Abstract
The major phosphoprotein in dentin is the aspartic acid and serine-rich protein called dentin phosphophoryn (DPP). DPP appears to be synthesized as a part of a larger compound protein, dentin sialophosphoprotein (DSPP). DSPP has never been isolated or detected in dentin extracts. It is now evident that DSPP is a chimeric protein composed of 3 parts: dentin sialoprotein (DSP), DPP, and dentin glycoprotein (DGP). Previous reports have suggested that the BMP1 protease is responsible for processing DSPP. However, unequal amounts of these products are present in the dentin matrix. Here, we provide evidence for an internal ribosome entry site in the DSPP gene that directs the synthesis of DPP. This mechanism would account for unequal amounts of intracellular DSP and DPP. The internal ribosomal entry site (IRES) activity varied in different cell types, suggesting the presence of additional regulatory elements during the translational regulation of DPP. Further, we provide evidence that DPP is transported to the extracellular matrix (ECM) through exosomes. Using tissue recombination and lentivirus-mediated gain-of-function approaches, we also demonstrate that DPP is essential for the formation of well-defined tooth structures with mineralized dentin matrix.
Keywords: extracellular matrix (ECM), cell biology, cell differentiation, dentin, matrix biology, odontoblast(s)
Introduction
Dentin sialophosphoprotein (DSPP) belongs to the SIBLING (small integrin-binding ligand, N-linked glycoprotein) family, which contains 5 genetically related members (Fisher et al., 2001). These genes are located as a tandem cluster on the chromosome among different species (Fisher, 2011). Initially deemed as an odontoblast-specific gene (Bègue-Kirn et al., 1998), DSPP has now been localized in many other tissues (Alvares et al., 2006). In humans, mutations in the DSPP gene result in different types of dentinogenesis-related diseases (reviewed by Bailleul-Forestier et al., 2008). The phenotype present in the DSPP knockout mouse shows defective dentin development, similar to human detinogenesis imperfecta type III (Sreenath et al., 2003).
The organic matrix of dentin consists predominantly of type I collagen and noncollagenous proteins (NCPs) (Linde, 1989). Dentin sialoprotein (DSP), the glycoprotein and proteoglycan forms, and dentin phosphophoryn (DPP) are the most abundant NCPs. Both DSP and DPP are encoded by a single-gene DSPP (MacDougall et al., 1997; Yamakoshi et al., 2008; Zhu et al., 2010). DSPP is a multidomain protein with DSP at the N terminus and DPP at the C terminus. The sequence in the middle encoded for a small protein called dentin glycoprotein (DGP) (Yamakoshi et al., 2005). It is believed that DSPP is synthesized in its entirety as a precursor protein and then proteolytically processed into DSP, DGP, and DPP by proteases like bone morphogenetic protein-1 (BMP1) (Yamakoshi et al., 2006; Sun et al., 2009; von Marschall and Fisher, 2010; Tsuchiya et al., 2011). Such a processing mechanism should generate a 1:1 ratio of DSP and DPP, in the absence of other processing mechanisms. However, published reports have demonstrated that DPP accounts for 50% of the non-collagenous proteins, while only 5% of DSP (glycoprotein form) is present in the dentin matrix (Qin et al., 2001; Yamakoshi et al., 2006). Moreover, DSPP holoprotein has not been detected during any stage of tooth development or in the dentin matrix. This observation implies that other alternative mechanisms might direct the synthesis of DPP.
During a search for mechanisms of translational regulation of DPP, we analyzed the DSPP sequence for the possibility of an internal ribosomal entry site (IRES). IRES is a cap-independent translational initiation mechanism. In this mechanism, the translation is initiated at an internal ribosome entry site sequence of specific mRNA. More recently, IRES activities have been detected in an increasing number of cellular mRNAS in several species, implying that the IRES process is far more extensive than previously thought. It is possible that IRES-mediated translational mechanism might play a crucial role in generating protein diversity (Touriol et al., 2003).
In this study we provide evidence for a unique internal expression of the carboxyl terminal domain of DSPP, mediated by a putative IRES element present within the coding region. Further, we demonstrate that DPP is transported to the extracellular matrix (ECM) through exosomes. Identifying the mechanism by which DPP is synthesized and transported is crucial, since failure of DPP translation is a potential cause of developmental dentin effects.
Materials & Methods
Cloning of DSPP in Double-labeling Vector
The full-length mouse DSPP cDNA without the signal peptide was cloned into the TOPO4 vector with the primers mentioned in the Appendix Table. DSPP plasmid was then excised with Spe1 and cloned into the engineered DsRed-GFP vector (a kind gift from Dr. Jantsch), and the insert was confirmed by sequencing.
