Abstract
IAA is a naturally occurring auxin that plays a crucial role in the regulation of plant growth and development. The endogenous concentration of IAA is spatiotemporally regulated by biosynthesis, transport and its inactivation in plants. Previous studies have shown that the metabolism of IAA to 2-oxindole-3-acetic acid (OxIAA) and OxIAA-glucoside (OxIAA-Glc) may play an important role in IAA homeostasis, but the genes involved in this metabolic pathway are still unknown. In this study, we show that UGT74D1 catalyzes the glucosylation of OxIAA in Arabidopsis. By screening yeasts transformed with Arabidopsis UDP-glycosyltransferase (UGT) genes, we found that OxIAA-Glc accumulates in the culture media of yeasts expressing UGT74D1 in the presence of OxIAA. Further, we showed that UGT74D1 expressed in Escherichia coli converts OxIAA to OxIAA-Glc. The endogenous concentration of OxIAA-Glc decreased by 85% while that of OxIAA increased 2.5-fold in ugt74d1-deficient mutants, indicating the major role of UGT74D1 in OxIAA metabolism. Moreover, the induction of UGT74D1 markedly increased the level of OxIAA-Glc and loss of root gravitropism. These results indicate that UGT74D1 catalyzes a committed step in the OxIAA-dependent IAA metabolic pathway in Arabidopsis.
Keywords: Arabidopsis, Auxin, Glucosyltransferase, Metabolism, Plant hormone
Introduction
The phytohormone auxin plays a key role in many aspects of a plant’s life cycle, including embryogenesis, organ formation and environmental responses (Woodward and Bartel 2005, Zhao 2010). Auxin regulates cell elongation and differentiation in a concentration-dependent manner. A naturally occurring auxin, IAA, is mainly synthesized from tryptophan via indole-3-pyruvate (IPA) by TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS (TAA) and YUCCA (YUC) flavin-containing monooxygenases in Arabidopsis (Fig. 1) (Mashiguchi et al. 2011, Stepanova et al. 2011, Won et al. 2011). The TAA and YUC family genes are regulated spatiotemporally and contribute to the uneven distribution of IAA in plants (Cheng et al. 2006, Cheng et al. 2007, Stepanova et al. 2008, Tao et al. 2008, Yamada et al. 2009). Plants modulate cellular IAA concentrations using membrane-localized proteins, including PIN efflux carriers, AUX1/LAX influx transporters and ABCB transporters (Hayashi 2012). PIN efflux carriers play a crucial role in the regulation of polar auxin transport (Vanneste and Friml 2009). A rapid change in PIN localization by vesicle trafficking determines the direction of auxin flow in plant organs (Grunewald and Friml 2010).
Fig. 1.
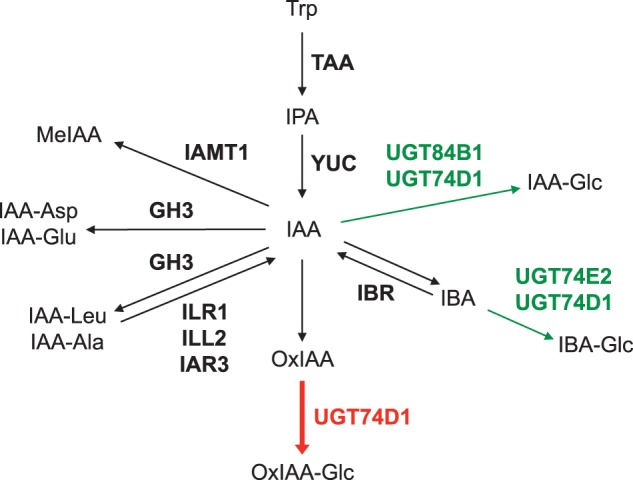
Main IAA biosynthetic pathway and proposed IAA metabolic pathways in Arabidopsis. Genes encoding IAA biosynthetic and metabolic enzymes identified in Arabidopsis are shown in bold. The red arrow indicates a novel reaction step catalyzed by UGT74D1. The green arrows indicate the proposed reaction steps catalyzed by previously characterized UGTs.
Previous studies indicate that various conjugation and degradation enzymes also play important roles in the regulation of IAA concentration (Fig. 1). Several GRETCHEN HAGEN 3 (GH3) genes encode enzymes that catalyze the formation of IAA–amino acid conjugates (Staswick et al. 2005). Each GH3 enzyme has a slightly different substrate specificity and probably contributes to the formation of several types of IAA–amino acid conjugates (Staswick et al. 2005). The overexpression of GH3 genes results in reduced auxin phenotypes, such as dwarfism, short hypocotyls and fewer lateral roots in Arabidopsis (Nakazawa et al. 2001, Takase et al. 2003, Takase et al. 2004). In contrast, gh3-deficient mutants of Arabidopsis show slightly increased sensitivity to IAA (Staswick et al. 2005). Previous studies using 14C-labeled IAA demonstrated that IAA is metabolized to IAA-Asp and IAA-Glu in Arabidopsis (Fig. 1) (Östin et al. 1998, LeClere et al. 2002). In addition, IAA-Asp and IAA-Glu levels are substantially increased in the IAA-overproducing supreroot1 and superroot2 mutants (Barlier et al. 2000, Mashiguchi et al. 2011, Novák et al. 2012). These results indicate that the GH3-mediated formation of IAA-Asp and IAA-Glu plays an important role in the inactivation of IAA.
Similar to IAA, treatment with some IAA–amino acid conjugates, such as IAA-Leu, IAA-Ala and IAA-Phe, inhibits root growth in Arabidopsis. Loss-of-function mutants of IAA-LEUCINE RESISTANT 1 (ILR1) and IAA-ALANINE RESISTSTANT 3 (IAR3) exhibit reduced sensitivity to IAA-Leu and IAA-Ala, respectively (Bartel and Fink 1995, Davies et al. 1999). ILR1, ILR1-LIKE 2 (ILL2) and IAR3 are endoplasmic reticulum (ER)-localized amidohydrolases that generate IAA from IAA–amino acid conjugates (Fig. 1) (LeClere et al. 2002). Previous studies demonstrated that ilr1 iar3 ill2 triple mutants exhibit low-auxin phenotypes, and the endogenous levels of IAA are reduced in these mutants (LeClere et al. 2002, Rampey et al. 2004). These reports suggest that IAA–amino acid conjugates function as auxin storage forms, and the GH3 and amidohydrolase families play a role in the regulation of IAA concentration.
IAA CARBOXYL METHYLTRANSFERASE 1 (IAMT1) catalyzes the methylation of the carboxyl group, which inactivates IAA to its methyl ester (MeIAA) (Fig. 1) (Zubieta et al. 2003). Ectopic expression of IAMT1 leads to the low-auxin phenotype, including upward curling leaves and disrupted gravitropism, and reduced IAA sensitivity in Arabidopsis. Suppression of IAMT1 by RNA interference (RNAi) results in epinastic small leaves and dwarfism. Analysis of a GUS (β-glucuronidase) reporter gene under the IAMT1 promoter suggests the spatiotemporal regulation of IAMT1 in Arabidopsis (Qin et al. 2005). IAMT1 may also play an important role in leaf morphogenesis (Qin et al. 2005).
Glucosylation is probably implicated in the inactivation of IAA, although its physiological roles are still unknown. UDP-glycosyltransferases (UGTs) are one of the largest families of glycosyltransferases in plants. There are 107 UGTs in Arabidopsis (Yonekura-Sakakibara and Saito 2009). Some UGTs catalyze glucosylation of plant hormones, including auxin, ABA, cytokinins and salicylic acid, using UDP-glucose as a co-substrate (Lim and Bowles 2004, Gachon et al. 2005, Yonekura-Sakakibara and Saito 2009). UGT84B1 catalyzes the conversion of IAA to 1-O-(indol-3-ylacetyl)-β-d-glucose (IAA-Glc) (Fig. 1) (Jackson et al. 2001), which widely exists in higher plants (Cohen and Bandurski 1982, Sitbon et al. 1993, Jakubowska and Kowalczyk 2004, Iyer et al. 2005). Overexpression of UGT84B1 leads to enhanced shoot branch formation, gravitropism defects and decreased sensitivity to auxin in Arabidopsis (Jackson et al. 2002, Lim and Bowles 2004). It has been shown that UGT74E2 catalyzes the formation of 1-O-(indol-3-ylbutanoyl)-β-d-glucose (IBA-Glc) from indole-3-butylic acid (IBA), which is probably generated from IAA in plants (Tognetti et al. 2010). Overexpression of UGT74E2 results in short stature, compressed rosette, enhanced shoot branching and high osmotic stress tolerance in Arabidopsis. A recent study demonstrated that UGT74D1 converts both IAA and IBA to their corresponding glucosides in vitro, implicating it in the glucosylation of both IAA and IBA in planta (Jin et al. 2013). However, the phenotypes of UGT74D1-overexpressing plants were different from those observed in UGT84B1- or UGT74E2-overexpressing plants (Jin et al. 2013), suggesting the distinct functions of UGT74D1 in Arabidopsis.
