Abstract
The hypothesis that domestication lead to a relaxation of purifying selection on mitochondrial (mt) genomes was tested by comparative analysis of mt genes from dog, pig, chicken, and silkworm. The three vertebrate species showed mt genome phylogenies in which domestic and wild isolates were intermingled, whereas the domestic silkworm (Bombyx mori) formed a distinct cluster nested within its closest wild relative (B. mandarina). In spite of these differences in phylogenetic pattern, significantly greater proportions of nonsynonymous SNPs than of synonymous SNPs were unique to the domestic populations of all four species. Likewise, in all four species, significantly greater proportions of RNA-encoding SNPs than of synonymous SNPs were unique to the domestic populations. Thus, domestic populations were characterized by an excess of unique polymorphisms in two categories generally subject to purifying selection: nonsynonymous sites and RNA-encoding sites. Many of these unique polymorphisms thus seem likely to be slightly deleterious; the latter hypothesis was supported by the generally lower gene diversities of polymorphisms unique to domestic populations in comparison to those of polymorphisms shared by domestic and wild populations.
Keywords: domestication, mitochondrial DNA, purifying selection, slightly deleterious mutations
1. Introduction
The availability of genomic data for domestic animals and plants has stimulated interest in discovering the genetic changes underlying the distinctive characteristics of domestic species (Amaral et al. 2011; Andersson 2001; Andersson and Georges 2004; MacEachern et al. 2009; Rubin et al. 2010). Some such genetic changes presumably involve the loci responsible for traits consciously or inadvertently subjected to artificial selection by humans in the process of domestication (Andersson and Georges 2004). In addition, certain neutral or even deleterious alleles may be fixed because they are linked to alleles selectively favored in the process of domestication (Fisher 1930). Other genetic changes may involve the relaxation of purifying selection after domestication, as a result of the less stringent selective pressures in the domestic environment (Björnerfeldt et al. 2006; Cruz et al. 2008; MacEachern et al. 2009). Because domestication may involve population bottlenecks, the effectiveness of purifying selection in eliminating slightly deleterious mutations may also be reduced in domesticated populations (Cruz et al. 2008).
Comparisons of the ratio of nonsynonymous to synonymous substitutions has been used to test for a relaxation of purifying selection on both mitochondrial and nuclear genes of the domestic dog (Björnerfeldt et al. 2006; Cruz et al. 2008) and on nuclear genes of domestic cattle (MacEachern et al. 2009). Mitochondrial (mt) genomes are expected to particularly prone to the accumulation of slightly deleterious mutations because of their high mutation rate and lack of recombination (Hasegawa et al. 1998; Hughes and Hughes 2007; Rand and Kann 1996).
Although some studies have claimed evidence of positive selection on mt protein-coding genes (Bazin et al.; Li et al. 2010; Mishmar et al. 2006) and RNA-encoding genes (Ruiz-Pesini and Wallace 2006), the methods used in these analyses are of questionable validity (Hughes 2007); Hughes and Friedman 2008; Hughes et al. 2008). Moreover, these studies have at best merely reported statistical patterns allegedly consistent with positive selection. Not even a single study has shown experimentally that any of the allegedly selectively favored substitutions is actually associated with an advantageous phenotype.
By contrast, there is strong evidence of purifying selection on mt protein-coding genes. First, mt protein coding genes almost always show higher numbers of synonymous substitutions per synonymous site (dS) than of nonsynonymous substitutions per nonsynonymous site (dN), a pattern indicating that purifying selection has acted to remove deleterious nonsynonymous mutations (Hughes and Hughes 2007; Kumar 1996). Moreover, there are numerous known diseases linked substitutions in mt protein-coding and RNA-encoding genes of humans and other mammals (Baranowska et al. 2009; Schaeffer et al. 2001; Wallace 1992). The existence of such mt-linked disorders supports the hypothesis of strong purifying selection by proving evidence of mutations that have a deleterious phenotypic effect (Kimura and Ohta 1973).
Phylogenies of mt genomes of domestic animal species and their closest wild relatives have revealed surprising complexity, with wild and domestic genomes not forming separate clades (Björnerfeldt et al. 2006; Giuffra et al. 2000; Larson et al. 2005; Liu et al. 2006). Such topologies may arise from independent domestication events, from domestication of populations involving several distinct matrilines, from post-domestication introgression, or from some combination of these processes. Nonetheless, on the hypothesis that domestication relaxes purifying selection present in the wild, one would expect to see the accumulation of slightly deleterious mutations in domesticated populations whatever their origin. Conversely one would expect to see intensified purifying selection in free-living populations, whether those represent the wild ancestral species or feral populations of domestic origin. Here I test this prediction using published sequences of complete mitochondrial genomes of the domestic dog, pig, chicken, and silkworm.
2. Methods
2.1. Sequence data
Complete or nearly complete mt genome sequences were obtained for the following taxa: 254 from the domestic dog (Canis lupus familiaris) and 19 from the wolf (Canis lupus); 59 from the domestic pig and 27 from wild boar (Sus scrofa); 41 from the domestic chicken (Gallus gallus domesticus) and 17 from the red junglefowl (Gallus gallus); and 33 from the domestic silkworm (Bombyx mori) and 15 from its closest known wild relative (Bombyx mandarina). Sequences were aligned by the CLUSTALX program (Thompson et al. 1997). When pairwise comparisons were made among a set of aligned sequences, any site at which the alignment postulated a gap in any of the sequences was excluded from the analysis so that a comparable set of sequences was available for each pairwise comparison. Phylogenetic trees were rooted with appropriate outgroup taxa: (1) for the dog, four sequences from the coyote (Canis latrans); (2) for the pig, a sequence from the common warthog (Phacochoerus africanus); for the chicken, a sequence from the green junglefowl (Gallus varius); and (4) for the silkworm, one sequence from the oriental fruit moth (Grapholita molesta) and one sequence from the gypsy moth (Lymantria dispar). For Genbank identifiers (gi numbers) of all sequences, see Supplementary Figures S1-S4.