Cloning of Putative IRES Region in Bicistronic Vectors
We amplified the speculated IRES fragment by PCR using Pfu DNA polymerase, and the template used was the cDNA obtained from mouse dental pulp cells. Four regions were amplified with the respective primers shown in the Appendix Table. Region 1 contained 72 bp of DSP and 24 bp of DGP. Region 2 contained 126 bp of DSP and 90 bp of DGP. Region 3 contained 126 bp of DSP. Region 4 contained 267 bp of DGP. The amplified fragments were cloned into TA cloning vector (Promega, Madison, WI, USA) and then cloned into the pRF vector, which is a bicistronic vector containing Renilla and Photinus luciferase as the first and second cistrons, and containing the simian virus 40 (SV40) promoter to transcribe the bicistronic mRNA. The positive control plasmid used, PVRF, contains polio virus IRES individually between Renilla and Photinus luciferase and was kindly provided by Dr. Anne E. Willis. Reporter constructs had the initial nomenclature as proposed earlier (Stoneley et al., 1998), which were named pRF.
The second bicistronic vector used to observe the localization of the processed fragments is the DsRed-GFP vector. The inserts cloned into this vector were the full-length DSPP (2784 bp) and a fragment containing only the DSP and DGP fragments (1302 bp).
Cell Culture and Transfection
Cell lines used were the mouse pre-osteoblast cell line MC3T3-E1 (ATCC catalog number CRL-2593), dental pulp stem cells (DPSCs) (Gronthos et al., 2002), T4-4 rat pre-odontoblast cells (Hao et al. 2002), and human embryonic kidney HEK293 cells. The MC3T3-E1 cells were cultured in DMEM/F12 media without ascorbic acid, purchased from Invitrogen (Carlsbad, CA, USA; Gibco code A10490-01). DPSC cells were grown in αMEM media, and HEK293 cells were incubated in DMEM media. All cell types except DPSCs were cultured in media supplemented with 10% fetal bovine serum (FBS) and antibiotics (100 U/mL penicillin and100 µg/mL streptomycin, Gibco Invitrogen). DPSCs were supplemented with 20% FBS. Cells were maintained at subconfluent conditions, in a humidified incubator at 37°C, with ambient oxygen and 5% CO2.
Reporter constructs and control vectors were transfected into the above-mentioned cells by means of the transfection kit FuGENE HD (Cat # 04709691001, Roche Applied Science, Indianapolis, IN, USA). Cells were harvested 24 hrs after transfection, and reporter gene activities were determined with the Dual-Luciferase Reporter Assay System (Promega E1960). Relative IRES activity was quantified as the ratio of the expression of Photinus luciferase to that of Renilla luciferase.
Confocal Microscopy
Cells expressing DsRed-DSPP-GFP plasmid were imaged with a Zeiss LSM 710 confocal microscope equipped with Zen image analysis software. The expression levels of DsRed or GFP were recorded by confocal images through a red or green channel. Prior to being imaged, the cells were fixed in 4% paraformaldehyde, permeabilized with 0.5% Triton X-100, and mounted on glass slides with Vectashield mounting medium containing DAPI nuclear stain (Vector Laboratories Inc., Burlingame, CA, USA).
Western Blot Analysis
Total proteins were isolated from HEK293 cells after transient transfections with pRGFP vectors containing either DSPP or DSP+DGP and subjected to SDS-PAGE followed by transfer onto nitrocellulose membranes. Detection of the antigen was performed with anti-DSP (1:500), anti-DPP (1:500), and anti-GFP (1/500) antibodies. The bands were visualized by the ECL-Western blot reagent (PerkinElmer Life Sciences, Waltham, MA, USA). The blots were stripped and probed with anti-tubulin antibody (1:10,000) (Sigma, St. Louis, MO, USA) to obtain loading control data. Total proteins from exosomes were processed and probed with anti-DPP antibody and CD-63 (1:500) (Santa Cruz Biotechnology, Santa Cruz, CA, USA) as above.
Cloning of DPP to Generate Lentivirus Particles
DPP was cloned into the expression vector p3X-FLAGS-CMB-13 (E4776, SIGMA) with the primers mentioned in the Table. The cloned DNA fragment was released by ECORI and SPEI from the p3X vector. A signal peptide was appended at the N-terminus, and 3 FLAG Tags were inserted at the C-terminal and then inserted into the SmaI site of pLVX-Puro (Lentivirus vector 632164, Clontech, Madison, WI, USA). The virus was packaged with the Lenti XHT packaging system (632160 Clontech), and the DPP lentivirus particles were collected as per the manufacturer’s protocol.
