct possibilities for GFR expression (low, both low and high, and high) depending on the number of GFR copies that were transfected, explaining why different experimental subclones exhibited these three different GFR profiles. (2) For a subclone with a bimodal GFR expression, the model analysis in Fig. 3B shows that E2 deprivation will force the cells into a high GFR state, as the low GFR state disappears, and re-adding E2 will return the cells to the bimodal state, in accord with experiment, Fig. 4. (3) Fig. 3B also shows why the transition of a bimodal sub-clone to a high GFR state upon E2 deprivation is fast, as there is only one stable state and no barrier to cross to get there, while reversing the transition by re-adding E2 is slow, as cells must transition across a barrier to regain the low GFR state. (4) For nontransfected MCF-7 cells, the model analysis in Fig. 3A shows that E2 deprivation will not result in a high GFR state, since there is a large barrier preventing state switching, even though such deprivation does result in a high GFR state for the bimodal subclones, Fig. 3B, as they have no barrier to cross. (5) The analysis in Fig. 5 shows that transient ER overexpression lowers the barrier to the high GFR state during a small time window and allows a few E2-deprived MCF-7 cells to cross the barrier. This explains why a population of E2 deprived MCF-7 cells will die out, but a similar population with a transient ER overexpression will eventually regrow, Fig. 6.
It should be emphasized that the model we present here, because it is an abstract and simplified version of reality, has some limitations. While a bistable switch is not unexpected, due to the presence of positive feedback loops in the ER–GFR crosstalk network, further experiments are required to confirm the exact mechanism. In particular, we emphasize epigenetic regulation of GFR promoters (EPI) in our current model, creating an epigenetic switch by assuming that EPI controls and is being controlled by GFR. The reality could be more complicated, since cell differentiation and de-differentiation processes are closely linked with both ER and GFR pathways. MicroRNAs, epigenetic regulation, and inter-cellular communications are implicated in differentiation/de-differentiation processes [1,37,51]; however, much is unknown at present. Moreover, cell-signaling networks may have as many steady states as there are physiological states, each steady state being represented in a subpopulation of cells within a cell culture or tissue. The low-GFR, E2-dependent and the high-GFR, E2-independent states are just two examples of many states that are possible.
Notwithstanding these limitations, the model of a survival-signaling switch presented here helps to understand certain dynamic behaviors of breast cancer cells that are difficult to comprehend by intuitive reasoning alone. Refining this model by obtaining data from primary breast cancers, as opposed to cell lines, will increase its clinical relevance and could aid in the search for new strategies to overcome breast cancer resistance.
Supplementary Material
Acknowledgements
This work was supported in part by the US National Institutes of Health Grant U54-CA149147 (to R.C., J.J.T. and W.T.B.), and by fellowships to C.C. provided by the Virginia Polytechnic Institute and State University graduate program in Genetics, Bioinformatics and Computational Biology.
Abbreviations
- AKT
a serine/threonine-specific protein kinase, also known as Protein Kinase B (PKB)
- CCS
charcoal-stripped fetal-calf serum
- CSC
cancer stem cell
- E2
17β-estradiol
- E2:ER
E2-bound estrogen receptor
- EGFR
epidermal growth factor receptor
- ER
estrogen receptor
- ER-P
phosphorylated estrogen receptor
- FCS
fetal calf serum
- GFR
growth factor receptor
- HER2
human epidermal growth factor receptor-2
- MAPK
mitogen activated protein kinases
- mTOR
mammalian target of rapamycin
- NFκB
nuclear factor kappa-light-chain-enhancer of activated B cells
- PI3K
phosphatidylinositide 3-kinases
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.febslet.2013.08.022.