2.2. Statistical methods
Phylogenetic trees were constructed on the basis of the entire mt genome DNA sequence by the maximum likelihood (ML) method in the MEGA program, version 5.05 (Tamura et al. 2011). The Model test function in MEGA was used to choose models for ML analyses by the Bayes Information Criterion (BIC). The reliability of branching patterns in ML trees was tested by bootstrapping (1000 samples). The following DNA sequence evolution models were used: (1) for the dog, GTR+I; (2) for the pig, TN93+G+I; (3) for the chicken, HKY+G+I; and for the silkworm, GTR+G+I.
The number of synonymous substitutions per synonymous site (dS) and the number of nonsynonymous substitutions per nonsynonymous site (dN) were estimated by Li’s (1993) method, which takes into account transitional bias (known to be marked in mitochondrial genomes). In coding regions, the mean for all pairwise comparisons of dS provided an estimate of nucleotide diversity at synonymous sites (πS); and the mean for all pairwise comparisons of dN provided an estimate of nucleotide diversity at nonsynonymous sites (πN) (Nei and Kumar 2000). The number of nucleotide substitutions per site (d) in non protein-coding regions was estimated by maximum composite likelihood method in MEGA (Tamura et al 2011); the nucleotide diversity (π) in non protein-coding regions was estimated by the mean of d in all pairwise comparisons. The computation of πS, πN, and π was equivalent to equation 10.6, p. 256, of Nei (1987). Standard errors of πS, πN, and π were estimated by the bootstrap method, which takes into account the non-independence of pairwise comparisons (Nei and Kumar 2000); and z-tests were used to test equality of nucleotide diversities in different genomic regions.
Gene diversity (Nei 1987, p. 177) was estimated separately at individual single nucleotide polymorphism (SNP) sites, using the PolyAna program (Hughes 2005); Hughes et al. 2003; Knapp et al. 2011); where xi is the frequency of the ith allele (nucleotide) at a given locus (site), the gene diversity is 1-Σxi2. Polymorphic (SNP) sites were classified as synonymous or nonsynonymous, based on the coding effect of the nucleotide change. There were certain SNP sites that could not be classified unambiguously as synonymous or nonsynonymous (ambiguous sites), either because both synonymous and nonsynonymous variants occurred at the same site or because, given polymorphic sites within a single codon, the coding effect of a given substitution depended on the pathway taken by evolution. Ambiguous sites constituted 15 of 1439 polymorphic sites in coding regions of the four domesticated taxa (1.0%); and 41 of 2055 (2.0%) of polymorphic sites in coding regions of the four wild taxa. Ambiguous sites were excluded from analyses of gene diversity at individual polymorphic sites.
3. Results
3.1. Phylogenetic Analyses
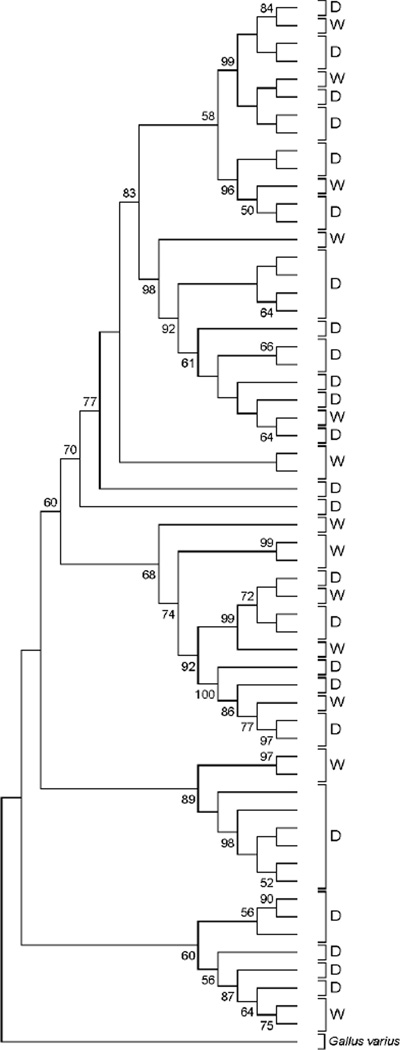
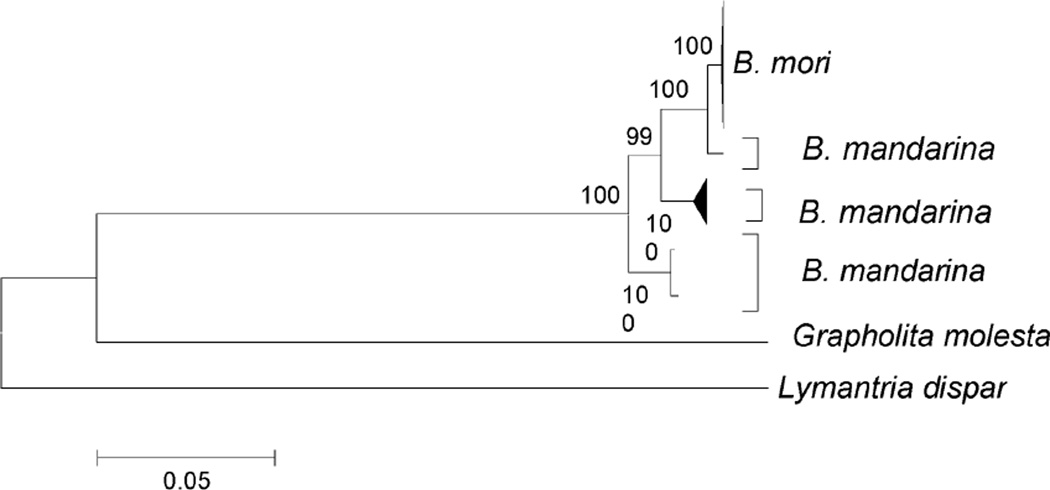
In rooted ML phylogenies of dog, pig, and chicken mt genomes, domestic and wild isolates were intermingled (Figure 1 and Supplementary Figures S1-S3). Often highly significant bootstrap support was obtained for clusters of sequences including one or more genomes of wild origin and one or more genomes of domestic origin (Figure 1 and Supplementary Figures S1-S3). For example, Figure 1 illustrates the ML tree of chicken mt genomes, showing topology only. Although in this tree, many of the deeper relationships did not receive strong bootstrap support, there were five clusters with 96% bootstrap support or better that included sequences of both wild and domestic origin (Figure 1). By contrast, in the case of the silkworm, all Bombyx mori sequences clustered together in a monophyletic group that received 100% boostrap support (Figure 2 and Supplementary Figure S4). The B. mori cluster fell within B. mandarina, suggesting that the latter taxon is paraphyletic (Figure 2 and Supplementary Figure S4).
