Abstract
The recent advancement of high-throughput genome sequencing technologies has resulted in a considerable increase in demands for large-scale genome annotation. While annotation is a crucial step for downstream data analyses and experimental studies, this process requires substantial expertise and knowledge of bioinformatics. Here we present MEGANTE, a web-based annotation system that makes plant genome annotation easy for researchers unfamiliar with bioinformatics. Without any complicated configuration, users can perform genomic sequence annotations simply by uploading a sequence and selecting the species to query. MEGANTE automatically runs several analysis programs and integrates the results to select the appropriate consensus exon–intron structures and to predict open reading frames (ORFs) at each locus. Functional annotation, including a similarity search against known proteins and a functional domain search, are also performed for the predicted ORFs. The resultant annotation information is visualized with a widely used genome browser, GBrowse. For ease of analysis, the results can be downloaded in Microsoft Excel format. All of the query sequences and annotation results are stored on the server side so that users can access their own data from virtually anywhere on the web. The current release of MEGANTE targets 24 plant species from the Brassicaceae, Fabaceae, Musaceae, Poaceae, Salicaceae, Solanaceae, Rosaceae and Vitaceae families, and it allows users to submit a sequence up to 10 Mb in length and to save up to 100 sequences with the annotation information on the server. The MEGANTE web service is available at https://megante.dna.affrc.go.jp/.
Keywords: Gene prediction, Plant genome annotation, Web service
Introduction
With the advent of high-throughput sequencing technologies, plant genome sequencing has been accelerated, and the data are being utilized for crop improvement (Bevan and Uauy 2013). The accumulation of the large amount of plant genome sequences led to constructions of comparative genomics databases (Mihara et al. 2010, Nagamura et al. 2011, Rouard et al. 2011, Goodstein et al. 2012) and development of a plant-specific controlled vocabulary for effective data integration (Cooper et al. 2013). However, the costs of the data management and analyses are increasing because of the need for high-spec computers, huge amounts of data storage and expertise in both computer science and molecular biology. In these data analyses, genome annotation is one of the most fundamental and indispensable steps (Yandell and Ence 2012), directly affecting further studies such as molecular evolutionary analyses, transposon tagging and microarray experiments. The annotation procedures require a higher level of bioinformatics skill, as several analysis programs must be conducted followed by the integration of the results to predict gene structures and assign gene functions. Thus, an easy-to-use annotation platform, which does not require any expertise in bioinformatics, would be essential for researchers to perform genome annotation and to visualize the results on a graphical viewer to interpret the annotation.
Currently, several types of analysis tools are available online for plant genome annotation. For example, online versions of ab initio gene prediction programs, such as AUGUSTUS (Stanke and Waack 2003), Fgenesh (Salamov and Solovyev 2000) and GeneMark.hmm (Lukashin and Borodovsky 1998), can be used to find open reading frames (ORFs) from genomic sequences. FPGP (Amano et al. 2010) aligns full-length cDNA (FLcDNA) sequences of dicot and monocot plants to a query sequence. Gramene (Youens-Clark et al. 2011) and PlantGDB (Duvick et al. 2008) provide a web service for a similarity search against plant nucleotide or protein databases. For graphical representation of the analysis results, WebGBrowse (Podicheti et al. 2009) is a good candidate. Although such web services are useful for genome annotation, it is time-consuming for researchers to access multiple web sites and interpret their results one by one. Moreover, such an annotation procedure is difficult for non-bioinformaticians to select the appropriate tools and parameter sets for the input sequences. Therefore, an integrated analysis tool to execute a series of analysis programs automatically is required to support genome analyses such as positional cloning of plant genes (Chen et al. 2009, Xu et al. 2011).
Several web-based annotation pipelines are available for plant genome sequences. Some of them are designed for specific plant genome annotation; RiceGAAS (Sakata et al. 2002) is for rice and TriAnnot (Leroy et al. 2012) is for wheat. There are also more versatile genome annotation tools that can be adapted not only for plants but also for other species. DNA subway (Goff et al. 2011) provides parameter sets for both animals and plants. MAKER (Cantarel et al. 2008) has a highly configurable web interface to select reference databases and parameters for analysis programs. However, there are few plant species that are supported in the existing annotation pipelines.
Here we describe a new plant genome annotation web service called MEGANTE that runs several analysis programs against query sequences, integrates the results and visualizes the annotation information on a genome browser. Compared with the existing tools, one of the notable features of MEGANTE is its simple interface, which is easy to use, even for non-experts. In addition, the service targets a wide variety of plant species and is able to accept large query sequences up to 10 Mb in length.
