Significance
The vascular tissues form a continuous network providing the long-distance transport of water and nutrients in all higher plants (tracheophytes). To incorporate separate organs into this network, it is essential that the position of different vascular cell types is tightly regulated. Several factors required for root vascular patterning (including hormones and gene products) have previously been identified in the model plant Arabidopsis. We have now established a mathematical model formulizing the interaction between these factors, allowing us to identify a minimal regulatory network capable of maintaining a stable vascular pattern in Arabidopsis roots. We envisage that this model will help future researchers understand how similar regulatory units can be applied to create alternative patterns in other species.
Keywords: mathematical modeling, plant development
Abstract
As multicellular organisms grow, positional information is continually needed to regulate the pattern in which cells are arranged. In the Arabidopsis root, most cell types are organized in a radially symmetric pattern; however, a symmetry-breaking event generates bisymmetric auxin and cytokinin signaling domains in the stele. Bidirectional cross-talk between the stele and the surrounding tissues involving a mobile transcription factor, SHORT ROOT (SHR), and mobile microRNA species also determines vascular pattern, but it is currently unclear how these signals integrate. We use a multicellular model to determine a minimal set of components necessary for maintaining a stable vascular pattern. Simulations perturbing the signaling network show that, in addition to the mutually inhibitory interaction between auxin and cytokinin, signaling through SHR, microRNA165/6, and PHABULOSA is required to maintain a stable bisymmetric pattern. We have verified this prediction by observing loss of bisymmetry in shr mutants. The model reveals the importance of several features of the network, namely the mutual degradation of microRNA165/6 and PHABULOSA and the existence of an additional negative regulator of cytokinin signaling. These components form a plausible mechanism capable of patterning vascular tissues in the absence of positional inputs provided by the transport of hormones from the shoot.
Plant vascular tissues (xylem and phloem) provide long distance transport between the root and the shoot. In Arabidopsis roots, the xylem and phloem are arranged in a bisymmetric pattern (Fig. 1A). There is a single xylem axis, with two protoxylem cells at the marginal positions and metaxylem cells in the central position. This axis is flanked by two domains of procambial cells and two phloem poles.
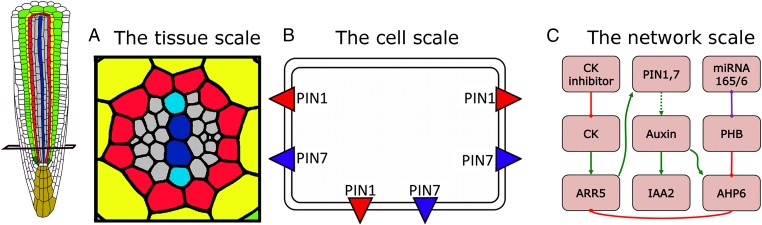
Fig. 1.
The multicellular model considers three levels of regulation. (A) Tissue scale. We use a cross-section through the Arabidopsis root as a template for our model. The cell lineages are color-coded as follows: yellow, endodermis; red, pericycle cells; gray, procambial cells and phloem initials; blue, the xylem axis (the protoxylem is shown in a lighter shade). (B) Cell scale. Each cell contains the auxin efflux proteins (PIN1, PIN3, and PIN7) positioned on the lateral membranes where they have been experimentally observed. (C) Network scale. The regulatory network has been embedded within each cell. Green arrows indicate transcriptional activation. The dotted green arrow indicates that auxin distribution is affected by polar auxin transport. Red bars indicate repression. The purple bar represents the mutual degradation of mRNAs.
Like many other developmental processes in roots, such as the positioning of lateral roots, the specification of the root pole, or the regulation of the root stem cell niche (1), the specification of a vascular pattern is the direct result of the heterogeneous distribution of the hormone auxin (2). Asymmetries in auxin distribution are largely controlled through the subcellular localization of the PIN-formed (PIN) class of auxin efflux carriers (3). Mathematical modeling of PIN-mediated auxin fluxes can generate auxin maxima at the root tip and the sites of organ initiation (4, 5). However, these models have only considered auxin flow within a 2D longitudinal root section and did not investigate the influence of the signaling network responsible for auxin perception at a subcellular level or its possible feedbacks on auxin levels. A 3D model of auxin transport was presented by Swarup et al. (6), but this model only considers the outer three cell layers (epidermis, cortex, and endodermis).
Mathematical models should account for the global effect of multiple hormonal pathways, consider where these hormones are synthesized, how they move between tissues, and how the pathways communicate within and between cells. Although studies investigating the role of auxin-cytokinin cross-talk in roots have identified nodes at which the hormonal pathways interact (2, 7), they have been unable to show the precise relationship between these two pathways or predict the effect that altering one component would have on the system as a whole.
