Abstract
Oscillatory processes in biological signal transduction have come under progressively increasing scrutiny in terms of their functional significance and mechanisms of emergence and regulation. Since oscillatory processes can be a by-product of rapid adaptation and can also easily emerge if the feedbacks underlying adaptive processes are inadvertently artificially enhanced, one needs to exercise caution in both claiming the existence of in vivo oscillations and in seeking to assign to them a specific functional significance. Nevertheless, oscillations can be a powerful means of encoding and transferring information both in time and in space, thus possessing important potential advantages for evolutionary selection and stabilization. Thus periodicity in the cell responses to diverse persistent external stimuli might become a more recognized and even expected feature of signaling processes.
The how
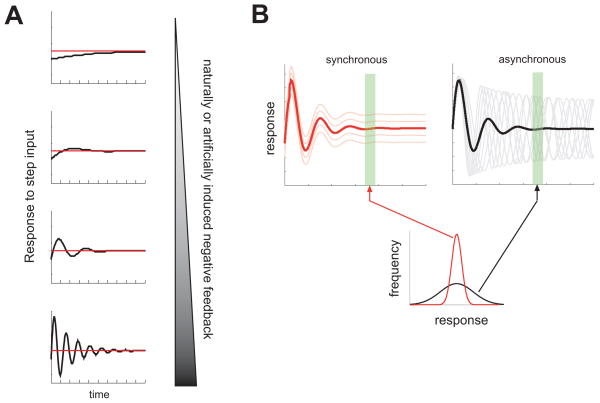
The requirement of appropriate responses to cues in the extracellular environment imposes important limitations in signal processing by intracellular biochemical networks. One such limitation is the frequent need to adapt to persistent signaling inputs. Adaptation, widely observed in biological processes, allows the cell to respond primarily to changes or variations in the microenvironment rather than the persistent environmental inputs, frequently a more physiologically important cue [1,2]. Adaptation can also allow the cell to respond over a wider range of the signaling doses, i.e., to have a higher dynamical range. However, the ability to adapt comes at a price [3]. If, as is frequently the case, adaptation is mediated by a negative feedback regulation [4,5], one can show that the need to adapt fast (mediated by strong negative feedback regulation processes) can lead to considerable overshooting in the adaptation process and, if the feedback is sufficiently strong, to oscillations (Fig. 1A). Oscillations can therefore be an emergent property of adaptive systems, and therefore can frequently arise spontaneously, without a specific need for periodicity in a regulatory process.
Figure 1.
(A) Schematic demonstrating that increased negative feedback, whether naturally or artificially induced, increases the propensity of oscillations. Plots show the response to a step input of a linear proportional-integral feedback system, thought to be commonly present in signaling pathways. In each case, the response eventually settles on the target steady state shown by the red line. As the strength of negative feedback is increased, the steady state is reached more rapidly, but at the cost of overshoot and oscillations. (B) Schematic showing how synchronous and asynchronous oscillations, though leading to similar average responses, may be distinguished experimentally. The response dynamics of individual cells are shown by the light red and light gray traces, leading to the same population-average dynamics as shown in red and black. However, when the population is sampled at a single time point(s) during the steady phase of the response (indicated by the green rectangle), the variance of the asynchronous population tends to be much larger than that of a synchronous population, enabling these possibilities to be distinguished by single-cell resolution experimental techniques not relying on live cell probes.
The availability of a mechanism to generate oscillations in ubiquitous biological processes creates an opportunity for evolutionary selection to recruit this particular form of biochemical kinetics for a variety of specific purposes, as addressed in this journal issue. Evolutionary selection of the oscillatory response as a functionally significant and therefore beneficial feature of a signaling output can also enhance its properties. Thus, although the oscillations arising from negative feedback only are usually subject to a gradual dampening of the amplitude of the response, emergence of additional (e.g., positive) feedback can dramatically increase the persistence of the oscillations, effectively making them permanent, subject only to regulatory or energy limitations [6,7]. Importantly, as discussed more in detail below, oscillatory processes are essentially dynamic, thus further evolutionary tinkering might be needed to match their kinetic properties with those of the processes regulated by the oscillations downstream in signaling networks. Nevertheless, from the evolutionary standpoint, emergence of oscillatory processes is easily envisioned based on the negative feedback mechanisms which in turn might have evolved to primarily regulate adaptation.