The oxidation of IAA to 2-oxindole-3-acetic acid (OxIAA) is one type of IAA inactivation reaction in plants (Fig. 1). This IAA inactivation reaction is irreversible, and OxIAA does not show growth-promoting activity (Reinecke and Bandurski 1983, Normanly 1997, Sztein et al. 1999, Chamarro et al. 2001, Kowalczyk and Sandberg 2001, Ljung et al. 2002, Woodward and Bartel 2005). In Zea mays tissue extracts, IAA was enzymatically converted to OxIAA under aerobic conditions (Reinecke and Bandurski 1988), and the levels of OxIAA increased significantly in plants that overproduce IAA (Novak et al. 2012, Sairanen et al. 2012). It has been demonstrated that IAA is metabolized to OxIAA and its glucoside in Arabidopsis seedlings (Östin et al. 1998). The endogenous levels of OxIAA glucoside, 1-O-(2-oxindol-3-ylacetyl)-β-d-glucose (OxIAA-Glc), are much higher than that of IAA-Asp, IAA-Glu, IAA-Glc and OxIAA in Arabidopsis (Kai et al. 2007), suggesting that the OxIAA pathway is an important metabolic pathway for IAA homeostasis in plants (Ljung et al. 2002, Rosquete et al. 2012, Ljung 2013). However, the genes involved in the OxIAA metabolic pathway are still unknown.
Although various metabolic pathways have been proposed for the regulation of the IAA level, the main IAA metabolic pathway in plants remains unclear. Östin et al. (1998) demonstrated that IAA was primarily metabolized to IAA-Asp and IAA-Glu by the application of a relatively high concentration of IAA in Arabidopsis. Interestingly, most of the IAA applied was metabolized to OxIAA and OxIAA-Glc and only a small portion was conjugated with Asp or Glu when a relatively low concentration of IAA was applied. These results suggest that the GH3 and OxIAA pathways may have critical but distinct roles in IAA homeostasis. Thus, identification of the genes involved in the OxIAA pathway may provide the key to elucidate the main IAA metabolic pathway in plants.
In this study, we show that UGT74D1 plays an important role in the OxIAA pathway in Arabidopsis (Fig. 1). UGT74D1 heterologously expressed in E. coli cells converted OxIAA to OxIAA-Glc in the presence of UDP-glucose. The endogenous level of OxIAA-Glc is dramatically reduced in ugt74d1-deficient mutants. Moreover, the induction of UGT74D1 resulted in increased level of OxIAA-Glc and loss of root gravitropism in Arabidopsis. In conclusion, UGT74D1 catalyzes a committed step in the OxIAA pathway, and contributes to IAA homeostasis in Arabidopsis.
Results
Isolation of an OxIAA glucosyltransferase gene in Arabidopsis
Arabidopsis UGTs are divided into 14 groups (groups A–N) (Li et al. 2001, Ross et al. 2001). Group L contained the UGTs (UGT84B1, UGT74E2 and UGT74D1) that catalyze the glucosylation of IAA and IBA (Fig. 1; Supplementary Fig. S1) (Jackson et al. 2001, Tognetti et al. 2010, Jin et al. 2013). In addition, UGT74F1, UGT74F2, UGT84A2 and UGT74B1 in group L mediate the glucosylation of various aromatic compounds: salicylic acid, anthranilate, sinapic acid and indole-glucosinolate (Lim et al. 2002, Messner et al. 2003, Quiel and Bender 2003, Grubb et al. 2004, Sinlapadech et al. 2007). From these results, we anticipated that OxIAA glucosyltransferases would also be present in group L.
To identify the genes that encode OxIAA glucosyltransferases in Arabidopsis, we used a screening system that involves budding yeast Saccharomyces cerevisiae transformed with each UGT gene in group L. Budding yeasts produce high amounts of UDP-glucose for the synthesis of cell wall components, β-1,3-glucan and β-1,6-glucan (Douglas et al. 1994, Shahinian and Bussey 2000, Oka and Jigami 2006). We expressed the 13 UGT genes individually in budding yeasts and carried out screening of the yeasts that produced OxIAA-Glc by supplementing the culture media with OxIAA (Table 1). Some UGTs (UGT74E1, UGT74E2, UGT75B2 and UGT84B2) are not tested in this study due to the limited availability of Arabidopsis full-length clones (Table 1; Supplementary Table S1). However, it has been shown previously that UGT75B2 and UGT84B2 do not convert OxIAA to OxIAA-Glc in vitro (Jackson et al. 2001). After incubation of the budding yeasts with OxIAA, we analyzed OxIAA metabolites in the yeast cells and the culture media using liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI-MS/MS). OxIAA-Glc was identified by using a product ion at m/z 190.1 (3.14 min) generated from the parent ion at m/z 352.1 on an MS/MS chromatogram (Fig. 2B). Our analysis showed accumulation of OxIAA-Glc in the culture media of yeasts transformed with UGT74D1 (Yeast-UGT74D1 + OxIAA), suggesting that UGT74D1 may convert OxIAA to OxIAA-Glc (Fig. 2A, B). It is important to note that a product ion peak at m/z 190.1 for OxIAA-Glc was not detected in the culture medium of yeast cells expressing UGT74D1 without supplementation of the OxIAA (Yeast-UGT74D1—OxIAA in Fig. 2B). Similarly, no OxIAA-Glc was detected in the culture of yeast cells transformed with the other UGT genes tested in this study (Table 1).
Table 1.
Screening of Arabidopsis OxIAA glucosyltransferases in S. cerevisiae in the presence of OxIAA
| Gene | AGI code | OxIAA-Glc |
|---|---|---|
| UGT74B1 | AT1G24100 | ND |
| UGT74F1 | AT2G43840 | ND |
| UGT74F2 | AT2G43820 | ND |
| UGT74C1 | AT2G31790 | ND |
| UGT74D1 | AT2G31750 | Detected |
| UGT74E1 | AT1G05675 | NT |
| UGT74E2 | AT1G05680 | NT |
| UGT75B1a | AT1G05560 | ND |
| UGT75B2a | AT1G05530 | NT |
| UGT75D1 | AT4G15550 | ND |
| UGT75C1 | AT4G14090 | ND |
| UGT84A1 | AT4G15480 | ND |
| UGT84A2 | AT3G21560 | ND |
| UGT84A3 | AT4G15490 | ND |
| UGT84A4 | AT4G15500 | ND |
| UGT84B1 | AT2G23260 | ND |
| UGT84B2a | AT2G23250 | NT |
a No conversion of OxIAA to OxIAA-Glc was detected in the previous study (Jackson et al. 2001).
ND, not detected. NT, not tested.
Fig. 2.