Figure 1.
Schematic illustration of ML tree of domestic chicken (D) and wild red junglefowl (W) mt genomes (topology only; for full tree and sequences used see Supplementary Figure S3). Numbers on branches represent the percentage of 1000 bootstrap samples supporting the branch; only values ≥ 50% are shown.
Figure 2.
Schematic illustration of ML tree of Bombyx mori and B. mandarina mt genomes (for full tree and sequences used see Supplementary Figure S4). Numbers on branches represent the percentage of 1000 bootstrap samples supporting the branch; only values ≥ 50% are shown.
3.2. Nucleotide diversity
Synonymous (πS) and nonsynonymous (πN) nucleotide diversities were estimated for the concatenated coding sequences of the 9 mt protein-coding genes; πS and πN were estimated separately for domestic and wild isolates in each of the four taxa (Table 1). In each case, πS was significantly greater than πN for both domestic and wild isolates (Table 1). However, the πN :πS ratio was in each case higher in the domestic population than in the corresponding wild population (Table 1), consistent with relaxation of purifying selection in domestic populations. However, the differences between the πN :πS ratios of domestic and wild populations were generally small (Table 1).
Table 1.
Synonymous (πS) and nonsynonymous (πN) nucleotide diversity (± S.E.) in protein-coding genes from mitochondrial genomes of domesticated and wild animals.
| Species | N | πS | πN | πN:πS |
|---|---|---|---|---|
| Dog | ||||
| Domestic | 254 | 0.00808 ± 0.00067 | 0.00116 ± 0.00018*** | 0.144 |
| Wild | 19 | 0.02706 ± 0.00140 | 0.00282 ± 0.00028*** | 0.104 |
| Pig | ||||
| Domestic | 59 | 0.01382 ± 0.00106 | 0.00235 ± 0.00026*** | 0.170 |
| Wild | 27 | 0.02150 ± 0.00123 | 0.00350 ± 0.00030*** | 0.163 |
| Chicken | ||||
| Domestic | 41 | 0.00435 ± 0.00059 | 0.00068 ± 0.00011*** | 0.155 |
| Wild | 17 | 0.00498 ± 0.00059 | 0.00071 ± 0.00013*** | 0.143 |
| Silkworm | ||||
| Domestic | 33 | 0.00151 ± 0.00042 | 0.00042 ± 0.00010*** | 0.278 |
| Wild | 15 | 0.03528 ± 0.00242 | 0.00528 ± 0.00050*** | 0.150 |
Z-test that πS= πN; P < 0.001.
Nucleotide diversity (π) in 12S rRNAs, 16S rRNAs, and tRNAs was significantly lower than the corresponding value of πS in both domestic and wild populations of dog, pig, and chicken (Table 2). The same was true of wild but not domestic silkworm (Table 2). The absence of this pattern in the silkworm can probably be explained by a much lower overall level of polymorphism in B. mori than in the other domestic population examined. For example, πS in B. mori was only about one third that in the domestic chicken, only about one fifth that in the domestic dog, and only about one tenth that in the domestic pig (Table 1).
Table 2.
Nucleotide diversity (π ± S.E.) in non-protein-coding regions of mitochondrial genomes of domesticated and wild animals.
| Species | Domestic | Wild |
|---|---|---|
| Dog | ||
| 12S rRNA | 0.00145 ± 0.00062*** | 0.00429 ± 0.00108*** |
| 16 S rRNA | 0.00243 ± 0.00077*** | 0.00559 ± 0.00122*** |
| tRNAs | 0.00135 ± 0.00082*** | 0.00609 ± 0.00120*** |
| D-loop | 0.06633 ± 0.01632*** | 0.05770 ± 0.08118 |
| Pig | ||
| 12S rRNA | 0.00591 ± 0.00126*** | 0.00944 ± 0.00220*** |
| 16 S rRNA | 0.00281 ± 0.00130*** | 0.00560 ± 0.00136*** |
| tRNAs | 0.00423 ± 0.00113*** | 0.00847 ± 0.00131*** |
| D-loop | 0.01436 ± 0.00313 | 0.00203 ± 0.00323 |
| Chicken | ||
| 12S rRNA | 0.00077 ± 0.00028*** | 0.00104 ± 0.00118*** |
| 16 S rRNA | 0.00166 ± 0.00057** | 0.00183 ± 0.00058*** |
| tRNAs | 0.00108 ± 0.00043*** | 0.00091 ± 0.00037*** |
| D-loop | 0.00810 ± 0.00145* | 0.00904 ± 0.00146*** |
| Silkworm | ||
| 12S rRNA | 0.00081 ± 0.00040 | 0.00945 ± 0.00269*** |
| 16 S rRNA | 0.00227 ± 0.00054 | 0.00630 ± 0.00125*** |
| tRNAs | 0.00128 ± 0.00060 | 0.00531 ± 0.00139*** |
| D-loop | 0.00176 ± 0.01078 | 0.01480 ± 0.02846 |
The pattern of reduced nucleotide diversity RNA-encoding genes in comparison to synonymous sites in protein-coding genes supported the hypothesis that the RNA-encoding genes are subject to stronger purifying selection than are synonymous sites in protein-coding genes. By contrast, π in the D-loop was either similar to or greater than the corresponding values of πS (Table 2). In fact, in the case of the domestic dog and both domestic and wild chicken, π in the D-loop was significantly greater than the corresponding value of πS (Table 2). Thus, sites in the D-loop were not subject to stronger purifying selection than synonymous sites in protein-coding genes, and in some cases were significantly less constrained than even synonymous sites in protein-coding genes.