Results
Features of the MEGANTE web service
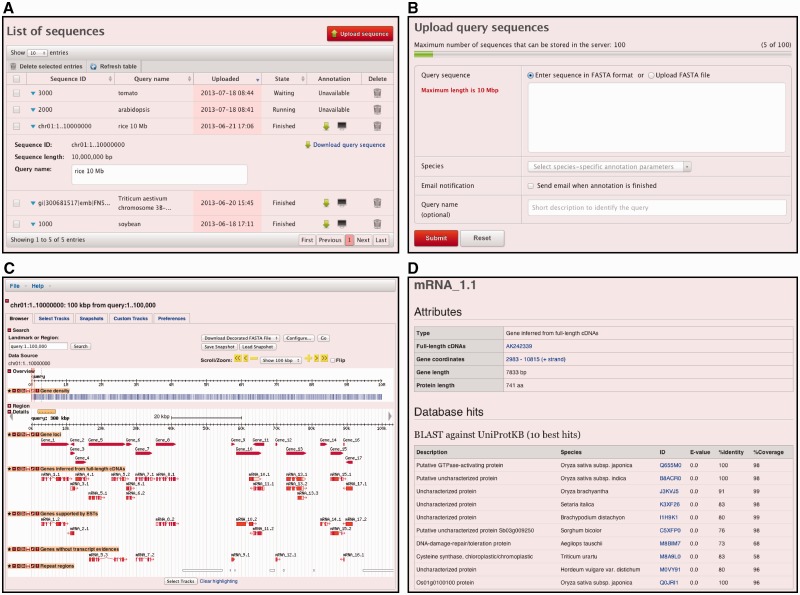
At the time of first use, MEGANTE requires an e-mail address and password to create an account. MEGANTE stores all data including query sequences submitted by users and the analysis results on the server side, and stores the data until users explicitly remove them via a web interface (Fig. 1A). Several analysis programs are automatically conducted in the system, but users do not need to specify any parameters or reference databases for the annotation process. Users can start annotation simply by copying and pasting a genomic sequence and then selecting the species of the query from a drop-down list (Fig. 1B). Currently, the service supports 24 species from the eight plant families shown in Table 1. Multiple sequences in FASTA format are acceptable. The length of each query sequence is limited to 10 Mb, and users can save up to 100 sequences in the server. These limitations are due to our current hardware resources that are available for this service.
Fig. 1.
Screenshots of the MEGANTE web interface. (A) Uploaded queries are listed. The list allows users to see the statuses of annotation jobs, download analysis results and jump to an annotation viewer. Clicking the sequence ID shows or hides detailed information about the query sequence. (B) Users can submit query sequences through this interface. (C) Annotation viewer with GBrowse. Users can select which data tracks to show or hide on the annotation map with the ‘Select Tracks’ tab. (D) Detailed annotation information of the predicted genes linked from the data tracks in GBrowse. ORF and amino acid sequences are also shown on this page.
Table 1.
Species supported in MEGANTE
| Families | Species |
|---|---|
| Brassicaceae | Arabidopsis thaliana |
| Brassica napus | |
| Brassica rapa | |
| Raphanus sativus | |
| Fabaceae | Glycine max |
| Lotus japonicus | |
| Medicago truncatula | |
| Vigna unguiculata | |
| Musaceae | Musa acuminata |
| Poaceae | Brachypodium distachyon |
| Hordeum vulgare | |
| Oryza sativa | |
| Phyllostachys edulis | |
| Sorghum bicolor | |
| Triticum aestivum | |
| Zea mays | |
| Salicaceae | Populus trichocarpa |
| Solanaceae | Nicotiana tabacum |
| Solanum lycopersicum | |
| Solanum melongena | |
| Solanum tuberosum | |
| Rosaceae | Malus × domestica |
| Prunus persica | |
| Vitaceae | Vitis vinifera |
The uploaded sequences are first queued, and then the queries are processed on an application server in a round-robin fashion to schedule the processes fairly. In the current system, five annotation processes can run in parallel. The whole annotation process can be completed within approximately 150 min for a 1 Mb sequence and approximately 15 h for a 10 Mb sequence. After finishing the annotation process, the following results are reported: repeat elements; alignments of transcript and protein sequences; predicted gene structures; similarities to known proteins; functional domains; and Gene Ontology (GO) terms (Ashburner et al. 2000). All the results are visualized with a widely used genome browser, GBrowse (Stein et al. 2002), which is integrated into the system. Furthermore, the system archives the annotation results in a single ZIP file for download. The file contains the annotation information in both Microsoft Excel and GFF3 (http://www.sequenceontoloty.org/gff3.html) formats. If users select an option for e-mail notification when submitting a query, an e-mail is sent to notify the users upon completion of the annotation. The data transfer between web browsers and the server is protected by SSL encryption.