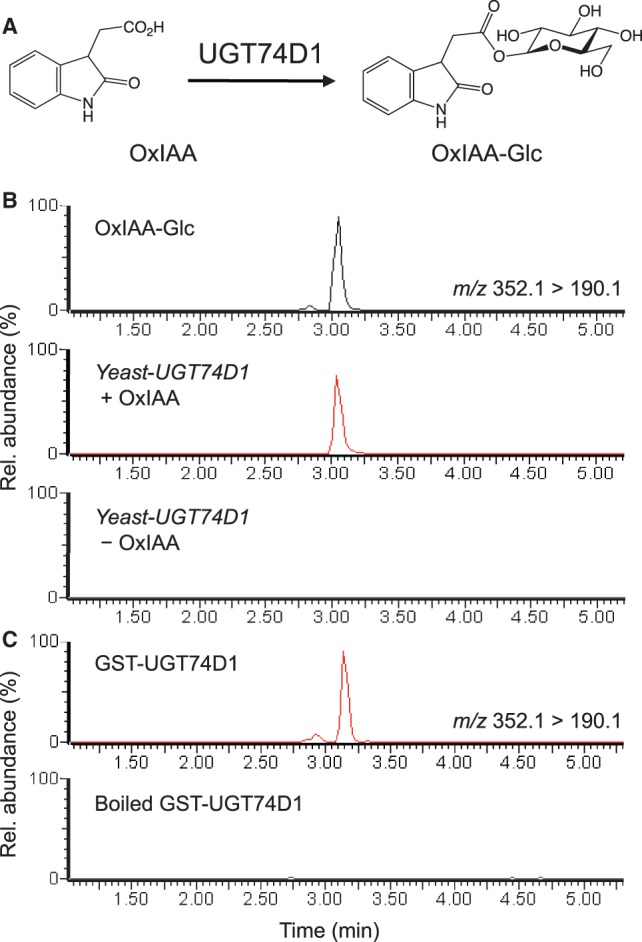
Conversion of OxIAA to OxIAA-Glc by UGT74D1. (A) Enzymatic reaction catalyzed by UGT74D1. (B) The MS/MS chromatogram for authentic OxIAA-Glc and metabolites of yeast expressing UGT74D1 in the culture medium with OxIAA (Yeast-UGT74D1 + OxIAA) or without OxIAA (Yeast-UGT74D1 - OxIAA). (C) The MS/MS chromatogram for the reaction products of GST–UGT74D1 and boiled GST–UGT74D1.
UGT74D1 catalyzes the conversion of OxIAA to OxIAA-Glc
Recently, Jin et al. (2013) demonstrated that UGT74D1 converts IBA, indole-3-propionic acid, IAA, 1-naphthalene acetic acid (NAA), 2,4-dichlorophenoxyacetic acid (2,4-D) and indole-3-carboxylic acid to their corresponding glucosides in vitro. However, OxIAA was not investigated as a naturally occurring substrate (Jin et al. 2013). To investigate directly if UGT74D1 exhibits glucosyltransferase activity against OxIAA, we expressed glutathione S-transferase (GST) fused with UGT74D1 (GST–UGT74D1) in E. coli for use in an enzyme assay. From the enzymatic reactions containing the GST–UGT74D1 fusion protein, we detected a product ion at m/z 190.1, representing OxIAA-Glc, on the MS/MS chromatogram (Fig. 2C), indicating that the GST–UGT74D1 protein efficiently converted OxIAA to OxIAA-Glc using UDP-glucose as a co-substrate. The formation of OxIAA-Glc was abolished in the absence of GST–UGT74D1 proteins (boiled GST–UGT74D1 in Fig. 2C). Consistent with the results of Jin et al. (2013), GST–UGT74D1 also catalyzed the conversion of IAA to IAA-Glc in our assay conditions (Supplementary Fig. S2). To determine the substrate preference of UGT74D1, we analyzed the Km values of GST–UGT74D1 for OxIAA and IAA (Table 2). The Km value of GST–UGT74D1 for OxIAA (15.98 µM) was 5.5 times lower than that for IAA (88.27 µM), indicating that UGT74D1 has higher affinity for OxIAA than IAA. The catalytic efficiency (kcat/Km) of GST–UGT74D1 for OxIAA (3.65 µM−1 min−1) was 6.6 times higher than that for IAA (0.55 µM−1 min−1) (Table 2). These results indicate that UGT74D1 has a higher specificity towards OxIAA than IAA.
Table 2.
Kinetic parameters for the glucosyltransferase activity of GST–UGT74D1 towards OxIAA and IAA
| Substrate | Km (µM) | kcat (min−1) | kcat/Km (µM−1min−1) |
|---|---|---|---|
| OxIAA | 15.98 | 58.31 | 3.65 |
| IAA | 88.27 | 48.56 | 0.55 |
OxIAA-Glc is mainly produced by UGT74D1 in Arabidopsis
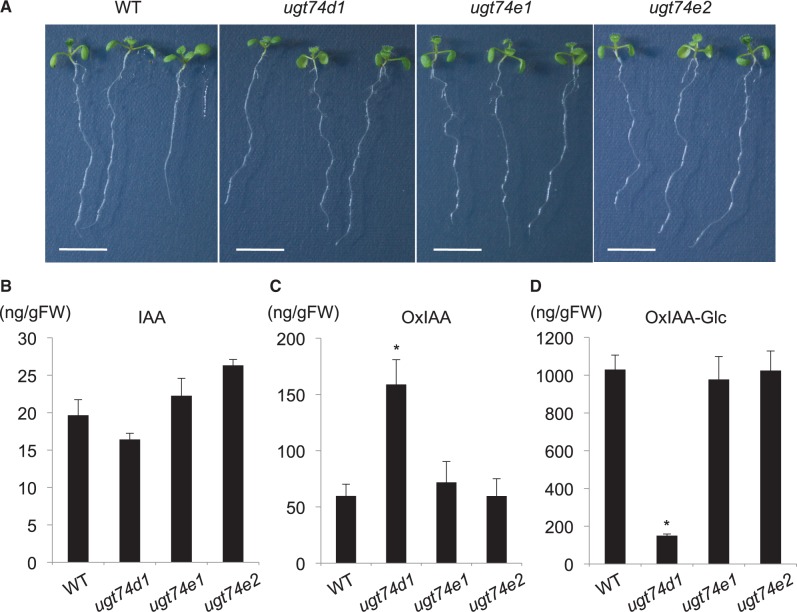
To investigate whether UGT74D1 mainly contributes to OxIAA metabolism in Arabidopsis, we analyzed the endogenous levels of IAA, OxIAA and OxIAA-Glc in the ugt74d1-deficient mutant. The transcripts of UGT74D1 were not detected in the T-DNA insertion mutant, ugt74d1 (SALK_004870) (Supplementary Fig. S3). The amounts of IAA metabolites in the wild type were found to be in the following order, OxIAA-Glc > OxIAA > IAA (Fig. 3). The levels of OxIAA-Glc were nearly two orders of magnitude higher than that of IAA. No significant phenotype or change in IAA concentration was observed in ugt74d1 (Jin et al. 2013; Fig. 3A, B). However, the endogenous level of OxIAA in the ugt74d1 mutant was 2.5-fold higher than that in the wild-type (Fig. 3C). Moreover, the level of OxIAA-Glc decreased by 85% in ugt74d1 when compared with that in the wild type (Fig. 3D). These results indicate that UGT74D1 mainly catalyzes the formation of OxIAA-Glc from OxIAA in Arabidopsis. To examine whether UGT74D1-related genes contribute to OxIAA-Glc formation, we further analyzed the endogenous levels of OxIAA and OxIAA-Glc in the knockout mutants of UGT74D1 homologs, ugt74e1 (SALK_045974) (Supplementary Fig. S3) and ugt74e2 (SALK_016116) (Tognetti et al. 2010). Our data showed that no significant change in the levels of OxIAA and OxIAA-Glc occurred in these mutants (Fig. 3B, C, D). From these results, we conclude that a large fraction of OxIAA-Glc is produced by UGT74D1, but other UGTs may play an auxiliary role in the formation of OxIAA-Glc in Arabidopsis.
Fig. 3.
Analysis of the OxIAA pathway in ugt74d1. (A) Phenotypes of the wild-type (WT), ugt74d1, ugt74e1 and ugt74e2. Bars indicate 5 mm. (B and C) Endogenous concentration of IAA, OxIAA and OxIAA-Glc in the WT, ugt74d1, ugt74e1 and ugt74e2. Values are mean ± SD (n = 3). *Differences between the WT and mutant are statistically significant at P < 0.05 (Student’s t-test).