3.3. Polymorphic sites
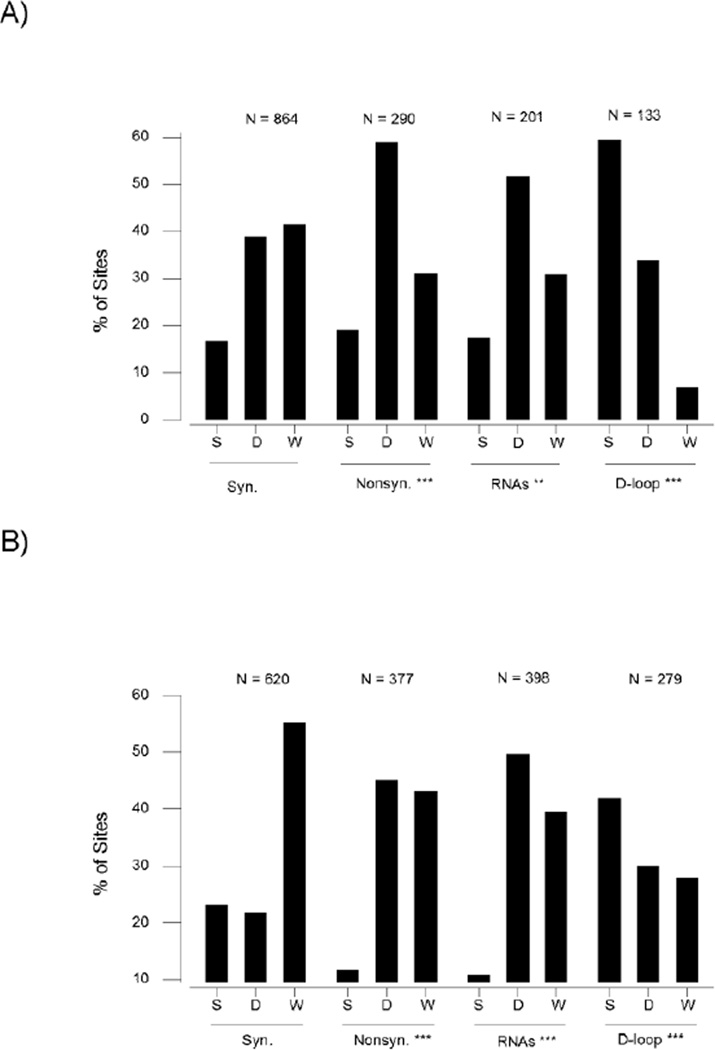
For each species, polymorphic nucleotide (SNP) sites were categorized as (1) unique to the domestic population; (2) unique to the wild population; or (3) polymorphic in both wild and domestic populations (shared polymorphic sites). These categories were compared among synonymous and non-synonymous sites in protein-coding genes; sites in 12S rRNA, 16S rRNA, and tRNA genes (RNA-encoding sites); and sites in the D-loop (Figure 3–4). In the dog, the proportions of nonsynonymous SNPs in the three categories differed significantly from that of synonymous SNPs (Figure 3A). 59.0% (171/290) nonsynonymous SNPs were unique to the domestic dog population and not found in the wolf population, whereas only 38.8% (335/864) of synonymous SNPs were unique to the domestic dog population (Figure 3A). Likewise, the proportions of RNA-encoding sites in the three categories differed significantly from that of synonymous sites (Figure 3A). 51.0% (104/201) of polymorphic RNA-encoding SNPs were unique to the domestic dog, again a higher proportion than in the case of synonymous SNPs (Figure 3A). On the other hand, in the case of D-loop SNPs, the majority (59.4% or 79/133) were shared between domestic dog and wolf, in marked contrast to the case of synonymous SNPs where only 16% (144/864) were shared (Figure 3A).
Figure 3.
Percentages of synonymous, nonsynonymous, RNA-encoding, and D-loop SNP sites shared between domestic and wild populations (S); unique to domestic population (D); and unique to wild population (W) in dog (A) and pig (B). Chi-square tests of the hypothesis that proportions in each category equal that of synonymous SNPs: ** P < 0.01; *** P < 0.001. In each case, the numbers of synonymous, nonsynonymous, RNA-encoding, and D-loop SNPs are indicated.