Reference databases used in the system and pre-processing for annotation
MEGANTE uses several reference databases for the genome annotation, which include FLcDNAs and expressed sequence tags (ESTs) obtained from INSDC (Nakamura et al. 2013), protein sequences from Swiss-Prot and the TrEMBL plant division of UniProtKB (Magrane and UniProt Consortium 2011), and a protein family and domain database, InterPro (Hunter et al. 2011). We update the databases on a regular basis. Up-to-date details of the databases, such as the number of sequences, are described on the MEGANTE web site. After retrieving FLcDNAs and ESTs for each species listed in Table 1, we run a SeqClean script (http://sourceforge.net/projects/seqclean/) to remove poly(A) tails, vectors, low complexities and short sequences from the transcripts.
Annotation workflow
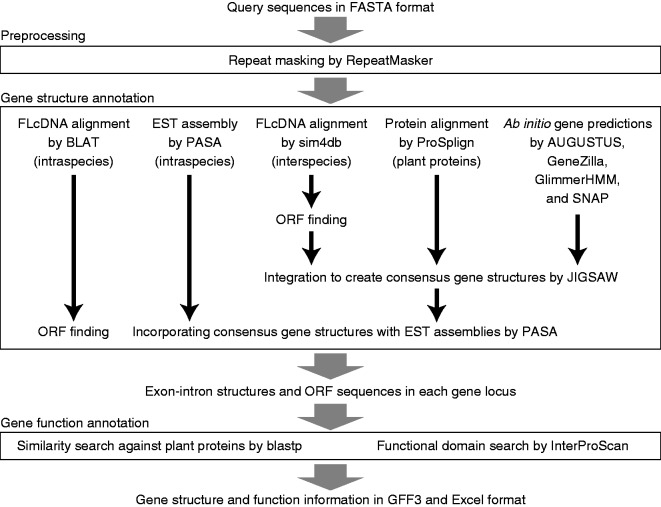
The overall annotation workflow is shown in Fig. 2. The annotation process begins with filtering out repeat elements detected by RepeatMasker (http://repeatmasker.org) with the MIPS Repeat Element Database (Nussbaumer et al. 2013). Next, to predict the exon–intron structures, the system aligns intraspecies FLcDNAs to a query sequence using BLAT (Kent 2002) with a cut-off of ≥98% identity and coverage. Although intraspecies FLcDNAs are effective for accurate gene prediction, in many cases the number of the sequences is not sufficient to cover entire genes. For this reason, we also use AUGUSTUS (Stanke and Waack 2003), GeneZilla (Allen et al. 2006), GlimmerHMM (Allen et al. 2006) and SNAP (Korf 2004) for ab initio gene prediction, ProSplign (Sayers et al. 2012) for protein alignment with SwissProt and TrEMBL, and sim4db (Walenz and Florea 2011) for interspecies FLcDNA alignment. In the sim4db alignment, only interspecies FLcDNAs from the same class (monocot or dicot) of plants are used to reduce the calculation time. The cut-off identity and coverage of the protein alignment are set to 90%. For gene prediction of the Musaceae and Rosaceae families, which have a relatively small number of sequences in protein databases, the values are relaxed to 80% to increase the number of proteins that could be mapped to the queried sequences. It was confirmed that the relaxed condition did not decrease the gene prediction accuracy (data not shown). The system runs sim4db with an identity and coverage cut-off of 50% and then finds the longest ORFs in each locus for a downstream analysis. All the results, which contain genes predicted by the four ab initio gene finders, protein alignments and ORFs generated from the sim4db alignments, are merged to create consensus gene structures using JIGSAW (Allen et al. 2006). Simultaneously, PASA (Haas et al. 2003) generates EST assemblies by mapping intraspecies ESTs to the query sequence. To achieve a more accurate prediction, the system runs PASA again to incorporate the EST assemblies into the consensus gene structures generated by JIGSAW. Consequently, predicted genes are classified into three categories: (i) genes inferred from intraspecies FLcDNAs; (ii) genes successfully incorporated with EST assemblies; and (iii) genes that did not overlap with EST assemblies or genes that failed to be incorporated with EST assemblies because of inconsistencies between the exon–intron structures. ORFs of class (i) genes are determined by selecting the longest ORFs. This process is not required for classes (ii) and (iii) because they already contain ORF information. If no ORFs ≥100 bp are found, those sequences are treated as non-protein-coding genes. Lastly, sequence similarity against SwissProt and TrEMBL proteins is examined with blastp in BLAST+ (Camacho et al. 2009), and InterProScan (Quevillon et al. 2005) is conducted to identify functional domains and assign GO terms to the ORFs.