Mouse Kidney Capsule Surgery Protocol
Sub-renal culture was performed in adult male DSPP null mice as reported previously (Song et al., 2006). After 30 days, the host mice were sacrificed, and implanted samples were analyzed by x-ray (Faxitron Analysis, Tucson, AZ, USA) and histological stains. All experiments were performed as per UIC protocol Animal Assurance number 10-221.
Results
Localization of DSP and DPP in the ECM Matrices Secreted by T4-4 Pre-odontoblasts
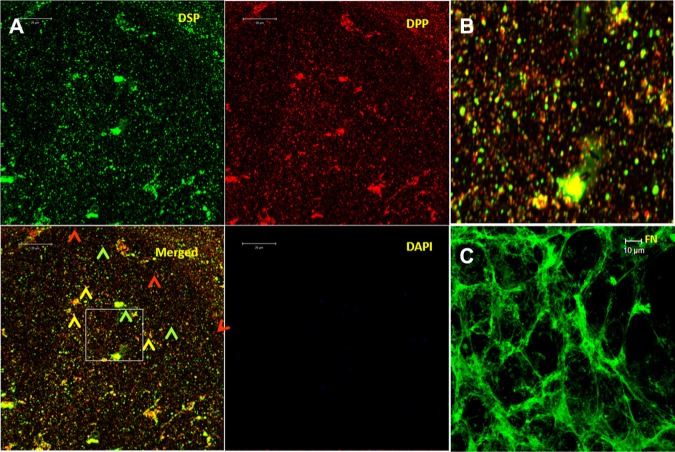
Immunohistochemical analysis performed on the ECM matrix showed areas of distinct localization of DSP (green) and DPP (red), and in some regions they appeared to be in close proximity (yellow) (Figs. 1A, 1B). Fibronectin was used as a positive control (Fig. 1C).
Figure 1.
Localization of DSP and DPP in the ECM secreted by T4-4 cells. (A) ECM was isolated from T4-4 cells grown to confluence as stated in “Materials & Methods”. Anti-DSP and anti-DPP antibodies were used to stain the matrix according to published protocols. Red, green, and yellow arrowheads depict DPP, DSP, and areas where both DSP and DPP are present in close proximity. (B) An enlarged portion of the boxed area. (C) Fibronectin (positive control) in the ECM matrix. (The figure can be viewed in color online.)
In silico Analysis of the DSPP Gene for Secondary Structures
To identify a mechanism by which DSPP is processed intracellularly, we examined the DSPP gene for the presence of IRES elements. Analysis of the DSPP gene indicated GC-rich repeats, suggesting a complex secondary structure at the RNA level. Therefore, RNA secondary structure was analyzed at various regions between the DSP and DPP portions of DSPP mRNA by the available RNA-fold software at http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi. In silico analysis showed the presence of various hairpin-loop secondary structures in regions 1-4 (Figs. 2A, 2B). The most complex secondary structure was observed in Region 2. This suggested that the IRES spanning domain might reside within 126 bp of DSP and 90 bp of DGP. Similar stem and Y structures are characteristic of viral IRES structures.
Figure 2.