In root vascular tissues, there is an auxin response maximum throughout the xylem axis (2, 8). This response domain is flanked by two domains of high cytokinin (ck) activity within the procambial and phloem cells (2, 9). High ck signaling promotes the expression and localization on the lateral membranes of PIN1, PIN3, and PIN7. High auxin signaling promotes the expression of the ck signaling inhibitor AHP6. PHABULOSA (PHB) acts redundantly with other members of the class III homeodomain-leucine zipper (Hd-zip) transcription factor gene family to repress AHP6 expression in a dose-dependent manner (10). In turn, PHB levels are determined through the degradation of PHB mRNA by microRNA165/6 (miRNA165/6) (10, 11). The asymmetric input of hormones into the root meristem also affects root vascular patterning. During embryogenesis, there is a migration of auxin response from the cotyledons to the root pole (2), and in the growing root, both auxin and ck are transported through the phloem (12–14).
Here, we present a model for investigating auxin fluxes within the radial root cross-section (Fig. 1). We show that interplay between these elements is capable of generating realistic patterns and that both hormonal signaling and mobile miRNA are required to specify the correct domain of auxin response.
Results
Localization of PINs on the Lateral Membranes Is Sufficient to Generate High Auxin Response in the Xylem Axis.
In addition to regulating the rootward flux of auxin in the root meristem (4, 15, 16), PIN1, PIN3, and PIN7 act in consort to channel a radial flow of auxin to the xylem axis (2).To test whether the radial transport of auxin alone offers a plausible mechanism to generate an auxin response maximum in the xylem axis, we developed a 2D computational model of the root vascular tissues based on realistic cell geometries taken from a cross-section through the root proximal meristem (SI Appendix, Fig. S1). Ordinary differential equations were embedded in each cell within this tissue geometry, and as in ref. 17, the temporal evolution of auxin concentration 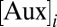 in a given cell i is given by
in a given cell i is given by
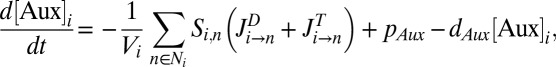 |
where 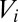 is the cell volume,
is the cell volume, 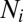 is the set of neighboring cells of cell i,
is the set of neighboring cells of cell i, 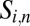 is the exchange surface between cells i and n,
is the exchange surface between cells i and n, 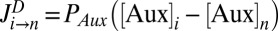 represents the passive auxin diffusion at rate
represents the passive auxin diffusion at rate 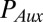 using Fick’s First Law, and
using Fick’s First Law, and 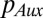 and
and 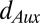 are the respective intracellular biosynthesis and degradation rates of auxin. Active transport of auxin depends on the efficiency of PIN transporters, and therefore,
are the respective intracellular biosynthesis and degradation rates of auxin. Active transport of auxin depends on the efficiency of PIN transporters, and therefore, 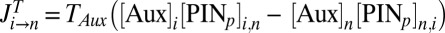 is the flux of auxin caused by active transport at rate
is the flux of auxin caused by active transport at rate 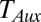 proportional to PIN protein concentration
proportional to PIN protein concentration 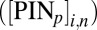 on
on 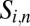 . Individual parameters were selected based on the current literature or reasonable estimates. Specific parameters are discussed further in SI Appendix.
. Individual parameters were selected based on the current literature or reasonable estimates. Specific parameters are discussed further in SI Appendix.
We used the CellSeT software (18) to determine the polarity of PIN:GFP from confocal images and examined 3D reconstructions of immunolocalizations for plants labeled with α-PIN1 or α-GFP antibodies (SI Appendix, Figs. S2–S5).
PIN proteins were incorporated onto specific cell membranes in our multicellular geometry (Fig. 1B and SI Appendix, Fig. S6); the total concentration of PIN proteins in each cell was set to zero or one and proportionally divided on any given membrane, resulting in a concentration of 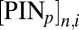 . We modeled auxin flux through the root and predicted the response by simulating the activity of the primary auxin response gene IAA2. Under these conditions, our model showed that the experimentally observed localization of the PINs on lateral membranes was capable of producing an auxin signaling maximum in the xylem axis closely resembling the observed pattern of IAA2 (Fig. 2 A and B and Movie S1). The positioning of this auxin signaling maximum persisted, regardless of whether auxin biosynthesis was assumed to be distributed uniformly across the root or localized in the phloem. We investigated the effect that PIN1, PIN3, and PIN7 exerted individually by running simulations with just one of three PINs (SI Appendix, Fig. S6). Simulations that considered only PIN1 or PIN7 did not show significant differences from the previous simulation, suggesting that these proteins play redundant roles in directing the auxin maximum (Fig. 2C and Movies S2 and S3). PIN3 activity alone was insufficient to reproduce the auxin signaling maximum seen in WT plants (Movie S4). To simplify our subsequent simulations and construct the minimum network required for vascular patterning, from this point, we only consider PIN7. Collectively, the simulations described above highlight that the observed localization of PINs on the lateral membranes is sufficient to direct auxin response to the xylem axis. We then applied our model to determine the minimum set of components that can operate alongside PIN7 during vascular patterning.