Fig. 2.
Overview of the genome annotation workflow in MEGANTE.
Visualization of annotation results
After all the annotation procedures are completed on an application server, the results are returned back to a web server for graphical representation with GBrowse. In addition to the three classes of predicted genes that were previously mentioned, gene loci and repeat regions are displayed on the annotation map in GBrowse (Fig. 1C). MEGANTE also provides other data tracks as follows: (i) protein alignments of the Swiss-Prot and TrEMBL plant division generated by ProSplign; (ii) intraspecies FLcDNA aligned by BLAT; (iii) EST assemblies generated by PASA; (iv) interspecies FLcDNA aligned by sim4db; and (v) repetitive elements detected by RepeatMasker. Users can select the tracks with the ‘Select Tracks’ tab in the window. Details of gene attributes and function annotation, such as top 10 BLAST hits to Swiss-Prot and TrEMBL, InterPro domains and GO terms, are linked from the predicted gene tracks (Fig. 1D). ORF and protein sequences can also be retrieved from the same page. For secure data management, we added authentication and authorization mechanisms into the original GBrowse so that user data are not disclosed to any others.
Application to plant genome sequences
To show the efficiency of MEGANTE, we applied this web service to genomic sequences from Arabidopsis thaliana, Glycine max, Musa acuminate, Oryza sativa, Populus trichocarpa, Solanum lycopersicum, Musca × domestica and Vitis vinifera. Each sequence consisted of 1,000 fragments of a genome sequence, and each one contained one transcript sequence. Details of the data sets are described in the Materials and Methods. To evaluate the performance of MEGANTE, we examined the predictive accuracies for the coding sequence (CDS) coordinates by sensitivity (Sn) and specificity (Sp) that are commonly used for the evaluation of gene prediction programs (Pavy et al. 1999, Rogic et al. 2001, Yao et al. 2005). Sn is defined as the proportion of actual positives that are correctly predicted, and Sp is defined as the proportion of predicted positives that are true positives. We calculated the Sn and Sp at both the exon and gene levels. The exon level means that the start and end positions of a CDS are checked at each exon. At gene level evaluation, it is necessary to identify all CDS coordinates in a transcript correctly. All of the results are summarized in Table 2. For comparison, the results of individual ab initio gene predictions employed in the system are also shown in the table. Differences in predictive accuracies can be observed among the species. For instance, approximately ≥80% of CDSs for A. thaliana and O. sativa were correctly identified, while the Sns at gene level for M. acuminate and M.×domestica were much lower, approximately 10–20%. However, MEGANTE exhibited higher Sn and Sp in almost all categories in comparison with the ab initio gene finders.
Table 2.
