Abstract
Malignant gliomas are highly invasive tumors with an almost invariably rapid and lethal outcome. Surgery and chemoradiotherapy fail to remove resistant tumor cells that disperse within normal tissue, which are a major cause for disease progression and therapy failure. Infiltration of the neural parenchyma is a distinctive property of malignant gliomas compared to other solid tumors. Thus, glioma cells are thought to produce unique molecular changes that remodel the neural extracellular matrix and form a microenvironment permissive for their motility. Here we describe the unique expression and pro-invasive role of fibulin-3, a mesenchymal matrix protein specifically upregulated in gliomas. Fibulin-3 is downregulated in peripheral tumors and thought to inhibit tumor growth. However, we found fibulin-3 highly upregulated in gliomas and cultured glioma cells, although the protein was undetectable in normal brain or cultured astrocytes. Overexpression and knockdown experiments revealed that fibulin-3 did not seem to affect glioma cell morphology or proliferation, but enhanced substrate-specific cell adhesion and promoted cell motility and dispersion in organotypic cultures. Moreover, orthotopic implantation of fibulin-3-overexpressing glioma cells resulted in diffuse tumors with increased volume and rostrocaudal extension compared to controls. Tumors and cultured cells overexpressing fibulin-3 also showed elevated expression and activity of matrix metalloproteases, such as MMP-2/9 and ADAMTS-5. Taken together, our results suggest that fibulin-3 has a unique expression and pro-tumoral role in gliomas, and could be a potential target against tumor progression. Strategies against this glioma-specific matrix component could disrupt invasive mechanisms and restrict dissemination of these tumors.
Keywords: EFEMP-1, fibulin, proteoglycans, extracellular matrix, glioma invasion
Malignant gliomas are the most common tumors originating within the CNS and account for over 13,000 deaths annually in the US (1). A hallmark of these highly aggressive tumors is their diffuse infiltration in the normal neural tissue with subsequent dispersion of isolated tumor cells far from the tumor core (2). Invasive glioma cells remain embedded in the CNS after tumor removal and are thought to be resistant to adjuvant chemoradiotherapy (3, 4), thus causing inevitable dissemination and recurrence of the disease, and failure of current therapeutic strategies in the long term (5).
The brain parenchyma is highly refractory to invasion by metastatic tumors, which essentially grow in the brain by displacing neural tissue aside rather than infiltrating it, even when those tumors may aggressively invade their tissues of origin (6). In contrast, glioma cells invade the central nervous tissue extensively, despite the absence of a supporting stroma (5) and the presence of inhibitory substrates for cell motility (5, 7). Interestingly, gliomas very rarely metastasize outside the CNS even after widespread dissemination within neural tissue, and do not disperse when implanted in peripheral tissues (8, 9). Thus, glioma cells seem to be highly specialized and selective to colonize the neural environment, which suggests the existence of mechanisms of invasion adapted for the particular composition and architecture of the CNS (10, 11).
One of the major barriers to cell dispersion in the adult CNS is the ubiquitous extracellular matrix (ECM), which is rich in hyaluronic acid and negatively charged proteoglycans but lacks most fibrillar proteins that support cell adhesion and motility (12, 13). Motile glioma cells can remodel this matrix by local degradation of ECM molecules and synthesis of a novel pericellular scaffold (11). The glioma ECM contains molecules that predominate during neural development (hyaluronic acid, phosphacan, SPARC) (14, 15), mesenchymal proteins found in peripheral stroma and basal lamina (collagens, fibronectin) (16), and chondroitin sulfate proteoglycans that limit cell motility in the adult CNS (versican, brevican) (17, 18) but instead promote glioma cell migration (7, 19, 20). Recent evidence from our and other laboratories has suggested that the presence of both mesenchymal and neural-specific matrix proteins in gliomas may result in novel molecular interactions that specifically promote glioma cell motility in the CNS (19, 21).
In an attempt to identify matricellular signals that could contribute to the unique invasive ability of glioma cells, we stimulated glioma cell adhesion and motility with a combination of mesenchymal (fibronectin) and neural-specific (brevican) matrix proteins, and analyzed the resulting upregulated genes using a microarray approach (Suppl. Fig. 1 and Suppl. Table I). From this analysis, we identified the protein fibulin-3, a recently identified member of the fibulin family, as a novel ECM component upregulated in motile glioma cells.
The fibulins are a family of ECM glycoproteins characterized by a tandem of epidermal-growth factor (EGF)-like repeats followed by a C-terminal fibulin-type domain common to all members of the family (22). Fibulin-1 and -2 are the prototypical, multimeric, large members of the family, located in the basal laminae of epidermal, muscle and connective tissues, whereas fibulin-3, 4 and 5 are smaller, monomeric proteins expressed in the ECM of blood vessels and elastic tissues (22). The small fibulins are expressed at low levels in the CNS, and fibulin-3 in particular has the lowest expression levels in rodent brain compared to the other members of the family (22, 23). Previous research has shown that fibulin-3 is downregulated in several types of solid tumors and its overexpression could limit tumor growth (24). However, we show here that fibulin-3 follows a strikingly different profile in gliomas, is highly and specifically upregulated in these tumors, and promotes glioma cell migration and tumor progression. Our results suggest that expression of fibulin-3 is differentially regulated in gliomas compared to other tumors, and that this protein may play a unique promoting role in glioma invasion.
EXPERIMENTAL PROCEDURES
Meta-analysis of microarray data
Expression of fibulin-3 mRNA in solid tumors, including gliomas, and their corresponding control tissues was obtained from 68 independent studies stored in the microarray database Oncomine Research™ (http://www.oncomine.org, Suppl. Table II), and compared by a two-sided t-test as described (25). Normalized data from Oncomine reveal the direction and significance of changes in gene expression but not their direct magnitude; therefore, we used the value of the Student’s t statistic from each study to represent changes in fibulin-3 expression (25). Expression of mRNA for the different members of the fibulin family in gliomas was obtained from 275 grade II-IV glioma specimens and 28 controls stored in the NCI Repository for Molecular Brain Neoplasia Data (REMBRANDT, http://rembrandt.nci.nih.gov, Suppl. Table III). Expression values were collected for ‘unified gene’ probesets corresponding to the different splice-forms of each fibulin gene (26), and plotted as the fold-level (i.e., log2 ratio) of each tumor to control samples. The expression of each fibulin mRNA was analyzed independently by one-way ANOVA, followed by post-hoc pairwise Dunnet’s test to compare the expression in each type of glioma against the controls.
Cell cultures and antibodies
The human glioma cell lines U87-MG, U251-MG and U373-MG (American Type Culture Collection, Manassas, VA), and the mouse glioma cell lines GL-261 and KR-158, were grown at 5% CO2 in DMEM supplemented with 10% fetal calf serum (FCS). The rat glioma cell line CNS-1 was grown in RPMI-1640 equally supplemented with 10% FCS. The recently described cell lines X12/GBM12 and X14/GBM14 (27, 28) were kindly provided by Dr. E. A. Chiocca (Department of Neurological Surgery, The Ohio State University). These cells were maintained as subcutaneous xenografts in nude mice and subcultured in DMEM supplemented with 1% FCS for one to two passages between in vivo implantations. Glioma initiating cells, prepared as neurospheres from clinical specimens (29), were characterized and kindly provided by Dr. Yoshinaga Saeki (Department of Neurological Surgery, The Ohio State University) (30). Glioma neurospheres were cultured in DMEM/F-12 supplemented with 2 μM glutamine, 20 ng/ml EGF, 20 ng/ml basic FGF and 1x B27 supplement (Invitrogen, Carlsbad, CA). Culture medium in all cases was supplemented with 50 U/ml penicillin and 50 μg/ml streptomycin.
Human fibulin-3 was detected with a mouse monoclonal antibody (mAb5-3, 0.4 μg/ml, Santa Cruz Biotechnology, Santa Cruz, CA) characterized and kindly provided by Dr. Lihua Marmorstein (Department of Ophthalmology and Vision Science, University of Arizona) (31). Preliminary characterization of this antibody suggests that it recognizes a short N-terminal sequence (90Pro-115Gly) containing several O-glycosylation sites. The antibody has low affinity for the native protein and may not recognize all potential glycoforms of fibulin-3. Subcellular fractionation markers were detected with monoclonal antibodies against proliferating cell nuclear antigen (PCNA, 0.2 μg/ml, Santa Cruz Biotechnology), microsomal ribophorin (0.25 μg/ml, Santa Cruz Biotechnology) and soluble α-tubulin (0.1 μg/ml, Invitrogen). Metalloproteases were detected with rabbit polyclonal antibodies against MMP2 (0.4 μg/ml, Santa Cruz Biotechnology) and MMP9 (1 μg/ml, Abcam, Cambridge, MA). Additional protein controls were detected with monoclonal antibodies against globular actin (1.5 μg/ml, Sigma-Aldrich) and the V5-epitope tag (1/5000, Invitrogen).
Human tissue processing
All studies involving human tissue specimens were performed in compliance with the guidelines of the Human Investigations Committee at The Ohio State University College of Medicine. Pathologically graded fresh-frozen surgical specimens of high-grade adult gliomas (patient age range: 37 to 74 years old) were obtained through the NCI Cooperative Human Tissue Network. Brain cortex tissue from age-matched controls was obtained from the NICHD Brain and Tissue Bank for Developmental Disorders at the University of Maryland, Baltimore MD. Tissues were individually homogenized in 20 mM TrisHCl, pH 7.4, containing 320 mM sucrose and a cocktail of protease inhibitors (Complete, Roche Applied Science, Indianapolis, IN). Total homogenates were subjected to subcellular fractionation as previously described (32) and further processed for protein electrophoresis.
