Abstract
The accumulation of sequenced genomes has expanded the already sizeable population of cysteine peptidases from parasites. Characterization of a few of these enzymes has ascribed key roles to peptidases in parasite life cycles and also shed light on mechanisms of pathogenesis. Here, we discuss recent observations on the physiological activities of cysteine peptidases of parasitic organisms, paired with a global view of all cysteine peptidases from the MEROPS database grouped by similarity. This snapshot of the landscape of parasite cysteine peptidases is complex and highly populated, which suggests that expansion of research beyond the few ‘model’ parasite peptidases is now timely.
Peptidases have central roles in parasite biology and pathogenesis
All parasites must infect their host(s) to survive and propagate, and peptidases are essential components of these processes [1,2]. Historically, peptidases have been referred to collectively as proteases (Box 1), and they are associated with a large and significant body of research in parasitology that spans many decades. Peptidases enable many disparate biological activities in parasitic organisms: they allow parasites to bore through cellular and tissue barriers [3] and to degrade host proteins for nutrition [4], and they are used to manipulate the host immune system to elude the immune response [3]. In addition, parasite peptidases are involved in processing secondary protein modifications [5,6] and are used as immunodiagnostic markers of infection. Parasite peptidases can also trigger allergies in humans and other hosts [7].
Box 1. What is a peptidase?
Peptidases – also known as proteases, proteinases, and peptide hydrolases – are enzymes that are best known for catabolizing proteins and polypeptides through the cleavage of peptide bonds (Figure I). Peptidases can be divided into two broad subtypes: The first subtype uses the activated oxygen of a water molecule as a nucleophile and active site residues with (metallopeptidase) or without (aspartyl peptidases) the aid of a metal cation; these enzymes do not form a transient covalent bond with the peptidase. The second type of peptidase forms a transient covalent bond between the substrate and a catalytic oxygen of serine (serine peptidases) or threonine (threonine peptidases), or the sulfur of an essential cysteine (cysteine peptidases). Cysteine peptidases are also referred to thiol-peptidases, sulfhydryl peptidases and cysteinyl peptidases. In the MEROPS peptidase database, cysteine peptidases are classified into nine classes referred to as clans, and one unclassified clan [9]. The clans have no common ancestor between them but are each comprised of family members with a common progenitor. The clans are: clan CA (type example, lysosomal cathepsins), clan CD (type example, caspase), clan CE (type example, adenain), clan CF (type example, pyroglutamyl peptidase), clan CH (type example, hedgehog protein), clan CL (type example, sortase B), clan CM (type example, hepatitis clan C virus peptidase), clan CN (type example, sindbis virus-type nsP2 peptidase) and clan CO (type example, dipeptidyl peptidase VI).
This review focuses on cysteine peptidases from medically important parasitic protozoa and helminths; an overview of the cysteine peptidase clans that contain proteins from parasites is shown in Figure 1. The many recently sequenced genomes from these organisms – including Babesia bovis, Leishmania infantum, Leishmania braziliensis, Plasmodium knowelsi, and Plasmodium vivax, which were all released within the past two years – prompt a new census of cysteine peptidases in parasites. Here, sequence similarity networks [8] are used to visualize all members of these groups that are found in the MEROPS peptidase database [9] and some old assumptions about parasite peptidases are revisited and updated. The hand-curated MEROPS database supplies a high quality snapshot of known peptidases and homologs, and the populations and relative similarities of parasite peptidases and model peptidases can then be viewed using sequence similarity networks. This representation provides a useful context for new work on parasite peptidases. Recent research has disproved a number of popular views, namely that parasitic protozoan cysteine peptidases are primarily catabolic and restricted to clan CA and clan CD and that all ‘cysteine peptidases’ have a nucleophilic cysteine. Many of these misconceptions are due, in part, to the tendency to concentrate research on proteins and systems for which there are existing techniques and resources, which is logical and yet limiting. The number of well-characterized parasite cysteine peptidases is now dwarfed by the number of unexamined proteins, and many of these proteins are predicted to have important biological roles. A number of these cysteine peptidases will likely be excellent drug targets. A global atlas based on sequence similarity of all cysteine peptidases highlights unappreciated relationships, which can be investigated with tools and techniques developed for nearby cysteine peptidase homologs.
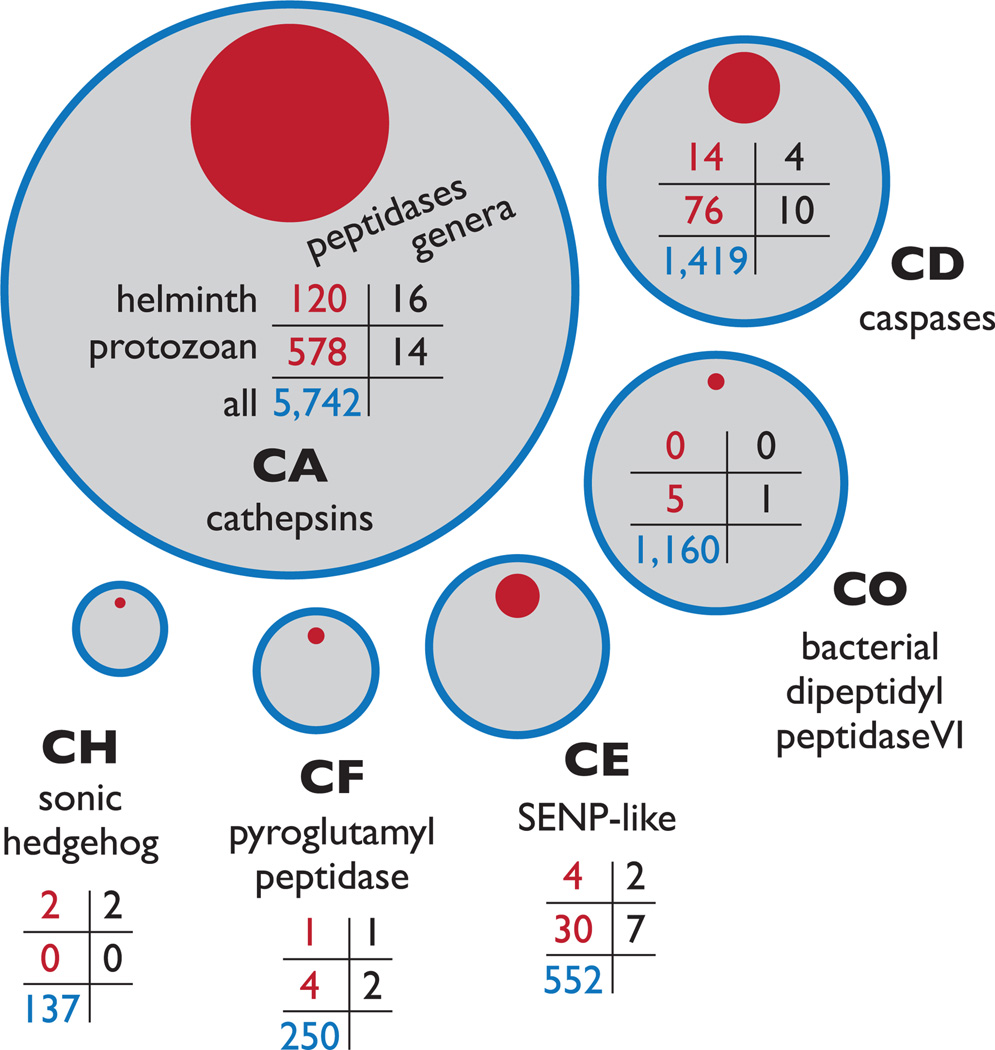
Figure 1.