The ease with which oscillations can emerge also should increase our caution about both the natural tendency to ascribe functional significance to all observed oscillatory processes and the possibility of inadvertent artifactual induction of oscillations in experimental analysis of adaptive signaling systems. For instance, mathematical modeling strongly suggests that over expression of the components of the signaling circuit that are parts of feedback loop at the expense of the components that are not, can enhance the feedback loop activity and lead to an increased probability of emergence of oscillatory responses (Fig. 1A). This might then lead to observations of potentially artifactual oscillatory processes, as has been suggested to be the case in the analysis of NF-kappaB signaling [8,9]. Whether an artifactual augmentation of the oscillation propensity indeed takes place should be determined by independent control experiments not involving genetic or other perturbations of the cell. For instance, if the oscillatory responses are synchronized across the cell population, biochemical methods can and have been used to detect them, e.g., common Western blots [10–13]. If, on the other hand, the oscillations are asynchronous, and thus would be masked in the average cell population response, one can resort to the analysis, such as flow cytometry or immunocytochemistry, that would allow taking snapshots of responses of single cells within a population at different times [14]. Considerable asynchronous oscillatory activity would be expected to lead to high variance of cell responses within a population at (almost) all time points (Fig. 1B), which can be easily either confirmed or argued against by these techniques.
Even if there is a substantial degree of certainty about the natural occurrence of oscillatory processes, their emergence as by-products of adaptive processes can mean that they have no easily distinguished functionality, as is still the case, e.g., for the oscillatory dynamics of p53 activation by DNA damage [15,16] or glycolytic oscillations in yeast [17]. Of course, proving absence of a functionality is frequently infinitely harder than proving its existence, and therefore it can be safely predicted that the search for possible functional significance of any oscillatory activity will persistently occupy a large set of interested researchers.
The why
If oscillations do naturally occur, they can present a wealth of possible functional advantages over simple steady inputs into signaling events. One of the key potential benefits provided by an oscillatory process is the possibility of entraining to another periodic activity. A simple example is the entraining of circadian clocks to the night-day periodicity [18]. Potentially more complex relationships might exist between e.g., the circadian oscillations primarily entrained by natural changes in light luminosity and the oscillations in the cell cycle or redox states of cells [19,20]. In this regard, it is of interest to investigate whether the coincidence of the duration of cell cycle in fast growing mammalian cells and the period of circadian clock (both approximately 24 hours) is merely coincidental.
Even if the interplay between different oscillatory events is not suspected, oscillations can be a powerful way to transmit signals. Indeed, signaling processes are primarily about transfer of information, and one can arguably encode considerably more information into an oscillatory process than into a steady one. Indeed, the information can be present both in the form of the average amplitude of the oscillations (their steady component) and the oscillation frequency. Mathematically, this is evident from the analysis called Fourier transform [21,22], with the oscillation amplitude and frequency defining its first two components for simple, sinusoid-like oscillations. For more complex oscillations, multiple frequencies can be ‘hidden’ within the oscillatory time course, which can be revealed by further terms of the Fourier transform and potentially decoded by the cells. All this is in contrast with just a single piece of information (amplitude) characterizing the steady inputs.