Induction of UGT74D1 reduces the root gravitropism in Arabidopsis
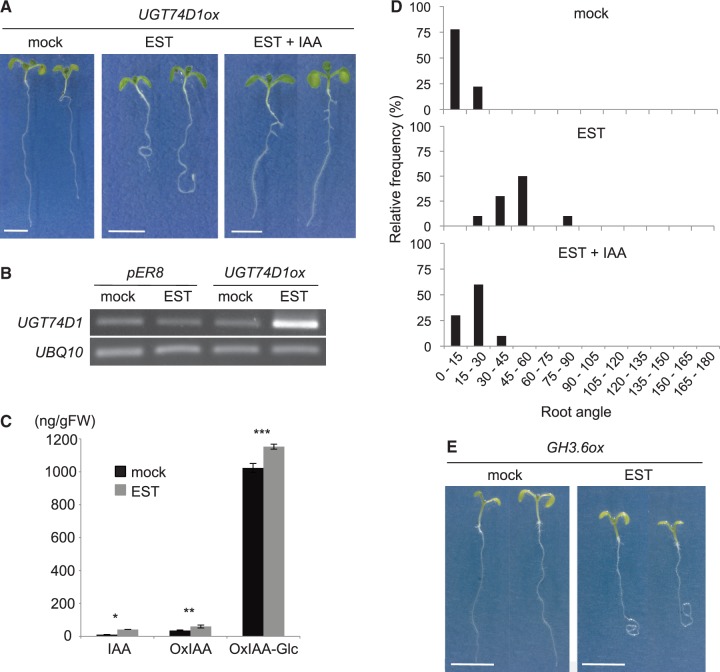
Previous studies clearly showed that the endogenous levels of IAA-Glc and IBA-Glc significantly increased in transgenic plants overexpressing UGT84B1 and UGT74E2, respectively (Jackson et al. 2002, Tognetti et al. 2010). To demonstrate further the biological function of UGT74D1 as an OxIAA glucosyltransferase in vivo, we generated Arabidopsis plants transformed with β-estradiol (EST)-inducible UGT74D1 (UGT74D1ox) (Fig. 4A). As shown in Fig. 4B, the expression of UGT74D1 was markedly increased in EST-treated UGT74D1ox plants when compared with non-EST-treated (mock) or vector control plants (pER8). We found that the endogenous levels of OxIAA-Glc increased by 13% in UGT74D1ox treated with EST (Fig. 4C), supporting that UGT74D1 mediates the formation of OxIAA-Glc in Arabidopsis. We also noted that the levels of IAA and OxIAA increased in UGT74D1ox plants (Fig. 4C), suggesting that enhanced glucosylation of OxIAA promotes the formation of IAA and OxIAA.
Fig. 4.
Analysis of the OxIAA pathway in UGT74D1ox. (A) Phenotypes of UGT74D1ox without EST (mock), with 10 µM EST (EST) and with 10 µM EST and 20 nM IAA (EST + IAA). Bars indicate 5 mm. (B) Semi-quantitative RT–PCR analysis of UGT74D1 expression in the vector control (pER8) and UGT74D1ox without EST (mock) or with 10 µM EST. (C) IAA, OxIAA and OxIAA-Glc concentrations in UGT74D1ox without (mock) or with 10 µM EST. Values are mean ± SD (n = 3). *, ***Differences between UGT74D1ox with and without EST are statistically significant at P < 0.01 (Student’s t-test). **Differences between UGT74D1ox with and without EST are statistically significant at P < 0.05 (Student’s t-test). (D) Histogram displaying the range of the root angles from the gravity vector for UGT74D1ox plants without EST (mock), with 10 µM EST and with 10 µM EST and 20 nM IAA (EST + IAA) are shown (n = 9–10). (E) Phenotypes of GH3.6ox without EST (mock) or with 10 µM EST. Bars indicate 5 mm.
Intriguingly, the induction of UGT74D1 remarkably reduced root gravitropism in Arabidopsis seedlings (Fig. 4A, D), indicating that UGT74D1 catalyzes a committed step in the OxIAA pathway. We note that the loss of root gravitropism in UGT74D1ox plants was similar to that observed in the EST-inducible GH3.6 (GH3.6ox) plants (Fig. 4E). GH3.6 encodes IAA–amino acid conjugate synthase, which catalyzes a committed step in the GH3 pathway, and ectopic expression of GH3.6 displays the low-auxin phenotype in Arabidopsis (Staswick et al. 2005). To examine if the decrease in auxin level led to loss of root gravitropism, we grew the UGT74D1ox plants in Murashige and Skoog (MS) agar medium containing 20 nM IAA. We found that root gravitropism in UGT74D1ox plants was recovered by supplementation with IAA (Fig. 4A, D). These results demonstrate that UGT74D1 catalyzes the glucosylation of OxIAA.
Responsiveness of UGT74D1 and GH3 genes to auxin
We analyzed the regulation of UGT74D1 by auxin in Arabidopsis through applying IAA. We treated 1-week-old seedlings of Arabidopsis with 1 µM IAA for from 1 to 48 h and analyzed the expression levels of UGT74D1 by reverse transcription–PCR (RT–PCR). We also analyzed the expression levels of GH3.2, GH3.3 and GH3.5 as early auxin-responsive genes (Staswick et al. 2005). As shown in Fig. 5, the expression of GH3 genes was strongly induced by IAA treatment within 1 h. The expression of UGT74D1 is different from that of GH3 in that it showed a slight increase only after 3 h of IAA treatment. This result demonstrates that both UGT74D1 and GH3 genes are implicated in IAA metabolism, but they are likely to be regulated by distinct mechanisms in Arabidopsis.
Fig. 5.
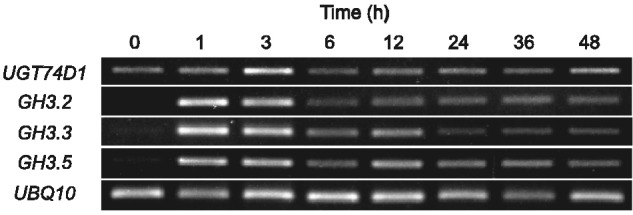
Auxin responsiveness of UGT74D1 and GH3 genes in Arabidopsis. Semi-quantitative RT–PCR analysis of UGT74D1, GH3.2, GH3.3 and GH3.5 expression levels in Arabidopsis treated with 1 µM IAA for 1–48 h. UBQ10, ubiquitin control.
Discussion
Biological function of UGT74D1
Herein, we provide genetic, biochemical and metabolite-based evidence that UGT74D1 plays a major role in the conversion of OxIAA to OxIAA-Glc in Arabidopsis. Recently, Jin et al. (2013) showed that UGT74D1 catalyzes the glucosylation of various auxin-related compounds in the following order of substrate preference: IBA, indole-3-propionic acid, IAA, NAA, 2,4-D and indole-3-carboxylic acid. In the present study, we demonstrated that UGT74D1 catalyzes the glucosylation of OxIAA and IAA to their corresponding glucosides (Table 2). These results taken together indicate that UGT74D1 has low substrate specificity towards auxin-related compounds. However, several lines of evidence indicate that UGT74D1 may actually have relatively high substrate specificity towards OxIAA in plants. First, the Km value of UGT74D1 for OxIAA was 5.5 times lower than that for IAA and the catalytic efficiency (kcat/Km) value of the enzyme for OxIAA was 6.6 times higher than that for IAA, indicating that OxIAA is a more preferred substrate in vitro (Table 2). Secondly, the endogenous concentration of OxIAA-Glc in plants varies with the expression level of UGT74D1; the OxIAA-Glc level decreased by 85% in the ugt74d1 mutant and increased by 13% in the UGT74D1ox plants (Figs. 3D, 4C). Moreover, previous studies have demonstrated that UGT84B1 and UGT74E2 catalyze the glucosylation of IAA and IBA, respectively, and ectopic expression of these genes in Arabidopsis results in similar phenotypes (e.g. compressed rosette, dwarf stature and a loss of apical dominance) (Jackson et al. 2002, Tognetti et al. 2010). On the other hand, UGT74D1 catalyzes the glucosylation of both IAA and IBA in vitro, but ectopic expression of UGT74D1 results in a curling leaf phenotype in Arabidopsis (Jin et al. 2013). In summary, these results suggest that the biological function of UGT74D1 is distinct from those of UGT84B1 and UGT74E2 in plants. However, we cannot exclude the possibility that UGT74D1 plays an auxiliary role in the glucosylation of IAA and IBA.