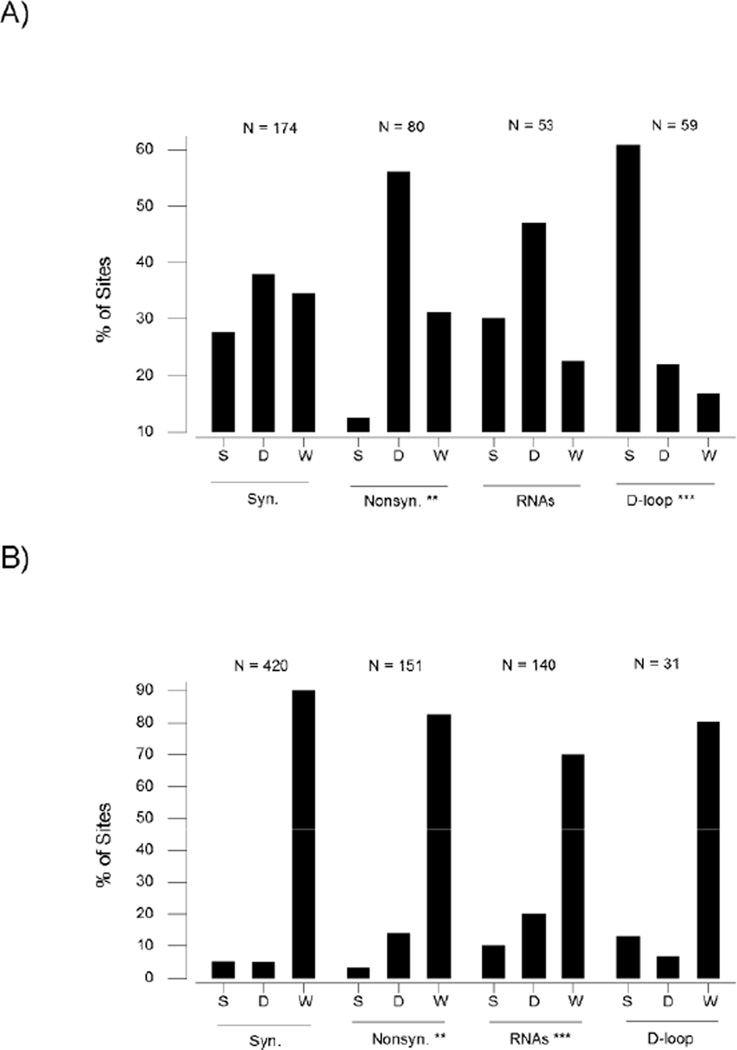
Figure 4.
Percentages of synonymous, nonsynonymous, RNA-encoding, and D-loop SNP sites shared between domestic and wild populations (S); unique to domestic population (D); and unique to wild population (W) in chicken (A) and silkworm (B). Chi-square tests of the hypothesis that proportions in each category equal that of synonymous SNPs: ** P < 0.01; *** P < 0.001. In each case, the numbers of synonymous, nonsynonymous, RNA-encoding, and D-loop SNPs are indicated.
In the other three taxa studied, the overall patterns were similar to those seen in the dog. In every case, a significantly greater proportion of nonsynonymous SNPs than of synonymous SNPs were unique to the domestic population (Figure 3–4). In the pig, 45.1% (170/377) of nonsynonymous SNPs were unique to the domestic population, whereas only 21.8% (135/620) of synonymous SNPs were unique to the domestic population (Figure 3B). In the chicken, 56.3% (45/80) of nonsynonymous SNPs were unique to the domestic population, whereas only 37.9% (66/174) of synonymous SNPs were unique to the domestic population (Figure 4A). In spite of the low level of polymorphism in B. mori, the pattern was similar in the silkworm. 13.9% (21/151) of nonsynonymous SNPs were unique to B. mori, whereas only 4.8% (20/420) of synonymous SNPs were unique to B. mori (Figure 4B).
Similarly, in every case, a significantly greater proportion of RNA-encoding SNPs than of synonymous SNPs were unique to the domestic population (Figure 3–4). In the pig, 49.7% (198/398) of RNA-encoding SNPs were unique to the domestic population, as opposed to 21.8% of synonymous SNPs (Figure 3B). In the chicken, 47.2% (25/53) of RNA-encoding SNPs were unique to the domestic population, as opposed to 37.9% of synonymous SNPs (Figure 4A). In the case of the silkworm, 20.0% (28/140) of RNA-encoding SNPs were unique to B. mori, as opposed to 4.8% of synonymous SNPs (Figure 4B).
In both the pig (Figure 3B) and the chicken (Figure 4A), the proportion of shared D-loop SNPs was significantly higher than that of shared synonymous SNPs. In the pig, 41.9% (117/279) of D-loop SNPs were shared, as opposed to 23.1% (143/620) of synonymous SNPs (Figure 3B). In the chicken, 61.0% (36/59) of D-loop SNPs were shared, as opposed to 27.6% (48/174) of synonymous SNPs (Figure 4A). In the case of the silkworm, 12.9% (4/51) of D-loop SNPs were shared, as opposed to 5.0% (21/420) of synonymous SNPs (Figure 4B). However, in the silkworm, the difference was not significant, since the number of polymorphic sites in the D-loop of silkworm was small (Figure 4B).
3.4. Gene diversity at polymorphic sites
Within each domestic population, the gene diversity at nonsynonymous SNP sites was compared between polymorphisms unique to the domestic population and those shared between domestic and wild populations (Table 3). In every case, median gene diversity at shared nonsynonymous SNPs was higher than that at nonsynonymous SNPs unique to the domestic population; and this difference was statistically significant in all cases but the dog (Table 3). In each species, the median gene diversity of unique nonsynonymous SNPs was the lowest possible value given the number of domestic genomes sampled, implying that in each domestic population more than half of the unique SNPs represented singletons (variants found in just one genome). In every domestic population, the proportion of singletons was greater among unique nonsynonymous polymorphisms than among shared nonsynonymous polymorphisms; and the difference was statistically significant in every case but the dog (Table 3).
Table 3.