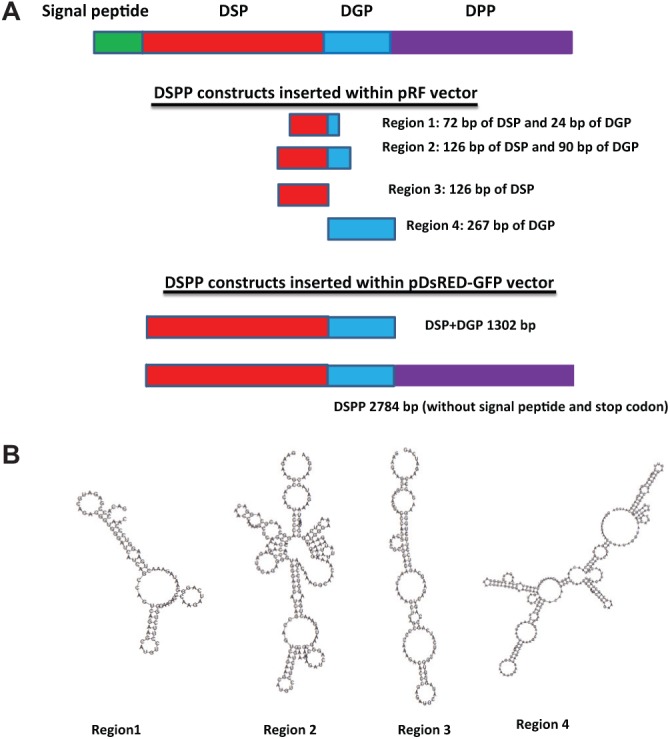
Identification of the IRES region in DSPP. (A) Four regions were identified for potential IRES domain in DSPP. They were inserted into the pRF vector between Renilla luciferase and Firefly luciferase using SpeI and EcoRI cutting sites. In the DsRed-GFP vector, the entire DSPP cDNA and a 1302-bp fragment containing DSP and DGP domain were inserted between the Spe1 and BamH1 sites. (B) In silico analysis of the putative ~400-bp region between the end of DSP and the start of DPP. This site was arbitrarily divided into 4 regions. Region 1 contains 72 bp of DSP and 24 bp of DGP and shows the characteristic stem-loop structure; Region 2 consists of 126 bp of DSP and 90 bp of DGP and shows a more complicated secondary structure containing stems and loops; Region 3 is comprised of 126 bp of DSP, and Region 4 consists of 267 bp of entire DGP.
Functional Activity of the IRES Domain in DSPP
To test if this domain possesses IRES activity, we cloned each of the 4 regions shown in Fig. 2 in-frame into the bicistronic luciferase vector. Here, the SV40 promoter drives Renilla luciferase in the first cistron, followed by the various DSPP-containing regions linked to Firefly luciferase in the second cistron. Results (Fig. 3A) depict the luciferase activity after transient transfections in HEK293 (1), MC3T3-E1 (2), and T4-4 (3) cells, assayed as a surrogate for translational activity. Luciferase activity indicated a substantial overall increase of IRES activity in pRF vectors containing Region 2. The increase over control levels was ~ 10-fold in HEK293 cells, ~ 15-fold in MC3T3-E1 cells, and ~13-fold in T4-4 cells. The activity within this region corroborates well with the complex secondary structure predicted for this region. Region 4, which contained 267 bp of DGP, contained the lowest activity. These findings provide strong evidence that the 216 bp in DSPP mRNA likely harbors bona fide IRES elements.
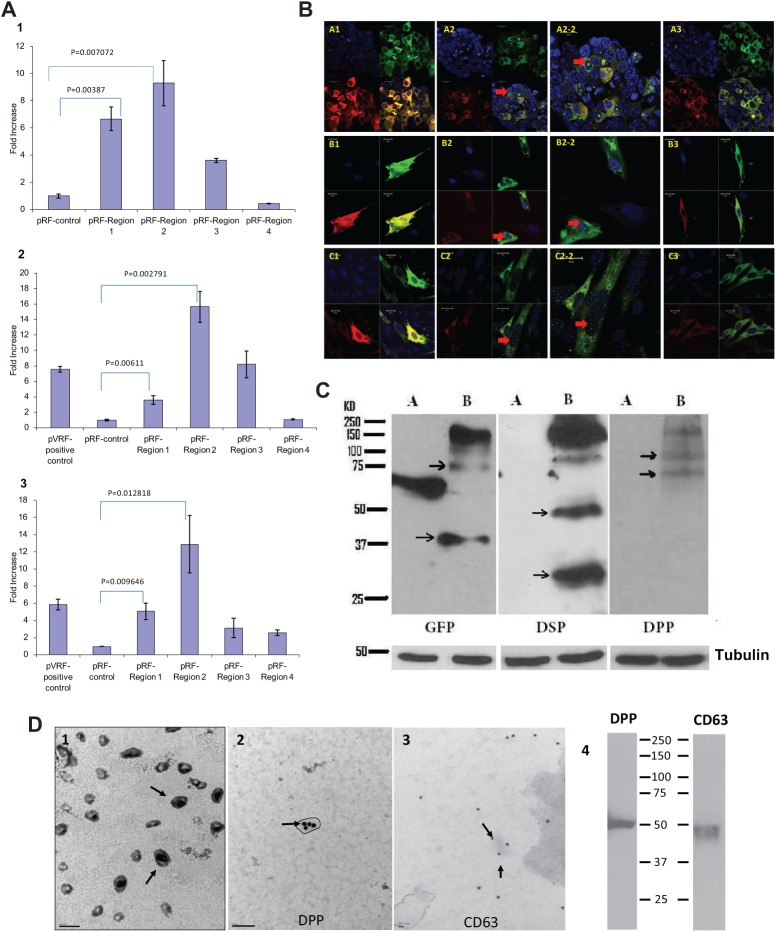
Figure 3.