Metabolic regulation of IAA concentration
OxIAA-Glc is one of the major IAA metabolites in terms of endogenous concentration, and the level of OxIAA-Glc was nearly two orders of magnitude higher than that of IAA, IAA-Asp, IAA-Glu, IAA-Glc and OxIAA in Arabidopsis (Kai et al. 2007). Herein, we showed that the OxIAA-Glc level is reduced by 85% in ugt74d1 mutants (Fig. 3D). However, the ugt74d1 mutant still accumulates a wild-type level of IAA and does not show any IAA-related morphological phenotype (Fig. 3A, B). This result suggests that the IAA concentration in ugt74d1 is probably compensated to wild-type levels by the regulation of IAA biosynthesis and/or metabolism. Detailed analysis of the expression levels for the IAA biosynthetic genes (e.g. TAA, YUC and CYP79B genes) and IAA metabolic genes (e.g. GH3 genes, UGT84B1 and IAMT1) in the ugt74d1 mutant may shed light on understanding the mechanisms regulating IAA homeostasis in plants.
Although previous studies suggest that the OxIAA pathway operates in plants, the rate-limiting step of this pathway remains unknown. Herein, we demonstrated that the induction of UGT74D1 resulted in the loss of root gravitropism in Arabidopsis (Fig. 4A, D), indicating that UGT74D1 catalyzes a committed step in the OxIAA pathway and that an increase in the glucosylation of OxIAA may promote the conversion of IAA to OxIAA. In agreement with this, an increase in OxIAA level was observed in the UGT74D1ox plants (Fig. 4C). Furthermore, treatment of UGT74D1ox plants with IAA led to recovery of root gravitropism (Fig. 4A, D), suggesting that the level of IAA in the root apex of UGT74D1ox plants decreased below the level required for gravitropic response. Intriguingly, the IAA level did not decrease but rather slightly increased in the UGT74D1ox plants when the whole plant was used for IAA analysis (Fig. 4C). A similar phenomenon was observed in Arabidopsis plants overexpressing UGT84B1 (Jackson et al. 2002), in which the overexpression of UGT84B1 resulted in the low-auxin phenotype, although an increase in IAA level was evident. Likewise, an increase in IBA level was observed in Arabidopsis plants overexpressing UGT74E2 (Tognetti et al. 2010). These results suggest that plants might overcompensate the decreases in IAA and OxIAA levels due to overexpression of UGT74D1 by enhancing their formation to maintain cellular homeostasis (Tognetti et al. 2010).
Östin et al. (1998) previously demonstrated that IAA was mainly metabolized to OxIAA and OxIAA-Glc and only a minor portion was conjugated with aspartate or glutamate when a relatively low level of IAA (0.5 µM) was applied to Arabidopsis. In contrast, IAA was mainly metabolized to IAA-Asp and IAA-Glu when a relatively high concentration of IAA (5 µM) was used (Östin et al. 1998). The levels of OxIAA (approximately 60 ng g−1 FW) and OxIAA-Glc (approximately 1,000 ng g−1 FW) were relatively higher than that of IAA (approximately 20 ng g−1 FW) in Arabidopsis seedlings (Fig. 3B–D). On the other hand, the levels of IAA-Asp and IAA-Glu (<10 ng g−1 FW) were much lower than that of IAA, but the co-induction of TAA1 and YUC genes in the IPA-dependent IAA biosynthesis pathway remarkably increased the levels of these metabolites (approximately 600–6,000 ng g−1 FW) (Mashiguchi et al. 2011). In accordance with these results, our RT–PCR data demonstrated that UGT74D1 was constitutively expressed and not markedly affected by the application of IAA, whereas GH3 genes were induced rapidly and strongly (Fig. 5). These results suggest that the OxIAA and GH3 pathways are major pathways that may have distinct roles in IAA homeostasis. The OxIAA pathway may function constitutively to maintain the basal levels of IAA concentration in plants. In contrast, the GH3 pathway may play an important role in cases where plant cells rapidly reduce a relatively large amount of IAA in response to developmental and environmental changes. Identification of the IAA oxidase that catalyzes the conversion of IAA to OxIAA is critical to elucidate further the role of the OxIAA pathway in the metabolism of IAA in Arabidopsis.
Materials and Methods
Plant materials
Arabidopsis thaliana ecotype Columbia-0 was used as the wild type in this study. Seeds of ugt74d1 (SALK_004870), ugt74e1 (SALK_045974) and ugt74e2 (SALK_016116) were obtained from the Arabidopsis Biological Resource Center (ABRC). Sterilized seeds were germinated on MS agar medium (pH 5.7) supplemented with thiamine hydrochloride (3 µg ml−1), nicotinic acid (5 µg ml−1), pyridoxine hydrochloride (0.5 µg ml−1), myo-inositol (100 µg ml−1) and sucrose (1%, w/v) after imbibition at 4°C. Seedlings were grown at 21°C under continuous white light (30–50 µmol m−2 s−1). For RT–PCR and LC-ESI-MS/MS analyses, 7-day-old seedlings grown on MS medium were used. For EST treatment, 4-day-old seedlings of pER8, UGT74D1ox and GH3.6ox were transferred to MS medium containing 10 µM EST and grown for a further 3 d. For recovery of root gravitropism in UGT74D1ox by IAA, 4-day-old seedlings were transferred to MS medium containing 10 µM EST or 10 µM EST with 20 nM IAA and grown for 1–3 d.
Generation of transgenic plants
A cDNA of UGT74D1 in the pDONR207 vector was transferred into the pMDC7 vector by an LR recombination reaction using LR clonase II™ (Invitrogen). A cDNA of GH3.6 was transferred into the pMDC7 vector from the entry vector pENTR/D-TOPO (Invitrogen) by an LR recombination reaction using LR clonase II™ (Invitrogen). Arabidopsis wild-type plants were transformed with the resulting construct pMDC7::UGT74D1 or pMDC7::GH3.6 by the floral dip method using the Agrobacterium tumefaciens GV3101 strain (Clough and Bent 1998).
Expression analysis
Total RNA was isolated from plants using the RNeasy Plant Mini Kit (Qiagen). The PrimeScript RT reagent Kit with gDNA Eraser (Takara) was used for the generation of first-strand cDNA. PCR was carried out with the primers listed in Supplementary Table S2. The cDNA (1 µl) was amplified by PCR using the UGT74D1 or GH3 gene-specific primers. Ubiquitin was used as a reference gene.
Chemicals
[phenyl-13C6]OxIAA, [phenyl-13C6]OxIAA-Glc, OxIAA, OxIAA-Glc and IAA-Glc were synthesized as described in the Supplementary Methods.