Cell transfection and transduction
A clone containing the complete coding sequence of human fibulin-3 (gene EFEMP-1, Genbank # BC098561) was subcloned by PCR into the vector pcDNA4.V5.6xHis (Invitrogen) to produce V5-tagged fibulin-3 isoforms. Constructs were transiently transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. Additionally, the cDNA for full-length fibulin-3 was subcloned into the lentiviral carrier vector pCDH-EF1-copGFP (System Biosciences, Mountain View, CA). Lentiviruses were produced in HEK293 cells using the ViraPower packaging system (Invitrogen) and titrated as described (33). Cells were transduced at different multiplicities of infection and tested by quantitative RT-PCR (qRT-PCR) and Western blotting to identify cultures overexpressing fibulin-3 within a physiological range comparable with that observed in clinical specimens compared against normal brain, as described (20).
For RNA interference experiments, predesigned siRNA oligonucleotides against human and rat fibulin-3 were purchased from QIAGEN (Valencia, CA). Control siRNAs from the same manufacturer included scrambled versions of the fibulin-3 siRNAs and a validated, non-silencing siRNA (“AllStars negative control”). siRNAs were transiently transfected at the rate of 100 pmol / (1.106 cells x 3ml culture medium) using Lipofectamine 2000. Cells were collected 48h post-transfection to quantify the levels of fibulin-3 and for further testing in adhesion and motility assays (list of siRNA sequences is provided in Suppl. Table IV).
Cell proliferation, adhesion and dispersion assays
Transduced cells were grown in 96-well plates at an initial density of 1,000 cells/well in 200 μl of culture medium. Proliferation was quantified by measuring the reduction of a soluble tetrazolium salt (CellTiter kit, Promega, Fitchburg, WI) according to the manufacturer’s instructions. In parallel plates, cells were stained with 0.5 μg/ml propidium iodide (apoptotic cells) and 2 μg/ml Hoechst-33258 (total cells) and the ratio of apoptotic/total cells was quantified by automated nuclei count using the software ImageJ.
For cell-adhesion assays, 48-well plates were precoated for 2 hours at room temperature with the following substrates: human fibronectin (5 μg/mL, Becton-Dickinson, San Jose, CA), mouse laminin (5 μg/mL, Invitrogen), high-Mw poly-L-lysine (50 μg/mL, Sigma-Aldrich, St Louis, MO) and high-Mw hyaluronic acid (200 μg/mL, Calbiochem, La Jolla, CA). Non-specific binding sites were subsequently blocked with 1% bovine serum albumin in Dulbecco’s PBS. Stably transduced glioma cells were gently resuspended and dissociated in DPBS + 2 mM EDTA, transferred to fresh culture medium, and plated at 50,000 cells/well on the precoated plates. After 30 minutes the cells were washed, fixed and quantified by crystal violet staining as described (21). To test direct binding of cells to fibulin-3, we repeated the same procedure using 1 μg/ml of purified human fibulin-3 (His-tagged) or commercially available fibulin-3 (GST-tagged, NovusBiologicals, Littleton, CO). All experiments were repeated at least three times with 4 to 6 replicates per experimental condition, and further analyzed by 2-way ANOVA.
Organotypic cultures of brain slices were prepared as previously described (20). Briefly, coronal brain slices (300-μm thick) from postnatal day 1 CD-1 mice were cultured on MilliCel membranes (0.4 μm-pore size, Millipore, Temecula, CA) over slice culture medium Neurobasal-A/HBSS, (70/30 ratio), supplemented with 1mM L-glutamine, 1mM sodium-pyruvate, 0.5% FCS, 1X B27 supplement, 1X G5 supplement (Invitrogen), 100 U/mL penicillin and 100 μg/mL streptomycin. Glioma cells were cultured in suspension at 1.105 cells/mL for 48h to form spherical aggregates, individually “seeded” on the brain slices, and imaged in culture during five days. Experiments were performed in triplicate, using at least 10 aggregates per experimental condition, and analyzed by 2-way ANOVA for repeated measures. To further analyze cell motility within the slices, 60–80 independent cells/condition were imaged overnight by time-lapse microscopy. Cell tracks were manually reconstructed with image software and migration parameters (average cell velocity and net migrated distance) were compared by two-tailed Mann-Whitney’s U-test. To test the effect of fibulin-3 siRNA on glioma cell dispersion, cells were transfected with siRNAs during the suspension culture, 48h before applying the aggregates on the brain slices.
Animal studies
All studies involving animals were approved by the institutional Animal Care and Use Committee at The Ohio State University. Glioma cells were harvested at 80% confluence and resuspended at 2.5x104 cells/μl in Hanks’ buffered saline solution supplemented with 0.1 % w/v glucose. The cell suspension (3 μl) was injected stereotactically into the right thalamus of 90-day old female Lewis rats (180–210 g), over a 5 min period, as previously described (34). Animals were euthanized and tumors harvested 15 days after tumor implantation.
Tumor histology
Brains were cryo-sectioned coronally at 25 μm and every fourth section stained with 0.1 % w/v cresyl violet. Tumor mass and detached cell clusters were identified by microscopy and delineated by a researcher blinded to the experimental conditions, as previously described (20). Imaging software (ImageJ) was used for image registration and alignment, and to calculate, for each section, the area of the tumor mass and detached cell clusters (35). Total tumor volume was calculated using Cavalieri’s estimator of morphometric volume as described (20). To represent antero-posterior spread of the tumor, the tumor origin (zero) was made to coincide with the section with largest tumor area, which matched the injection site in all cases.
Immunohistochemistry
Coronal brain tissue sections (20-μm thick) were subjected to antigenic recovery in 10mM sodium citrate buffer, pH 6.0, for 30 min at 65 °C, and subsequently blocked for non-specific binding with 100 mM sodium phosphate buffer, pH 7.4, containing 0.25% w/v Triton X-100 and 3% w/v bovine serum albumin. Sections were probed overnight at 4 °C with an antibody against the proliferation marker Ki67 (1/50, Thermo-Fisher, Waltham, MA), and counterstained with 4′,6-diamino-2-phenylindole (DAPI). Ten random sections per tumor (n=3 tumors for each condition) were probed for Ki67 antigen expression. The proliferative index (% of Ki67-positive nuclei relative to total number of DAPI-stained nuclei in each field) was measured in three fields per section and calculated by automated scoring of at least 300 nuclei per field (using ImageJ software), as described (20).
Western blotting, zymography, and RT-PCR
Cells processed for protein electrophoresis were lysed in 25 mM TrisHCl, pH 7.4, containing 150 mM NaCl, 1% w/v CHAPS and a cocktail of protease inhibitors (Complete). Conditioned, serum-free, culture medium was filtered and concentrated as previously described (32). Cell and tissue samples containing 15–20 μg total protein were electrophoresed on reducing 7.5% SDS-polyacrylamide gels and analyzed by Western blotting (32). To quantify metalloprotease activity, concentrated samples of conditioned medium were prepared in non-reducing electrophoresis buffer and processed for gelatin zymography as described (36).
Animals selected for tumor mRNA analysis were anesthetized and perfused transcardially with 100 mM sodium phosphate buffer, pH 7.4, followed by tissue dissection and snap freezing in liquid nitrogen. Frozen tissues and cultured glioma cells were extracted in Trizol (Invitrogen), residual DNA was degraded using Turbo-DNA Free (Applied Biosystems, Foster City, CA) and total cDNA was prepared with M-MLV reverse transcriptase (Invitrogen), following the instructions of the manufacturers. cDNA was processed for RT-PCR or quantitative RT-PCR using iQ SYBR Green Supermix (Bio-Rad, Hercules, CA) and the following parameters suggested by the manufacturer: denaturation step (95 °C, 30 seconds), annealing step (58 °C, 30 seconds), and elongation step (72 °C, 30 seconds), for a total of 45 cycles. A list of all primers used for RT-PCR is provided in Suppl. Table IV.
RESULTS
Fibulin-3 is highly upregulated in high-grade gliomas and secreted by glioma cells
Fibulin-3 is a poorly studied member of the fibulin family, usually associated with the basal lamina of non-neural tissues. It is downregulated in peripheral solid tumors and thought to impair tumor progression (24), making its detection and possible role in malignant brain tumors intriguing.