Overview of the six cysteine peptidase clans containing parasite proteins Cysteine peptidases of parasitic organisms are found in six clans: CA, CD, CE, CH, CF and CO. The areas of the red circles are proportional to the populations of helminth and protozoan parasite peptidase sequences in MEROPS, whereas the grey circles are proportional to the total population of each clan. The tables indicate the number of peptidases in each clan associated with helminths, parasitic protozoa and associated genera; table cells are defined in the clan CA table. No relationship is implied by the spacing between the different clans. Examples of parasite peptidases are: CA: Fasciola hepatica cathepsin L1; CD: Trypanosoma cruzi metacaspase 2; CO: Trichomonas vaginalis PgpA peptidase; CE: SENP-like protein from Theileria parva; CF: Trypanosoma brucei pyroglutamyl peptidase 1; CH: hedgehog-like protein from Brugia malayi.
Most of the parasite cysteine peptidase population is contained within clan CA
Of the 24 families in clan CA, 12 families include peptidases from parasitic helminths and protozoa, and collectively these families encompass most of the known parasite cysteine peptidase sequences (84%; see Figure 1). The remaining families are primarily composed of viral and bacterial peptidases. However, that there has been much more progress in sequencing the genomes of protozoan parasites than for those of in helminths, and this bias affects the contents of the MEROPS database, which was used to calculate these statistics. The global relationships among these families can be displayed as a sequence similarity network (Box 2), illustrating species-specific representation (Figure 2b). Most of the clan CA families that contain proteins from parasites are associated with deubiquitinating activity (DUB) [10], and there is evidence of a ubiquitin-linked proteasome pathway in several parasites [11]. There is a notable paucity of literature on DUBs in parasites, although in Trypanosoma cruzi, the etiologic agent of Chagas disease, ubiquitin-dependent degradation of cytoskeletal proteins has been linked to the transition between morphologically dissimilar life-cycle stages [5]. Most DUBs are cysteine peptidases. Two major classes contain many members from protozoan parasites: (i) ubiquitin C-terminal hydrolases (family C12), which act mainly in the recycling of ubiquitin when it is erroneously conjugated to intracellular nucleophiles, and (ii) ubiquitin-specific proteases (family C19), which oppose and co-evolve with ubiquitin E3 ligases [10]. The network in Figure 2b shows that DUB-associated families include many members from parasites. Family C2, which is associated with the calcium-binding calpains, contains a Plasmodium falciparum peptidase that is required for cell-cycle progression [12], but like DUBs, this class is otherwise poorly studied in parasites.
Box 2. Using sequence similarity networks to view MEROPS peptidase classes.
Sequence similarity networks (SSN) allow very large groups of homologous proteins to be viewed together, clustered by similarity, as shown here with different classes of cysteine peptidase. An SSN is a graphical description of a group of proteins and is used here to display characteristics, such as species type, of large numbers of proteins so that the trends that are correlated with protein structural similarity are exposed [8]. Protein sequences are symbolized as nodes (e.g. circles or squares) connected by edges (lines) that represent pairwise sequence alignments the scores of which are better than a given threshold. In the displayed SSNs, proteins with similar amino acid sequences cluster together because the most similar subsets of proteins are connected and pulled together by many alignments better than the threshold; we report statistics (median percentage identity and alignment length) that report the quality of the least significant alignments included that are assigned the threshold score. (More specifically, this score is the BLAST E-value; see Glossary). Different degrees of sequence similarity can be emphasized by varying the threshold score, for example in Figure 2b, distant relationships are included, emphasizing family-level groupings. In Figure 2c, a subset of the sequences from Figure 2b are shown as an inset, but the threshold is more stringent; therefore sequences must be more similar to one another to share a cluster than in Figure 2b. In general, the shorter the edge connecting two proteins, the more similar the pair of proteins. Disconnected proteins and clusters might be related by detectable sequence similarity at levels below the selected threshold score. These disconnected proteins typically appear in rows at the bottom of an SSN-based figure, and their location relative to other groups is arbitrary, except where noted as in Figure 2c. (For more information on interpreting SSNs, see Ref.[8].)
The SSNs and peptidase statistics in this review are calculated from sets of proteins downloaded from the MEROPS database (http://merops.sanger.ac.uk) [9] on 6 February 2009 (Release 8.10). This database is a hand-curated repository of information about peptidases; the different classes of peptidase in the networks include only proteins that were in the database by that date. The snapshot of the parasite cysteine peptidase landscape that these networks provide will evolve as new genomes are sequenced and new peptidases are discovered. To create the networks, the sequence similarity between each pair of protein sequences classified within a MEROPS clan or family is calculated using the sequence alignment program BLAST [64]; all pairwise similarities better than the specified threshold are used to create a network, which is then visualized using a variant of a force-directed graph layout algorithm via Cytoscape [65].
Once groups of similar proteins are exposed using the network structure, the network can then be overlaid with additional information such as pathogen class (Figure 2b), substrate specificity (Figure 3a), and the nature of the catalytic residue (as calculated from sequence alignments to a statistical model of the family [66] in Figure 3c) to create customized atlases of protein families. The networks displayed in the figures can be searched, explored and used to extract and view sets of protein attributes tailored to specific interests using network visualization software such as Cytoscape and are available for download from http://babbittlab.compbio.ucsf.edu/resources/TIPs09.
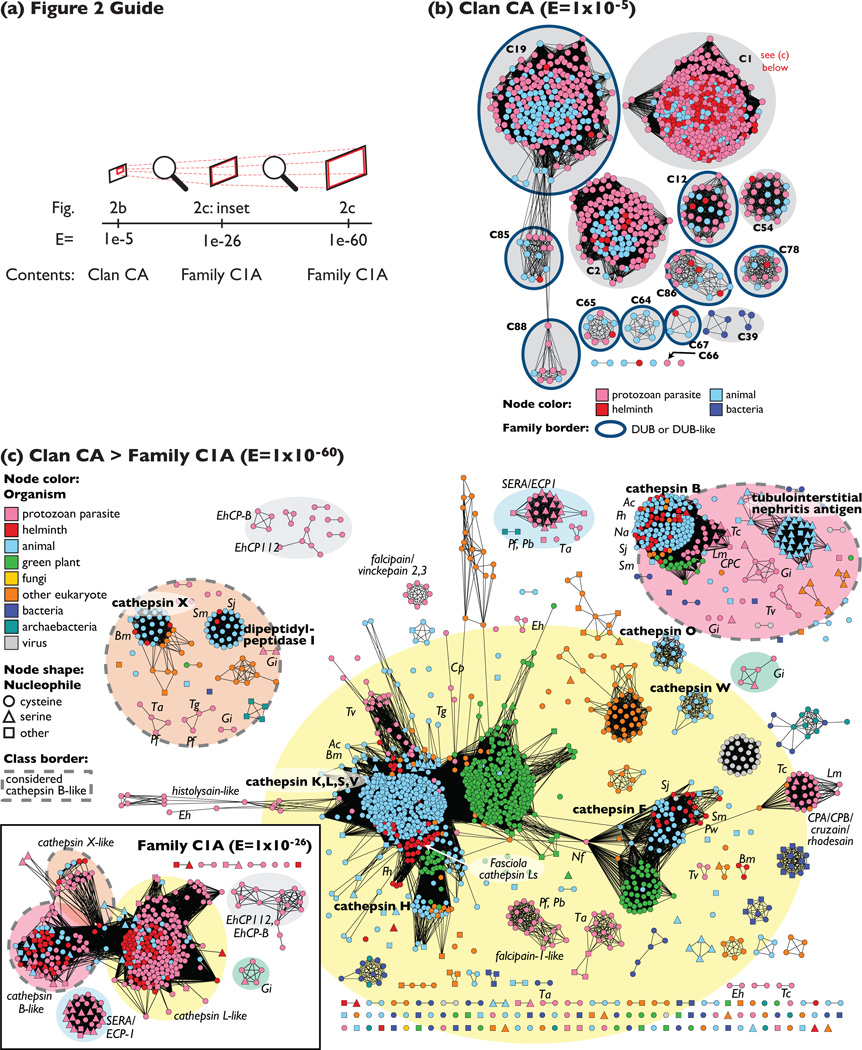
Figure 2.