Predictive accuracies of MEGANTE and individual gene prediction programs used in the system
| Test gene sets | Evaluation categories | MEGANTE | AUGUSTUS | GeneZilla | GlimmerHMM | SNAP | |
|---|---|---|---|---|---|---|---|
| A. thaliana | Exon level | Sn (%) | 95.2 | 84.4 | 71.8 | 80.9 | 75.5 |
| Sp (%) | 87.9 | 75.6 | 66.5 | 72.7 | 59.2 | ||
| Gene level | Sn (%) | 84.2 | 58.4 | 43.6 | 49.1 | 38.7 | |
| Sp (%) | 58.8 | 43.4 | 28.0 | 35.2 | 22.4 | ||
| G. max | Exon level | Sn (%) | 78.8 | 75.3 | 57.5 | 56.3 | 61.8 |
| Sp (%) | 86.7 | 71.1 | 54.2 | 59.1 | 54.3 | ||
| Gene level | Sn (%) | 51.4 | 35.9 | 22.1 | 28.2 | 18.2 | |
| Sp (%) | 48.8 | 32.5 | 13.6 | 16.0 | 12.1 | ||
| M. acuminata | Exon level | Sn (%) | 46.1 | 48.3 | 19.9 | 34.7 | 30.5 |
| Sp (%) | 62.3 | 54.4 | 28.7 | 31.5 | 36.4 | ||
| Gene level | Sn (%) | 12.8 | 12.2 | 5.9 | 7.6 | 5.8 | |
| Sp (%) | 12.7 | 11.2 | 4.1 | 3.4 | 4.0 | ||
| O. sativa | Exon level | Sn (%) | 91.9 | 52.9 | 57.0 | 74.0 | 45.4 |
| Sp (%) | 86.7 | 67.4 | 46.4 | 59.0 | 50.6 | ||
| Gene level | Sn (%) | 78.0 | 29.5 | 21.9 | 37.8 | 19.7 | |
| Sp (%) | 57.1 | 29.3 | 12.2 | 21.6 | 15.6 | ||
| P. trichocarpa | Exon level | Sn (%) | 76.7 | 73.7 | 60.8 | 55.0 | 63.4 |
| Sp (%) | 81.1 | 71.0 | 57.8 | 59.3 | 57.4 | ||
| Gene level | Sn (%) | 32.3 | 26.8 | 19.6 | 21.4 | 14.4 | |
| Sp (%) | 32.0 | 25.8 | 13.4 | 13.2 | 10.7 | ||
| S. lycopersicum | Exon level | Sn (%) | 85.7 | 69.4 | 49.0 | 49.3 | 57.0 |
| Sp (%) | 91.1 | 74.4 | 49.1 | 52.4 | 49.3 | ||
| Gene level | Sn (%) | 62.6 | 29.5 | 22.0 | 26.8 | 19.3 | |
| Sp (%) | 60.0 | 34.0 | 13.0 | 14.5 | 12.3 | ||
| M.×domestica | Exon level | Sn (%) | 59.0 | 59.1 | 47.1 | 49.8 | 42.3 |
| Sp (%) | 68.5 | 62.5 | 47.3 | 50.1 | 46.1 | ||
| Gene level | Sn (%) | 22.1 | 22.5 | 13.2 | 19.1 | 11.2 | |
| Sp (%) | 19.0 | 18.4 | 7.7 | 9.7 | 7.8 | ||
| V. vinifera | Exon level | Sn (%) | 61.0 | 51.4 | 46.8 | 35.8 | 41.7 |
| Sp (%) | 83.7 | 51.2 | 38.5 | 31.0 | 36.0 | ||
| Gene level | Sn (%) | 22.7 | 10.5 | 7.1 | 6.6 | 5.0 | |
| Sp (%) | 27.3 | 7.9 | 3.5 | 2.6 | 2.6 | ||
Gene prediction parameters we used for each target species are described in the Materials and Methods.
Furthermore, we ran MEGANTE against 13 genome contigs from wheat chromosome 3B (Choulet et al. 2010), which were used in the evaluation of a wheat genome annotation pipeline, TriAnnot (Leroy et al. 2012). The overall size of the contigs is approximately 18 Mb, and they contain 172 CDSs. We used the same evaluation as previously described. The results revealed that the Sn and Sp were 77.6% and 88.2% at the exon level and 64.5% and 63.5% at the gene level, respectively. The results could not be directly compared with the values described in the study on TriAnnot because the numbers of contigs and genes used for the evaluation were not identical between these two. However, both the Sn and Sp of MEGANTE were comparable with those of TriAnnot.
Discussion
In this article, we introduced MEGANTE, a web service for integrated plant genome annotation. The interface of MEGANTE is designed mainly for non-bioinformatics researchers. Complex configurations for annotation procedures are not required; therefore, users can perform genome annotation simply by copying and pasting genomic sequences and selecting the species they want to query. Graphical representation is important for quickly interpreting the analysis results. We utilized GBrowse (Stein et al. 2002) for data visualization because this viewer is widely used in several plant genome databases (Goodstein et al. 2012, Lamesch et al. 2012, Sakai et al. 2013) and users should be familiar with its interface.