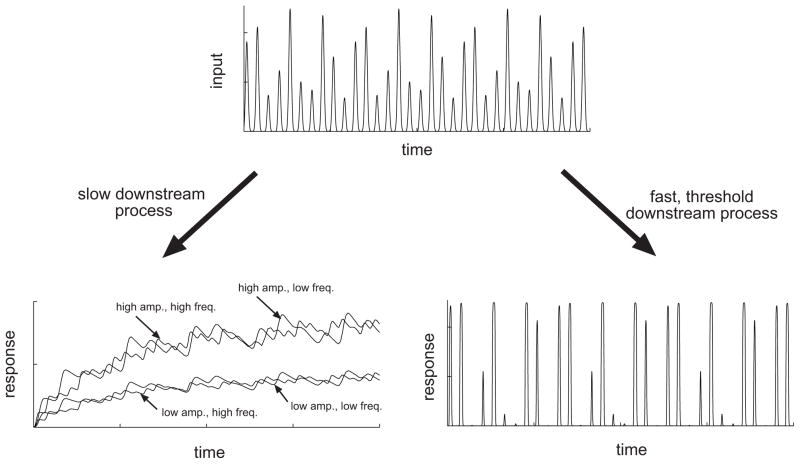
Can cells decode different pieces of information embedded in the oscillatory inputs? The answer is most likely ‘yes’. A simple example of how this can occur within a cell follows below. Essentially, this example makes a very simple assumption: oscillatory signaling inputs can impinge on either relatively fast or relatively slow biochemical processes (Fig. 2). If the process controlled by the oscillatory input is slow compared to the oscillation period, it will not be able to oscillate effectively, responding rather to the oscillation amplitude only. If on the other hand the process controlled by the oscillatory input is fast and furthermore responds only if the input has reached a certain threshold, the output would be less sensitive to the exact oscillation amplitude (reacting only to whether it is above or below a threshold), but extremely sensitive and responsive to the oscillation frequency, i.e., how many times the threshold is exceeded over the signaling duration. By contrast, in decoding steady inputs, the dynamical features of the downstream processes are not as important suggesting that the responses of fast and slow downsteam processes can be very similar. As oscillatory inputs can be coupled to many target processes, the same oscillation can thus be decoded in different ways and can therefore be richer in the information content vs. more steady inputs.
Figure 2.
Extraction of information from an oscillatory input. An example oscillatory input is shown at the top. (Lower left) When the input impinges on a response element with slow kinetics, only the amplitude of the input is extracted. This effect is revealed by the varying the amplitude and frequency of the input. The response amplitude differs when the input amplitude is varied (compare high vs. low amp.), but not when the input frequency is varied (compare high vs. low freq.); (Lower right) When the input impinges on a response element with rapid kinetics, especially with thresholding capability, the frequency is primarily extracted. The resulting response amplitude is mostly held fixed, but the response tends to oscillate at a frequency similar to that of the input.
Interestingly, the high information content of oscillatory inputs has been used in reverse engineering of man-made systems, and more recently, in biological analyses, in unraveling the dynamic properties, or even composition of signaling networks. In this situation, although there might not be a natural oscillatory process occurring, the biological system of interest may be stimulated by the artificially oscillatory inputs and the corresponding outputs then assayed [22–28]. As a result, it can be established how the network of interest decodes different frequencies and whether it may be essentially blind to signals in certain frequency ranges. In this fashion, the architectures of natural signaling and metabolic networks have been explored, and differential gene expression capabilities detected as a function of different input frequencies.
It is of interest to hypothesize that the ability of cells to respond to oscillations only at certain frequencies might serve as a way to couple them with the rest of the population of similarly responsive cells. The resulting ‘resonance’ in responses might serve as a way to establish collective behavior and a certain pass-key for cells to participate in this behavior. Indeed, it has been recently observed that oscillations might mediate a quorum sensing-like response in D. discoideum populations allowing cells to enhance the signaling at a resonance frequency, selecting the area with the highest local cAMP concentration as the ‘resonance’ area, where ultimately most cells are attracted by chemotaxis [29]. Whether other collectively oscillating multi-cellular ensembles are organized in a similar fashion, e.g., in pancreatic islets of Langerhans [30], remains to be determined.