A global map of clan CA places most parasite peptidases into family C1 (a) Guide to Figure 2(b) and (c) showing the relationship between the different networks. (b) All 698 parasite peptidase sequences from clan CA are displayed as a sequence similarity network, along with reference CA peptidases from Homo sapiens, Anopheles gambiae, Caenorhaobditis elegans, and Escherichia coli. The sequences cluster by sequence similarity; the least significant sequence similarities included have a BLAST E-value of 1 × 10−5, or a median of 27% identity over alignments of 110 amino acids. Sequences are colored according to type of organism. Each cluster corresponding to a MEROPS family is labeled and highlighted with a grey oval. If the family is associated with deubiquitinating (DUB) or DUB-like activity, the family is circled with a dark blue border. Family examples are: C19: ubiquitin-specific protease; C1: papain-like enzyme; C85: DUBA deubiquitinating enzyme; C2: calpain; C12: ubiquitin C-terminal hydrolase; C54: autophagin-1; C86: ataxin-3; C78: ubiquitin-fold modifier 1-specific protease 1 (UfSP1) peptidase; C88: ovarian tumor protease; C65: otubain-1; C64: Cezanne de-ubiquitinylating peptidase; C67: cylindromatosis-related deubiquitinating enzyme (CylD); C39: colicin V processing peptidase; C66: immunoglobulin G-degrading enzyme of Streptococcus pyogenes (IdeS)-like protein from T. vaginalis. CA families that do not include parasite or reference organism peptidases are not shown. (c) Family C1A (subset of family C1, member of Clan CA) peptidases are displayed as networks. The inset shows clustering of representatives at an intermediate threshold level of sequence similarity (E = 1 × 10−26), and colored ovals highlighting clusters and single proteins correspond between the inset and the main panel. The main panel includes C1A peptidases from all organisms in a sequence similarity network at the threshold of at E = 1 × 10−60 (50% identity over 220 amino acids; 1779 sequences). Sequence classes considered part of the ‘cathepsin-B-like’ group have a dashed grey border. Sequences are colored according to type of organism, and the shape of the node corresponds to the predicted residue at the nucleophile position for that peptidase. Bold labels indicate the human protein; italics indicate the general location of selected species and classes of parasite enzyme. Species abbreviations: Ac, Ancylostoma caninum; Bm, Brugia malayi; Cp, Cryptosporidium parvum; Eh, Entamoeba histolytica; Fh, Fasciola hepatica; Gi, Giardia intestinalis; Lm, Leishmania mexicana; Nf, Naegleria fowleri; Na, Necator americanus; Pw, Paragonimus westermani; Pf, Plasmodium falciparum; Pb, Plasmodium berghei; Sj, Schistosoma japonicum; Sm, Schistosoma mansoni; Ta, Theileria annulata; Tg, Toxoplasma gondii; Tv, Trichomonas vaginalis; Tc, Trypanosoma cruzi.
Proteins from family C1, found within clan CA, are the most-studied parasite cysteine peptidases and include enzymes that are bona fide targets for drug development, namely, hemoglobinolytic falcipains from Plasmodium [4,13] and cruzain from T. cruzi [1]. Perhaps because it is well known that these enzymes are associated with degradation of proteins for nutrition, parasite peptidases have acquired a reputation for being involved solely in catabolism. However, cysteine peptidases are known to participate in many aspects of the parasite life cycle, and as most parasite proteins, in family C1 and in other classes, have not been examined experimentally, it is likely that the true range of functional roles for peptidases will only broaden with time. Family C1 encompasses 80% of helminth CA peptidases and 45% of protozoan parasite CA peptidases and is large and diverse. At a very basic level, this family splits into two broad classes: a smaller class associated with bleomycin hydrolase activity, which contains no parasite proteins (family C1B in the MEROPS database), and a second much larger class typified by the catabolic human cathepsin proteins found in the lysosome (family C1A). Within this cathepsin-like group (shown in Figure 2c), proteins can be further divided into two complementary subclasses based on specific regions of sequence similarity around the catalytic residues: the cathepsin-L-like group and the cathepsin-B-like group [14]. As shown by the distribution of species in the network in Figure 2c, the division between cathepsin L- and B-like enzymes is quite ancient. Although little can be said about the functional implications of this distinction, peptidases are commonly classified as cathepsin-L- or cathepsin-B-like. Drug development targeting parasite cathepsin-L-like and cathepsin-B-like proteins has benefited enormously from existing tools and structures related to human proteins [15]; an example is human cathepsin K, which is successfully targeted in therapies for osteoporosis [16]. Some of these screening library and kinetic data have been released by pharmaceutical companies and are now freely available through the Collaborative Drug Discovery database [17] to be re-appropriated for use with parasite targets.
In protozoa, there is increasing evidence for the involvement of family C1 in evasion of the immune system in addition to nutrition. Cathepsin-L-like peptidases of the cysteine protease B-locus from Leishmania mexicana were shown to inhibit the host T helper cell type 1 response, and the corresponding increased cysteine peptidase activity in L. mexicana might explain some of the differences between leishmaniasis caused by L. mexicana and L. major [18]. In people with suppressed immune systems, a diagnosis of Chagas disease is particularly devastating, but an inhibitor targeting cruzain was shown to rescue immunodeficient mice infected by T. cruzi [19].
Family C1 is quite large in part because many members – particularly, in protozoa – are the result of gene duplication; some C1 peptidases are found in tandem arrays of similar isoforms (e.g., see Refs [20,21]) and occasionally located on different chromosomes [22]. The biological consequences of increased numbers of active enzymes have proven difficult to recognize or to explain [20], because gene expansions are not uniform among phylogenetic groups. Among the trypanosomatids, the T. brucei genome includes only two C1 peptidases (a cathepsin-L-like and a cathepsin-B-like peptidase) [23] (Figure S1 in the supplementary material online), one of which is necessary for the parasite to cross the blood–brain barrier of the host [24], whereas T. congolense has 13 cathepsin-B-like peptidases [25]. A third trypanosomatid, L. mexicana, expresses at least 19 cathepsin-L-like peptidases and a single cathepsin B [21]; as mentioned earlier, increased cysteine peptidase activity in comparison with L. major might be the cause of differing disease outcomes [18]. Differential gene expansion is also seen in family C2, in which L. major contains 22 calpain-like proteins [26], and P. falciparum contains only one [13].