MEGANTE has unique features that are not found in similar services. For instance, the service is able to accept a query sequence with a length of 10 Mb, which is larger than the other services. Another prominent feature of MEGANTE is that it targets a wide variety of plant species including 24 species from eight families. This was made possible by adapting common parameter sets for gene structure prediction for all species in the same family. For example, the system creates consensus gene structures for the Poaceae family by using JIGSAW (Allen et al. 2006) with a parameter matrix generated from a reference gene set from O. sativa. Although it is generally preferable to optimize gene prediction parameters for a particular species, the number of reference genes with high-quality annotation is not large enough for parameter optimization, and enriched annotation of closely related species shows much better performance for this application. In fact, our evaluation of wheat genome sequence annotation using MEGANTE revealed that the prediction parameters for O. sativa were sufficient for wheat in our annotation workflow.
One important point to be considered is the updates of data, including reference databases and parameter files for gene prediction in the system. We plan to update the transcript and protein databases at least twice a year, and regenerate gene prediction parameters when new reference annotation data are released. Most of the methods used to generate parameter files for gene prediction are automated in the system, and thus the overall procedure for one species can be completed within a week. Furthermore, it is possible to adapt MEGANTE to any other species by collecting transcript sequences or optimizing gene prediction parameters with reference annotation data for the species of interest.
Materials and Methods
Compiling reference gene sets for gene prediction
To optimize gene prediction parameters for each species, annotated genes are required as reference gene sets. We use the term [training] to refer to the optimization. We collected A. thaliana gene sets from the Brassicaceae family, G. max from Fabaceae, M. acuminate from Musaceae, O. sativa from Poaceae, P. trichocarpa from Salicaceae, S. lycopersicum from Solanaceae, M.×domestica from Rosaceae, and V. vinifera from Vitaceae. The system uses the same parameter sets for gene prediction of all species in the same family, while transcript sequences used for alignment are distinct from each other. The A. thaliana gene set was retrieved from representative genes in TAIR10 (Lamesch et al. 2012); G. max and P. trichocarpa from Phytozome v9.0 (Goodstein et al. 2012); M. acuminate from The Banana Genome Hub version 1 (Droc et al. 2013); O. sativa from representative genes in RAP-DB IRGSP 1.0 (Sakai et al. 2013); S. lycopersicum from ITAG2.3 in SGN (Bombarely et al. 2011); M.×domestica from v1.0p assembly and annotation in GDR (Jung et al. 2008); and V. vinifera from the 12X version of genome assembly and annotation in Grape Genome Browser (Jaillon et al. 2007). First, we excluded genes that did not begin with an initiation codon or did not end with a stop codon in each gene set. Then, we randomly selected 1,000 genes for training of ab initio gene prediction programs, 10,000 for training of JIGSAW and 1,000 for an evaluation of the overall performance of MEGANTE. The data sets for training and evaluation did not overlap with each other so that valid evaluation between independent sets was possible. All of the gene sequences extracted from genome assemblies contain CDSs and their 1 kb upstream and downstream sequences.
Training of gene prediction programs
We initially trained ab initio gene finders, AUGUSTUS (Stanke and Waack 2003), GeneZilla (Allen et al. 2006), GlimmerHMM (Allen et al. 2006) and SNAP (Korf 2004), with each gene set. However, the gene finders had pre-trained parameter files for some plant species; thus, we did not train the programs for those species. The pre-trained parameters we used were AUGUSTUS for Arabidopsis, maize and tomato; GlimmerHMM for Arabidopsis and rice; and SNAP for Arabidopsis and rice. AUGUSTUS for maize is used for the Poaceae family. For GeneZilla, we employed an automatic training program, GRAPE (Majoros and Salzberg 2004). To train the other programs, we followed the instructions provided by the software developer. Subsequently, training of JIGSAW with all of the ab initio gene finders was conducted using 10,000 CDSs. In addition, interspecies FLcDNA alignment with sim4db and protein alignment with ProSplign were also conducted against the same data sets. These procedures were the same as previously described for the annotation workflow. All the results were fed to JIGSAW as sources of evidence. The training of JIGSAW was performed by the train_jigsaw.pl script in the package with default options.
Availability and implementation of the system
MEGANTE was implemented in a web application framework, Catalyst, with MySQL as the backend database. The frontend web interface was built with HTML5, CSS3 and JavaScript, and was tested on the following web browsers: Safari 6, Chrome 28, Firefox 23 and Internet Explorer 9 and 10. This service is available for free at https://megante.dna.affrc.go.jp/.
Funding
This work was supported by the Ministry of Agriculture, Forestry and Fisheries of Japan [Genomics for Agricultural Innovation, grant GIR1001; Development of Genome Information Database System for Innovation of Crop and Livestock Production].
Disclosures
The authors have no conflicts of interest to declare.