Functional characterization of the IRES region in DSPP. (A) IRES activities of different constructs in various cell lines. cDNAs (shown in Fig. 2A) were generated by PCR and introduced between the upstream Renilla luciferase cistron and the downstream Firefly luciferase cistron. The plasmids were transfected in HEK293 (1), MC3T3-E1 cells (2), and T4-4 (3) cells. Forty-eight hrs after transient transfection of the various constructs, Firefly luciferase and Renilla luciferase activities of the lysates were determined by the Dual-Luciferase Reporter Assay system (Promega). The Firefly luciferase values were normalized with the corresponding values for Renilla luciferase. We determined the fold increase in IRES activity by dividing the normalized luciferase value by the value obtained from control pRF transfection. Plasmid PVRF was used as a positive control. Note that Region 2 showed the highest IRES activity in all the cell lines tested. Each experiment was done in triplicate. Data are expressed as mean ± SEM. (B) Spatial localization of DSP and DPP with fluorescently tagged DSPP: DsRED-GFP plasmid with or without the full-length DSPP or DSP-DGP (containing the IRES domain) construct was transiently transfected in HEK293, MC3T3-E1, and DPSC cells. Images were captured with the Zeiss LSM 710 confocal microscope equipped with Zen image analysis software. Transfection with green and red fluorescent protein fusion (pDsRED-GFP) shows equal amounts of DsRED and GFP expression in all cell types (A1, B1, C1). Transfection with the pDsRED-DSPP-GFP shows more GFP expression than DsRED (A2, B2, C2) and localization in the nucleus (A2-2, B2-2, C2-2), implying more DPP-translated products. Transfection with the pDsRED-DSP-DGP-GFP plasmid, which contains the IRES domain, shows more GFP expression, indicating the presence of an IRES element in the construct (A3, B3, C3). Note the absence of GFP expression in the nucleus. (C) Presence of intracellular DSP and DPP. Total proteins were isolated from HEK293 cells transfected with either RFP-GFP plasmid (A) or RFP-DSPP-GFP plasmid (B). Western blot analysis was performed with either anti-DSP, anti-DPP, or anti-GFP antibody. Arrows denote the presence of GFP-DPP or RFP-DSP and their processed forms. Tubulin was used as loading control. (D) DPP is transported to the secretome in exosomes: (1) TEM showing the presence of exosomes in the secretome of T4-4 cells; (2) exosomes were processed for immuno-TEM as described in “Materials & Methods”, with sections stained for DPP followed by 20-nm gold secondary antibody [Note the presence of DPP in the isolated exosomes.]; (3) sections were stained for CD-63, a marker for exosomes; and (4) Western blot performed on total proteins isolated from the exosome with anti-DPP (1:200) and anti-CD63 (1:250) antibodies. (The figure can be viewed in color online.)
Spatial and Differential Translational Regulation of DSPP Gene Products in vitro
To address the question of IRES-dependent translation of DPP, we generated a bicistronic vector containing DSPP and also the DSP-DGP region in the DsRed-GFP vector to visualize sites of DSP and DPP synthesis in HEK293, DPSC, and MC3T3-E1 cells. Results in Fig. 3B show that the control plasmid containing the DsRed and GFP reporters showed equal distribution of DsRed and GFP expression when viewed under confocal microscopy in all 3 cell types (A1, B1, C1). Interestingly, cells transfected with full-length DSPP displayed abundant GFP-positive cells with consistent nuclear localization (A2, B2, C2) and in the enlarged view (A2-2, B2-2, C2-2). The DSP-DGP construct showed more GFP expression, localized predominantly in the cytoplasm. These results provide evidence that an IRES-mediated molecular process may account for an additional mechanism by which DPP is translated and has the potential to translocate to the nucleus.
Total Intracellular Proteins Contain DSP and DPP
To investigate the possibility that DSP and DPP exist intracellularly, we transiently transfected full-length mouse DSPP cDNA cloned in the DsRed-GFP vector in HEK293 cells. Western blot analysis (Fig. 3C), performed on total proteins, showed the intact and processed forms of DPP with anti-DPP antibody, while the DSP-RFP protein and its processed forms were identified by the anti-DSP antibody. The GFP antibody confirmed the presence of ~75 kDa DPP-GFP chimeric protein, confirming that DSP and DPP exist intracellularly as separate proteins.