Within the helminth parasites, gene expansion has led to new roles for the cathepsins, complementing the general catabolic role of cathepsins in protein turnover for nutrition. In Fasciola, the cathepsin Ls are secreted and allow the flukes to burrow through host intestinal and liver tissue by degrading matrix proteins such as fibronectin and collagen and also to counter the host immune system by cleaving CD4 proteins from T lymphocytes, a process that is thought to be responsible for the suppression of lymphocyte proliferation [3]. The cathepsin Ls of the lung fluke Paragonimus, which are derived from nine peptidase genes, are also used in tissue invasion and evasion of the immune system as well as in excystment [27]. In filarial nematodes, cathepsin L activity is important in developmental processes: for example, this cathepsin is required in cuticle molting of the L3 stage in Onchocerca volvulus [28] and in embryogenesis in Brugia malayi [29].
Clan CD
Clan CD proteins from protozoa and helminths have been investigated only during the past decade [30–33]. Clan CD has attracted the attention of parasitologists in part because this clan contains the enzyme that adds a glycosylphosphatidylinositol (GPI) lipid moiety to the plasma membrane of a plethora of parasitic organisms; a secondary modification has been linked to virulence in T. brucei [34]. Second, the role of metacaspases as regulators of the populations of certain parasitic protozoa has recently been bought to light [35]. Unlike degradative peptidases with relatively broad substrate specificity, members of clan CD are associated with tightly defined substrate specificities; characterized clan members are tuned to prefer a specific amino acid at the P1 position (see Figure 3a and Box 1). Although most parasite members of clan CD have poorly defined functions, a few are known to play important roles in interactions with the host and in cell division. Three clan CD families contain a significant number of parasite cysteine peptidases: the family including the GPI:protein transamidases and asparaginyl endopeptidases/legumains (C13); the caspase family (C14); and the separase family (C50).
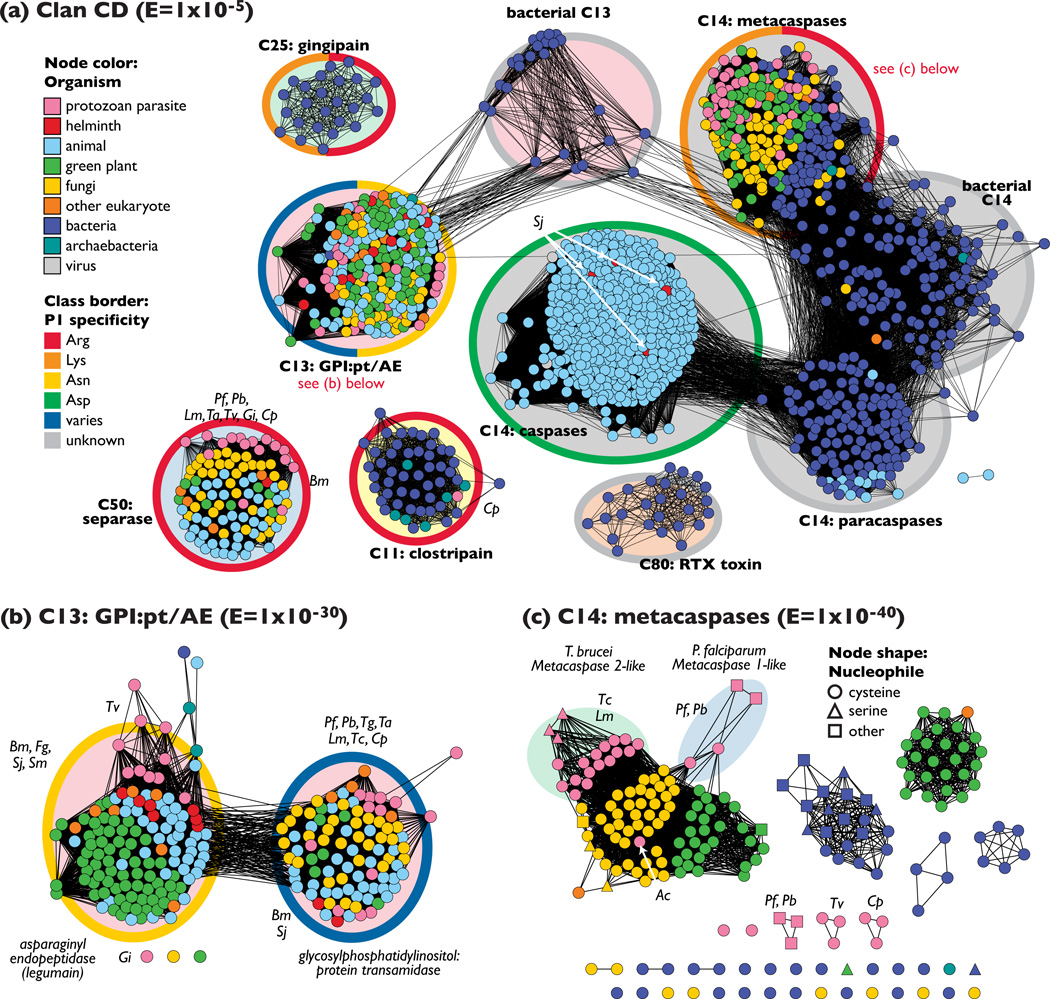
Figure 3.
Clan CD features an uneven distribution of peptidases from parasites (a) All 1419 peptidase sequences in clan CD are displayed as a sequence similarity network; the least significant similarities included have an E-value of 1 × 10−5 (27% identity over 130 amino acids). Sequences are colored according to type of organism, and the P1 substrate specificity is highlighted with thick class borders. Labels in bold indicate MEROPS family membership. Groups that are part of the same family have the same class background color. (b) Family C13 peptidases are displayed as a sequence similarity network at the threshold E-value of 1 × 10−30 (30% identity over 200 amino acids; 235 sequences) colored and labeled as in (a). (c) Metacaspases and similar proteins from family C14 displayed as a sequence similarity network at a threshold of E = 1 × 10−40 (37% identity over 250 amino acids; 198 sequences). The shape of the symbol denotes the predicted residue at the nucleophile position. Species abbreviations: Bm, Brugia malayi; Cp, Cryptosporidium parvum; Eh, Entamoeba histolytica; Fg, Fasciola gigantica; Gi, Giardia intestinalis; Lm, Leishmania mexicana; Pf, Plasmodium falciparum; Pb, Plasmodium berghei; Sj, Schistosoma japonicum; Sm, Schistosoma mansoni; Ta, Theileria annulata; Tg, Toxoplasma gondii; Tv, Trichomonas vaginalis; Tc, Trypanosoma cruzi. Sequences from many more species are present than are labeled here; interactive networks are available for download from http://babbittlab.compbio.ucsf.edu/resources/TIPs09.
Family C13 provides an interesting example that illustrates the need to view cysteine peptidases as enzymes that have evolved to catalyze a collection of different overall reactions, rather than being limited to protease (peptide hydrolase) activity. As shown by the network in Figure 3b, family C13 clearly separates into two groups that share about 30% sequence identity; these two groups correspond to two quite distinct enzyme activities. One activity entails the transfer of a nascent protein to a GPI anchor, which results in the reformation of a peptide bond between the protein and the anchor [30]. Many pathogens have surface-bound GPI-anchored proteins. These surface proteins are the first antigens to come into contact with the host immune system, and can trigger disease symptoms if recognized as foreign by the immune system. GPI-anchored proteins can also bind to host cell-surface receptors, allowing entry into the cells [6]; examples of these proteins include glycoprotein 63 (GP63) in Leishmania [36] and merozoite surface protein-1 in Plasmodium [37]. In addition, the C13 family includes a separate group of enzymes that catalyze asparaginyl endopeptidase reactions (also known as legumains). In Schistosoma mansoni, asparaginyl endopeptidases can proteolytically process and activate cathepsin-B-like gut peptidases in vitro [38]. As seen in Figure 3b, the known asparaginyl endopeptidases are phylogenetically diverse and are represented by all phylogenetic classes of organism except fungi and viruses.