. We modeled auxin flux through the root and predicted the response by simulating the activity of the primary auxin response gene IAA2. Under these conditions, our model showed that the experimentally observed localization of the PINs on lateral membranes was capable of producing an auxin signaling maximum in the xylem axis closely resembling the observed pattern of IAA2 (Fig. 2 A and B and Movie S1). The positioning of this auxin signaling maximum persisted, regardless of whether auxin biosynthesis was assumed to be distributed uniformly across the root or localized in the phloem. We investigated the effect that PIN1, PIN3, and PIN7 exerted individually by running simulations with just one of three PINs (SI Appendix, Fig. S6). Simulations that considered only PIN1 or PIN7 did not show significant differences from the previous simulation, suggesting that these proteins play redundant roles in directing the auxin maximum (Fig. 2C and Movies S2 and S3). PIN3 activity alone was insufficient to reproduce the auxin signaling maximum seen in WT plants (Movie S4). To simplify our subsequent simulations and construct the minimum network required for vascular patterning, from this point, we only consider PIN7. Collectively, the simulations described above highlight that the observed localization of PINs on the lateral membranes is sufficient to direct auxin response to the xylem axis. We then applied our model to determine the minimum set of components that can operate alongside PIN7 during vascular patterning.
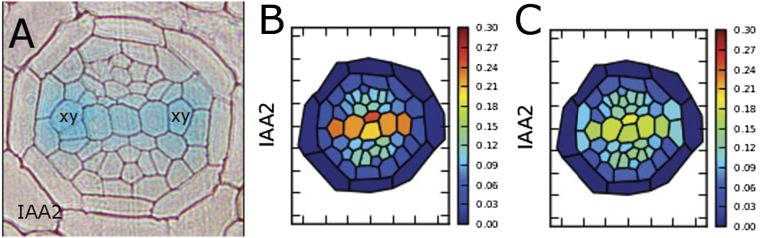
Fig. 2.
Coordination of PINs in specifying the auxin response. (A) pIAA2::GUS marks high auxin response in the xylem axis (xy). (B) Predicted auxin response based on the collective output of PIN1, PIN3, and PIN7. (C) A similar auxin response is predicted when only PIN7 is active.
Integrating Signaling Networks into a Multicellular Model of Vascular Patterning.
We have incorporated the regulatory pathways that are known to determine vascular patterning (2, 10) in the model by including the following processes (Fig. 1C): (i) The transcription of IAA2 and AHP6 is promoted by auxin; (ii) AHP6 transcription is negatively regulated by PHB; (iii) PHB mRNA and miRNA165/6 mutually degrade each other; (iv) miRNA165/6 is produced in the endodermis but diffuses throughout the vascular tissues; (v) ARR5 transcription is activated by ck and repressed by AHP6; and (vi) PIN7 transcription is activated by ck response (ARR5).
We derived a system of ordinary differential equations governing the dynamics of the interaction network that was embedded in our multicellular geometry. Based on previous subcellular models (19, 20), the transcription of all genes was modeled using Hill Kinetics, with the mRNA 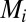 produced through the transcription of gene i in response to transcriptional regulator
produced through the transcription of gene i in response to transcriptional regulator 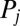 given by
given by
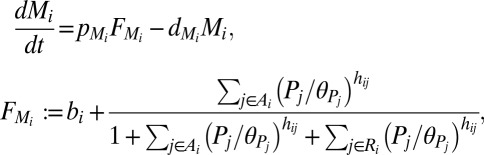 |
where 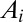 and
and 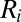 are the sets of activators and repressors acting on gene i, respectively,
are the sets of activators and repressors acting on gene i, respectively, 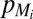 and
and 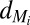 are production and degradation rates of
are production and degradation rates of 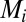 , respectively,
, respectively, 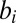 is a dimensionless parameter regulating basal transcription,
is a dimensionless parameter regulating basal transcription, 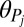 is a protein–DNA binding threshold, and
is a protein–DNA binding threshold, and 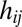 is the Hill coefficient of protein
is the Hill coefficient of protein 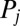 when acting on gene i. Translation of the protein
when acting on gene i. Translation of the protein 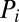 from
from 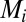 is then governed by the equation
is then governed by the equation
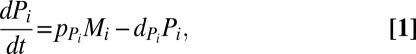 |
where 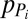 and
and 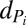 are the production and degradation rates of
are the production and degradation rates of 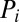 , respectively. Mutual degradation of PHB mRNA
, respectively. Mutual degradation of PHB mRNA 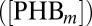 and miRNA
and miRNA 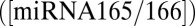 is governed by the reaction
is governed by the reaction  at rate
at rate 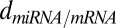 , which in the evolution equations of
, which in the evolution equations of 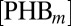 and
and 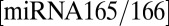 , is described by the negative term
, is described by the negative term 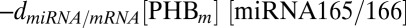 (21). Passive diffusion of ck, miRNA165/6, and SHORT ROOT (SHR) are governed by the same rules as auxin. The full set of reactions constituting this signaling network is listed in SI Appendix, section 1.