Median gene diversity and % singleton at individual nonsynonymous SNP sites unique to domestic populations and at those shared between domestic and wild populations.
| SNP category | Species | Unique to Domestic | Shared | ||||
|---|---|---|---|---|---|---|---|
| Nonsynonymous | N | Gene Diversity |
% Singleton |
N | Gene Diversity |
% Singleton |
|
| Dog | 175 | 0.0078 | 50.9% | 28 | 0.0156 | 39.3% | |
| Pig | 170 | 0.0333 | 91.2% | 44 | 0.3703*** | 38.6%††† | |
| Chicken | 45 | 0.0476 | 93.2% | 10 | 0.2701*** | 10.0%††† | |
| Silkworm | 21 | 0.0588 | 81.0% | 5 | 0.1653* | 20.0%† | |
| RNA-encoding | |||||||
| Dog | 104 | 0.0078 | 63.5% | 35 | 0.0233** | 37.1%†† | |
| Pig | 165 | 0.0333 | 92.1% | 57 | 0.2586*** | 33.3%††† | |
| Chicken | 25 | 0.0476 | 80.0% | 16 | 0.2130*** | 6.3%††† | |
| Silkworm | 28 | 0.1139 | 46.4% | 14 | 0.1653*** | 7.1%† | |
Mann-Whitney tests of the hypothesis that median gene diversity for shared SNPs equals that for unique SNPs:
P < 0.05;
P < 0.01;
P < 0.001.
Fisher’s exact tests of the hypothesis that the proportion singleton in unique SNPs equals that in shared SNPs:
P < 0.05;
P < 0.01;
P < 0.001.
A similar pattern was seen in the case of RNA-encoding SNP sites. In each of the four taxa, median gene diversity was significantly higher at shared RNA-encoding SNPs than at those unique to the domestic population (Table 3). In each species, the proportion of singletons was significantly greater among unique RNA-encoding polymorphisms than among shared nonsynonymous polymorphisms (Table 3).
3.5. Divergence vs. polymorphism in silkworm
Because the domestic silkworm isolates clustered together in the phylogeny it was possible to compare polymorphism and divergence at synonymous and nonsynonymous sites. Compared with the nearest B. mandarina sequence (gi 210062303; Supplementary Figure S4), there were 55 synonymous and 12 nonsynonymous changes (ratio S:N = 4.6) in mt coding regions in B. mori. By contrast, there were 43 synonymous polymorphisms and 26 nonsynonymous polymorphisms in B. mori (ratio S:N = 1.7). The difference between these proportions was significant (χ2 = 6.60; 1 d.f. P = 0.01). This result shows an excess of nonsynonymous polymorphism over divergence.
4. Discussion
In protein-coding genes of mt genomes of both domestic and wild populations of dog, pig, chicken, and silkworm, nonsynonymous nucleotide diversity (πN) was significantly lower than synonymous nucleotide diversity (πS), supporting the hypothesis that nonsynonymous sites are subject to strong purifying selection. Likewise, nucleotide diversities (π) in RNA-encoding genes were consistently lower than πS, supporting the hypothesis that RNA-encoding sites are also subject to purifying selection. The πN :πS ratios were higher in domestic populations than in wild populations, suggesting a slight relaxation of purifying selection on protein-coding genes of domestic populations; but this effect was modest.
Much more striking evidence of relaxed purifying selection on domestic populations was obtained by comparing SNPs unique to domestic populations with those unique to wild populations and those shared between wild and domestic populations. In all four species, significantly greater proportions of nonsynonymous SNPs than of synonymous SNPs were unique to the domestic populations. Likewise, in all four species, significantly greater proportions of RNA-encoding SNPs than of synonymous SNPs were unique to the domestic populations. Thus, domestic populations were characterized by an excess of unique polymorphisms in two categories generally subject to purifying selection: nonsynonymous sites and RNA-encoding sites. Many of these unique polymorphisms seem likely to be slightly deleterious. The latter interpretation is supported by their generally lower gene diversities in comparison to those of polymorphisms shared by domestic and wild populations (Table 3).
The phylogeny of silkworm showed a different pattern from that seen the three vertebrates, with all domestic (B. mori) isolates forming a cluster distinct from, though nested within, the wild silkworm (B. mandarina; Figure 2). Given this phylogeny, it was possible to compare synonymous and nonsynonymous divergence and polymorphism in the silkworm. This analysis showed an excess of nonsynonymous polymorphisms over nonsynonymous divergence, a pattern consistent with relaxed purifying selection in B. mori. This pattern was the opposite of the result reported byLi et al. (2010) on the basis of a smaller B. mandarina sample, which did not include the B. mandarina sequence that in the present phylogeny formed the closest sister group to B. mori (gi 210062303; Supplementary Figure S4).Li et al. (2010) claimed that the pattern they observed was evidence of positive selection leading to fixation of certain amino acid substitutions in mt protein-coding genes as a result of domestication, following the reasoning of the McDonald-Kreitman (MK) test (McDonald and Kreitman 1991). The MK test is not really a valid test of positive selection because strong within-population purifying selection can lead to an excess of nonsynonymous divergence relative to polymorphism, thereby giving a false signal of positive selection (Hughes et al. 2008). The present analysis suggests a further problem with the MK test; namely, that it is very sensitive to the choice of outgroup.