Intracellular DPP is Transported to the ECM through Exosomes
IRES-mediated translated DPP does not contain a signal peptide; therefore, we sought to determine if DPP is associated with exosomes. The isolated exosomes from the secretome of T4-4 cells are shown in Fig. 3D-1. Immuno-TEM images showed that CD-63-positive exosomes (Fig. 3D-3) contained DPP (Fig. 3D-2). Western blot confirmed the presence of CD-63 and DPP in the exosomes (Fig. 3D-4). However, no DSP was detected in the exosomes.
Expression of DPP in DSPP Null Mouse Tooth Germs Restores Mineralized Matrix Formation
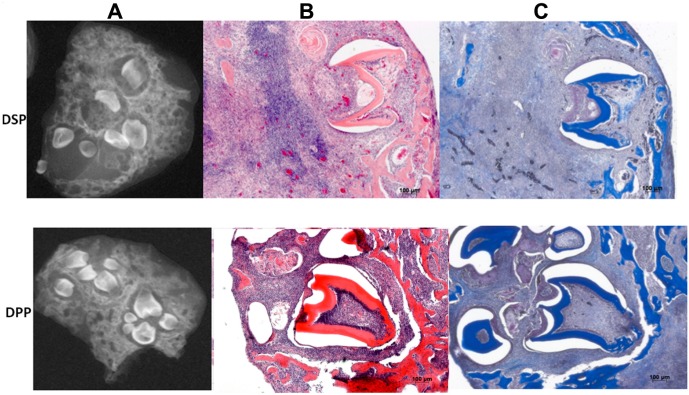
To demonstrate that the independently translated DPP is functional in vivo, we overexpressed DPP in dental mesenchymal cells isolated from DSPP knockout mice using lentivirus expressing DPP, to recapitulate the in vivo function of the cleaved products of DSPP. After 30 days of sub-renal culture, Faxitron imaging results (Fig. 4) showed the formation of several tooth-like structures with mineralized dentin matrix. In contrast, DSP overexpression showed fewer tooth-like structures with unmineralized dentin matrix. This clearly indicates that the translated products of DSPP have mutually exclusive functions in dentinogenesis.
Figure 4.
DSPP gene products have distinct functions during dentinogenesis. (A) X-ray analysis of dental mesenchyme overexpressing DPP shows formation of mineralized tooth-like structures when recombined with mouse dental epithelium, after sub-renal culture at 30 days in a DSPP null mouse, while cells overexpressing DSP show less mineralized matrix. (B) Histological analysis by H& E. (C) Masson Trichrome staining shows the tooth architecture with the organic matrix and mineralized dentin synthesized by the differentiated odontoblasts.
Discussion
Eukaryotic cells can generate different proteins from a single gene through mechanisms like alterative RNA splicing, alternative promoter at 5′ UTR, and IRES. While many reports have focused on transcriptional regulation, there is little support for translational regulation.
In the present study, we demonstrate that DSP and DPP can exist as separate proteins both intracellularly and in the ECM. However, the absence of equal amounts of DSP and DPP lead us to search for translational regulation mechanisms. There are several mechanisms that can be envisaged by which DPP is present in greater amounts than DSP. First, there is a possibility that the holoprotein DSPP is secreted into the matrix, where it is cleaved by proteases like BMP-1 and the cell preferentially uptakes DPP by endocytosis. However, analysis of the data shown here indicates more cellular distribution of DPP tagged with GFP when compared with DsRed-DSP, and this evidence does not support this possibility. The second scenario is that proteases responsible for processing DSPP exist intracellularly. In this context, DSP must be subjected to a further degradation process, and this has yet to be identified. However, this possibility cannot be excluded. In the third scenario, alternative splicing of DSPP mRNA is a possibility. However, full-length DSPP cDNA was used, and this can be excluded. Recently, DSPP was shown to be cleaved by BMP1 in the ECM, but this does not explain the unequal distribution of DSP and DPP (Tsuchiya et al., 2011; Yang et al., 2013). Therefore, we examined whether DPP can be translated by an alternative translational mechanism. The linker region in the DSPP gene (i.e., between DSP and DPP) is conserved in several species, and therefore this region was analyzed for possible IRES elements that might regulate the translation of DPP. In silico analysis predicted a folding structure that displayed a complex hairpin-loop structure for the region which encompasses 126 bp of DSP and 90 bp of DGP. It is believed that the unique secondary structure of IRES elements collaborates in the recruitment of the translational machinery; specifically, such structures are necessary for mediating direct interaction with the 40S ribosomal subunit. This characteristic stem and Y loop structure is present in cellular IRES in BiP and FGF-2 (Le and Maizel, 1997). Classification of viral IRES is partially based on such a characteristic secondary structure (reviewed by Fitzgerald and Semler, 2009). Other studies show that preservation of such a structure may not be necessary to retain the IRES activity (Baird et al., 2006). Overall, because of the lack of conserved primary sequence or structural similarities, cellular IRES remains elusive for detection.