(21). Passive diffusion of ck, miRNA165/6, and SHORT ROOT (SHR) are governed by the same rules as auxin. The full set of reactions constituting this signaling network is listed in SI Appendix, section 1.
Mutual Degradation of miR165/6 and PHB Generates Sharp Boundaries of Gene Expression.
We first considered mobile miRNA165/6. The fate of miRNA after it has regulated its target is not well-understood. However, a previous study has shown that the degradation of miRNA223 is accelerated when its target is present (22). Additionally, a previous modeling study has postulated that the mutual degradation of target and miRNA can sharpen the boundary of its mRNA target’s spatial distribution (21). Levine et al. (21) developed a general model based on reaction–diffusion equations in one spatial dimension, in which small RNA molecules interact with a target gene and move from cell to cell through diffusion. Although diffusion generally smoothes spatial expression patterns, Levine et al. (21) found that intercellular mobility of small RNAs sharpens the boundaries between target expression domains in a robust manner, and this sharpening was because of the codegradation of both small RNAs and their targets. The codegradation means that, when the small RNAs diffuse into areas of low gene expression, they can eliminate the expression of their targets in these cells, whereas they are less able to affect the levels of target genes in cells in which the target is expressed abundantly. Carlsbecker et al. (10) proposed that a similar mechanism may operate during vascular patterning as endodermally produced miRNA165/6 moves into the vascular cylinder and encounters its target mRNA (PHB); however, they did not test this prediction with a mathematical model and did not explore whether the mutual degradation of miRNA165/6 and PHB mRNA is required to produce the observed expression patterns.
We introduced the molecular circuitry into our model to compare scenarios in which the degradation of both PHB and miRNA occurs mutually. In the former scenario, we assume that binding of miRNA and PHB mRNA targets both molecules for degradation, and in the latter scenario, miRNA accelerates the degradation of PHB but is not itself consumed. In simulations without codegradation of miRNA and target, we observed a diffuse gradient of PHB, resulting in accumulation of AHP6 throughout the xylem axis and ectopically in the procambium (Movie S5). By including the mutual degradation, we observed a much sharper boundary of PHB with high expression in the central cells and low expression in the outer cell layers, resulting in a much greater repression of AHP6 in the central cells. These simulations suggest that, under our parameter set, the mutual degradation of these components provides a suitable mechanism to generate distinct domains of expression of both components, where the expression of PHB and other network components are remarkably similar to the experimentally observed data (Fig. 3 A and B and Movie S6). We investigated the effect that doubling or halving these parameters had on this gradient and observed that, in almost every scenario, incorporating a mutual degradation between these two components resulted in a sharper gradient of PHB expression (SI Appendix, section 2).
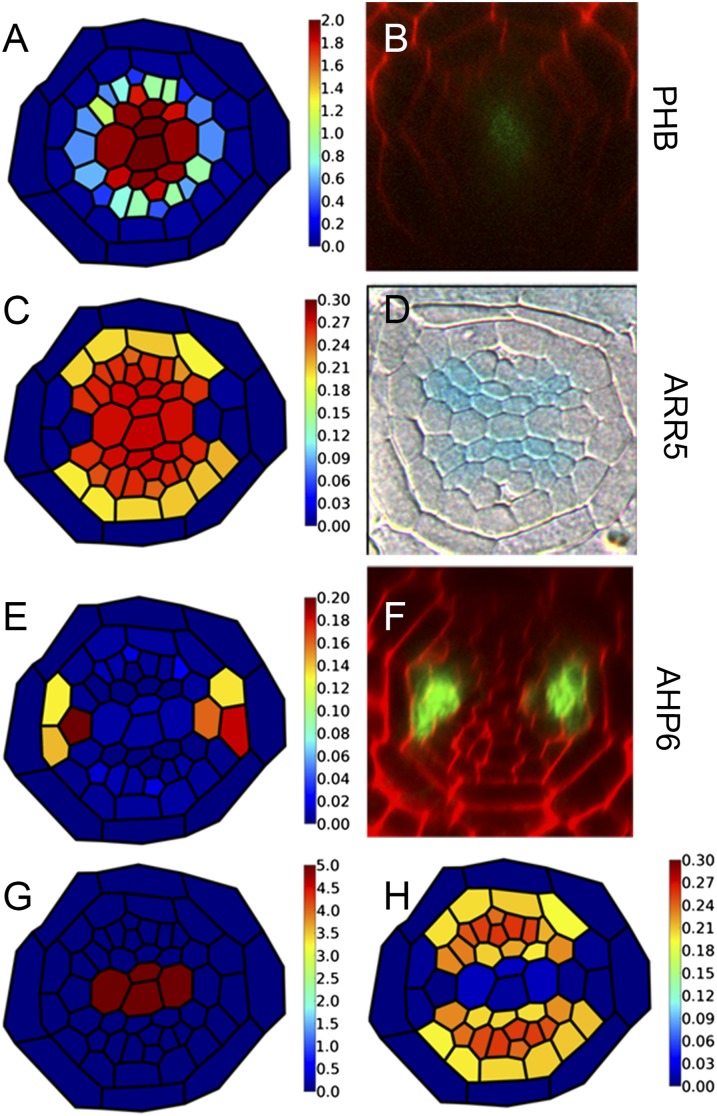
Fig. 3.