LC-ESI-MS/MS analysis of IAA and its metabolites
IAA and OxIAA were analyzed as described in the Supplementary Methods. For analysis of OxIAA-Glc in Arabidopsis seedlings, 30–70 mg of fresh plants were quickly weighed, frozen with liquid nitrogen and stored at −80°C. Plant material was homogenized in 80% acetone/H2O (0.2–1 ml) containing 20–40 ng of [phenyl-13C6]OxIAA-Glc with ceramic beads (3 mm) using a Tissue Lyser (Qiagen) for 3 min. The supernatants were centrifuged at 15,000×g for 3 min at 4°C and transferred to test tubes. The extraction was repeated twice using 80% acetone/H2O (0.2–1 ml) without the internal standard. The supernatants were combined and evaporated by nitrogen gas and centrifuged at 15,000×g for 5 min after the volume was decreased to <200 µl. The supernatant was applied to a 5 µm, 4.6 × 150 mm Symmetry shield C18 column (Waters) coupled to a 5 µm, 4.6 × 10 mm C18 guard column (Senshu Pak) connected to an HPLC system equipped with a 2475 multi λ-fluorescence detector (Waters). The samples were eluted at a flow rate of 1 ml min−1 with 0.01 M ammonium acetate (solvent A) and 100% methanol (solvent B) by using 10% solvent B for 1 min and a gradient of 10–50% solvent B over 30 min. HPLC fractions eluted at the retention time of OxIAA-Glc (16.5–17.5 min) were collected and evaporated using a SpeedVac (Thermo). The dried OxIAA-Glc fraction was redissolved with 1% acetic acid/H2O (1 ml) and applied to an Oasis HLB column (Waters). The column was washed with 10% methanol/H2O containing 1% acetic acid (1 ml) before eluting the OxIAA-Glc with 60% methanol/H2O containing 1% acetic acid (1 ml). The eluate was evaporated to dryness using a SpeedVac. OxIAA-Glc was analyzed by an ACQUITY Ultra Performance Liquid Chromatography (UPLC)-MS/MS Q-Tof-premier (Waters). The chromatography was performed on an ACQUITY UPLC BEH C18 1.7 µm, 2.1 × 50 mm column (Waters). The OxIAA-Glc fraction was redissolved in 50% acetonitrile/H2O (20 µl) and injected into the UPLC column. Elution of the samples was carried out with 0.05% acetic acid (solvent A2) and acetonitrile with 0.05% acetic acid (solvent B2) using 2% solvent B2 for 0.1 min and a gradient ranging from 2% to 30% of solvent B2 for 7.4 min at a flow rate of 0.2 ml min−1. The temperature of the UPLC column was 40°C. The retention time of OxIAA-Glc and [phenyl-13C6]OxIAA-Glc was 3.14 min. OxIAA-Glc and [phenyl-13C6]OxIAA-Glc were analyzed in the negative ion mode. MS/MS analysis conditions were as follows: capillary = 2.65 kV, source temperature = 100°C, desolvation temperature = 400°C, collision energy = 9.4 V, sampling cone voltage = 28 V, scan time = 0.6 s per scan (delay = 0.05 s) and the MS/MS transition (m/z) = 352.1/190.1 for unlabeled OxIAA-Glc and 358.1/196.1 for [phenyl-13C6]OxIAA-Glc. Quantification was carried out using the extracted ion chromatogram of OxIAA-Glc and [phenyl-13C6]OxIAA-Glc. A standard curve was generated using OxIAA-Glc and [phenyl-13C6]OxIAA-Glc as described above, except for the omission of the HPLC purification step.
Isolation of OxIAA glucosyltransferase using budding yeast
Full-length cDNAs of UGTs were amplified by PCR with the primers listed in Supplementary Table S2 using RIKEN Arabidopsis full-length clones as templates (Supplementary Table S1). To express the UGT proteins in yeast, UGT genes were subcloned into the pDONR207 vector by the BP recombination reaction using BP clonase II™ (Invitrogen) and transferred into the modified pYES-DEST52 vector described in Kanno et al. (2012) by an LR recombination reaction using LR clonase II™ (Invitrogen). The primer design and recombination reactions were performed following the manufacturer’s instructions. The S. cerevisiae w303-1a strain was transformed and incubated in medium for 16 h at 30°C. The culture was added to the medium when the OD600 was 0.1. After incubation with 10 µM OxIAA for 24 h at 30°C, the cell culture was centrifuged at 3,000×g for 5 min. The supernatant was filtered by a 0.22 µm filter and fractionated by HPLC. The OxIAA-Glc fraction was applied to an Oasis HLB cartridge and analyzed by LC-ESI-MS/MS.
Preparation of recombinant GST–UGT74D1
To express UGT74D1 as a GST-fused protein (GST–UGT74D1), a cDNA of UGT74D1 was amplified using UGT74D1_F and UGT74D1_R primers (Supplementary Table S2), cloned into the pDONR207 vector and transferred into the pDEST15 vector (Invitrogen) by an LR recombination reaction. The E. coli BL21 Star (DE3) strain transformed with the resulting construct pDEST15::UGT74D1 was incubated at 18°C in Terrific-broth medium containing 100 µg ml−1 carbenicillin. For induction of GST–UGT74D1 expression, Isopropyl β-d-1-thiogalactopyranoside (1 mM, final concentration) was added to the culture when the OD600 was approximately 0.6. After incubation of cells for 2 d at 18°C, the GST–UGT74D1 protein was purified using glutathione–Sepharose 4B (GE Healthcare) following the manufacturer’s instructions. The purified proteins were kept at −80°C with 20% (w/v) glycerol until enzyme assays.
Assay for glucosyltransferase activity
OxIAA glucosyltransferase activity was determined by measuring OxIAA-Glc formation. Each assay consisted of HEPES buffer (50 mM, pH 7.5), UDP-glucose (1 mM) and OxIAA (100 µM) in a total volume of 100 µl. The enzyme reaction was initiated by the addition of the GST–UGT74D1 protein (1 µg). The reactions were carried out at 30°C for 4 min. The reactions were terminated by adding 100 µl of acetonitrile. After centrifugation at 15,000×g, the supernatant was diluted 10-fold with 50% acetonitrile/H2O and quickly injected into the LC-ESI-MS/MS apparatus for OxIAA-Glc analysis.
IAA glucosyltransferase activity was determined by measuring IAA-Glc formation. Each assay consisted of HEPES buffer (50 mM, pH 7.5), UDP-glucose (1 mM) and IAA (100 µM) in a total volume of 100 µl. The enzyme reaction was initiated by the addition of the GST–UGT74D1 protein (1 µg). The reactions were carried out at 30°C for 8 min. The reactions were terminated by adding 100 µl of acetonitrile. After centrifugation at 15,000×g, the supernatant was injected into the HPLC system. IAA-Glc was applied to a Symmetry shield C18 column coupled to a C18 guard column connected to an HPLC system and detected by a fluorescence detector. The samples were eluted at a flow rate of 1 ml min−1 with 0.01 M ammonium acetate (solvent A) and 100% methanol (solvent B) using 10% solvent B for 1 min and a gradient ranging from 10% to 50% of solvent B over 30 min.
Steady-state kinetic parameters
The OxIAA glucosyltransferase assay based on the formation of OxIAA-Glc was employed for the steady-state kinetic studies of UGT74D1. Each assay consisted of HEPES buffer (50 mM, pH 7.5), UDP-glucose (2.5 mM) and OxIAA in a total volume of 50 µl. The concentration of OxIAA varied (2.5, 5, 7.5, 10, 20, 30 and 40 µM). The enzyme reaction was initiated by the addition of the GST–UGT74D1 protein. The reactions were carried out at 30°C for 4 min. The reactions were terminated by adding 50 µl of acetonitrile containing [phenyl-13C6]OxIAA-Glc. The reaction mixtures were centrifuged at 15,000×g for 10 min at 4°C. The supernatants were diluted 10-fold with 50% acetonitrile/H2O and quickly injected into the LC-ESI-MS/MS system for OxIAA-Glc analysis. The initial velocity was determined from the slope of a plot of OxIAA-Glc concentration vs. incubation time. Steady-state kinetic parameters were calculated from a Lineweaver–Burk plot.
An IAA glucosyltransferase assay based on the formation of IAA-Glc was employed for the steady-state kinetic studies of UGT74D1. Each assay consisted of HEPES buffer (50 mM, pH 7.5), UDP-glucose (2.5 mM) and IAA in a total volume of 50 µl. The concentration of IAA varied (10, 20, 30, 40, 60, 80, 100, 150 and 200 µM). The enzyme reaction was initiated by the addition of the GST–UGT74D1 protein, and the reaction was carried out at 30°C for 8 min. The reaction was terminated by adding 50 µl of acetonitrile. The reaction mixtures were centrifuged at 15,000×g for 10 min at 4°C. The supernatant was injected into the HPLC system for IAA-Glc analysis. Initial velocity was determined from the slope of a plot of the concentration of IAA-Glc vs. incubation time. Steady-state kinetic parameters were calculated from a Lineweaver–Burk plot.
Supplementary data
Supplementary data are available at PCP online.
Funding
This study was supported by the Japan Society for the Promotion of Science [KAKENHI Grant 24370027 (to H.K.), Research Fellowship for Young Scientists 256665 (to K.T.) and JST, PRESTO (to H.K.)].
Supplementary Material
Acknowledgements
We thank Dr. Kiyoshi Mashiguchi, Ms. Yumiko Takebayashi and Dr. Belay Ayele for their helpful comments on LC-ESI-MS/MS analysis, enzyme assays and the critical reading of the manuscript. We thank Professor Nam-Hai Chua for providing the pMDC7 vector, the RIKEN Bio Resource Center for providing the Arabidopsis full-length cDNA clones, and the ABRC for providing the T-DNA insertion mutant seeds of ugt74d1, ugt74e1 and ugt74e2. We are grateful to Ms. Aya Ide for assistance in preparing plant materials.