In the three vertebrate species analyzed, domestic and wild isolates were intermingled in the phylogenies (Supplementary Figures S1-S3). Factors that may have given rise to such topologies include independent domestication events, initial domestication of a diverse population including several distinct matrilines, and post-domestication introgression (Björnerfeldt et al. 2006; Giuffra et al. 2000; Larson et al. 2005; Liu et al. 2006). From the point of view of the present study, it is not important which of these factors is responsible for the observed topology in a given case. What is striking is that all four domesticated species, in spite of differences in phylogeny, showed an essentially similar pattern with respect to the relaxation of purifying selection on protein-coding and RNA-encoding genes in domestic populations, and more stringent selection in wild populations. Note that such a pattern is expected if the observed pattern is due to increased intensity of purifying selection in wild populations. Even if a wild population has a feral origin (that is domestic individuals that escaped from domestication; Larson et al. 2005), it is expected that they will become subject to more stringent purifying selection once they become free-living; and this prediction is supported by the current results. The fact that feral populations of domestic species show evidence of selection favoring wild-like phenotypes is evidence for the fundamentally deleterious character of many phenotypes that have arisen in domestication (Brisbin et al. 1977; Sol 2008).
Population bottlenecks during domestication may lead to inbreeding, which is known to occur in the event of a prolonged and severe bottleneck (Nei et al. 1975). Inbreeding in turn is expected to lead to the purging of deleterious recessives, especially those with a strongly deleterious effects, from the nuclear genome of a diploid organism (Lacy and Ballou 1998). However, this effect will not be observed in mt genomes of vertebrates or insects, because mt genomes are haploid and inherited in the maternal line only. Rather, it might be predicted that in the case of the mt genome, the major effect of a bottleneck will be the relaxation of purifying selection due to decreased effective population size. The results of the present analyses are consistent with this prediction. Future studies might compare nuclear and mitochondrial genomes of domesticated animal species in order to test whether there is evidence that the purging of strongly deleterious recessive variants by inbreeding has occurred in the former. In addition, computer simulations of various bottleneck scenarios might provide refined predictions regarding potential differences between nuclear and mitochondrial genes with respect to the effects of selection and drift on the fate of deleterious variants during the process of domestication.
Supplementary Material
Highlights.
Domestic animals show different patterns of mitochondrial DNA variation than wild relatives.
Domestic animals show evidence of an excess of slightly deleterious variants
Domestication relaxes purifying selection on animal mitochondrial genomes
Abbreviations
- ML
maximum likelihood
- mt
mitochondrial
- SNP
single nucleotide polymorphism
- πS
nucleotide diversity at synonymous sites
- πN
nucleotide diversity at nonsynonymous sites
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amaral AJ, Ferretti L, Megens H-J, Crooijmans RP, Nie H, Ramos-Onsins SE, Perez-Enciso M, Schook LB, Groenen MA. Genome-wide footprints of pig domestication and selection revealed through massive parallel sequencing of pooled DNA. PLoS ONE. 2011;6(4):e14782. doi: 10.1371/journal.pone.0014782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson L. How selective sweeps in domestic animals provide new insight into biological mechanisms. J. Intern. Med. 2011;271:1–14. doi: 10.1111/j.1365-2796.2011.02450.x. [DOI] [PubMed] [Google Scholar]
- Andersson L, Georges M. Domestic-animal genomics: deciphering the genetics of complex traits. Nature Rev. Genet. 2004;5:202–212. doi: 10.1038/nrg1294. [DOI] [PubMed] [Google Scholar]
- Baranowska I, Jäderund KH, Nennesmo I, Holmqvist E, Heidrich N, Larsson N-G, Andersson G, Wagner EG, Hedhammar Å, Wibom R, Andersson L. Sensory ataxic neuropathy in Golden Retriever dogs is caused by a deletion in the mitochondrial tRNATyr gene. PLoS Genet. 2009;5(5):e1000499. doi: 10.1371/journal.pgen.1000499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazin E, Glémin S, Galtier N. Population size does not influence mitochondrial genetic diversity in animals. Science. 2006;312:570–572. doi: 10.1126/science.1122033. [DOI] [PubMed] [Google Scholar]
- Björnerfeldt S, Webster MT, Vilà C. Relaxation of selective constraint on dog mitochondrial DNA following domestication. Genome Res. 2006;16:990–994. doi: 10.1101/gr.5117706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisbin IL, Jr, Geiger RA, Graves HB, Pinder JE, III, Sweeney JM, Sweeney JR. Morphological characterizations of two populations of feral swine. Acta Theriol. 1977;22:75–85. [Google Scholar]
- Cruz F, Vilà C, Webster MT. The legacy of domestication: accumulation of deleterious mutations in the dog genome. Mol. Biol. Evol. 2008;25:2331–2336. doi: 10.1093/molbev/msn177. [DOI] [PubMed] [Google Scholar]
- Fisher RA. The genetical theory of natural selection. Oxford: Oxford University Press; 1930. [Google Scholar]
- Giuffra E, Kijas JM, Amarger V, Carlborg Ö, Jeon J-T, Andersson L. The origin of the domestic pig: independent domestication and subsequent introgression. Genetics. 2000;154:1785–1791. doi: 10.1093/genetics/154.4.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M, Cao Y, Yang Z. Preponderance of slightly deleterious polymorphism in mitochondrial DNA: nonsynonymous/synonymous substitution rate is much higher within species than between species. Mol. Biol. Evol. 1998;15:1499–1505. doi: 10.1093/oxfordjournals.molbev.a025877. [DOI] [PubMed] [Google Scholar]