The metacaspases are a subset of the caspase-associated family C14 that were originally identified using bioinformatics techniques in [31]. As shown in Figure 3a, these enzymes provide an alternative to the canonical caspases found in animals, and are expressed in parasitic protozoa, fungi, plants, and bacteria. Initial reports indicate specificity for arginine, lysine, or both amino acids at P1 rather than the aspartic acid specificity for which caspases are named [39,40]. The metacaspases are homologs of caspases, which mediate apoptosis in animals [41], and indeed, processes similar to programmed cell death have been observed in a number of parasitic protozoa. Although the occurrence of programmed cell death in single-celled organisms is somewhat counterintuitive, a possible rationale for selective suicide in parasites is the avoidance of levels of parasitemia that would kill the host and cause the death of all of the descendants of the founder of the infection [42]. However, whether metacaspases are involved in these processes is quite controversial [43]. Metacaspase-dependent apoptosis has been well characterized in yeast [44], but most published work investigating a link between programmed cell death and metacaspases expressed in kinetoplastids and apicoplexans has been inconclusive (e.g., T. brucei [45,46]; P. berghei [47]). Little is known about additional potential roles for these proteins in parasites, but the metacaspase from Leishmania major is considered essential for correct segregation of the nucleus and kinetoplast during cell division [35]. Helminths have no metacaspases, but four canonical caspase homologs from both the liver fluke Schistosoma japonicum and the blood fluke S. mansoni have been identified and presumably function in a similar way to caspases in humans [48].
Separase activity, provided by family C50, is responsible for the separation of the sister chromatids during eukaryotic cell division at the metaphase–anaphase transition [49]. This family of peptidases is predicted to occur in all eukaryotes and members have been identified in the helminth Brugia malayi and a number of protozoan parasites, such as the trypanosomatids and the Plasmodium separases. Although targeting separase activity should be detrimental to parasite replication and therefore inhibit survival, no parasite C50 enzymes have been characterized functionally or validated as drug targets.
Other classes of parasite cysteine peptidase are mostly uncharacterized
A search of the MEROPS peptidase database [9] affirms the presence of parasite CPs in four clans in addition to CA and CD (Figure 1). These peptidases, a few of which are mentioned here, are mostly unexamined and might have unusual roles in parasite life cycles. First, five Trichomonas vaginalis peptidases are present in the otherwise bacterial clan CO in MEROPS. Bacterial clan CO peptidases are associated with cell-wall disassembly via hydrolysis, although very distant relatives have been reported in eukaryotes that act as acyl transferases to the rhodopsin coenzyme retinal and form a subdomain of kinetoplastid glutathionylspermidine synthase [50].
Clan CE, composed of sentrin-specific protease (SENP)-like proteins that interact with the small ubiquitin-like modifier protein (SUMO) [51], contains proteins from B. malayi and S. japonicum, as well as from an array of protozoan parasites. SUMOylation might have a role in host-cell invasion and cyst genesis in Toxoplasma [52] and has also been found in numerous parasitic protozoa [5], including Plasmodium [53] and Trypanosoma [54].
Clan CF is known for pyroglutamyl peptidase activity, and the MEROPS clan includes a protein from B. malayi as well as from T. vaginalis and two trypanosomatids; this peptidase is also expressed in Leishmania [55]. This activity is used to remove the N-terminal pyroglutamyl residues that are thought to discourage degradation of neuropeptides by aminopeptidases. The expression of a pyroglutamyl peptidase from T. brucei might contribute to endocrine dysfunction in people with sleeping sickness [56].
Clan CH – associated with hedgehog protein and other oddly named animal developmental proteins – includes proteins from the helminths B. malayi and Trichinella spiralis. Hedgehog protein exhibits a unique variant of peptidase chemistry involving an essential cysteine residue; the peptidase domain of the protein undergoes a chemical rearrangement in a non-catalytic self-cleaving mechanism [57].
Functional divergence of cysteine peptidases in parasites
A global survey emphasizes the considerable functional breadth and diversity of parasite cysteine peptidases. Certain emerging classes of functional divergence, including loss of the canonical cysteine peptidase active site and the unexpected presence of parasite proteins in bacteria-associated peptidase classes are particularly pertinent in parasites.
By definition, a cysteine peptidase will require a catalytic cysteine residue for canonical activity. However, in a number of expressed cysteine peptidase-like enzymes, different amino acids replace the conventional catalytic residues. Some of these proteins still appear to be enzyme-like, and the active site might be catalytically active, as in the case of the cysteine nucleophile mutated to a serine (Box 1), whereas other proteins are not capable of catalyzing peptidase-type chemistry. In fact, of all 259 proteins from parasitic protozoa in family C1 (“cathepsin-like”), 16% do not appear to have the classic Cys-His catalytic dyad. 8% of family C1 helminths have substitutions at these positions (Box 2). (Humans have two C1 peptidases out of a total of 13 in which serines replace the cysteine nucleophile; these proteins appear to be involved in some cases of renal failure [58].) Family C1 classes containing these unusual proteins include the ECP1/SERA proteins from Plasmodium, which are key to host cell egress through an unknown mechanism [59], and isoforms of the T. congolense cathepsin-B-like peptidases and Theileria annulata cathepsin-L-like enzymes [2] (Figure 2c). In several metacaspases from clan CD, the catalytic cysteine has been substituted with a serine or threonine (e.g. T. brucei homologs of metacaspase 2) (see Figure 3c and Figure S2 in the supplementary material online at http://babbittlab.compbio.ucsf.edu/resources/TIPs09), but the prediction of these nucleophiles is complicated by the divergence of these proteins from a solved structure. Many of these cysteine-to-serine substitutions occur in parasites that reside in an oxygen-rich environment, such as blood, and could be a means to moderate oxidation of the essential catalytic cysteine, although this remains to be shown experimentally. The functional consequences of these amino acid replacements are unknown. Because only a small number of enzymes from these diverse families have been characterized in depth, the relationship between the mechanistic differences that these replacements might represent and their biological roles in the parasites in which they are found remains a mystery.
When there is a nonfunctional active site, cysteine peptidase homologs have been described that have regulatory roles, for example human c-FLIPL contains a catalytically inactive caspase-like domain (from clan CD), and is thought to form a heterodimer with procaspase-8, resulting in allosteric activation of procaspase-8 and leading to apoptotic signaling [60]. Suggested roles for some apparently inactivated cysteine peptidases of the scabies mite Sarcoptes scabiei include competitive inhibition of the host blood coagulation cascade and antagonization of cleavage of protease-activated receptors to reduce the inflammation response [61].