Glossary
Abbreviations
- EST
β-estradiol
- GH3
GRETCHEN HAGEN 3
- GST
glutathione S-transferase
- GUS
β-glucuronidase
- IAA-Asp
IAA-aspartate
- IAA-Glc
1-O-(indol-3-ylacetyl)-β-d-glucose
- IAA-Glu
IAA-glutamate
- IBA
indole-3-butylic acid
- IBA-Glc
1-O-(indol-3-ylbutanoyl)-β-d-glucose
- LC-ESI-MS/MS
liquid chromatography-electrospray ionization-tandem mass spectrometry
- MS
Murashige and Skoog
- MS/MS
tandem mass spectrometry
- OxIAA
2-oxindole-3-acetic acid
- OxIAA-Glc
OxIAA-glucoside
- RT–PCR
reverse transcription–PCR
- UGT
UDP-glycosyltransferase
- UPLC
ultra performance liquid chromatography
Disclosures
The authors have no conflicts of interest to declare.
References
- Barlier I, Kowalczyk M, Marchant A, Ljung K, Bhalerao R, Bennett M, et al. The SUR2 gene of Arabidopsis thaliana encodes the cytochrome P450 CYP83B1, a modulator of auxin homeostasis. Proc. Natl Acad. Sci. USA. 2000;97:14819–14824. doi: 10.1073/pnas.260502697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel B, Fink GR. ILR1, an amidohydrolase that releases active indole-3-acetic-acid from conjugates. Science. 1995;268:1745–1748. doi: 10.1126/science.7792599. [DOI] [PubMed] [Google Scholar]
- Chamarro J, Östin A, Sandberg G. Metabolism of indole-3-acetic acid by orange (Citrus sinensis) flavedo tissue during fruit development. Phytochemistry. 2001;57:179–187. doi: 10.1016/s0031-9422(01)00023-1. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 2006;20:1790–1799. doi: 10.1101/gad.1415106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y. Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell. 2007;19:2430–2439. doi: 10.1105/tpc.107.053009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cohen JD, Bandurski RS. Chemistry and physiology of the bound auxins. Annu. Rev. Plant Physiol. 1982;33:403–430. [Google Scholar]
- Davies RT, Goetz DH, Lasswell J, Anderson MN, Bartel B. IAR3 encodes an auxin conjugate hydrolase from Arabidopsis. Plant Cell. 1999;11:365–376. doi: 10.1105/tpc.11.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas CM, Foor F, Marrinan JA, Morin N, Nielsen JB, Dahl AM, et al. The Saccharomyces cerevisiae FKS1 (ETG1) gene encodes an integral membrane protein which is a subunit of 1,3-β-d-glucan synthase. Proc. Natl Acad. Sci. USA. 1994;91:12907–12911. doi: 10.1073/pnas.91.26.12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachon CM, Langlois-Meurinne M, Saindrenan P. Plant secondary metabolism glycosyltransferases: the emerging functional analysis. Trends Plant Sci. 2005;10:542–549. doi: 10.1016/j.tplants.2005.09.007. [DOI] [PubMed] [Google Scholar]
- Grubb CD, Zipp BJ, Ludwig-Müller J, Masuno MN, Molinski TF, Abel S. Arabidopsis glucosyltransferase UGT74B1 functions in glucosinolate biosynthesis and auxin homeostasis. Plant J. 2004;40:893–908. doi: 10.1111/j.1365-313X.2004.02261.x. [DOI] [PubMed] [Google Scholar]
- Grunewald W, Friml J. The march of the PINs: developmental plasticity by dynamic polar targeting in plant cells. EMBO J. 2010;29:2700–2714. doi: 10.1038/emboj.2010.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K. The interaction and integration of auxin signaling components. Plant Cell Physiol. 2012;53:965–975. doi: 10.1093/pcp/pcs035. [DOI] [PubMed] [Google Scholar]
- Iyer M, Slovin JP, Epstein E, Cohen JD. Transgenic tomato plants with a modified ability to synthesize indole-3-acetyl-β-1-O-d-glucose. J. Plant Growth Regul. 2005;24:142–152. [Google Scholar]
- Jackson RG, Kowalczyk M, Li Y, Higgins G, Ross J, Sandberg G, et al. Over-expression of an Arabidopsis gene encoding a glucosyltransferase of indole-3-acetic acid: phenotypic characterisation of transgenic lines. Plant J. 2002;32:573–583. doi: 10.1046/j.1365-313x.2002.01445.x. [DOI] [PubMed] [Google Scholar]
- Jackson RG, Lim EK, Li Y, Kowalczyk M, Sandberg G, Hoggett J, et al. Identification and biochemical characterization of an Arabidopsis indole-3-acetic acid glucosyltransferase. J. Biol. Chem. 2001;276:4350–4356. doi: 10.1074/jbc.M006185200. [DOI] [PubMed] [Google Scholar]
- Jakubowska A, Kowalczyk S. The auxin conjugate 1-O-indole-3-acetyl-β-d-glucose is synthesized in immature legume seeds by IAGlc synthase and may be used for modification of some high molecular weight compounds. J. Exp. Bot. 2004;55:791–801. doi: 10.1093/jxb/erh086. [DOI] [PubMed] [Google Scholar]
- Jin SH, Ma XM, Han P, Wang B, Sun YG, Zhang GZ, et al. UGT74D1 is a novel auxin glycosyltransferase from Arabidopsis thaliana. PLoS One. 2013;8:e61705. doi: 10.1371/journal.pone.0061705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai K, Horita J, Wakasa K, Miyagawa H. Three oxidative metabolites of indole-3-acetic acid from Arabidopsis thaliana. Phytochemistry. 2007;68:1651–1663. doi: 10.1016/j.phytochem.2007.04.030. [DOI] [PubMed] [Google Scholar]
- Kanno Y, Hanada A, Chiba Y, Ichikawa T, Nakazawa M, Matsui M, et al. Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proc. Natl Acad. Sci. USA. 2012;109:9653–9658. doi: 10.1073/pnas.1203567109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk M, Sandberg G. Quantitative analysis of indole-3-acetic acid metabolites in Arabidopsis. Plant Physiol. 2001;127:1845–1853. [PMC free article] [PubMed] [Google Scholar]
- LeClere S, Tellez R, Rampey RA, Matsuda SP, Bartel B. Characterization of a family of IAA–amino acid conjugate hydrolases from Arabidopsis. J. Biol. Chem. 2002;277:20446–20452. doi: 10.1074/jbc.M111955200. [DOI] [PubMed] [Google Scholar]
- Li Y, Baldauf S, Lim EK, Bowles DJ. Phylogenetic analysis of the UDP-glycosyltransferase multigene family of Arabidopsis thaliana. J. Biol. Chem. 2001;276:4338–4343. doi: 10.1074/jbc.M007447200. [DOI] [PubMed] [Google Scholar]
- Lim EK, Bowles DJ. A class of plant glycosyltransferases involved in cellular homeostasis. EMBO J. 2004;23:2915–2922. doi: 10.1038/sj.emboj.7600295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim EK, Doucet CJ, Li Y, Elias L, Worrall D, Spencer SP, et al. The activity of Arabidopsis glycosyltransferases toward salicylic acid, 4-hydroxybenzoic acid, and other benzoates. J. Biol. Chem. 2002;277:586–592. doi: 10.1074/jbc.M109287200. [DOI] [PubMed] [Google Scholar]
- Ljung K. Auxin metabolism and homeostasis during plant development. Development. 2013;140:943–950. doi: 10.1242/dev.086363. [DOI] [PubMed] [Google Scholar]
- Ljung K, Hull AK, Kowalczyk M, Marchant A, Celenza J, Cohen JD, et al. Biosynthesis, conjugation, catabolism and homeostasis of indole-3-acetic acid in Arabidopsis thaliana. Plant Mol. Biol. 2002;50:309–332. doi: 10.1023/a:1016024017872. [DOI] [PubMed] [Google Scholar]
- Mashiguchi K, Tanaka K, Sakai T, Sugawara S, Kawaide H, Natsume M, et al. The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl Acad. Sci. USA. 2011;108:18512–18517. doi: 10.1073/pnas.1108434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messner B, Thulke O, Schaffner AR. Arabidopsis glucosyltransferases with activities toward both endogenous and xenobiotic substrates. Planta. 2003;217:138–146. doi: 10.1007/s00425-002-0969-0. [DOI] [PubMed] [Google Scholar]