Glossary
Abbreviations
- CDS
coding sequence
- EST
expressed sequence tag
- FLcDNA
full-length cDNA
- GO
gene ontology
- ORF
open reading frame
- Sn
sensitivity
- Sp
specificity
References
- Allen JE, Majoros WH, Pertea M, Salzberg SL. JIGSAW, GeneZilla, and GlimmerHMM: puzzling out the features of human genes in the ENCODE regions. Genome Biol. 2006;7(Suppl. 1):S9.1–S9.13. doi: 10.1186/gb-2006-7-s1-s9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano N, Tanaka T, Numa H, Sakai H, Itoh T. Efficient plant gene identification based on interspecies mapping of full-length cDNAs. DNA Res. 2010;17:271–279. doi: 10.1093/dnares/dsq017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan MW, Uauy C. Genomics reveals new landscapes for crop improvement. Genome Biol. 2013;14:206. doi: 10.1186/gb-2013-14-6-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bombarely A, Menda N, Tecle IY, Buels RM, Strickler S, Fischer-York T, et al. The Sol Genomics Network (solgenomics.net): growing tomatoes using Perl. Nucleic Acids Res. 2011;39:D1149–D1155. doi: 10.1093/nar/gkq866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel BL, Korf I, Robb SM, Parra G, Ross E, Moore B, et al. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008;18:188–196. doi: 10.1101/gr.6743907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Zhang Y, Zhao L, Zhu Z, Lin J, Zhang S, et al. Fine mapping and candidate gene analysis of a green-revertible albino gene gra(t) in rice. J. Genet. Genomics. 2009;36:117–123. doi: 10.1016/S1673-8527(08)60098-3. [DOI] [PubMed] [Google Scholar]
- Choulet F, Wicker T, Rustenholz C, Paux E, Salse J, Leroy P, et al. Megabase level sequencing reveals contrasted organization and evolution patterns of the wheat gene and transposable element spaces. Plant Cell. 2010;22:1686–1701. doi: 10.1105/tpc.110.074187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper L, Walls RL, Elser J, Gandolfo MA, Stevenson DW, Smith B, et al. The plant ontology as a tool for comparative plant anatomy and genomic analyses. Plant Cell Physiol. 2013;54:e1. doi: 10.1093/pcp/pcs163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droc G, Larivière D, Guignon V, Yahiaoui N, This D, Garsmeur O, et al. The banana genome hub. Database (Oxford) 2013;2013:bat035. doi: 10.1093/database/bat035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvick J, Fu A, Muppirala U, Sabharwal M, Wilkerson MD, Lawrence CJ, et al. PlantGDB: a resource for comparative plant genomics. Nucleic Acids Res. 2008;36:D959–D965. doi: 10.1093/nar/gkm1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SA, Vaughn M, McKay S, Lyons E, Stapleton AE, Gessler D, et al. The iPlant Collaborative: cyberinfrastructure for plant biology. Front. Plant Sci. 2011;2:34. doi: 10.3389/fpls.2011.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Delcher AL, Mount SM, Wortman JR, Smith RK, Hannick LI, et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 2003;31:5654–5666. doi: 10.1093/nar/gkg770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, Bateman A, et al. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 2012;40:D306–D312. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- Jung S, Staton M, Lee T, Blenda A, Svancara R, Abbott A, et al. GDR (Genome Database for Rosaceae): integrated web-database for Rosaceae genomics and genetics data. Nucleic Acids Res. 2008;36:D1034–D1040. doi: 10.1093/nar/gkm803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I. Gene finding in novel genomes. BMC Bioinformatics. 2004;5:59. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, et al. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2012;40:D1202–D1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy P, Guilhot N, Sakai H, Bernard A, Choulet F, Theil S, et al. TriAnnot: a versatile and high performance pipeline for the automated annotation of plant genomes. Front. Plant Sci. 2012;3:5. doi: 10.3389/fpls.2012.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukashin AV, Borodovsky M. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 1998;26:1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magrane M, UniProt Consortium UniProt Knowledgebase: a hub of integrated protein data. Database (Oxford) 2011;2011:bar009. doi: 10.1093/database/bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majoros WH, Salzberg SL. An empirical analysis of training protocols for probabilistic gene finders. BMC Bioinformatics. 