The where
Oscillatory processes can be distributed in space both within multi-cellular organisms and within single cells. This can have interesting consequences for the control of signaling events both in space and in time. A now classical example of such a process is the establishment of somites in vertebrate embryos by the means of the fixation in space of the results of oscillations in time in the Notch signaling pathway in differentiating tissue [31], not unlike the way heart beats get registered and recorded in a spatially unfolding way in a cardiogram. Speaking of the heart, there too, the pacemaker oscillations propagate through the tissue triggering waves of functional contraction [32]. However, it might be worth examining less obvious but potentially no less exciting instances of functional uses of spatially regulated oscillatory activities.
One such instance is in the control of the effective random search of the mating partner in the default response of the yeast mating pathway. When unable to sense the gradient of the pheromone secreted by the cells of the opposite mating type, but sensing the presence of the pheromone in the immediate environment, yeast cells attempt to find the mating partner by trying to grow several projections sequentially in random directions. This periodic morphogenesis processes, that may ultimately leave a cell resembling a starfish, is controlled by a periodic activation and expression of a MAPK molecule regulating a multitude of the requisite genes [10]. The oscillation thus can control the duration of sequential phases of cell growth, with the MAPK activity localized to each new projection in preparation for a potential mating event.
Another instance of oscillatory inputs affecting spatial signaling patterns is the modulation of localization of signaling molecules to different intracellular compartments. For instance, the nuclear localization of the transcription factor NFAT is controlled by calcium mediated input that can be oscillatory. The probability of NFAT localization to the nucleus is directly affected by the frequency of the calcium oscillations [33]. Thus input frequency can be translated into the initiation and extent of gene transcription in this and possibly other pathways, through the spatial control of the transcription regulators [34].
Conclusions and Future Directions
Oscillatory processes in mammalian cells continue to hold fascination for most scientists due to their essential dynamism and patent non-randomness. There is a natural, and, for the most part, productive tendency to search for and ascribe meaning and significance to oscillations, even if no evidence of their importance for cell function exists. Arguably, oscillations can arise as, and, in many systems, continue to be mere by-products of the more physiologically pressing need for the systems to adapt to persistent stimuli, even though, in many cases, this by-product process can potentially be recruited to play important additional information processing roles through evolutionary selection. As oscillatory signals can be much more information rich versus their more steady counterparts, the evolutionary pressure for selection and stabilization of initially accidental oscillatory responses can be very high. As a result, there is currently a co-existence of oscillatory phenomena in which the functionality of the oscillatory processes can be easily interpreted, with a set of pulsatile processes that seems to hold no particular consequences for the cell or organism function. Looking ahead, one can fully expect that more examples of oscillatory events in cell signaling will be increasingly found. Interpreting them with appropriate caution, it would be of interest to determine whether oscillations are indeed a hallmark of signaling events, allowing cells to organize and coordinate a variety of processes on both intracellular and intercellular levels, or whether they would remain curious but rare and potentially inconsequential specimens in the inexhaustible boutique of biological processes.
Acknowledgments
R.C. and A.L. acknowledge support from the National Institutes of Health (GM072024, GM084332). R.C. also acknowledges support from the Medical Scientist Training Program at Johns Hopkins University.