- Nakazawa M, Yabe N, Ichikawa T, Yamamoto YY, Yoshizumi T, Hasunuma K, et al. DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. Plant J. 2001;25:213–221. doi: 10.1046/j.1365-313x.2001.00957.x. [DOI] [PubMed] [Google Scholar]
- Normanly J. Auxin metabolism. Physiol. Plant. 1997;100:431–442. [Google Scholar]
- Novák O, Hényková E, Sairanen I, Kowalczyk M, Pospíšil T, Ljung K. Tissue-specific profiling of the Arabidopsis thaliana auxin metabolome. Plant J. 2012;72:523–536. doi: 10.1111/j.1365-313X.2012.05085.x. [DOI] [PubMed] [Google Scholar]
- Oka T, Jigami Y. Reconstruction of de novo pathway for synthesis of UDP-glucuronic acid and UDP-xylose from intrinsic UDP-glucose in Saccharomyces cerevisiae. FEBS J. 2006;273:2645–2657. doi: 10.1111/j.1742-4658.2006.05281.x. [DOI] [PubMed] [Google Scholar]
- Östin A, Kowalyczk M, Bhalerao RP, Sandberg G. Metabolism of indole-3-acetic acid in Arabidopsis. Plant Physiol. 1998;118:285–296. doi: 10.1104/pp.118.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin G, Gu H, Zhao Y, Ma Z, Shi G, Yang Y, et al. An indole-3-acetic acid carboxyl methyltransferase regulates Arabidopsis leaf development. Plant Cell. 2005;17:2693–2704. doi: 10.1105/tpc.105.034959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiel JA, Bender J. Glucose conjugation of anthranilate by the Arabidopsis UGT74F2 glucosyltransferase is required for tryptophan mutant blue fluorescence. J. Biol. Chem. 2003;278:6275–6281. doi: 10.1074/jbc.M211822200. [DOI] [PubMed] [Google Scholar]
- Rampey RA, LeClere S, Kowalczyk M, Ljung K, Sandberg G, Bartel B. A family of auxin-conjugate hydrolases that contributes to free indole-3-acetic acid levels during Arabidopsis germination. Plant Physiol. 2004;135:978–988. doi: 10.1104/pp.104.039677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinecke DM, Bandurski RS. Oxindole-3-acetic acid, an indole-3-acetic acid catabolite in Zea mays. Plant Physiol. 1983;71:211–213. doi: 10.1104/pp.71.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinecke DM, Bandurski RS. Oxidation of indole-3-acetic acid to oxindole-3-acetic acid by an enzyme preparation from Zea mays. Plant Physiol. 1988;86:868–872. doi: 10.1104/pp.86.3.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosquete MR, Barbez E, Kleine-Vehn J. Cellular auxin homeostasis: gatekeeping is housekeeping. Mol. Plant. 2012;5:772–786. doi: 10.1093/mp/ssr109. [DOI] [PubMed] [Google Scholar]
- Ross J, Li Y, Lim E, Bowles DJ. Higher plant glycosyltransferases. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-2-reviews3004. reviews 3004.1–3004.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sairanen I, Novák O, Pěnčík A, Ikeda Y, Jones B, Sandberg G, et al. Soluble carbohydrates regulate auxin biosynthesis via PIF proteins in Arabidopsis. Plant Cell. 2012;24:4907–4916. doi: 10.1105/tpc.112.104794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahinian S, Bussey H. β-1,6-Glucan synthesis in Saccharomyces cerevisiae. Mol. Microbiol. 2000;35:477–489. doi: 10.1046/j.1365-2958.2000.01713.x. [DOI] [PubMed] [Google Scholar]
- Sinlapadech T, Stout J, Ruegger MO, Deak M, Chapple C. The hyper-fluorescent trichome phenotype of the brt1 mutant of Arabidopsis is the result of a defect in a sinapic acid:UDPG glucosyltransferase. Plant J. 2007;49:655–668. doi: 10.1111/j.1365-313X.2006.02984.x. [DOI] [PubMed] [Google Scholar]
- Sitbon F, Östin A, Sundberg B, Olsson O, Sandberg G. Conjugation of indole-3-acetic acid (IAA) in wild-type and IAA-overproducing transgenic tobacco plants, and identification of the main conjugates by frit-fast atom bombardment liquid chromatography-mass spectrometry. Plant Physiol. 1993;101:313–320. doi: 10.1104/pp.101.1.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, Maldonado MC, et al. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell. 2005;17:616–627. doi: 10.1105/tpc.104.026690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie DY, Dolezal K, et al. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell. 2008;133:177–191. doi: 10.1016/j.cell.2008.01.047. [DOI] [PubMed] [Google Scholar]
- Stepanova AN, Yun J, Robles LM, Novak O, He W, Guo H, et al. The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell. 2011;23:3961–3973. doi: 10.1105/tpc.111.088047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztein AE, Cohen JD, de la Fuente IG, Cooke TJ. Auxin metabolism in mosses and liverworts. Amer. J. Bot. 1999;86:1544–1555. [PubMed] [Google Scholar]
- Takase T, Nakazawa M, Ishikawa A, Kawashima M, Ichikawa T, Takahashi N, et al. ydk1-D, an auxin-responsive GH3 mutant that is involved in hypocotyl and root elongation. Plant J. 2004;37:471–483. doi: 10.1046/j.1365-313x.2003.01973.x. [DOI] [PubMed] [Google Scholar]
- Takase T, Nakazawa M, Ishikawa A, Manabe K, Matsui M. DFL2, a new member of the Arabidopsis GH3 gene family, is involved in red light-specific hypocotyl elongation. Plant Cell Physiol. 2003;44:1071–1080. doi: 10.1093/pcp/pcg130. [DOI] [PubMed] [Google Scholar]
- Tao Y, Ferrer JL, Ljung K, Pojer F, Hong F, Long JA, et al. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell. 2008;133:164–176. doi: 10.1016/j.cell.2008.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognetti VB, Van Aken O, Morreel K, Vandenbroucke K, van de Cotte B, De Clercq I, et al. Perturbation of indole-3-butyric acid homeostasis by the UDP-glucosyltransferase UGT74E2 modulates Arabidopsis architecture and water stress tolerance. Plant Cell. 2010;22:2660–2679. doi: 10.1105/tpc.109.071316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste S, Friml J. Auxin: a trigger for change in plant development. Cell. 2009;136:1005–1016. doi: 10.1016/j.cell.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Won C, Shen X, Mashiguchi K, Zheng Z, Dai X, Cheng Y, et al. Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc. Natl Acad. Sci. USA. 2011;108:18518–18523. doi: 10.1073/pnas.1108436108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward AW, Bartel B. Auxin: regulation, action, and interaction. Ann. Bot. 2005;95:707–735. doi: 10.1093/aob/mci083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M, Greenham K, Prigge MJ, Jensen PJ, Estelle M. The TRANSPORT INHIBITOR RESPONSE2 gene is required for auxin synthesis and diverse aspects of plant development. Plant Physiol. 2009;151:168–179. doi: 10.1104/pp.109.138859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Saito K. Functional genomics for plant natural product biosynthesis. Nat. Prod. Rep. 2009;26:1466–1487. doi: 10.1039/b817077k. [DOI] [PubMed] [Google Scholar]
- Zhao Y. Auxin biosynthesis and its role in plant development. Annu. Rev. Plant Biol. 2010;61:49–64. doi: 10.1146/annurev-arplant-042809-112308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubieta C, Ross JR, Koscheski P, Yang Y, Pichersky E, Noel JP. Structural basis for substrate recognition in the salicylic acid carboxyl methyltransferase family. Plant Cell. 2003;15:1704–1716. doi: 10.1105/tpc.014548. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.