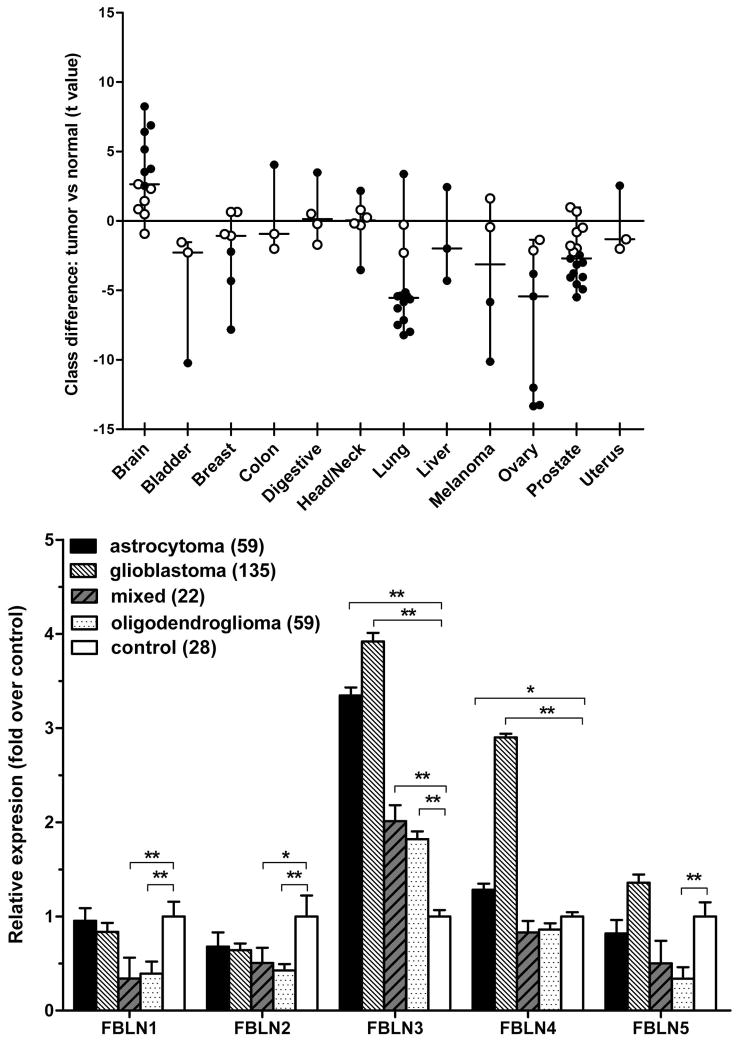
To study the expression of fibulin-3 in solid tumors, we first analyzed microarray data from two curated databases that have been used to identify specific patterns of gene expression in cancer. Analysis of fibulin-3 expression in the studies provided by Oncomine Research™ confirmed that this protein was mostly downregulated in non-neural solid tumors when compared against normal tissue (Fig. 1A), in agreement with previous findings (24). However, fibulin-3 expression in gliomas showed a surprisingly distinct pattern, with moderate to strong upregulation in most cases. To corroborate this result we compared the expression of fibulin-1 to -5 in primary brain tumors, using microarray data from the NCI Repository for Molecular Brain Neoplasia Data. Our results (Fig. 1B) confirmed that fibulin-3 was strongly upregulated in gliomas and, in addition, revealed that this protein was the most upregulated member of the family in all classes of glioma analyzed. Taken together, these results strongly suggested that fibulin-3 expression was likely under a different type of regulation in gliomas compared to peripheral tumors.
Figure 1.
Fibulin-3 is specifically upregulated in primary brain tumors. A) Relative expression of fibulin-3 mRNA in solid tumors, according to meta-analysis of microarray data from the database Oncomine Research™. Each dot represents a study that compared a class of solid tumors and their respective control tissue. The t value corresponds to the Student’s t-test used for comparison in each study; larger absolute values of t indicate greater difference between the two classes compared. Black and white dots indicate significant and non-significant differences at p<0.05, respectively. Note the consistently upregulated expression of fibulin-3 mRNA in primary brain tumors. A list of all studies and the number of control and tumor specimens per study is provided in Suppl. Table II. B) The mRNA expression levels for the members of the fibulin (FBLN) family were compared in different groups of gliomas versus control brain tissue, using microarray data from the NCI Repository for Molecular Brain Neoplasia Data. Bars indicate the level of expression (mean ± S.E.M.) for each tumor type or normal brain tissue. Data for each fibulin member were analyzed by one-way ANOVA followed by post-hoc Dunnet’s test to compare each type of glioma against control (* p<0.05; ** p<0.001). Fibulin-3 was consistently upregulated in all types of gliomas analyzed.
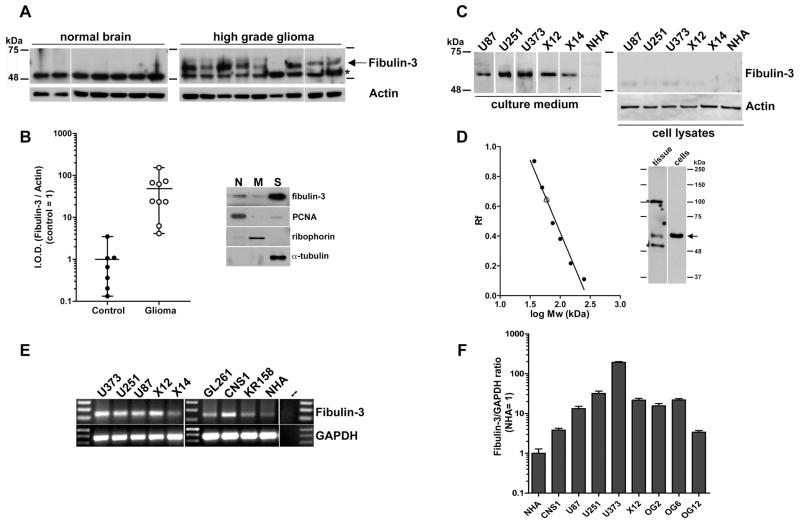
To validate the expression of fibulin-3 in gliomas at the protein level, we processed fresh-frozen specimens of grade III anaplastic astrocytoma and grade IV glioblastoma, as well as age-matched controls, for Western blotting. Fibulin-3 was absent in total homogenates from normal brain tissue, but could be detected in almost all glioma samples (Fig. 2A–B). Further fractionation of homogenates and analysis of subcellular fractions (Fig 2B) suggested that fibulin-3 was largely soluble and had weak association with the nuclear- and membrane-containing fractions.
Figure 2.
Fibulin-3 is highly expressed in glioma tissue and cells. A) Total homogenates from nine high-grade gliomas (two grade III astrocytomas and seven grade IV glioblastomas) and seven age-matched controls were probed for fibulin-3 (arrow). The asterisk indicates non-specific cross-reactivity of the antibodies used. B) Quantification of results from (A); notice that fibulin-3 expression is virtually absent in normal brain compared to gliomas. I.O.D., integrated optical density. The accompanying western blot shows a representative subcellular fractionation of glioma tissue revealing the distribution of fibulin-3. Nuclear antigen PCNA, microsomal ribophorin and soluble alpha-tubulin were used as specific markers for the nuclei-enriched (N), membrane-enriched (M) and soluble (S) subcellular fractions, respectively. C) Expression of fibulin-3 in the conditioned medium and total lysates of glioma cell lines (U87, U251, U373), primary cultures of glioma xenografts (X12, X14) and cultured normal human astrocytes (NHA). Equal protein loading for culture media was determined by protein measurement and by comparable amido-black staining of the blot membranes prior to incubation with antibody. D) Analysis of relative migration of fibulin-3 from soluble fraction of glioma tissue (tissue, open circle) or medium from U251-MG glioma cells (cells, open triangle) indicated a single form of Mw~54-kDa, as expected. No other fibulin-3 bands were observed. Asterisks indicate antibody cross-reactivity as indicated above. E) Detection of fibulin-3 mRNA in human glioma cell lines, rodent glioma cell lines (rat CNS-1; mouse GL-261 and KR-158) and NHA. The rightmost lane corresponds to RT-PCR in the absence of template. E) Results from qRT-PCR show the comparative expression of fibulin-3 mRNA (mean ± S.E.M.) in NHA (control=1) versus glioma cell lines and glioma neurospheres prepared from patient specimens (OG2, OG6, OG12) as indicated in the methods section. Gliomaspheres cultured in serum-free conditions express fibulin-3 at levels comparable to those of typical glioma cell lines, suggesting that expression of fibulin-3 in culture is not caused by serum-induced mesenchymal drift.
To determine if glioma cells were effectively a source of fibulin-3 in the tumor, we analyzed mRNA and protein samples from glioma cell lines, primary cultures of glioma cells and neurospheres of glioma initiating cells prepared from clinical specimens. We detected fibulin-3 in the conditioned medium of all human glioma cell lines analyzed, including U87MG, U251MG and U373MG (Fig 2C) as well as U118MG and A172 (not shown). Only one isoform of Mw ~54 kDa was observed both in glioma cells and tissue, with perfectly matching migration profiles (Fig. 2D). Fibulin-3 was not detected in total cell lysates, probably due to low association of the secreted protein to cell membranes, and the inability of the antibody to detect incompletely glycosylated intracellular fibulin-3 (BH and MSV, unpublished observations). We also detected fibulin-3 mRNA in rat (CNS-1) and mouse (GL-261, KR-158) glioma cell lines (Fig. 2E), although we could not use anti-fibulin-3 antibody to detect the protein because of species-specific recognition.
To test the expression of fibulin-3 in primary rather than long-established glioma cells, we analyzed the medium of low-passage GBM12/X12 and GBM14/X14 glioma cells, two models of invasive human glioma that are maintained as xenografts to prevent serum-dependent culture adaptation (27). We also measured, by qRT-PCR, the expression of fibulin-3 mRNA in primary glioma-derived neurospheres cultured in serum-free conditions (Fig. 2F). Fibulin-3 was highly expressed in all cases analyzed. However, using equivalent procedures and detection conditions we could not find fibulin-3 in the conditioned medium of low-passage normal human astrocytes. Fibulin-3 mRNA levels were also very low in these cells, four- to five-fold lower than the lowest level observed in glioma cells. (Fig. 2F).