References
- 1.Alberts B. Garland Science. 4th ed xxxiv. New York: 2002. Molecular biology of the cell; p. 1548. [Google Scholar]
- 2.Dickson RB, Lippman ME. Growth factors in breast cancer. Endocr. Rev. 1995;16(5):559–589. doi: 10.1210/edrv-16-5-559. [DOI] [PubMed] [Google Scholar]
- 3.Rimawi MF, Osborne CK. Breast cancer blocking both driver and escape pathways improves outcomes. Nature Reviews: Clinical Oncology. 2012;9(3):133–134. doi: 10.1038/nrclinonc.2012.9. [DOI] [PubMed] [Google Scholar]
- 4.Patani N, Martin LA, Dowsett M. Biomarkers for the clinical management of breast cancer: international perspective. Int. J. Cancer. 2013;133(1):1–13. doi: 10.1002/ijc.27997. [DOI] [PubMed] [Google Scholar]
- 5.Musgrove EA, Sutherland RL. Biological determinants of endocrine resistance in breast cancer. Nat. Rev. Cancer. 2009;9(9):631–643. doi: 10.1038/nrc2713. [DOI] [PubMed] [Google Scholar]
- 6.Arteaga CL, et al. Treatment of HER2-positive breast cancer: current status and future perspectives. Nat Rev Clin Oncol. 2012;9(1):16–32. doi: 10.1038/nrclinonc.2011.177. [DOI] [PubMed] [Google Scholar]
- 7.Clarke R, et al. Progression of human breast cancer cells from hormone-dependent to hormone-independent growth both in vitro and in vivo. Proc Natl Acad Sci USA. 1989;86(10):3649–3653. doi: 10.1073/pnas.86.10.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clarke R, et al. Hormonal carcinogenesis in breast cancer: cellular and molecular studies of malignant progression. Breast Cancer Res. Treat. 1994;31(2–3):237–248. doi: 10.1007/BF00666157. [DOI] [PubMed] [Google Scholar]
- 9.Santen RJ, et al. Adaptive hypersensitivity to estrogen: mechanism for sequential responses to hormonal therapy in breast cancer. Clin. Cancer Res. 2004;10(1 Pt 2):337S–345S. doi: 10.1158/1078-0432.ccr-031207. [DOI] [PubMed] [Google Scholar]
- 10.Saji S, Sato F, Ueno NT. Fuel, electricity, ER and HER2–a hybrid-car model of breast cancer. Nat Rev Clin Oncol. 2012;9(7) doi: 10.1038/nrclinonc.2012.9-c1. [DOI] [PubMed] [Google Scholar]
- 11.Sainsbury JR, et al. Epidermal-growth-factor receptors and oestrogen receptors in human breast cancer. Lancet. 1985;1(8425):364–366. doi: 10.1016/s0140-6736(85)91385-6. [DOI] [PubMed] [Google Scholar]
- 12.Clarke R, et al. Antiestrogen resistance in breast cancer and the role of estrogen receptor signaling. Oncogene. 2003;22(47):7316–7339. doi: 10.1038/sj.onc.1206937. [DOI] [PubMed] [Google Scholar]
- 13.Nicholson RI, et al. Growth factor signalling in endocrine and anti-growth factor resistant breast cancer. Rev Endocr Metab Disord. 2007;8(3):241–253. doi: 10.1007/s11154-007-9033-5. [DOI] [PubMed] [Google Scholar]
- 14.Gee JMW, et al. Antihormone induced compensatory signalling in breast cancer: an adverse event in the development of endocrine resistance. Hormone Molecular Biology and Clinical Investigation. 2011;5(2):2. doi: 10.1515/HMBCI.2011.009. [DOI] [PubMed] [Google Scholar]
- 15.Hiscox S, Gee J, Nicholson RI. Therapeutic resistance to anti-hormonal drugs in breast cancer: new molecular aspects and their potential as targets. Vol. 9. Springer; Dordrecht: 2009. p. 198. [Google Scholar]
- 16.Bayliss J, et al. Reversal of the estrogen receptor negative phenotype in breast cancer and restoration of antiestrogen response. Clin. Cancer Res. 2007;13(23):7029–7036. doi: 10.1158/1078-0432.CCR-07-0587. [DOI] [PubMed] [Google Scholar]