2004;5:206. doi: 10.1186/1471-2105-5-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihara M, Itoh T, Izawa T. SALAD database: a motif-based database of protein annotations for plant comparative genomics. Nucleic Acids Res. 2010;38:D835–D842. doi: 10.1093/nar/gkp831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamura Y, Antonio BA, Sato Y, Miyao A, Namiki N, Yonemaru J, et al. Rice TOGO Browser: a platform to retrieve integrated information on rice functional and applied genomics. Plant Cell Physiol. 2011;52:230–237. doi: 10.1093/pcp/pcq197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Cochrane G, Karsch-Mizrachi I, The International Nucleotide Sequence Database Collaboration The International Nucleotide Sequence Database Collaboration. Nucleic Acids Res. 2013;41:D21–D24. doi: 10.1093/nar/gks1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussbaumer T, Martis MM, Roessner SK, Pfeifer M, Bader KC, Sharma S, et al. MIPS PlantsDB: a database framework for comparative plant genome research. Nucleic Acids Res. 2013;41:D1144–D1151. doi: 10.1093/nar/gks1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavy N, Rombauts S, Déhais P, Mathé C, Ramana DV, Leroy P, et al. Evaluation of gene prediction software using a genomic data set: application to Arabidopsis thaliana sequences. Bioinformatics. 1999;15:887–899. doi: 10.1093/bioinformatics/15.11.887. [DOI] [PubMed] [Google Scholar]
- Podicheti R, Gollapudi R, Dong Q. WebGBrowse—a web server for GBrowse. Bioinformatics. 2009;25:1550–1551. doi: 10.1093/bioinformatics/btp239. [DOI] [PubMed] [Google Scholar]
- Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, et al. InterProScan: protein domains identifier. Nucleic Acids Res. 2005;33:W116–W120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogic S, Mackworth AK, Ouellette FB. Evaluation of gene-finding programs on mammalian sequences. Genome Res. 2001;11:817–832. doi: 10.1101/gr.147901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouard M, Guignon V, Aluome C, Laporte MA, Droc G, Walde C, et al. GreenPhylDB v2.0: comparative and functional genomics in plants. Nucleic Acids Res. 2011;39:D1095–D1102. doi: 10.1093/nar/gkq811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H, Lee SS, Tanaka T, Numa H, Kim J, Kawahara Y, et al. Rice Annotation Project Database (RAP-DB): an integrative and interactive database for rice genomics. Plant Cell Physiol. 2013;54:e6. doi: 10.1093/pcp/pcs183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata K, Nagamura Y, Numa H, Antonio BA, Nagasaki H, Idonuma A, et al. RiceGAAS: an automated annotation system and database for rice genome sequence. Nucleic Acids Res. 2002;30:98–102. doi: 10.1093/nar/30.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salamov AA, Solovyev VV. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 2000;10:516–522. doi: 10.1101/gr.10.4.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2012;40:D13–D25. doi: 10.1093/nar/gkr1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Waack S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics. 2003;19(Suppl. 2):ii215–ii225. doi: 10.1093/bioinformatics/btg1080. [DOI] [PubMed] [Google Scholar]
- Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walenz B, Florea L. Sim4db and Leaff: utilities for fast batch spliced alignment and sequence indexing. Bioinformatics. 2011;27:1869–1870. doi: 10.1093/bioinformatics/btr285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Wang B, Wu Y, Du P, Wang J, Wang M, et al. Fine mapping and candidate gene analysis of ptgms2-1, the photoperiod-thermo-sensitive genic male sterile gene in rice (Oryza sativa L.) Theor. Appl. Genet. 2011;122:365–372. doi: 10.1007/s00122-010-1452-0. [DOI] [PubMed] [Google Scholar]
- Yandell M, Ence D. A beginner’s guide to eukaryotic genome annotation. Nat. Rev. Genet. 2012;13:329–342. doi: 10.1038/nrg3174. [DOI] [PubMed] [Google Scholar]
- Yao H, Guo L, Fu Y, Borsuk LA, Wen TJ, Skibbe DS, et al. Evaluation of five ab initio gene prediction programs for the discovery of maize genes. Plant Mol. Biol. 2005;57:445–460. doi: 10.1007/s11103-005-0271-1. [DOI] [PubMed] [Google Scholar]
- Youens-Clark K, Buckler E, Casstevens T, Chen C, Declerck G, Derwent P, et al. Gramene database in 2010: updates and extensions. Nucleic Acids Res. 2011;39:D1085–D1094. doi: 10.1093/nar/gkq1148. [DOI] [PMC free article] [PubMed] [Google Scholar]