Fibulin-3 secreted by glioma cells has a premoninant isoform
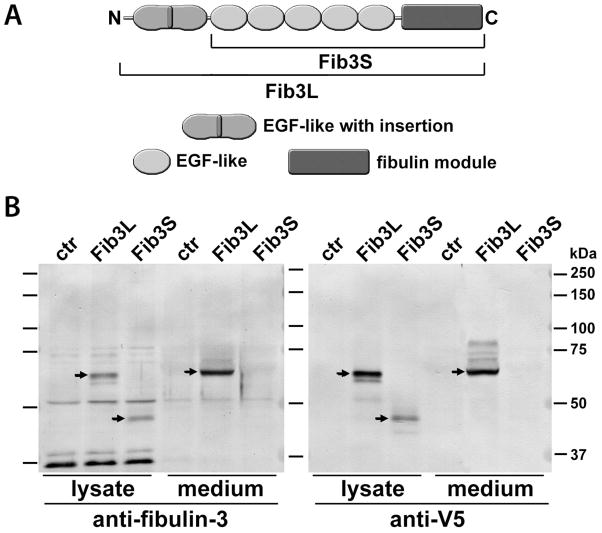
The first study of fibulin-3 gene (37) predicted two possible isoforms for this protein, generated by differential initiation of translation: a ‘long’ form of 53–55 kDa, with an N-terminal Ca++-binding EGF-like repeat, and a ‘short’ form of 40–43 kDa without that domain (Fig. 3A). However, in all our experiments we only detected the long form of fibulin-3 in glioma cells and tissues, suggesting that the putative short isoform was either expressed at extremely low levels or not at all. These results agreed with the original description of endogenous fibulin-3 protein in retinal cells (31) and with the observation that recombinant 43-kDa fibulin-3 is likely degraded and does not exist in substantial amounts in the extracellular environment (38).
Figure 3.
Fibulin-3 has a predominant isoform in glioma cells. A) Two possible isoforms of human fibulin-3 have been proposed: Fib3L (Long, 53–55 kDa) and Fib3S (Short, 43–45 kDa). B) Both isoforms were created by PCR, tagged with a C-terminal V5 epitope and expressed in rat CNS-1 glioma cells. Blots were probed with anti-V5 antibody and with antibody against human fibulin-3 that does not cross-react with endogenous rat fibulin-3. Results obtained with both antibodies strongly suggest that recombinant Fib3S is produced much less efficiently than Fib3L and secreted at very low or undetectable amounts to the culture medium. Note that the V5 antibody detected a number of faint bands above the position of Fib3L in the culture medium, which may correspond to glycoforms not detected by the anti-fibulin-3 mAb3-5 antibody.
To confirm these observations, we transfected glioma cells with the cDNA corresponding to each possible fibulin-3 isoform and observed that the long form of fibulin-3 was largely secreted to the culture medium, whereas the short form was expressed at lower levels, accumulated in the total cell lysate, and was virtually absent from the culture medium (Fig. 3B). Together, our results suggested that only the long form of fibulin-3 was expressed in vivo, and prompted us to utilize this isoform for all subsequent experiments.
Fibulin-3 enhances glioma cell adhesion and migration
Because we had identified fibulin-3 in an assay designed to detect genes upregulated in adherent glioma cells (Suppl. Fig. 1), we hypothesized that this protein could play a role in regulating glioma cell adhesion or motility. To test this hypothesis we analyzed the effect of fibulin-3 overexpression and knockdown in adhesion and migration assays.
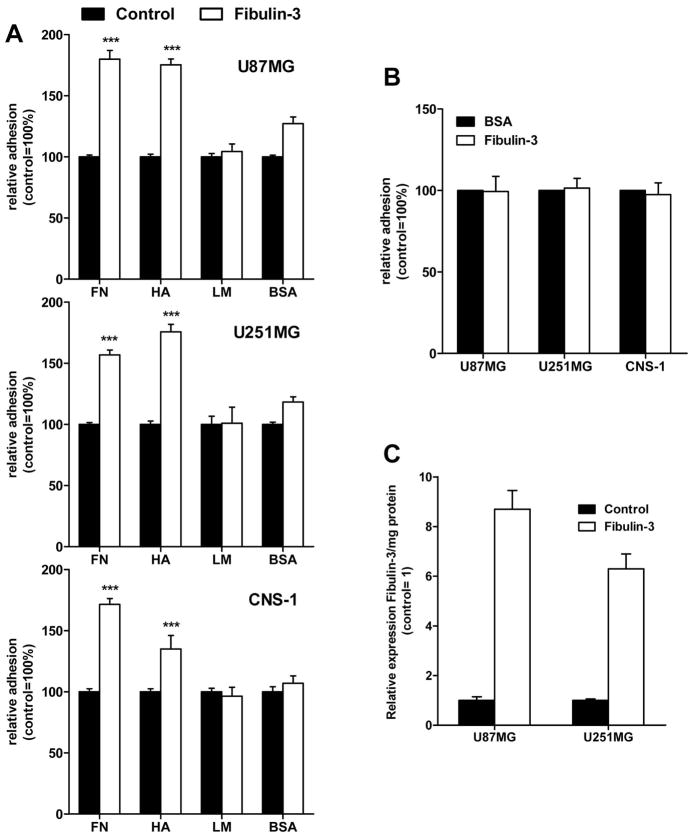
Physiological overexpression of fibulin-3 stimulated cell adhesion to ECM molecules abundant in the glioma matrix, such as fibronectin and hyaluronic acid, but not to uncoated plates or those coated with type-I laminin or poly-L-lysine (Fig. 4A,C). However, glioma cells did not show increased adhesion to wells coated with purified fibulin-3 (Fig. 4B), suggesting that fibulin-3 was not acting as an adhesive substrate by itself but had indirect effect on cell adhesion.
Figure 4.
Expression of fibulin-3 increases substrate-dependent glioma cell adhesion. A) Human (U87MG, U251MG) and rat (CNS-1) glioma cells stably overexpressing fibulin-3 were plated and quantified on multiwell plates coated with fibronectin (FN), type-I laminin (LN), high-MW hyaluronic acid (HA), or left uncoated and blocked with bovine albumin (BSA). B) Non-transfected cells were plated as above on multiwell plates coated with purified fibulin-3 (white bars) or left uncoated (BSA, black bars). Fibulin-3 did not act as a pro-adhesive substrate for any of the cell lines tested. All experiments were repeated at least three times with 3–6 replicates per condition. Data (mean ± S.E.M.) were analyzed by two-way ANOVA (*** p<0.001). Results on poly-L-lysine-coated wells were undistinguishable from those on uncoated surfaces (not shown). C) Relative expression levels of fibulin-3 in conditioned medium from control and fibulin-3-overexpressing U87MG and U251MG glioma cells were determined by Western blotting and confirmed to be within the physiological range observed in clinical specimens, as described in the methods section. Overexpression of fibulin-3 in transfected rat CNS-1 cells was quantified by qRT-PCR (not shown) and found comparable with mRNA overexpression levels in transfected human glioma cells.
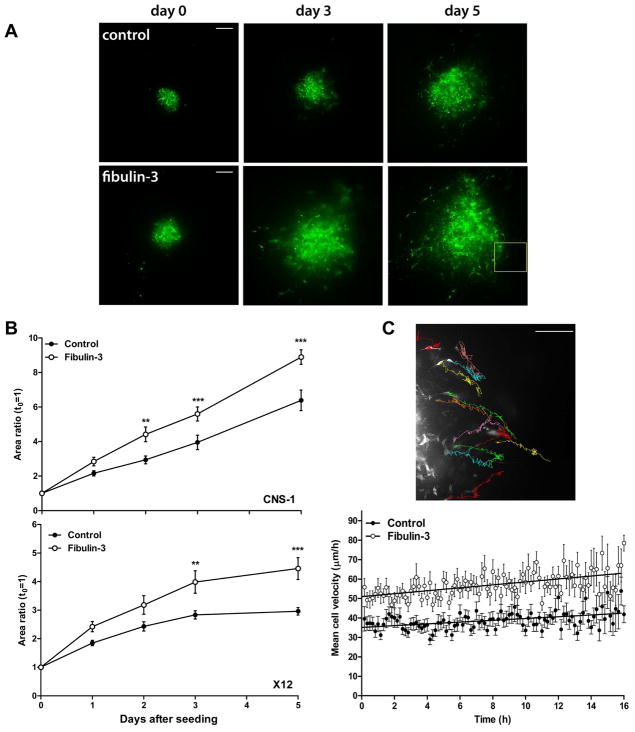
Next, we tested the effect of fibulin-3 on glioma cell invasion in cultured brain slices that mimic the cytoarchitecture and natural barriers of the brain (20, 21) (Fig. 5A). Overexpression of fibulin-3 in the invasive cell lines CNS-1 and X12 consistently enhanced glioma cell dispersion in cultured neural tissue (Fig. 5B). Further analysis of the fast-migrating CNS-1 cells by time-lapse microscopy revealed that fibulin-3-overexpressing cells had significantly higher migration velocity than controls (55.95 ± 2.14 versus 38.16 ± 1.58 μm/h, p<0.001_) and migrated a longer net distance from their points of origin (229.92 ± 12.60 versus 142.20 ± 8.94 μm over 16h, p<0.001), suggesting increased efficiency of migration (Fig. 5C).
Figure 5.