- 17.Xia W, et al. A model of acquired autoresistance to a potent ErbB2 tyrosine kinase inhibitor and a therapeutic strategy to prevent its onset in breast cancer. Proc. Natl. Acad. Sci. USA. 2006;103(20):7795–7800. doi: 10.1073/pnas.0602468103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Curtis C, et al. The genomic and transcriptomic architecture of 2000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koboldt DC, et al. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Osborne CK, Schiff R. Mechanisms of endocrine resistance in breast cancer. Annu. Rev. Med. 2011;62:233–247. doi: 10.1146/annurev-med-070909-182917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lannigan DA. Estrogen receptor phosphorylation. Steroids. 2003;68(1):1–9. doi: 10.1016/s0039-128x(02)00110-1. [DOI] [PubMed] [Google Scholar]
- 22.Nicholson RI, et al. Growth factor-driven mechanisms associated with resistance to estrogen deprivation in breast cancer: new opportunities for therapy. Endocr. Relat. Cancer. 2004;11(4):623–641. doi: 10.1677/erc.1.00778. [DOI] [PubMed] [Google Scholar]
- 23.Pratt MAC, et al. Estrogen withdrawal-induced NF-kappa B activity and Bcl-3 expression in breast cancer cells: Roles in growth and hormone independence. Mol. Cell. Biol. 2003;23(19):6887–6900. doi: 10.1128/MCB.23.19.6887-6900.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalaitzidis D, Gilmore TD. Transcription factor cross-talk: the estrogen receptor and NF-kappaB. Trends Endocrinol. Metab. 2005;16(2):46–52. doi: 10.1016/j.tem.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 25.Gionet N, et al. NF-kappaB and estrogen receptor alpha interactions: differential function in estrogen receptor-negative and -positive hormone-independent breast cancer cells. J. Cell. Biochem. 2009;107(3):448–459. doi: 10.1002/jcb.22141. [DOI] [PubMed] [Google Scholar]
- 26.Zhou Y, et al. The NFkappaB pathway and endocrine-resistant breast cancer. Endocr. Relat. Cancer. 2005;12(Suppl. 1):S37–46. doi: 10.1677/erc.1.00977. [DOI] [PubMed] [Google Scholar]
- 27.Bates NP, Hurst HC. An intron 1 enhancer element mediates oestrogen-induced suppression of ERBB2 expression. Oncogene. 1997;15(4):473–481. doi: 10.1038/sj.onc.1201368. [DOI] [PubMed] [Google Scholar]
- 28.Chrysogelos SA, et al. Mechanisms of EGF receptor regulation in breast cancer cells. Breast Cancer Res. Treat. 1994;31(2–3):227–236. doi: 10.1007/BF00666156. [DOI] [PubMed] [Google Scholar]
- 29.Hutcheson IR, et al. Oestrogen receptor-mediated modulation of the EGFR/MAPK pathway in tamoxifen-resistant MCF-7 cells. Breast Cancer Res. Treat. 2003;81(1):81–93. doi: 10.1023/A:1025484908380. [DOI] [PubMed] [Google Scholar]
- 30.Nicholson RI, et al. Nonendocrine pathways and endocrine resistance: observations with antiestrogens and signal transduction inhibitors in combination. Clin. Cancer Res. 2004;10(1 Pt 2):346S–354S. doi: 10.1158/1078-0432.ccr-031206. [DOI] [PubMed] [Google Scholar]
- 31.Nicholson RI, et al. Growth factor signalling and resistance to selective oestrogen receptor modulators and pure anti-oestrogens: the use of anti-growth factor therapies to treat or delay endocrine resistance in breast cancer. Endocr. Relat. Cancer. 2005;12(Suppl. 1):S29–S36. doi: 10.1677/erc.1.00991. [DOI] [PubMed] [Google Scholar]
- 32.Zilli M, et al. Molecular mechanisms of endocrine resistance and their implication in the therapy of breast cancer. Biochim. Biophys. Acta – Rev. Cancer. 2009;1795(1):62–81. doi: 10.1016/j.bbcan.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 33.Biswas DK, et al. Crossroads of estrogen receptor and NF-kappaB signaling. Sci STKE. 2005;2005(288):pe27. doi: 10.1126/stke.2882005pe27. [DOI] [PubMed] [Google Scholar]