References
- 1.Parent CA, Devreotes PN. A cell’s sense of direction. Science. 1999;284:765–770. doi: 10.1126/science.284.5415.765. [DOI] [PubMed] [Google Scholar]
- 2.Luo DG, Xue T, Yau KW. How vision begins: an odyssey. Proc Natl Acad Sci U S A. 2008;105:9855–9862. doi: 10.1073/pnas.0708405105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3•.Csete ME, Doyle JC. Reverse engineering of biological complexity. Science. 2002;295:1664–1669. doi: 10.1126/science.1069981. A clear exposition of the routes to the emergence of oscillations through adaptive processes and ways they can be controlled. [DOI] [PubMed] [Google Scholar]
- 4.Yi TM, Huang Y, Simon MI, Doyle J. Robust perfect adaptation in bacterial chemotaxis through integral feedback control. Proc Natl Acad Sci U S A. 2000;97:4649–4653. doi: 10.1073/pnas.97.9.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levchenko A, Iglesias PA. Models of eukaryotic gradient sensing: application to chemotaxis of amoebae and neutrophils. Biophys J. 2002;82:50–63. doi: 10.1016/S0006-3495(02)75373-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng P, Yang Y, Liu Y. Interlocked feedback loops contribute to the robustness of the Neurospora circadian clock. Proc Natl Acad Sci U S A. 2001;98:7408–7413. doi: 10.1073/pnas.121170298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferrell JE, Jr, Pomerening JR, Kim SY, Trunnell NB, Xiong W, Huang CY, Machleder EM. Simple, realistic models of complex biological processes: positive feedback and bistability in a cell fate switch and a cell cycle oscillator. FEBS Lett. 2009;583:3999–4005. doi: 10.1016/j.febslet.2009.10.068. [DOI] [PubMed] [Google Scholar]
- 8.Barken D, Wang CJ, Kearns J, Cheong R, Hoffmann A, Levchenko A. Comment on “Oscillations in NF-kappaB signaling control the dynamics of gene expression”. Science. 2005;308:52. doi: 10.1126/science.1107904. author reply 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheong R, Hoffmann A, Levchenko A. Understanding NF-kappaB signaling via mathematical modeling. Mol Syst Biol. 2008;4:192. doi: 10.1038/msb.2008.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hilioti Z, Sabbagh W, Jr, Paliwal S, Bergmann A, Goncalves MD, Bardwell L, Levchenko A. Oscillatory phosphorylation of yeast Fus3 MAP kinase controls periodic gene expression and morphogenesis. Curr Biol. 2008;18:1700–1706. doi: 10.1016/j.cub.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoffmann A, Levchenko A, Scott ML, Baltimore D. The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation. Science. 2002;298:1241–1245. doi: 10.1126/science.1071914. [DOI] [PubMed] [Google Scholar]
- 12.Maeda M, Lu S, Shaulsky G, Miyazaki Y, Kuwayama H, Tanaka Y, Kuspa A, Loomis WF. Periodic signaling controlled by an oscillatory circuit that includes protein kinases ERK2 and PKA. Science. 2004;304:875–878. doi: 10.1126/science.1094647. [DOI] [PubMed] [Google Scholar]
- 13.Shankaran H, Ippolito DL, Chrisler WB, Resat H, Bollinger N, Opresko LK, Wiley HS. Rapid and sustained nuclear-cytoplasmic ERK oscillations induced by epidermal growth factor. Mol Syst Biol. 2009;5:332. doi: 10.1038/msb.2009.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheong R, Wang CJ, Levchenko A. High content cell screening in a microfluidic device. Mol Cell Proteomics. 2009;8:433–442. doi: 10.1074/mcp.M800291-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lahav G, Rosenfeld N, Sigal A, Geva-Zatorsky N, Levine AJ, Elowitz MB, Alon U. Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat Genet. 2004;36:147–150. doi: 10.1038/ng1293. [DOI] [PubMed] [Google Scholar]
- 16.Lev Bar-Or R, Maya R, Segel LA, Alon U, Levine AJ, Oren M. Generation of oscillations by the p53-Mdm2 feedback loop: a theoretical and experimental study. Proc Natl Acad Sci U S A. 2000;97:11250–11255. doi: 10.1073/pnas.210171597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richard P. The rhythm of yeast. FEMS Microbiol Rev. 2003;27:547–557. doi: 10.1016/S0168-6445(03)00065-2. [DOI] [PubMed] [Google Scholar]
- 18.Boivin DB, Duffy JF, Kronauer RE, Czeisler CA. Dose-response relationships for resetting of human circadian clock by light. Nature. 1996;379:540–542. doi: 10.1038/379540a0. [DOI] [PubMed] [Google Scholar]