Fibulin-3 enhances radial glioma cell dispersion and individual cell migration. A) Representative image of control and fibulin-3-expressing cell aggregates (CNS-1 cells) seeded on brain slices and cultured during five days. Bars= 200 μm. B) The dispersion of the invasive cell lines CNS-1 and X12 on brain slices was analyzed by 2-way ANOVA for repeated measures followed by post-hoc Bonferroni’s test (**p<0.01; ***p<0.001). Area ratio= area occupied by cells at each time point relative to the original area (mean ± S.E.M.). C) Semiautomated tracking of CNS-1 cells was used to calculate mean cell velocity (average of frame-to-frame velocity for each cell) and net migrated distance (net difference in cell position between first and last frame). The image shows representative tracks of individual cells imaged overnight. Mean cell velocities remained stable but showed a trend to increase toward the end of the cell-tracking experiment, probably due to cumulative tissue disruption by migrating cells. Bar= 200 μm
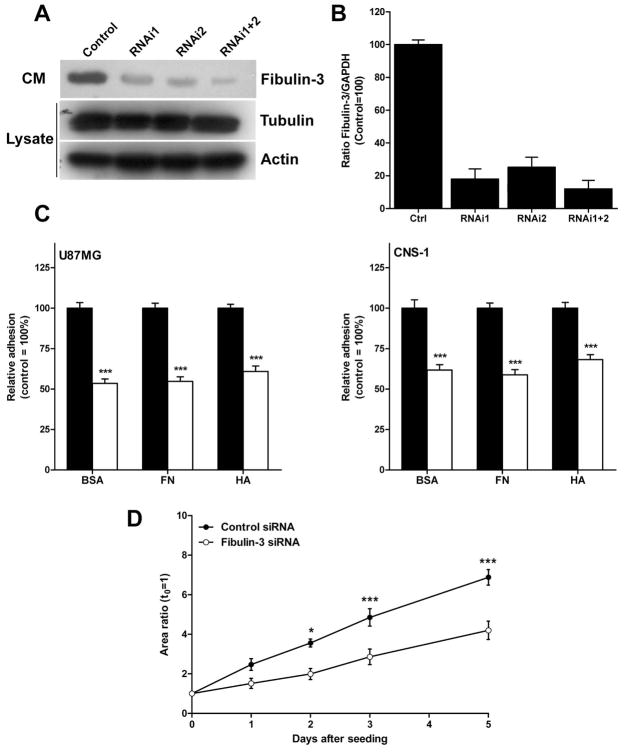
To confirm the specificity of the pro-adhesive and pro-motility effects of fibulin-3, we next decreased this protein in two cell lines (U87MG and CNS-1) by means of RNA interference. Efficient transient knockdown of fibulin-3 in both cell lines was achieved 48h after transfection of specific siRNAs (Fig. 6A–B). Using this technique we did not see any obvious changes on cell morphology (not shown), but confirmed a significant reduction in cell adhesion to all substrates tested (Fig. 6C). Further testing with CNS-1 cells (Fig. 6D) and X12 cells (not shown) revealed a strong decrease in dispersion area when these cells were seeded on cultured brain slices as before.
Figure 6.
Knockdown of fibulin-3 reduces glioma cell adhesion and motility. A) Reduction of fibulin-3 in the conditioned medium of the cell line U87MG after transient transfection with two independent siRNA sequences. Housekeeping control proteins were quantified in cell lysates. Total protein content was 15 μg/lane in all cases; samples were processed 48h after transfection. B) Knockdown of fibulin-3 in the cell line CNS-1, monitored by qRT-PCR, following the same procedure as in (A). C) Transient knockdown of fibulin-3 reduces cell adhesion to all substrates tested. Transfected cells were analyzed as indicated in Figure 4. Black bars: control siRNA; white bars: fibulin-3 siRNA. (***p<0.001 by two-way ANOVA). Results were virtually identical with the cell line U251MG (not shown). D) CNS-1 cells were transiently transfected with fibulin-3 siRNAs 48h before seeding on cultured brain slices. Follow-up over five days, as indicated in Figure 5, showed a significant reduction in the total area occupied by dispersed cells (*p<0.025, *** p<0.001 by two-way ANOVA for repeated measures).
Fibulin-3 increases tumor invasion
To test the effects of fibulin-3 expression in vivo, we implanted stably transduced CNS-1 cells intracranially in Lewis rats and allowed them to grow for 15 days. This syngeneic tumor model was selected by its ability to reproduce accurately the phenotype of clinical gliomas, including nuclear atypia, pleomorphism, vascularization, necrotic foci and tumor invasion (39, 40).
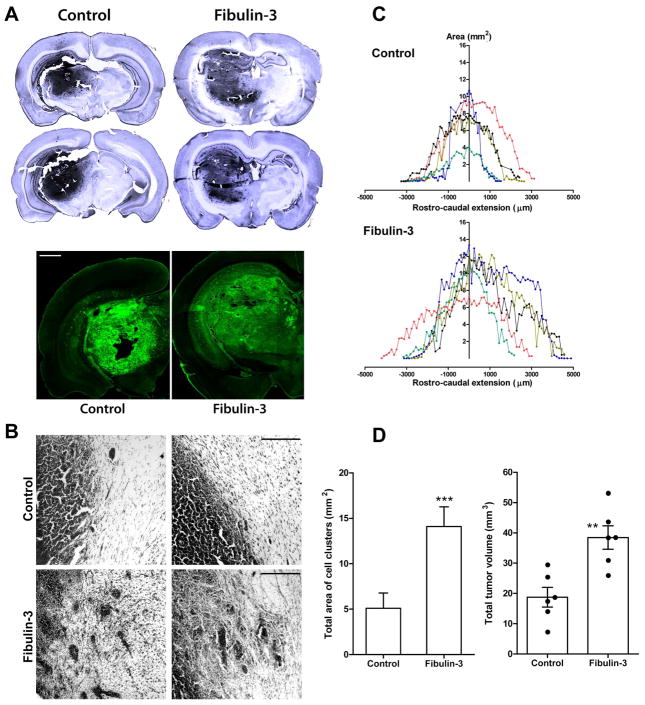
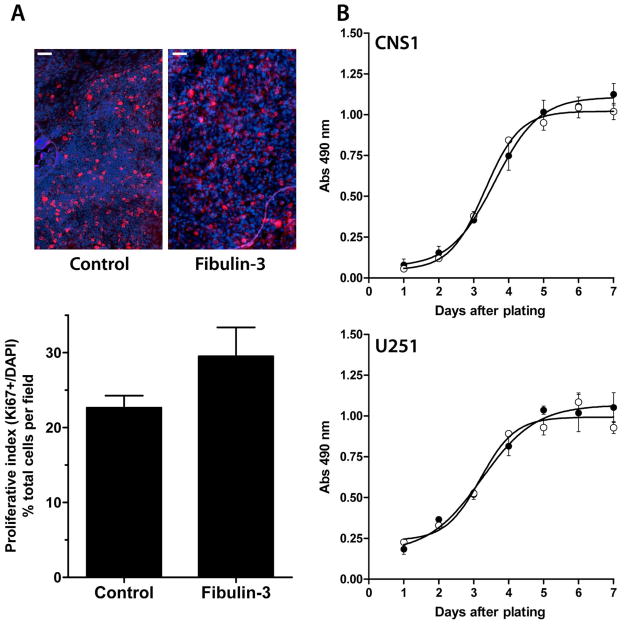
We found that fibulin-3-overexpressing tumors appeared visually less compact and more diffuse than controls (Fig. 7A) and presented larger cell clusters separated from the tumor mass (Figs. 7B,D). Morphometric analysis disclosed that fibulin-3-overexpressing tumors had larger transversal areas than controls, extended over a longer rostro-caudal distance, and covered a larger total volume (Figs. 7C–E). Interestingly, analysis of cell proliferation, measured by the proportion of Ki67 positive cells in the tumor, did not reveal significant differences between control and fibulin-3-overexpressing cells (Fig. 8A). In agreement, overexpression of fibulin-3 in vitro did not affect total cell proliferation (Fig. 8B) or apoptosis (not shown).
Figure 7.
Fibulin-3 increases tumor dispersion in vivo. A) Representative control and fibulin-3-overexpressing tumors 15 days after intracranial injection of CNS-1 cells, revealed by Nissl staining (upper images) or detection of GFP-fluorescent tumor cells (lower images, bar = 1 mm). Fibulin-3 tumors appear larger and less compact than controls. B) Higher magnification of Nissl-stained tumors. Fibulin-3-overexpressing tumors show more diffuse borders and large clusters separated from the tumor mass (bar= 200 μm). C) Tumor area (in mm2) was quantified every fourth section and plotted against the rostro-caudal extension of the tumor, centered on the injection site (20). Notice the increase in caudal invasion of fibulin-3-expressing tumors, probably due to invasion of thalamo-reticular tracts. D) Cell clusters were counted and measured every fourth section as described (35) and their cumulative area quantified using image analysis software. (*** p<0.001 by Students’ t-test). E) Total tumor volumes were calculated using Cavalieri’s estimator of morphometric volume (20), and compared by Student’s t-test (** p<0.01).
Figure 8.
Expression of fibulin-3 does not affect cell proliferation. A) Representative staining for the proliferation marker Ki67 around the core of control or fibulin-3-overexpressing tumors formed by CNS-1 cells. The proliferative index (% of Ki67-positive nuclei relative to total number of DAPI-stained nuclei in each field, mean ± S.E.M.) was calculated by automated scoring of at least 300 nuclei per field (using ImageJ software), and compared by Student’s t-test (p=0.228, no significant differences). Note the higher cell density in control tumors. Bars=50 μm. B) Proliferation rates of CNS-1 cells stably transduced with fibulin-3 (black circles) or a control (white circles) cDNA were analyzed using a metabolic assay for reduction of tetrazolium (21). Proliferation curves were repeated twice using triplicates for each day. The absence of significant differences was verified in a second glioma cell line (U251MG).