Supporting data for an IRES-mediated translation of DPP were confirmed by luciferase assays within Region 2. This region was comprised of 126 bp of the C-terminal end of DSP and 90 bp of the N-terminal portion of DGP. The IRES-mediated translation of DPP could explain the physiological levels of DPP in the matrix.
Many cellular IRES are activated in response to various stress conditions (Stoneley et al., 1998). Since odontoblasts synthesize a mineralized matrix and are involved in calcium transport, it is possible that the intracellular stress conditions might activate IRES-mediated DPP translation in a robust manner. Our results show, for the first time, that, in addition to regulation at the transcriptional and splicing levels, DPP generation is also controlled at the translational level. Using immuno-TEM, we showed that only DPP and not DSP is associated with exosomes, and this finding suggests that intracellular DPP could be transported to the ECM through exosomes. Exosomes have the property of transporting various proteins to the ECM (Keller et al., 2006).
The biological significance of the 2 protein products of DSPP was demonstrated by lentiviral-mediated overexpression of DSP and DPP in DSPP null mice, since lentivirus can infect both dividing and non-dividing cells as well as become integrated into the chromosome. Tissue recombination and sub-renal culture of the recombinants showed the formation of more tooth-like structures and mineralized dentin with DPP.
Overall, this study shows that the alternative IRES translational mechanism may permit DPP expression to continue under conditions when cap-dependent translation is impaired, and the large amounts of DPP secreted to the ECM are an essential component for the formation of mineralized dentin.
Supplementary Material
Acknowledgments
The authors thank Dr. Karthikeyan Narayanan (IBN, Singapore) for useful discussions.
Footnotes
A supplemental appendix to this article is published electronically only at http://jdr.sagepub.com/supplemental.
This work was supported by the National Institute of Dental and Craniofacial Research, National Institutes of Health, US Department of Health and Human Services (R01 DE 19633), and by the Brodie Endowment Fund.
The authors declare no potential conflicts of interest with respect to the authorship and/or publications of this article.
References
- Alvares K, Kanwar YS, Veis A. (2006). Expression and potential role of dentin phosphophoryn (DPP) in mouse embryonic tissues involved in epithelial-mesenchymal interactions and branching morphogenesis. Dev Dyn 235:2980-2990. [DOI] [PubMed] [Google Scholar]
- Baird SD, Turcotte M, Korneluk RG, Holcik M. (2006). Searching for IRES. RNA 12:1755-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailleul-Forestier I, Molla M, Verloes A, Berdal A. (2008). The genetic basis of inherited anomalies of the teeth. Part 1: Clinical and molecular aspects of non-syndromic dental disorders. Eur J Med Genet 51:273-291. [DOI] [PubMed] [Google Scholar]
- Bègue-Kirn C, Krebsbach PH, Bartlett JD, Butler WT. (1998). Dentin sialoprotein, dentin phosphoprotein, enamelysin and ameloblastin: tooth-specific molecules that are distinctly expressed during murine dental differentiation. Eur J Oral Sci 106:963-970. [DOI] [PubMed] [Google Scholar]
- Fisher LW. (2011). DMP1 and DSPP: evidence for duplication and convergent evolution of two SIBLING proteins. Cells Tissues Organs 194:113-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher LW, Torchia DA, Fohr B, Young MF, Fedarko NS. (2001). Flexible structures of SIBLING proteins, bone sialoprotein, and osteopontin. Biochem Biophys Res Commun 280:460-465. [DOI] [PubMed] [Google Scholar]
- Fitzgerald KD, Semler BL. (2009). Bridging IRES elements in mRNAs to the eukaryotic translation apparatus. Biochim Biophys Acta 1789:518-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronthos S, Brahim J, Li W, Fisher LW, Cherman N, Boyde A, et al. (2002). Stem cell properties of human dental pulp stem cells. J Dent Res 81:531-535. [DOI] [PubMed] [Google Scholar]