Integration of hormonal signaling and Hd-zip activity is sufficient to create a bisymmetric auxin response. (A–F) Model predictions and experimental observations of key marker genes are shown side by side. (A and B) Simulated PHB mRNA levels are shown alongside the experimentally observed pattern of pPHB::PHB:GFP, (C and D) predicted ARR5 output is shown alongside pARR5::GUS, and (E and F) predicted AHP6 output is shown alongside pAHP6::GFP activity. These simulations are based on the original regulatory network lacking an additional inhibitor of ck. Although the simulations recover the observed pattern of PHB and AHP6, they fail to recover the observed pattern of ARR5. (G) By including the repressive effect of CKIN in the metaxylem, (H) our model reproduces the experimental pattern of pARR5::GUS.
Additional Component Is Required to Position the ck Signaling Maximum Correctly.
When PHB regulation by miRNA165/6 is incorporated into our model, our simulations recreate the experimentally observed responses of some but not all markers. The predicted outputs of IAA2 and AHP6 match the data; IAA2 expression has been observed throughout the xylem axis, and AHP6 has only been observed in the marginal positions and the protoxylem-associated pericycle cells (Fig. 3 E and F and Movie S6). However, whereas in our experimental observations, ARR5, PIN7, and the synthetic cytokinin reporter TCSn (23) were always absent throughout the xylem axis (Fig. 3D and SI Appendix, Fig. S7), our simulations predict that both ARR5 and ck are present in the metaxylem (Fig. 3 C and D and Movie S6), suggesting that an additional factor may be required to restrict ck homeostasis/signaling in these cells. Likely candidates include type A ARRs, which are known to inhibit ck signaling (24), or a member of the CKX family of proteins, which is known to degrade ck (25). Inclusion of either a CKX gene or an inhibitor of ck signaling driven in the metaxylem (CKIN) into our model (Fig. 3G) alters the pattern of ck response, such that all network components become similar to experimental observations (Fig. 3H and Movie S7).
Phloem Transport Does Not Provide an Essential Source of Positional Information During Root Vascular Patterning.
Transport of both auxin and ck through the phloem provides an important source of hormones in the root meristem (12, 13). Plants with impaired phloem transport show unstable patterns of auxin response in the root meristem (14). However, it is unclear whether these unstable patterns are purely the result of decreased levels of auxin and ck in the vascular tissues or whether the phloem is required to provide a positional bias in the input of these hormones. We ran simulations to investigate the effect that introducing a spatial bias in the input of hormones had on vascular patterning. In the first simulation, auxin and ck were produced uniformly in all cells, and in subsequent simulations, we allowed synthesis of auxin or ck in the phloem at two times the rate in other cells. We found that both scenarios were able to produce robust domains of hormonal output (Movies S7 and S8). However, when we specified the phloem as the main source of ck, we saw a nonuniform gradient of ARR5 response, with the highest response close to the phloem (Movie S8). We have never observed such a distribution in our experimental data. Together, our simulations suggest that phloem-mediated ck transport is unlikely to act as a source of positional information but rather, ensures that there is a sufficient supply of ck in the root meristem.
Auxin influx carriers also control auxin transport in the vascular tissues (12). As previously published (26), we also observed localization of AUX1::AUX1:YFP on the lateral membranes of protophloem cells (SI Appendix, Fig. S7). To test whether such a component would have a significant effect on vascular patterning, we incorporated a phloem-localized auxin importer into our model and observed only modest changes in the predicted patterns, with a very slight increase in auxin response in protophloem and no change in the expression of key components, such as AHP6 (Movie S9).
Evaluation of Model Sensitivity.
By encapsulating the interaction network and the experimentally observed localization of PIN proteins, our model can reproduce the observed expression patterns of many network components in each cell. As in most models of signaling networks based on differential equations, the predicted outcome of the model is reliant on the choice of its parameters (production and degradation rates, protein–DNA binding thresholds, and Hill coefficients) (27, 28). We have based certain parameters, such as the rates of auxin transport and permeability, on parameters used in previous models (4, 5), whereas we have had to estimate others. To explore the degree to which our choice of parameters affects the outcome of our model, we performed local and global sensitivity analyses of a 1D subcellular network model (SI Appendix, section 3). These analyses identified the parameters to which the model is most sensitive. These parameters were mainly associated with auxin and ck levels and, specifically in the metaxylem, the cooperativity of CKIN. We investigated the effect of perturbing all of these parameters alongside transport and permeability rates in the spatial model. Although there was some variation in intensities, the pattern of key network components was maintained in all these simulations (SI Appendix, section 3).
Maintenance of Steady State Vascular Pattern.