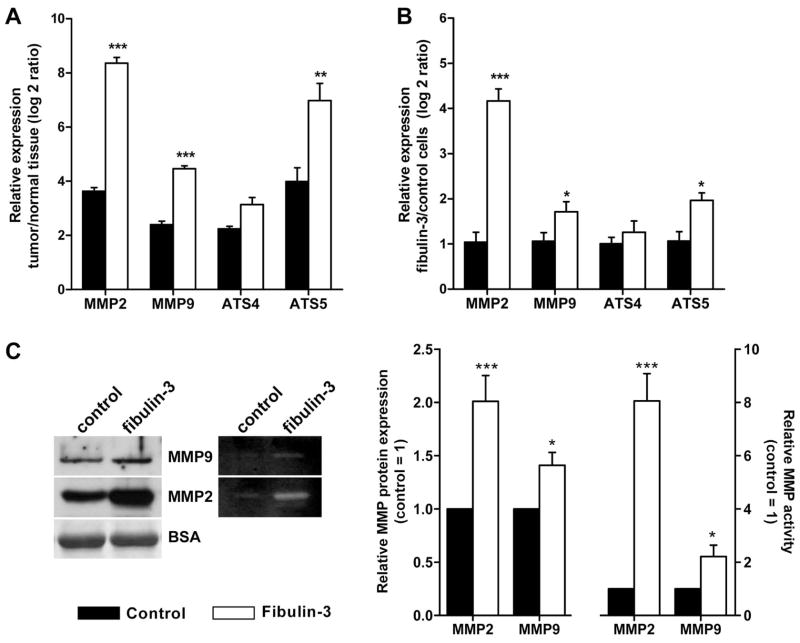
Taken together, our results suggested that fibulin-3 could act as a signal to promote cell invasion and intracranial dispersion of the tumor. In agreement with this pro-invasive role, we observed that CNS-1 tumors overexpressing fibulin-3 had significantly upregulated mRNA levels for several matricellular proteases (Fig. 9A): matrix metalloprotease (MMP)-2, MMP-9, and the disintegrin and metalloprotease with thrombospondin motifs (ADAMTS)-5, all of which are involved in pericellular ECM degradation and glioma invasion. Moreover, cultured CNS-1 cells overexpressing fibulin-3 exhibited significant upregulation of the same metalloproteases at the mRNA and protein levels (Fig. 9B,C) as well as a strong increase of metalloprotease activity (Fig. 9C and data not shown). This suggested a direct regulatory effect of fibulin-3 on metalloprotease expression and activity in glioma cells.
Figure 9.
Fibulin-3 promotes metalloprotease expression and activity. A) Tumor tissue (microdissected from the brain) and contralateral, normal tissue from three control and three fibulin-3-expressing CNS-1 tumors were individually collected and processed for qRT-PCR. mRNA expression for the metalloproteases MMP-2, MMP-9, ADAMTS-4 (ATS4) and ADAMTS-5 (ATS5) was compared in each tumor tissue to its contralateral control. Values (mean ± S.E.M.) represent the log2 ratio of tumor to normal brain expression. B) Expression of the same metalloproteases was also compared by qRT-PCR in cultured control (baseline expression = 1) and fibulin-3-overexpressing CNS-1 cells. GAPDH was used as internal normalizing control in all cases. (***p<0.001; **p< 0.01; *p<0.025, by two-way ANOVA). C) Increased expression and activity of metalloproteases in the medium of transfected CNS-1 cells was verified by Western blotting for MMP2 and MMP9, and by gelatin-zymography. Total amount of protein and detection of serum albumin (BSA, external reference) in the conditioned medium were used as normalization controls to calculate relative MMP expression and activity (20). Analysis of proteoglycan cleavage in these cells also suggested increased activity of the proteoglycan-degrading protease ADAMTS-5 (BH and MSV, in preparation).
DISCUSSION
The invasion of malignant gliomas within the brain parenchyma involves a number of coordinated processes, including cell detachment from the tumor mass, receptor-mediated adhesion to the surrounding matrix, local matrix remodeling, and chemo- and haptotaxis to guide cell motility (11, 41). These processes are common to all invasive tumors; however, the effective and selective dispersion of glioma cells within the neural environment suggests that they may also have unique mechanisms of invasion adapted for the particular composition and structure of the CNS (11). We and others have observed that the ECM produced in gliomas contains mesenchymal proteins absent from the matrix in the surrounding neural parenchyma, as well as neural-specific components that are not produced by peripheral tumor cells (11, 16, 42). We have hypothesized that glioma-specific matrix components may underlie in part the unique invasive abilities of these cells (21). Here, we have identified fibulin-3 as a novel, pro-invasive molecule specific to the glioma ECM and absent from the microenvironment of the brain and most peripheral tumors.
The fibulins form a family of matrix-associated glycoproteins that are thought to act as intermolecular bridges in the ECM, forming supra-molecular structures that regulate cell adhesion, motility and proliferation (43, 44). Several members of this family have been implicated in the pathobiology of solid tumors, including ovarian (fibulin-1), breast (fibulin-1) and colorectal (fibulin-4) cancers (43, 44). However, the role of the fibulins in cancer initiation and progression has been difficult to define because their expression is variable in different tumors (45–47), and they have been shown to act as positive or negative regulators of tumor growth in different models (43, 44). For example, the splice variants of fibulin-1 are hypothesized to have opposite roles in tumor formation (48). Similarly, fibulin-5 enhances cell motility in vitro (49) and promotes epithelial-to-mesenchymal transition (50), but reduces angiogenesis and bulk growth in fibrosarcomas (24). Thus, additional information from experimental models is required to establish more clearly the relevance of these proteins in cancer. Importantly, because the expression of all fibulins in the CNS is very low compared to other tissues (22), no members of this family have been assessed in primary brain tumors. This is therefore the first account of the expression and functional relevance of a fibulin member in malignant gliomas.
Fibulin-3, together with fibulin-4 and -5, forms a subgroup of ‘small’ fibulins that lack the anaphylotoxin motif from fibulin-1 and -2 and other large N-terminal domains present in fibulin-2 and -6 (51). Although upregulation of this protein was originally linked to cell senescence (37), the only known pathological consequences of altered fibulin-3 expression are some dominant forms of macular degeneration caused by its misfolding and accumulation in the retinal epithelium (31, 52). On the other hand, absence of fibulin-3 causes connective tissue deficiencies that result in multiple hernias, organ atrophy and early aging phenotype (53), in agreement with the association of this protein to the basal lamina of elastic tissues.
A previous investigation of fibulin-3 in several solid, non-neural tumors demonstrated that fibulin-3 mRNA was downregulated in most of the specimens analyzed (24), in agreement with our analysis of microarray data (Fig. 1). In stark contrast, we found that fibulin-3 was consistently upregulated in malignant glioma tissue, in agreement with a previous mention of this gene (EFEMP1) as one of the mesenchymal genes highly expressed in primary brain tumors (42). Moreover, we observed a strong expression of fibulin-3 in all glioma cell types tested (Fig. 2). Although this result does not preclude other possible sources of fibulin-3 in the tumor mass, the complete absence of fibulin-3 in the brain (22, 54) strongly suggest the tumor cells as major source of this protein in gliomas. In agreement, gene expression profiling of human cell lines and the NCI-60 panel of human tumor lines (https://biogps.gnf.org) has shown the highest levels of fibulin-3 mRNA among glioma cell lines, whereas peripheral tumor, endothelial and myeloid cells express this gene at much lower, or undetectable, levels. Our results thus suggest that fibulin-3 is a constitutive component of the glioma ECM but absent from the same compartment in the brain and other solid tumors. This, in turn, suggests that fibulin-3 could be specifically involved in the progression of malignant gliomas.
Expression of fibulin-3 had a significant enhancing effect on cell adhesion (Fig. 4), which was most notorious on cells plated on fibronectin or hyaluronic acid, two ECM components abundantly secreted by glioma cells. In a previous study (21), we described the same substrate-dependent, pro-adhesive effect for the proteoglycan brevican, another ECM protein secreted by glioma cells. However, brevican’s effects were likely mediated by upregulation and binding of soluble fibronectin to the surface of glioma cells (21), whereas we did not see any of these effects on cells overexpressing fibulin-3 (unpublished observations). Additionally, fibulin-3 itself did not act as an adhesive substrate, suggesting that overexpression of this protein promoted adhesion indirectly, likely through increase of cell-surface adhesion molecules. A similar pro-adhesive effect of fibulin-3 has been recently observed in peripheral olfactory glia (55), and the highly homologous fibulin-5 has also been shown to promote adhesion and migration in cancer cells (49, 50). Importantly, expression of fibulin-3 not only increased cell adhesion but also acted as a motogenic signal, increasing individual glioma cell motility and dispersion in cultured neural tissue (Fig. 5). Similar results obtained with matrigel transmigration assays (unpublished observations) suggest that the pro-migratory effect of fibulin-3 could have been partly due to upregulation of metalloprotease activity (see below).
In agreement with the pro-migratory effects in vitro, overexpression of fibulin-3 in a rat glioma model (CNS-1 glioblastoma) resulted in invasive tumors with larger cross-sectional area, longer rostro-caudal extension, increased formation of cell clusters, and more overall diffuse appearance than controls (Fig. 7). These tumors showed increased expression of the matricellular metalloproteases MMP2, MMP9 and ADAMTS-5 (Fig. 9A). MMP2 and MMP9 have been extensively shown to promote brain tumor growth and invasion through local matrix degradation and release of trophic factors (56, 57), whereas ADAMTS-5 is thought to promote invasion through cleavage of ECM proteoglycans (58) and release of pro-migratory signals (20, 21). The expression and activity of these proteases were also found upregulated in cultured CNS-1 cells (Fig. 9B,C), suggesting that fibulin-3 levels could regulate these enzymes in an autocrine/paracrine manner. In agreement, we have observed that MMP2 and MMP9 are downregulated following fibulin-3 knockdown in cultured cells (BH and MSV, unpublished observations).