- Hughes AL. Evidence for abundant slightly deleterious polymorphisms in bacterial populations. Genetics. 2005;169:553–558. doi: 10.1534/genetics.104.036939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL. Looking for Darwin in all the wrong places: the misguided quest for positive selection at the nucleotide sequence level. Heredity. 2007;99:364–373. doi: 10.1038/sj.hdy.6801031. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Friedman R. Codon-based tests of positive selection, branch lengths, and the evolution of mammalian immune system genes. Immunogenetics. 2008;60:495–506. doi: 10.1007/s00251-008-0304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Hughes MA. Coding sequence polymorphism in avian mitochondrial genomes reflects population histories. Mol. Ecol. 2007;16:1369–1376. doi: 10.1111/j.1365-294X.2007.03242.x. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Packer B, Welsch R, Bergen AW, Chanock SJ, Yeager M. Widespread purifying selection at polymorphic sites in human protein-coding loci. Proc. Natl. Acad. Sci. USA. 2003;100:15754–15757. doi: 10.1073/pnas.2536718100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Friedman R, Rivailler P, French JO. Synonymous and nonsynonymous polymorphisms and divergences in bacterial genomes. Mol. Biol. Evol. 2008;25:2199–2209. doi: 10.1093/molbev/msn166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimua M, Ohta T. Mutation and evolution at the molecular level. Genetics Suppl. 1973;73:19–35. [PubMed] [Google Scholar]
- Knapp EW, Irausquin SJ, Friedman R, Hughes AL. PolyAna: analyzing synonymous and nonsynonymous polymorphic sites. Conservation Genet. Resources. 2011;3:429–431. doi: 10.1007/s12686-010-9372-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S. Patterns of nucleotide substitution in mitochondrial protein coding genes of vertebrates. Genetics. 1996;143:537–548. doi: 10.1093/genetics/143.1.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacy RC, Ballou JD. Effectiveness of selection in reducing genetic load in populations of Peromyscus polionotus during generations of inbreeding. Evolution. 1998;52:900–909. doi: 10.1111/j.1558-5646.1998.tb03715.x. [DOI] [PubMed] [Google Scholar]
- Larson G, Dobney K, Albarella U, Fang M, Matisoo-Smith E, Robins J, Lowden S, Finlayson H, Brand T, Willerslev E, Rowley-Conwy P, Andersson L, Cooper A. Worldwide phylogeography of wild boar reveals multiple centers of pig domestication. Science. 2005;307:1618–1621. doi: 10.1126/science.1106927. [DOI] [PubMed] [Google Scholar]
- Li D, Guo Y, Shao H, Tellier LC, Wang J, Xiang Z, Xia Q. Genetic diversity, molecular phylogeny and selection evidence of the silkworm mitochondria implicated by complete resequencing of 41 genomes. BMC Evol. Biol. 2010;2010:10–81. doi: 10.1186/1471-2148-10-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W-H. Unbiased estimates of the rates of synonymous and nonsynonymous substitution. J. Mol. Evol. 1993;36:96–99. doi: 10.1007/BF02407308. [DOI] [PubMed] [Google Scholar]
- Liu Y-P, Wu G-S, Yao Y-G, Miao Y-W, Luikart G, Baig M, Beja-Pereira A, Ding Z-L, Palinchamy MG, Zhang Y-P. Multiple maternal origins of chickens: out of Asian jungles. Mol.Phyl.Evol. 2006;38:12–19. doi: 10.1016/j.ympev.2005.09.014. [DOI] [PubMed] [Google Scholar]
- MacEachern S, McEwan J, McCulloch A, Mather A, Savin K, Goddard M. Molecular evolution of the Bovini tribe (Bovidae, Bovinar): is there evidence of rapid evolution or reduced selective constraint in domestic cattle? BMC Genomics. 2009;2009:10–179. doi: 10.1186/1471-2164-10-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishmar D, Ruiz-Pesini E, Mondragon-Palomino M, Procaccio V, Gaut B, Wallace DC. Adaptive selection of mitochondrial complex I subunits during primate radiation. Gene. 2006;378:11–18. doi: 10.1016/j.gene.2006.03.015. [DOI] [PubMed] [Google Scholar]
- Nei M. Molecular Evolutionary Genetics. New York: Columbia University Press; 1987. [Google Scholar]
- Nei M, Kumar S. Molecular Evolution and Phylogenetics. New York: Oxford University Press; 2000. [Google Scholar]
- Nei M, Maruyama T, Chakraborty R. The bottleneck effect and genetic variability in populations. Evolution. 1975;29:1–10. doi: 10.1111/j.1558-5646.1975.tb00807.x. [DOI] [PubMed] [Google Scholar]
- Rand DM, Kann LM. Excess amino acid polymorphism in mitochondrial DNA: contrats among genes from Drosophila, mice, and humans. Mol. Biol. Evol. 1996;13:734–748. doi: 10.1093/oxfordjournals.molbev.a025634. [DOI] [PubMed] [Google Scholar]
- Rubin C-J, Zody MC, Ericksson J, Meadows JR, Sherwood E, Webster MT, Jiang L, Ingman M, Sharpe T, Ka S, Hallböök F, Besnier F, Carlborg Ö, Beh’hom B, Tixier-Boichard M, Jensen P, Siegel P, Lindblad-Toh K, Andersson L. Whole-genome resequencing reveals loci under selection during chicken domestication. Nature. 2010;464:587–593. doi: 10.1038/nature08832. [DOI] [PubMed] [Google Scholar]
- Ruiz-Pesini E, Wallace DC. Evidence for adaptive selection acting on the tRNA and rRNA genes of human mitochondrial DNA. Human Mut. 2006;27:1072–1081. doi: 10.1002/humu.20378. [DOI] [PubMed] [Google Scholar]
- Schaeffer AM, Taylor RW, Turnbull DM. The mitochondrial genome and mitochondrial muscle disorders. Curr. Opin. Pharmacol. 2001;1:288–293. doi: 10.1016/s1471-4892(01)00051-0. [DOI] [PubMed] [Google Scholar]
- Sol D. Artificial selection, naturalization, and fitness: Darwin’s pigeons revisited. Biol. J. Linn. Soc. 2008;93:657–665. [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Diggins DG. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. Diseases of the mitochondrial DNA. Annu. Rev. Biochem. 1992;61:1175–1212. doi: 10.1146/annurev.bi.61.070192.005523. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.