We next investigated whether the model was robust to small changes in the multicellular geometry. We repeated the simulations in new templates based on root cross-sections taken at ∼40 μm from the quiescent center (QC), representing smaller vascular cylinders in which the cellular pattern is less developed. Although these alterations in geometry inevitably brought some small variation in the output of individual markers, the key patterning events were maintained in all these simulations (SI Appendix, Fig. S8). Vascular pattern is maintained robustly in living roots; therefore, small changes in hormone input have negligible effects, and only extreme changes (such as treatments with very high levels of ck) have dramatic effects on vascular patterning (9).
We next tested whether our model conferred a similar robustness. To test robustness, we moved away from simulations where the expression of the PINs were fixed based on experimental observations and allowed every cell the potential to express PIN7, meaning that all vascular cells could potentially express any network component (with the exception of miRNA165/6 and CKIN). We used the output from our previous simulation (Movie S7) as a set of initial conditions that closely resembled WT. We then ran this simulation to steady state and observed that, despite the fact that each cell had a similar potential to express both ck and auxin marker genes and the same auxin and cytokinin production rates, the initial vascular pattern was maintained robustly (Fig. 4 A, B, F, and G and Movie S10). For patterning to be maintained, adjacent cells of similar size and shape must be able show significantly different levels of steady state gene expression in response to a uniform auxin and cytokinin signal.
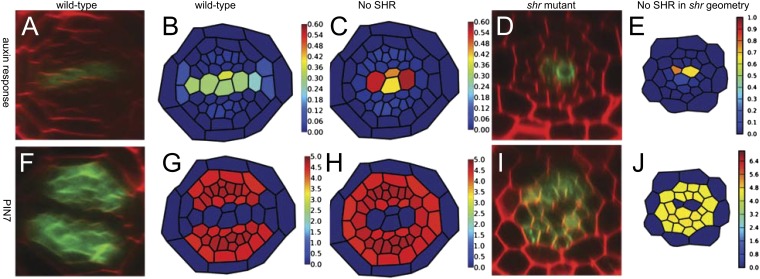
Fig. 4.
Mobile miRNA165/6 is required to maintain stable vascular pattern. Simulations and experimental observations of (A–E) auxin output and (F–J) PIN7 activity. (B and G) When all network components are present, the auxin response in the xylem axis is robustly maintained in accordance with experimental data. The synthetic pDR5rev::GFP reporter line (used here) and pIAA2::GFP line are primary auxin targets and have near-identical expression in vascular tissues. (C and H) In simulations where there is no SHR, the expression domain of PIN7 expands into the position normally occupied by protoxylem, and auxin response becomes restricted to the central cells. (D and I) We observed the same effect in shr mutants and when we ran the simulations using the geometry of an shr mutant.
Robust patterning under homogenous conditions can be explained by the existence of multiple steady states of gene expression within each cell in the model, conferring a switch-like property in which different subsets of vascular cells can have either high AHP6 expression (protoxylem) or high ARR5 and PIN7 expression (procambium). This possibility was tested by finding the steady states of a simplified version of the model in a tissue structure consisting of two cells of equal size with a single shared cell wall (SI Appendix, section 4). The analysis shows that, using the default parameter set, at very low and very high auxin production rates, there is a single stable steady state for the model system that is equal in both cells. However, for a broad range of intermediate values for auxin production, there are two possible steady states, in which one cell has high ARR5 and PIN7 and low AHP6 and auxin and vice versa in the other cell. This bistability is also seen for a range of intermediate values of ck and sufficiently low PHB expression, suggesting that, at least in the two-cell model, after an asymmetry between neighboring cells has been established, it should be robust to small changes in auxin, ck, and PHB; however, for sufficiently large changes, the asymmetry is lost, and gene expression is equalized in both cells.
Maintenance of Stable Vascular Pattern Requires Integration of Mobile miRNA165/6 with Hormonal Signaling.
The apparent affect of ck and PHB levels on pattern maintenance was additionally tested using the full model in the realistic tissue structure. The effect of exogenous treatment of ck was simulated by repeating the simulation shown in Movie S10 but increasing the rate of ck synthesis in all vascular cells. In a similar manner to previous experimental observations (2, 9), we observed loss of AHP6 expression and restriction of auxin response to the metaxylem (Movie S11). We next considered whether miRNA165/6 is required to maintain a stable auxin response in the xylem axis. The total levels of miRNA165/6 are dramatically reduced in the shr mutant. SHR is transcribed in the stele, but the protein is actively transported into the endodermis (29, 30). SHR is sequestered to the nucleus by the transcription factor SCR, where it promotes the expression of miRNA165/6 (10, 11).