For a number of parasite cysteine peptidases, it is possible that horizontal gene transfer explains why a class of cysteine peptidases previously associated solely with a different kingdom of organisms would include proteins from parasites. However, horizontal gene transfer (in which genes are transferred through an asexual interaction between different species) is notoriously difficult to establish, and there are few examples in eukaryotes (for example, see Ref. [62]). Alternatively, these instances may be a symptom of biases and sampling issues in sequencing projects and experimental characterization. As mentioned earlier, there are five Trichomonas clan CO peptidases; all other members of that clan are found in bacteria (or extremely distantly related as reported in Ref. [50]). Additional protozoan peptidases found in otherwise bacterial and archaebacterial groups are the Cryptosporidium peptidase in the clostripains (clan CD, family C11), and the T. vaginalis protein in clan CA, family C66; this family had previously been associated exclusively with bacteria that use member peptidases to evade the human immune system through cleavage of immunoglobulin G [63].
Concluding remarks
This is an opportune time to investigate the range of cysteine peptidases and their activities in pathogenic parasites. These enzymes are crucial to many aspects of the parasite life cycle, and databases such as MEROPS and the recently released S. mansoni genome [48] provide convenient lists of new, unexamined parasite peptidases the characterization of which may lead to the development of novel therapies. It is clear that most of our knowledge of parasite cysteine peptidases is confined to a small subset of highly studied peptidases from specific organisms, whereas the current landscape of parasite CPs is crowded and steadily growing, leaving the field with many open questions (see Box 3). Sequence similarity networks provide one way to navigate this landscape because they confirm and extend existing knowledge of cysteine peptidase classes. With an eye toward the development of new therapies, it is important to move beyond the identification of molecular function and associate parasite peptidases with their physiological roles, and to use a broader perspective in considering where existing techniques can be adapted and brought to bear on unexamined proteins. Resources such as MEROPS database and the sequence-based atlases of parasite cysteine peptidases presented here can be used as a starting point. A better understanding of how the many parasite cysteine parasites contribute to different parasite life cycles could help to mitigate the public health burden of parasitic diseases.
Box 3. Open questions.
Why do parasites have so many different types of de-ubiquitinating enzymes and calpains?
What are the functional implications of the differences in the numbers of isoforms of many clan CA peptidases in parasites?
What is the relationship between programmed cell death in animals and similar processes in protozoan parasites? Are metacaspases involved in these processes?
What are the physiological roles of parasite cysteine peptidases with absent or non-standard catalytic residues? Do any of these proteins still mediate catalysis?
Is horizontal gene transfer responsible for the collection of parasite cysteine peptidases found in classes previously associated with other phylogenetic groups? How do these proteins participate in the parasite life cycle?
Are there any undiscovered parasite cysteine peptidases with novel structural folds?
Supplementary Material
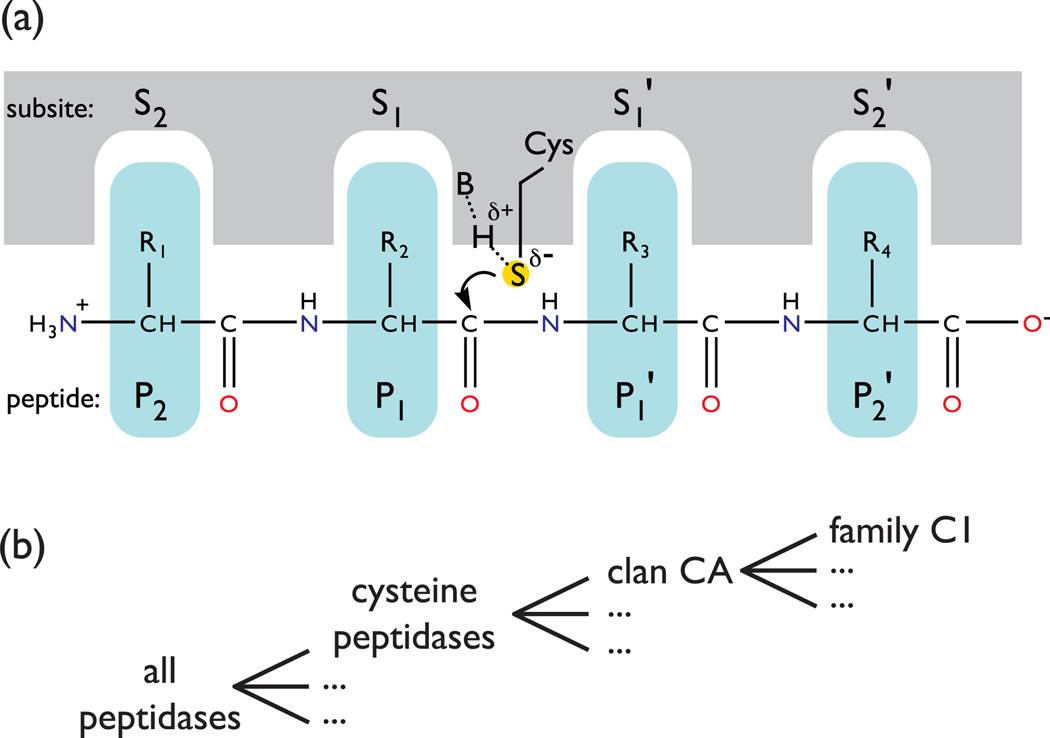
Figure I.
Peptidase conventions and taxonomy (a) The binding of a peptide substrate to the peptidase subsites (active site pockets) positions the sulfur of cysteine near the carbonyl of the peptide bond to be cleaved. The sulfur of the active site cysteine is particularly nucleophilic due to the presence of an indispensable chemical base (e.g. histidine). The peptide amino acid residues and subsites are numbered and designated prime or non-prime based on their position relative to the targeted peptide bond [67]. Key: S, subsite; P, peptide amino acid residue; R, amino acid side chain; S (within a yellow circle) is the catalytic sulfur of cysteine; B, catalytic base. (b) In the hierarchical taxonomy defined by the MEROPS database [9], all peptidases are assigned to families constituting all groups of peptidases that share statistically significant sequence similarity; families are then grouped into the larger clans that each has a common ancestral progenitor. Members of different clans are not homologous (i.e., they are not thought to be evolutionarily related).
Acknowledgements
Tools used for visualization of sequence similarity networks were created in part by the UCSF Resource for Biocomputing, Visualization, and Informatics, supported by NIH P41 RR-01081. Support for this work was provided in part by NIH GM60595 (P.C.B.) and the Sandler Foundation (M.S.). In addition, we thank Conor R. Caffrey and James H. McKerrow of the Sandler Center for Basic Research in Parasitic Diseases, UCSF, for their helpful feedback on this manuscript.
Glossary of terms
- BLAST E-value (E)
a statistic describing the significance of the scored protein sequence similarity between two sequences aligned using the sequence comparison program BLAST [64]. An expectation value (E-value) is the number of different alignments expected to have an equal or better score in the current database of sequences. Smaller E-values correspond to better alignments.
- Clan
a group of MEROPS families that are thought to share a common ancestor, based on evidence from structural similarity or the same ordering of active site residues within the protein sequences [9].
- DUB
deubiquitinating peptidase, i.e. a peptidase with isopeptidases activity that can catalytically remove the ubiquitin covalently attached to specific lysine side chain.
- E3 ligase
an enzyme catalyzing the final step of a three-step process that serves to activate and transfer ubiquitin (or ubiquitin-like moleules) to a protein destined for a various intracellular targets, the most studies of which is the proteasome.
- Family
a group of MEROPS peptidase species with sequences that are related at a statistically significant level of similarity [9].
- Peptidase species
in the MEROPS database, a group of peptidases, each of which is homologous to a sequenced and biochemically characterized type example peptidase [9].