- 34.Shostak K, Chariot A. NF-kappaB, stem cells and breast cancer: the links get stronger. Breast Cancer Res. 2011;13(4):214. doi: 10.1186/bcr2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baud V, Karin M. Is NF-kappaB a good target for cancer therapy? Hopes and pitfalls. Nat. Rev. Drug Discov. 2009;8(1):33–40. doi: 10.1038/nrd2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat. Rev. Cancer. 2013;13(2):97–110. doi: 10.1038/nrc3447. [DOI] [PubMed] [Google Scholar]
- 37.Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010;29(34):4741–4751. doi: 10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mishra SK, et al. Dynamic chromatin remodeling on the HER2 promoter in human breast cancer cells. FEBS Lett. 2001;507(1):88–94. doi: 10.1016/s0014-5793(01)02951-9. [DOI] [PubMed] [Google Scholar]
- 39.Massarweh S, Schiff R. Unraveling the mechanisms of endocrine resistance in breast cancer: new therapeutic opportunities. Clin. Cancer Res. 2007;13(7):1950–1954. doi: 10.1158/1078-0432.CCR-06-2540. [DOI] [PubMed] [Google Scholar]
- 40.Gee JMW, et al. Deciphering antihormone-induced compensatory mechanisms in breast cancer and their therapeutic implications. Endocr. Relat. Cancer. 2006;13:S77–S88. doi: 10.1677/erc.1.01274. [DOI] [PubMed] [Google Scholar]
- 41.Tyson JJ, et al. Dynamic modelling of oestrogen signalling and cell fate in breast cancer cells. Nat. Rev. Cancer. 2011;11(7):523–532. doi: 10.1038/nrc3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tolhurst RS, et al. Transient over-expression of estrogen receptor-alpha in breast cancer cells promotes cell survival and estrogen-independent growth. Breast Cancer Res. Treat. 2011;128(2):357–368. doi: 10.1007/s10549-010-1122-6. [DOI] [PubMed] [Google Scholar]
- 43.Liu Y, et al. MCF-7 breast cancer cells overexpressing transfected cerbB-2 have an in vitro growth advantage in estrogen-depleted conditions and reduced estrogen-dependence and tamoxifen-sensitivity in vivo. Breast Cancer Res. Treat. 1995;34(2):97–117. doi: 10.1007/BF00665783. [DOI] [PubMed] [Google Scholar]
- 44.Miller DL, et al. Emergence of MCF-7 cells overexpressing a transfected epidermal growth factor receptor (EGFR) under estrogen-depleted conditions: evidence for a role of EGFR in breast cancer growth and progression. Cell Growth Differ. 1994;5(12):1263–1274. [PubMed] [Google Scholar]
- 45.Tyson JJ, Novak B. Functional motifs in biochemical reaction networks. Annu. Rev. Phys. Chem. 2010;61:219–240. doi: 10.1146/annurev.physchem.012809.103457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palmari J, et al. Distribution of estrogen receptor heterogeneity in growing MCF-7 cells measured by quantitative microscopy. Cytometry. 1997;27(1):26–35. [PubMed] [Google Scholar]
- 47.Brock A, Chang H, Huang S. OPINION Non-genetic heterogeneity - a mutation-independent driving force for the somatic evolution of tumours. Nat. Rev. Genet. 2009;10(5):336–342. doi: 10.1038/nrg2556. [DOI] [PubMed] [Google Scholar]
- 48.O'Brien CS, et al. Resistance to Endocrine Therapy in Breast Cancer: Are It Breast Cancer Stem Cells Implicated? Cancer Stem Cells in Solid Tumors. 2011:381–402. [Google Scholar]
- 49.Shipitsin M, et al. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11(3):259–273. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 50.Zhou J, et al. NF-kappaB pathway inhibitors preferentially inhibit breast cancer stem-like cells. Breast Cancer Res. Treat. 2008;111(3):419–427. doi: 10.1007/s10549-007-9798-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Melo SA, Esteller M. Dysregulation of microRNAs in cancer: playing with fire. FEBS Lett. 2011;585(13):2087–2099. doi: 10.1016/j.febslet.2010.08.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.