To allow us to compare our simulation results with subsequent experimental data, we introduced SHR into our model. Guided by a previous modeling study (31), we allowed SHR transcription and protein synthesis within the stele and for SHR protein (but not mRNA) to move throughout the stele and into the endodermis through passive diffusion. Our model does not explicitly include SCR, but we have simulated its effect by allowing the production of miRNA165/6 by SHR only in the endodermis and assigning a Hill coefficient that reflects this cooperative binding. This mechanism reproduced the WT expression of all marker genes and produced similar results to our previous model (Movie S12). We then investigated whether vascular pattern could be maintained without SHR using the same conditions as shown in Movie S10 but setting SHR transcription to zero. Under these conditions, we predicted that PHB mRNA is present throughout the vascular tissues, and AHP6 is repressed in all cells. Uniformly low levels of AHP6 expression causes up-regulation of ck response at the protoxylem position and approximately homogeneous levels of PIN7 in all tissues except for the metaxylem. As a consequence, auxin is transported from the cells with high PIN7 concentration to the central cells in the xylem axis (Fig. 4 C and H and Movie S13). We then imported our entire regulatory network into the geometry of an shr mutant and ran another simulation from zero initial conditions, which predicted a highly similar pattern to the previous simulation (Fig. 4 E and J and Movie S14).
We tested these predictions by analyzing the expression of both PIN7 and the auxin response marker DR5rev::GFP in the shr mutant. Consistent with model predictions, we observed that the domain of auxin response was confined to the central cells of the xylem axis, and PIN7 was present in a radially symmetric pattern in all but the central vascular cells (Fig. 4 D and I). Together our results show that, in addition to the documented roles of SHR in specifying cell identity, stem cell function, and lateral root growth (10, 29, 32), SHR also directs hormonal responses during vascular patterning.
Discussion
We have used a multicellular mathematical model to probe whether the mutually inhibitory interaction between the transport/signaling of two hormones can act as a plausible vascular patterning mechanism. Previous models based exclusively on auxin transport in roots have analyzed the flux of auxin in the longitudinal sections (4, 5, 33). We have extended this concept to account for the lateral transport of auxin and shown that it determines radial patterning.
Our multicellular model also incorporates auxin and ck signaling networks together with PHB, miRNA165/6, and SHR. It has previously been suggested that small RNA species could repress their targets with a tunable threshold to create sharp boundaries of gene expression (21). We have modeled this interactive mechanism at the organ scale and showed that it is capable of creating the sharp gradient of PHB seen in WT roots. Previously, it has been shown that miRNA165/6 can regulate PHB through ARR1 to control root growth (34). However, because ARR1 is absent in the domain that we have modeled, it suggests that similar components can regulate the same targets in a developmentally specific context. Our study provides insights into how these species may be regulated in vascular tissues, and it will be interesting to see whether these concepts can be applied more widely to the role of PHB in controlling root growth as well as other developmental contexts.
Our simulations show that the restriction of PHB by miRNA165/6 is required in establishing a bisymmetric auxin response, and we have verified this prediction experimentally by showing that shr mutants (which lack miRNA165/6) are unable to generate a bisymmetric auxin response throughout the xylem axis. Our model also revealed that a missing component is required to explain the spatial expression of ARR5. By including such a component, our model yields gene expression patterns that are consistent with the experimental data, illustrating the power of computational modeling as a predictive tool for determining minimum network requirements. We have identified a minimum framework necessary for establishing vascular pattern in Arabidopsis roots. We recognize that, in biological systems, the minimal network is rarely used, and there is often a high degree of genetic redundancy. We have addressed this redundancy for the PINs and investigated the role of each PIN protein.
In conclusion, we have presented a multicellular computational model that embodies the concepts that mobile miRNAs and mutually inhibitory domains of hormonal signaling can act as mechanisms for generating pattern. This regulatory network for vascular patterning differs conceptually from other mechanisms that have been shown to generate pattern in plants based on either reaction–diffusion dynamics or reflux patterns (4, 35). Our model shows that the integration of movement by diffusion (miRNA165/6), the targeted transport of auxin, and the mutual degradation of PHB and miRNA165/6 offer a plausible way to generate vascular pattern.
Materials and Methods
The mathematical model is described in detail in the SI Appendix, section 1. Plant lines were all in the Columbia background and are described in SI Appendix, section 6. Confocal microscopy was performed on an inverted Leica SP5 confocal with propidium iodide used to counterstain the roots. Visualization of the GUS marker was performed as previously described (2).
Supplementary Material
Acknowledgments
We thank Leah Band for helpful comments. We acknowledge the Biotechnology and Biological Sciences Research Council and the Engineering and Physical Sciences Research Council for funding. H.M.B. was funded, in part, by King Abdullah University of Science and Technology (KAUST) Award KUK-013-04, J.R.K. was funded by the Royal Society and Wolfson Foundation, and A.B. was funded by a Royal Society University Research Fellowship. J.G and C.G. were funded by the Institut de Biologie Computationnelle de Montpellier and the Morphogenetics Inria Project-Lab.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. B.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1221766111/-/DCSupplemental.