- Hao J, Narayanan K, Ramachandran A, He G, Almushayt A, Evans C, et al. (2002). Odontoblast cells immortalized by telomerase produce mineralized dentin-like tissue both in vitro and in vivo. J Biol Chem 277:19976-19981. [DOI] [PubMed] [Google Scholar]
- Keller S, Sanderson MP, Stoeck A, Altevogt P. (2006). Exosomes: from biogenesis and secretion to biological function. Immunol Lett 107:102-108. [DOI] [PubMed] [Google Scholar]
- Le SY, Maizel JV., Jr (1997). A common RNA structural motif involved in the internal initiation of translation of cellular mRNAs. Nucleic Acids Res 25:362-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linde A. (1989). Dentin matrix proteins: composition and possible functions in calcification. Anat Rec 224:154-166. [DOI] [PubMed] [Google Scholar]
- MacDougall M, Simmons D, Luan X, Nydegger J, Feng J, Gu TT. (1997). Assignment of dentin sialophosphoprotein (DSPP) to the critical DGI2 locus on human chromosome 4 band q21.3 by in situ hybridization. J Biol Chem 272:835-842. [DOI] [PubMed] [Google Scholar]
- Qin C, Cook RG, Orkiszewski RS, Butler WT. (2001). Identification and characterization of the carboxyl-terminal region of rat dentin sialoprotein. J Biol Chem 276:904-909. [DOI] [PubMed] [Google Scholar]
- Song Y, Zhang Z, Yu X, Yan M, Zhang X, Gu S, et al. (2006). Application of lentivirus-mediated RNAi in studying gene function in mammalian tooth development. Devel Dyn 235:1334-1344. [DOI] [PubMed] [Google Scholar]
- Sreenath T, Thyagarajan T, Hall B, Longenecker G, D’Souza R, Hong S, et al. (2003). Dentin sialophosphoprotein knockout mouse teeth display widened predentin zone and develop defective dentin mineralization similar to human dentinogenesis imperfecta type III. J Biol Chem 278:24874-24880. [DOI] [PubMed] [Google Scholar]
- Stoneley M, Paulin FE, Le Quesne JP, Chappell SA, Willis AE. (1998). C-Myc 5′untranslated region contains an internal ribosome entry segment. Oncogene 16:423-428. [DOI] [PubMed] [Google Scholar]
- Sun Y, Lu Y, Chen S, Prasad M, Wang X, Zhu Q, et al. (2010). Key proteolytic cleavage site and full-length form of DSPP. J Dent Res 89:498-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touriol C, Bornes S, Bonnal S, Audigier S, Prats H, Prats AC, et al. (2003). Generation of protein isoform diversity by alternative initiation of translation at non-AUG codons. Biol Cell 95:169-178. [DOI] [PubMed] [Google Scholar]
- Tsuchiya S, Simmer JP, Hu JC, Richardson AS, Yamakoshi F, Yamakoshi Y. (2011). Astacin proteases cleave dentin sialophosphoprotein (Dspp) to generate dentin phosphoprotein (Dpp). J Bone Miner Res 26: 220-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Marschall Z, Fisher LW. (2010). Dentin sialophosphoprotein (DSPP) is cleaved into its two natural dentin matrix products by three isoforms of bone morphogenetic protein-1 (BMP1). Matrix Biol 29:295-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakoshi Y, Hu JC, Fukae M, Zhang H, Simmer JP. (2005). Dentin glycoprotein: the protein in the middle of the dentin sialophosphoprotein chimera. J Biol Chem 280:17472-17479. [DOI] [PubMed] [Google Scholar]
- Yamakoshi Y, Hu JC, Iwata T, Kobayashi K, Fukae M, Simmer JP. (2006). Dentin sialophosphoprotein is processed by MMP-2 and MMP-20 in vitro and in vivo. J Biol Chem 281:38235-38243. [DOI] [PubMed] [Google Scholar]
- Yamakoshi Y, Lu Y, Hu JC, Kim JW, Iwata T, Kobayashi K, et al. (2008). Porcine dentin sialophosphoprotein, length polymorphisms, glycosylations, phosphorylation and stability. J Biol Chem 283:14835-14844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang RT, Lim GL, Dong Z, Lee AM, Yee CT, Fuller RS, et al. (2013). The efficiency of dentin sialoprotein-phosphophoryn processing is affected by mutations both flanking and distant from the cleavage site. J Biol Chem 288:6024-6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Sun Y, Prasad M, Wang X, Yamoah AK, Li Y, et al. (2010). Glycosaminoglycan chain of dentin sialoprotein proteoglycan. J Dent Res 89:808-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.