- 19•.Yang Q, Pando BF, Dong G, Golden SS, van Oudenaarden A. Circadian gating of the cell cycle revealed in single cyanobacterial cells. Science. 327:1522–1526. doi: 10.1126/science.1181759. A particularly well argued example of the potential linkage between circadian and cell cycle rhythms. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirayama J, Cho S, Sassone-Corsi P. Circadian control by the reduction/oxidation pathway: catalase represses light-dependent clock gene expression in the zebrafish. Proc Natl Acad Sci U S A. 2007;104:15747–15752. doi: 10.1073/pnas.0705614104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oppenheim AV, Willsky AS. Signals & Systems. Vol. 2. Prentice Hall; 1996. [Google Scholar]
- 22.Geva-Zatorsky N, Dekel E, Batchelor E, Lahav G, Alon U. Fourier analysis and systems identification of the p53 feedback loop. Proc Natl Acad Sci U S A. 107:13550–13555. doi: 10.1073/pnas.1001107107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hersen P, McClean MN, Mahadevan L, Ramanathan S. Signal processing by the HOG MAP kinase pathway. Proc Natl Acad Sci U S A. 2008;105:7165–7170. doi: 10.1073/pnas.0710770105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lipan O, Wong WH. The use of oscillatory signals in the study of genetic networks. Proc Natl Acad Sci U S A. 2005;102:7063–7068. doi: 10.1073/pnas.0403790102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25••.Mettetal JT, Muzzey D, Gomez-Uribe C, van Oudenaarden A. The frequency dependence of osmo-adaptation in Saccharomyces cerevisiae. Science. 2008;319:482–484. doi: 10.1126/science.1151582. The first study to apply oscillatory inputs to not only understand the dynamics of the underlying system, but also attempt its reconstruction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paliwal S, Wang CJ, Levchenko A. Pulsing cells: how fast is too fast? Hfsp J. 2008;2:251–256. doi: 10.2976/1.2969901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ashall L, Horton CA, Nelson DE, Paszek P, Harper CV, Sillitoe K, Ryan S, Spiller DG, Unitt JF, Broomhead DS, et al. Pulsatile stimulation determines timing and specificity of NF-kappaB-dependent transcription. Science. 2009;324:242–246. doi: 10.1126/science.1164860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bennett MR, Pang WL, Ostroff NA, Baumgartner BL, Nayak S, Tsimring LS, Hasty J. Metabolic gene regulation in a dynamically changing environment. Nature. 2008;454:1119–1122. doi: 10.1038/nature07211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gregor T, Fujimoto K, Masaki N, Sawai S. The onset of collective behavior in social amoebae. Science. 328:1021–1025. doi: 10.1126/science.1183415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Porksen N, Hollingdal M, Juhl C, Butler P, Veldhuis JD, Schmitz O. Pulsatile insulin secretion: detection, regulation, and role in diabetes. Diabetes. 2002;51 (Suppl 1):S245–254. doi: 10.2337/diabetes.51.2007.s245. [DOI] [PubMed] [Google Scholar]
- 31.Jiang YJ, Aerne BL, Smithers L, Haddon C, Ish-Horowicz D, Lewis J. Notch signalling and the synchronization of the somite segmentation clock. Nature. 2000;408:475–479. doi: 10.1038/35044091. [DOI] [PubMed] [Google Scholar]
- 32.Vinogradova TM, Maltsev VA, Bogdanov KY, Lyashkov AE, Lakatta EG. Rhythmic Ca2+ oscillations drive sinoatrial nodal cell pacemaker function to make the heart tick. Ann N Y Acad Sci. 2005;1047:138–156. doi: 10.1196/annals.1341.013. [DOI] [PubMed] [Google Scholar]
- 33.Cooling MT, Hunter P, Crampin EJ. Sensitivity of NFAT cycling to cytosolic calcium concentration: implications for hypertrophic signals in cardiac myocytes. Biophys J. 2009;96:2095–2104. doi: 10.1016/j.bpj.2008.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34••.Dolmetsch RE, Xu K, Lewis RS. Calcium oscillations increase the efficiency and specificity of gene expression. Nature. 1998;392:933–936. doi: 10.1038/31960. The first study linking oscillations to specificity of signaling and suggesting that the same oscillatory signal can be interpreted differently by different target processes. [DOI] [PubMed] [Google Scholar]