- MEROPS
a curated database that classifies all known peptidases, and the data source for all figures and peptidase statistics in this review (http://merops.sanger.ac.uk) [9]. MEROPS also includes information on peptidase inhibitors, substrates, specificity and extensive summaries and links to the literature.
- Nucleophile
in this review, used as shorthand for the amino acid providing the functional group that makes an initial attack on the carbonyl carbon of a peptide bond. In canonical cysteine peptidases, the nucleophile is a cysteine.
References
- 1.McKerrow JH, et al. Proteases in parasitic diseases. Annu. Rev. Pathol. 2006;1:497–536. doi: 10.1146/annurev.pathol.1.110304.100151. [DOI] [PubMed] [Google Scholar]
- 2.Sajid M, et al. Proteases of parasitic protozoa - current status and validation. In: Selzer P, editor. Drug Discovery in Infectious Diseases: From Drug Targets to Drug Candidates. Wiley-VCH Verlag; 2009. pp. 177–208. [Google Scholar]
- 3.Dixit AK, et al. Immunodiagnostic/protective role of cathepsin L cysteine proteinases secreted by Fasciola species. Vet. Parasitol. 2008;154:177–184. doi: 10.1016/j.vetpar.2008.03.017. [DOI] [PubMed] [Google Scholar]
- 4.Goldberg DE. Hemoglobin degradation. Curr. Top. Microbiol. Immunol. 2005;295:275–291. doi: 10.1007/3-540-29088-5_11. [DOI] [PubMed] [Google Scholar]
- 5.Ponder EL, Bogyo M. Ubiquitin-like modifiers and their deconjugating enzymes in medically important parasitic protozoa. Eukaryotic Cell. 2007;6:1943–1952. doi: 10.1128/EC.00282-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zacks MA, Garg N. Recent developments in the molecular, biochemical and functional characterization of GPI8 and the GPI-anchoring mechanism. Mol. Membr. Biol. 2006;23:209–225. doi: 10.1080/09687860600601494. [DOI] [PubMed] [Google Scholar]
- 7.Sajid M, McKerrow JH. Cysteine proteases of parasitic organisms. Mol. Biochem. Parasitol. 2002;120:1–21. doi: 10.1016/s0166-6851(01)00438-8. [DOI] [PubMed] [Google Scholar]
- 8.Atkinson HJ, et al. Using sequence similarity networks for visualization of relationships across diverse protein superfamilies. PLoS ONE. 2009;4 doi: 10.1371/journal.pone.0004345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rawlings ND, et al. MEROPS: the peptidase database. Nucleic Acids Res. 2008;36:D320–D325. doi: 10.1093/nar/gkm954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nijman SM, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 11.Paugam A, et al. Characterization and role of protozoan parasite proteasomes. Trends Parasitol. 2003;19:55–59. doi: 10.1016/s1471-4922(02)00064-8. [DOI] [PubMed] [Google Scholar]
- 12.Russo I, et al. A calpain unique to alveolates is essential in Plasmodium falciparum and its knockdown reveals an involvement in pre-S-phase development. Proc. Natl. Acad. Sci. USA. 2009;106:1554–1559. doi: 10.1073/pnas.0806926106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenthal PJ. Cysteine proteases of malaria parasites. Int. J. Parasitol. 2004;34:1489–1499. doi: 10.1016/j.ijpara.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 14.Karrer KM, et al. Two distinct gene subfamilies within the family of cysteine protease genes. Proc. Natl. Acad. Sci. USA. 1993;90:3063–3067. doi: 10.1073/pnas.90.7.3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Renslo AR, McKerrow JH. Drug discovery and development for neglected parasitic diseases. Nat. Chem. Biol. 2006;2:701–710. doi: 10.1038/nchembio837. [DOI] [PubMed] [Google Scholar]
- 16.Stoch SA, Wagner JA. Cathepsin K inhibitors: a novel target for osteoporosis therapy. Clin. Pharmacol. Ther. 2008;83:172–176. doi: 10.1038/sj.clpt.6100450. [DOI] [PubMed] [Google Scholar]
- 17.Hohman M, et al. Novel web-based tools combining chemistry informatics, biology and social networks for drug discovery. Drug Discov. Today. 2009;14:261–270. doi: 10.1016/j.drudis.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 18.Buxbaum LU, et al. Cysteine protease B of Leishmania mexicana inhibits host Th1 responses and protective immunity. J. Immunol. 2003;171:3711–3717. doi: 10.4049/jimmunol.171.7.3711. [DOI] [PubMed] [Google Scholar]
- 19.Doyle PS, et al. A cysteine protease inhibitor cures Chagas' disease in an immunodeficient-mouse model of infection. Antimicrob. Agents Chemother. 2007;51:3932–3939. doi: 10.1128/AAC.00436-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.El-Sayed NM, et al. Comparative genomics of trypanosomatid parasitic protozoa. Science. 2005;309:404–409. doi: 10.1126/science.1112181. [DOI] [PubMed] [Google Scholar]
- 21.Mottram JC, et al. The multiple cpb cysteine proteinase genes of Leishmania mexicana encode isoenzymes that differ in their stage regulation and substrate preferences. J. Biol. Chem. 1997;272:14285–14293. doi: 10.1074/jbc.272.22.14285. [DOI] [PubMed] [Google Scholar]
- 22.Miller SK, et al. A subset of Plasmodium falciparum SERA genes are expressed and appear to play an important role in the erythrocytic cycle. J. Biol. Chem. 2002;277:47524–47532. doi: 10.1074/jbc.M206974200. [DOI] [PubMed] [Google Scholar]
- 23.Caffrey CR, Steverding D. Kinetoplastid papain-like cysteine peptidases. Mol. Biochem. Parasitol. 2009;167:12–19. doi: 10.1016/j.molbiopara.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 24.Nikolskaia OV, et al. Blood-brain barrier traversal by African trypanosomes requires calcium signaling induced by parasite cysteine protease. J. Clin. Invest. 2006;116:2739–2747. doi: 10.1172/JCI27798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mendoza-Palomares C, et al. Molecular and biochemical characterization of a cathepsin B-like protease family unique to Trypanosoma congolense . Eukaryot. Cell. 2008;7:684–697. doi: 10.1128/EC.00405-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Besteiro S, et al. Protein turnover and differentiation in Leishmania . Int. J. Parasitol. 2007;37:1063–1075. doi: 10.1016/j.ijpara.2007.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chung YB, et al. A 27 kDa cysteine protease secreted by newly excysted Paragonimus westermani metacercariae induces superoxide anion production and degranulation of human eosinophils. Korean J. Parasitol. 2008;46:95–99. doi: 10.3347/kjp.2008.46.2.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Craig H, et al. Unravelling the moulting degradome: new opportunities for chemotherapy? Trends Parasitol. 2007;23:248–253. doi: 10.1016/j.pt.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 29.Ford L, et al. Functional Analysis of the Cathepsin-Like Cysteine Protease Genes in Adult Brugia malayi Using RNA Interference. PLoS Negl. Trop. Dis. 2009;3 doi: 10.1371/journal.pntd.0000377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mottram JC, et al. Clan CD cysteine peptidases of parasitic protozoa. Trends Parasitol. 2003;19:182–187. doi: 10.1016/s1471-4922(03)00038-2. [DOI] [PubMed] [Google Scholar]
- 31.Uren AG, et al. Identification of paracaspases and metacaspases: two ancient families of caspase-like proteins, one of which plays a key role in MALT lymphoma. Mol. Cell. 2000;6:961–967. doi: 10.1016/s1097-2765(00)00094-0. [DOI] [PubMed] [Google Scholar]