The enhancing effect of fibulin-3 on cell adhesion and migration, and its apparent lack of effect on cell proliferation (Fig. 8), leads us to suggest that this protein could promote glioma progression by acting as a signal that enhances invasive ability. Current studies in our laboratory are further analyzing other mechanisms that could contribute to the progression of fibulin-3-expressing tumors (see below). It is important to remark that this pro-tumoral effect of fibulin-3 in gliomas contrasts markedly with the anti-tumoral effect proposed for this protein in peripheral tumors (24). Interestingly, that anti-tumoral effect was attributed to an anti-angiogenic effect of fibulin-3 in the tumor mass. However, our preliminary observations do not suggest changes in microvessel density after fibulin-3 expression (BH, HS and MSV, in preparation), in agreement with the fact that high-grade gliomas express large amounts of fibulin-3 and are at the same time some of the most highly vascularized solid tumors (59). It is also worth noting that the previous study on the anti-angiogenic role of fibulin-3 was performed with the short isoform of this protein (24), which we did not detect in gliomas and has not been observed in vivo.
Further studies will focus on the potential partners of fibulin-3 in gliomas, a task made difficult by the almost complete absence of molecular information about the fibulins in the nervous system. To date, the only identified partner of fibulin-3 in vitro is the tissue inhibitor of metalloproteases-3 (TIMP-3), which may bind to fibulin-3 in the ECM of the retinal epithelium (60). Interestingly, TIMP-3 is predominantly associated to ECM proteins and inhibits MMP2/MMP9 (61) and ADAMTS-4/5 (62). It is tempting to speculate whether interaction of fibulin-3 and TIMP-3 in gliomas could impair the inhibitory function of the latter to increase metalloprotease activity and promote glioma invasion.
In sum, our results suggest that fibulin-3 is an ECM component specifically upregulated in gliomas that may contribute to the distinctive invasive ability of these tumors. This protein is an example of potentially important, tumor-specific, highly accessible ECM targets for strategies aiming to disrupt the mechanisms of brain tumor dispersion.
Supplementary Material
Acknowledgments
This work was supported by seed grants (to MSV) from the American Cancer Society, the Dardinger Center Fund for Neuro-Oncology Research and the Program in Molecular Biology and Cancer Genetics (OSU-CCC), and the Joel A. Gringras fellowship from the American Brain Tumor Association (to BH). The authors would like to thank helpful discussions and valuable advice from Drs. E. Antonio Chiocca, Balveen Kaur and Sean Lawler, and the technical expertise of Dr. Kazuhiko Kurozumi, from the Department of Neurological Surgery, The Ohio State University.
BIBLIOGRAPHY
- 1.Central Brain Tumor Registry of the United States C. Statistical Report: Primary Brain Tumors in the United States 2000–2004. CBTRUS. 2007 [Google Scholar]
- 2.Louis DN. Molecular Pathology of Malignant Gliomas. Annual Review of Pathology: Mechanisms of Disease. 2006;1:97–117. doi: 10.1146/annurev.pathol.1.110304.100043. [DOI] [PubMed] [Google Scholar]
- 3.Lefranc F, Brotchi J, Kiss R. Possible future issues in the treatment of glioblastomas: special emphasis on cell migration and the resistance of migrating glioblastoma cells to apoptosis. J Clin Oncol. 2005;23:2411–22. doi: 10.1200/JCO.2005.03.089. [DOI] [PubMed] [Google Scholar]
- 4.Lamszus K, Kunkel P, Westphal M. Invasion as limitation to anti-angiogenic glioma therapy. Acta Neurochir Suppl. 2003;88:169–77. doi: 10.1007/978-3-7091-6090-9_23. [DOI] [PubMed] [Google Scholar]
- 5.Giese A, Bjerkvig R, Berens ME, Westphal M. Cost of migration: invasion of malignant gliomas and implications for treatment. J Clin Oncol. 2003;21:1624–36. doi: 10.1200/JCO.2003.05.063. [DOI] [PubMed] [Google Scholar]
- 6.Subramanian A, Harris A, Piggott K, Shieff C, Bradford R. Metastasis to and from the central nervous system--the ‘relatively protected site’. Lancet Oncol. 2002;3:498–507. doi: 10.1016/s1470-2045(02)00819-7. [DOI] [PubMed] [Google Scholar]
- 7.Viapiano MS, Matthews RT. From barriers to bridges: chondroitin sulfate proteoglycans in neuropathology. Trends Mol Med. 2006;12:488–96. doi: 10.1016/j.molmed.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 8.Pilkington GJ. The paradox of neoplastic glial cell invasion of the brain and apparent metastatic failure. Anticancer Res. 1997;17:4103–5. [PubMed] [Google Scholar]
- 9.Bolteus AJ, Berens ME, Pilkington GJ. Migration and invasion in brain neoplasms. Curr Neurol Neurosci Rep. 2001;1:225–32. doi: 10.1007/s11910-001-0022-x. [DOI] [PubMed] [Google Scholar]
- 10.Bernstein JJ. Local invasion and intraparenchymal metastasis of astrocytomas. Neuropathol Appl Neurobiol. 1996;22:421–4. doi: 10.1111/j.1365-2990.1996.tb00914.x. [DOI] [PubMed] [Google Scholar]
- 11.Bellail AC, Hunter SB, Brat DJ, Tan C, Van Meir EG. Microregional extracellular matrix heterogeneity in brain modulates glioma cell invasion. Int J Biochem Cell Biol. 2004;36:1046–69. doi: 10.1016/j.biocel.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 12.Ruoslahti E. Brain extracellular matrix. Glycobiology. 1996;6:489–92. doi: 10.1093/glycob/6.5.489. [DOI] [PubMed] [Google Scholar]
- 13.Novak U, Kaye AH. Extracellular matrix and the brain: components and function. J Clin Neurosci. 2000;7:280–90. doi: 10.1054/jocn.1999.0212. [DOI] [PubMed] [Google Scholar]
- 14.Delpech B, Maingonnat C, Girard N, et al. Hyaluronan and hyaluronectin in the extracellular matrix of human brain tumour stroma. Eur J Cancer. 1993;29A:1012–7. doi: 10.1016/s0959-8049(05)80214-x. [DOI] [PubMed] [Google Scholar]
- 15.Norman SA, Golfinos JG, Scheck AC. Expression of a receptor protein tyrosine phosphatase in human glial tumors. J Neurooncol. 1998;36:209–17. doi: 10.1023/a:1005840420136. [DOI] [PubMed] [Google Scholar]
- 16.Gladson CL. The extracellular matrix of gliomas: modulation of cell function. J Neuropathol Exp Neurol. 1999;58:1029–40. doi: 10.1097/00005072-199910000-00001. [DOI] [PubMed] [Google Scholar]
- 17.Rhodes KE, Fawcett JW. Chondroitin sulphate proteoglycans: preventing plasticity or protecting the CNS? J Anat. 2004;204:33–48. doi: 10.1111/j.1469-7580.2004.00261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crespo D, Asher RA, Lin R, Rhodes KE, Fawcett JW. How does chondroitinase promote functional recovery in the damaged CNS? Exp Neurol. 2007;206:159–71. doi: 10.1016/j.expneurol.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 19.Zheng PS, Wen J, Ang LC, et al. Versican/PG-M G3 domain promotes tumor growth and angiogenesis. FASEB J. 2004;18:754–6. doi: 10.1096/fj.03-0545fje. [DOI] [PubMed] [Google Scholar]
- 20.Viapiano MS, Hockfield S, Matthews RT. BEHAB/brevican requires ADAMTS-mediated proteolytic cleavage to promote glioma invasion. J Neurooncol. 2008;88:261–72. doi: 10.1007/s11060-008-9575-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu B, Kong LL, Matthews RT, Viapiano MS. The proteoglycan brevican binds to fibronectin after proteolytic cleavage and promotes glioma cell motility. J Biol Chem. 2008;283:24848–59. doi: 10.1074/jbc.M801433200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kobayashi N, Kostka G, Garbe JH, et al. A comparative analysis of the fibulin protein family. Biochemical characterization, binding interactions, and tissue localization. J Biol Chem. 2007;282:11805–16. doi: 10.1074/jbc.M611029200. [DOI] [PubMed] [Google Scholar]
- 23.Giltay R, Timpl R, Kostka G. Sequence, recombinant expression and tissue localization of two novel extracellular matrix proteins, fibulin-3 and fibulin-4. Matrix Biol. 1999;18:469–80. doi: 10.1016/s0945-053x(99)00038-4. [DOI] [PubMed] [Google Scholar]
- 24.Albig AR, Neil JR, Schiemann WP. Fibulins 3 and 5 antagonize tumor angiogenesis in vivo. Cancer Res. 2006;66:2621–9. doi: 10.1158/0008-5472.CAN-04-4096. [DOI] [PubMed] [Google Scholar]
- 25.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3. 0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–80. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Madhavan S, Zenklusen JC, Kotliarov Y, Sahni H, Fine HA, Buetow K. Rembrandt: helping personalized medicine become a reality through integrative translational research. Mol Cancer Res. 2009;7:157–67. doi: 10.1158/1541-7786.MCR-08-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giannini C, Sarkaria JN, Saito A, et al. Patient tumor EGFR and PDGFRA gene amplifications retained in an invasive intracranial xenograft model of glioblastoma multiforme. Neuro-Oncol. 2005;7:164–76. doi: 10.1215/S1152851704000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sarkaria JN, Carlson BL, Schroeder MA, et al. Use of an orthotopic xenograft model for assessing the effect of epidermal growth factor receptor amplification on glioblastoma radiation response. Clin Cancer Res. 2006;12:2264–71. doi: 10.1158/1078-0432.CCR-05-2510. [DOI] [PubMed] [Google Scholar]
- 29.Lee J, Kotliarova S, Kotliarov Y, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 30.Otsuki A, Patel A, Camilli S, et al. Oncolytic Virotherapy Using the HSV-Mutant rQNestin34. 5 towards CD133+ Brain Tumor Stem Cells. Mol Therapy. 2006;13:S167-S. [Google Scholar]
- 31.Marmorstein LY, Munier FL, Arsenijevic Y, et al. Aberrant accumulation of EFEMP1 underlies drusen formation in Malattia Leventinese and age-related macular degeneration. Proc Natl Acad Sci USA. 2002;99:13067–72. doi: 10.1073/pnas.202491599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Viapiano MS, Bi WL, Piepmeier J, Hockfield S, Matthews RT. Novel tumor-specific isoforms of BEHAB/brevican identified in human malignant gliomas. Cancer Res. 2005;65:6726–33. doi: 10.1158/0008-5472.CAN-05-0585. [DOI] [PubMed] [Google Scholar]
- 33.Tiscornia G, Singer O, Verma IM. Production and purification of lentiviral vectors. Nat Protoc. 2006;1:241–5. doi: 10.1038/nprot.2006.37. [DOI] [PubMed] [Google Scholar]
- 34.Jaworski DM, Kelly GM, Piepmeier JM, Hockfield S. BEHAB (brain enriched hyaluronan binding) is expressed in surgical samples of glioma and in intracranial grafts of invasive glioma cell lines. Cancer Res. 1996;56:2293–8. [PubMed] [Google Scholar]
- 35.Zhang H, Kelly G, Zerillo C, Jaworski DM, Hockfield S. Expression of a cleaved brain-specific extracellular matrix protein mediates glioma cell invasion In vivo. J Neurosci. 1998;18:2370–6. doi: 10.1523/JNEUROSCI.18-07-02370.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toth M, Fridman R. Assessment of Gelatinases (MMP-2 and MMP-9) by Gelatin Zymography. In: Brooks SA, Schumacher U, editors. Metastasis Research Protocols, part I (Analysis of Cells and Tissues) Totowa, NJ: Humana Press; 2001. pp. 163–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lecka-Czernik B, Lumpkin CK, Jr, Goldstein S. An overexpressed gene transcript in senescent and quiescent human fibroblasts encoding a novel protein in the epidermal growth factor-like repeat family stimulates DNA synthesis. Mol Cell Biol. 1995;15:120–8. doi: 10.1128/mcb.15.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kundzewicz A, Munier F, Matter JM. Expression and cell compartmentalization of EFEMP1, a protein associated with Malattia Leventinese. Adv Exp Med Biol. 2008;613:277–81. [PubMed] [Google Scholar]
- 39.Kruse CA, Molleston MC, Parks EP, Schiltz PM, Kleinschmidt-DeMasters BK, Hickey WF. A rat glioma model, CNS-1, with invasive characteristics similar to those of human gliomas: a comparison to 9L gliosarcoma. J Neurooncol. 1994;22:191–200. doi: 10.1007/BF01052919. [DOI] [PubMed] [Google Scholar]
- 40.Barth RF, Kaur B. Rat brain tumor models in experimental neuro-oncology: the C6, 9L, T9, RG2, F98, BT4C, RT-2 and CNS-1 gliomas. J Neurooncol. 2009 doi: 10.1007/s11060-009-9875-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Giese A, Westphal M. Glioma invasion in the central nervous system. Neurosurgery. 1996;39:235–50. doi: 10.1097/00006123-199608000-00001. [DOI] [PubMed] [Google Scholar]
- 42.Tso CL, Shintaku P, Chen J, et al. Primary glioblastomas express mesenchymal stem-like properties. Mol Cancer Res. 2006;4:607–19. doi: 10.1158/1541-7786.MCR-06-0005. [DOI] [PubMed] [Google Scholar]
- 43.Gallagher WM, Currid CA, Whelan LC. Fibulins and cancer: friend or foe? Trends Mol Med. 2005;11:336–40. doi: 10.1016/j.molmed.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 44.Argraves WS, Greene LM, Cooley MA, Gallagher WM. Fibulins: physiological and disease perspectives. EMBO Rep. 2003;4:1127–31. doi: 10.1038/sj.embor.7400033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Greene LM, Twal WO, Duffy MJ, et al. Elevated expression and altered processing of fibulin-1 protein in human breast cancer. Br J Cancer. 2003;88:871–8. doi: 10.1038/sj.bjc.6600802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gallagher WM, Greene LM, Ryan MP, et al. Human fibulin-4: analysis of its biosynthetic processing and mRNA expression in normal and tumour tissues. FEBS Lett. 2001;489:59–66. doi: 10.1016/s0014-5793(00)02389-9. [DOI] [PubMed] [Google Scholar]
- 47.Wlazlinski A, Engers R, Hoffmann MJ, et al. Downregulation of several fibulin genes in prostate cancer. Prostate. 2007;67:1770–80. doi: 10.1002/pros.20667. [DOI] [PubMed] [Google Scholar]
- 48.Bardin A, Moll F, Margueron R, et al. Transcriptional and posttranscriptional regulation of fibulin-1 by estrogens leads to differential induction of messenger ribonucleic acid variants in ovarian and breast cancer cells. Endocrinology. 2005;146:760–8. doi: 10.1210/en.2004-1239. [DOI] [PubMed] [Google Scholar]
- 49.Schiemann WP, Blobe GC, Kalume DE, Pandey A, Lodish HF. Context-specific effects of fibulin-5 (DANCE/EVEC) on cell proliferation, motility, and invasion. Fibulin-5 is induced by transforming growth factor-beta and affects protein kinase cascades. J Biol Chem. 2002;277:27367–77. doi: 10.1074/jbc.M200148200. [DOI] [PubMed] [Google Scholar]
- 50.Lee YH, Albig AR, Maryann R, Schiemann BJ, Schiemann WP. Fibulin-5 Initiates Epithelial-Mesenchymal Transition (EMT) and Enhances EMT Induced by TGF-beta in Mammary Epithelial Cells Via a MMP-Dependent Mechanism. Carcinogenesis. 2008 doi: 10.1093/carcin/bgn199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Vega S, Iwamoto T, Nakamura T, et al. TM14 is a new member of the fibulin family (fibulin-7) that interacts with extracellular matrix molecules and is active for cell binding. J Biol Chem. 2007;282:30878–88. doi: 10.1074/jbc.M705847200. [DOI] [PubMed] [Google Scholar]
- 52.Marmorstein LY, McLaughlin PJ, Peachey NS, Sasaki T, Marmorstein AD. Formation and Progression of Sub-Retinal Pigment Epithelium Deposits in Efemp1 Mutation Knock-in Mice: A Model for the Early Pathogenic Course of Macular Degeneration. Hum Mo lGenet. 2007 doi: 10.1093/hmg/ddm199. [DOI] [PubMed] [Google Scholar]
- 53.McLaughlin PJ, Bakall B, Choi J, et al. Lack of fibulin-3 causes early aging and herniation, but not macular degeneration in mice. Hum Mol Genet. 2007;16:3059–70. doi: 10.1093/hmg/ddm264. [DOI] [PubMed] [Google Scholar]
- 54.Ehlermann J, Weber S, Pfisterer P, Schorle H. Cloning, expression and characterization of the murine Efemp1, a gene mutated in Doyne-Honeycomb retinal dystrophy. Gene Expr Patterns. 2003;3:441–7. doi: 10.1016/s1567-133x(03)00084-x. [DOI] [PubMed] [Google Scholar]
- 55.Vukovic J, Ruitenberg MJ, Roet K, et al. The glycoprotein fibulin-3 regulates morphology and motility of olfactory ensheathing cells in vitro. Glia. 2008 doi: 10.1002/glia.20771. [DOI] [PubMed] [Google Scholar]
- 56.Rao JS. Molecular mechanisms of glioma invasiveness: the role of proteases. Nat Rev Cancer. 2003;3:489–501. doi: 10.1038/nrc1121. [DOI] [PubMed] [Google Scholar]
- 57.Levicar N, Nuttall RK, Lah TT. Proteases in brain tumour progression. Acta Neurochir(Wien) 2003;145:825–38. doi: 10.1007/s00701-003-0097-z. [DOI] [PubMed] [Google Scholar]
- 58.Held-Feindt J, Paredes EB, Blomer U, et al. Matrix-degrading proteases ADAMTS4 and ADAMTS5 (disintegrins and metalloproteinases with thrombospondin motifs 4 and 5) are expressed in human glioblastomas. Int J Cancer. 2006;118:55–61. doi: 10.1002/ijc.21258. [DOI] [PubMed] [Google Scholar]
- 59.Kaur B, Tan C, Brat DJ, Post DE, Van Meir EG. Genetic and hypoxic regulation of angiogenesis in gliomas. J Neurooncol. 2004;70:229–43. doi: 10.1007/s11060-004-2752-5. [DOI] [PubMed] [Google Scholar]
- 60.Klenotic PA, Munier FL, Marmorstein LY, nand-Apte B. Tissue inhibitor of metalloproteinases-3 (TIMP-3) is a binding partner of epithelial growth factor-containing fibulin-like extracellular matrix protein 1 (EFEMP1). Implications for macular degenerations. J Biol Chem. 2004;279:30469–73. doi: 10.1074/jbc.M403026200. [DOI] [PubMed] [Google Scholar]
- 61.Butler GS, Apte SS, Willenbrock F, Murphy G. Human tissue inhibitor of metalloproteinases 3 interacts with both the N- and C-terminal domains of gelatinases A and B. Regulation by polyanions. J Biol Chem. 1999;274:10846–51. doi: 10.1074/jbc.274.16.10846. [DOI] [PubMed] [Google Scholar]
- 62.Kashiwagi M, Tortorella M, Nagase H, Brew K. TIMP-3 is a potent inhibitor of aggrecanase 1 (ADAM-TS4) and aggrecanase 2 (ADAM-TS5) J Biol Chem. 2001;276:12501–4. doi: 10.1074/jbc.C000848200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.