- 32.Caffrey CR, et al. Blood 'n' guts: an update on schistosome digestive peptidases. Trends Parasitol. 2004;20:241–248. doi: 10.1016/j.pt.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 33.Götz MG, et al. Aza-peptidyl Michael acceptors. A new class of potent and selective inhibitors of asparaginyl endopeptidases (legumains) from evolutionarily diverse pathogens. J. Med. Chem. 2008;51:2816–2832. doi: 10.1021/jm701311r. [DOI] [PubMed] [Google Scholar]
- 34.Lillico S, et al. Essential roles for GPI-anchored proteins in African trypanosomes revealed using mutants deficient in GPI8. Mol. Biol. Cell. 2003;14:1182–1194. doi: 10.1091/mbc.E02-03-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ambit A, et al. An essential role for the Leishmania major metacaspase in cell cycle progression. Cell Death Differ. 2008;15:113–122. doi: 10.1038/sj.cdd.4402232. [DOI] [PubMed] [Google Scholar]
- 36.Tuon FF, et al. Toll-like receptors and leishmaniasis. Infect. Immun. 2008;76:866–872. doi: 10.1128/IAI.01090-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plattner F, Soldati-Favre D. Hijacking of host cellular functions by the Apicomplexa. Annu. Rev. Microbiol. 2008;62:471–487. doi: 10.1146/annurev.micro.62.081307.162802. [DOI] [PubMed] [Google Scholar]
- 38.Sajid M, et al. Functional expression and characterization of Schistosoma mansoni cathepsin B and its trans-activation by an endogenous asparaginyl endopeptidase. Mol. Biochem. Parasitol. 2003;131:65–75. doi: 10.1016/s0166-6851(03)00194-4. [DOI] [PubMed] [Google Scholar]
- 39.Watanabe N, Lam E. Two Arabidopsis metacaspases AtMCP1b and AtMCP2b are arginine/lysine-specific cysteine proteases and activate apoptosis-like cell death in yeast. J. Biol. Chem. 2005;280:14691–14699. doi: 10.1074/jbc.M413527200. [DOI] [PubMed] [Google Scholar]
- 40.González IJ, et al. Leishmania major metacaspase can replace yeast metacaspase in programmed cell death and has arginine-specific cysteine peptidase activity. Int. J. Parasitol. 2007;37:161–172. doi: 10.1016/j.ijpara.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 41.Salvesen GS, Riedl SJ. Caspase mechanisms. Adv. Exp. Med. Biol. 2008;615:13–23. doi: 10.1007/978-1-4020-6554-5_2. [DOI] [PubMed] [Google Scholar]
- 42.Bruchhaus I, et al. Protozoan parasites: programmed cell death as a mechanism of parasitism. Trends Parasitol. 2007;23:376–383. doi: 10.1016/j.pt.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 43.Vercammen D, et al. Are metacaspases caspases? J. Cell Biol. 2007;179:375–380. doi: 10.1083/jcb.200705193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mazzoni C, Falcone C. Caspase-dependent apoptosis in yeast. Biochim. Biophys. Acta. 2008;1783:1320–1327. doi: 10.1016/j.bbamcr.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 45.Helms MJ, et al. Bloodstream form Trypanosoma brucei depend upon multiple metacaspases associated with RAB11-positive endosomes. J. Cell Sci. 2006;119:1105–1117. doi: 10.1242/jcs.02809. [DOI] [PubMed] [Google Scholar]
- 46.Moss CX, et al. Metacaspase 2 of Trypanosoma brucei is a calcium-dependent cysteine peptidase active without processing. FEBS Lett. 2007;581:5635–5639. doi: 10.1016/j.febslet.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 47.Le Chat L, et al. The role of metacaspase 1 in Plasmodium berghei development and apoptosis. Mol. Biochem. Parasitol. 2007;153:41–47. doi: 10.1016/j.molbiopara.2007.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berriman M, et al. The genome of the blood fluke Schistosoma mansoni . Nature. 2009;460:352–358. doi: 10.1038/nature08160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gregan J, et al. Solving the shugoshin puzzle. Trends Genet. 2008;24:205–207. doi: 10.1016/j.tig.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anantharaman V, Aravind L. Evolutionary history, structural features and biochemical diversity of the NlpC/P60 superfamily of enzymes. Genome Biol. 2003;4 doi: 10.1186/gb-2003-4-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Drag M, Salvesen GS. DeSUMOylating enzymes – SENPs. IUBMB Life. 2008;60:734–742. doi: 10.1002/iub.113. [DOI] [PubMed] [Google Scholar]
- 52.Braun L, et al. The small ubiquitin-like modifier (SUMO)-conjugating system of Toxoplasma gondii . Int. J. Parasitol. 2009;39:81–90. doi: 10.1016/j.ijpara.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 53.Issar N, et al. Identification of a novel post-translational modification in Plasmodium falciparum: protein sumoylation in different cellular compartments. Cell Microbiol. 2008;10:1999–2011. doi: 10.1111/j.1462-5822.2008.01183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shang Q, et al. Solution structure of SUMO from Trypanosoma brucei and its interaction with Ubc9. Proteins. 2009;76:266–269. doi: 10.1002/prot.22409. [DOI] [PubMed] [Google Scholar]
- 55.Schaeffer M, et al. Differentiation of Leishmania major is impaired by over-expression of pyroglutamyl peptidase. I. Mol. Biochem. Parasitol. 2006;150:318–329. doi: 10.1016/j.molbiopara.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 56.Morty RE, et al. Pyroglutamyl peptidase type I from Trypanosoma brucei: a new virulence factor from African trypanosomes that de-blocks regulatory peptides in the plasma of infected hosts. Biochem. J. 2006;394:635–645. doi: 10.1042/BJ20051593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Varjosalo M, Taipale J. Hedgehog: functions and mechanisms. Genes Dev. 2008;22:2454–2472. doi: 10.1101/gad.1693608. [DOI] [PubMed] [Google Scholar]
- 58.Charonis A, et al. Basement membrane peptides: functional considerations and biomedical applications in autoimmunity. Curr. Med. Chem. 2005;12:1495–1502. doi: 10.2174/0929867054039071. [DOI] [PubMed] [Google Scholar]
- 59.Blackman MJ. Malarial proteases and host cell egress: an 'emerging' cascade. Cell Microbiol. 2008;10:1925–1934. doi: 10.1111/j.1462-5822.2008.01176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu JW, Shi Y. FLIP and the death effector domain family. Oncogene. 2008;27:6216–6227. doi: 10.1038/onc.2008.299. [DOI] [PubMed] [Google Scholar]
- 61.Holt DC, et al. A multigene family of inactivated cysteine proteases in Sarcoptes scabiei . J. Invest. Dermatol. 2004;123:240–241. doi: 10.1111/j.0022-202X.2004.22716.x. [DOI] [PubMed] [Google Scholar]
- 62.Sanders IR. Rapid disease emergence through horizontal gene transfer between eukaryotes. Trends. Ecol. Evol. (Amst.) 2006;21:656–658. doi: 10.1016/j.tree.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 63.von Pawel-Rammingen U, et al. IdeS, a novel streptococcal cysteine proteinase with unique specificity for immunoglobulin. G. EMBO J. 2002;21:1607–1615. doi: 10.1093/emboj/21.7.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bateman A, et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schechter I, Berger A. On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.