References
- 1.Vanstraelen M, Benková E. Hormonal interactions in the regulation of plant development. Annu Rev Cell Dev Biol. 2012;28:463–487. doi: 10.1146/annurev-cellbio-101011-155741. [DOI] [PubMed] [Google Scholar]
- 2.Bishopp A, et al. A mutually inhibitory interaction between auxin and cytokinin specifies vascular pattern in roots. Curr Biol. 2011;21(11):917–926. doi: 10.1016/j.cub.2011.04.017. [DOI] [PubMed] [Google Scholar]
- 3.Gälweiler L, et al. Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science. 1998;282(5397):2226–2230. doi: 10.1126/science.282.5397.2226. [DOI] [PubMed] [Google Scholar]
- 4.Grieneisen VA, Xu J, Marée AF, Hogeweg P, Scheres B. Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature. 2007;449(7165):1008–1013. doi: 10.1038/nature06215. [DOI] [PubMed] [Google Scholar]
- 5.Laskowski M, et al. Root system architecture from coupling cell shape to auxin transport. PLoS Biol. 2008;6(12):e307. doi: 10.1371/journal.pbio.0060307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swarup R, et al. Root gravitropism requires lateral root cap and epidermal cells for transport and response to a mobile auxin signal. Nat Cell Biol. 2005;7(11):1057–1065. doi: 10.1038/ncb1316. [DOI] [PubMed] [Google Scholar]
- 7.Dello Ioio R, et al. A genetic framework for the control of cell division and differentiation in the root meristem. Science. 2008;322(5906):1380–1384. doi: 10.1126/science.1164147. [DOI] [PubMed] [Google Scholar]
- 8.Dubrovsky JG, et al. Auxin minimum defines a developmental window for lateral root initiation. New Phytol. 2011;191(4):970–983. doi: 10.1111/j.1469-8137.2011.03757.x. [DOI] [PubMed] [Google Scholar]
- 9.Mähönen AP, et al. Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science. 2006;311(5757):94–98. doi: 10.1126/science.1118875. [DOI] [PubMed] [Google Scholar]
- 10.Carlsbecker A, et al. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature. 2010;465(7296):316–321. doi: 10.1038/nature08977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyashima S, Koi S, Hashimoto T, Nakajima K. Non-cell-autonomous microRNA165 acts in a dose-dependent manner to regulate multiple differentiation status in the Arabidopsis root. Development. 2011;138(11):2303–2313. doi: 10.1242/dev.060491. [DOI] [PubMed] [Google Scholar]
- 12.Swarup R, et al. Localization of the auxin permease AUX1 suggests two functionally distinct hormone transport pathways operate in the Arabidopsis root apex. Genes Dev. 2001;15(20):2648–2653. doi: 10.1101/gad.210501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirose N, et al. Regulation of cytokinin biosynthesis, compartmentalization and translocation. J Exp Bot. 2008;59(1):75–83. doi: 10.1093/jxb/erm157. [DOI] [PubMed] [Google Scholar]
- 14.Bishopp A, et al. Phloem-transported cytokinin regulates polar auxin transport and maintains vascular pattern in the root meristem. Curr Biol. 2011;21(11):927–932. doi: 10.1016/j.cub.2011.04.049. [DOI] [PubMed] [Google Scholar]
- 15.Vieten A, et al. Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development. 2005;132(20):4521–4531. doi: 10.1242/dev.02027. [DOI] [PubMed] [Google Scholar]
- 16.Blilou I, et al. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature. 2005;433(7021):39–44. doi: 10.1038/nature03184. [DOI] [PubMed] [Google Scholar]
- 17.Stoma S, et al. Flux-based transport enhancement as a plausible unifying mechanism for auxin transport in meristem development. PLOS Comput Biol. 2008;4(10):e1000207. doi: 10.1371/journal.pcbi.1000207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pound MP, French AP, Wells DM, Bennett MJ, Pridmore TP. CellSeT: Novel software to extract and analyze structured networks of plant cells from confocal images. Plant Cell. 2012;24(4):1353–1361. doi: 10.1105/tpc.112.096289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Middleton AM, King JR, Bennett MJ, Owen MR. Mathematical modelling of the Aux/IAA negative feedback loop. Bull Math Biol. 2010;72(6):1383–1407. doi: 10.1007/s11538-009-9497-4. [DOI] [PubMed] [Google Scholar]
- 20.Muraro D, et al. The influence of cytokinin-auxin cross-regulation on cell-fate determination in Arabidopsis thaliana root development. J Theor Biol. 2011;283(1):152–167. doi: 10.1016/j.jtbi.2011.05.011. [DOI] [PubMed] [Google Scholar]
- 21.Levine E, McHale P, Levine H. Small regulatory RNAs may sharpen spatial expression patterns. PLoS Comput Biol. 2007;3(11):e233. doi: 10.1371/journal.pcbi.0030233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baccarini A, et al. Kinetic analysis reveals the fate of a microRNA following target regulation in mammalian cells. Curr Biol. 2011;21(5):369–376. doi: 10.1016/j.cub.2011.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zürcher E, et al. A robust and sensitive synthetic sensor to monitor the transcriptional output of the cytokinin signaling network in planta. Plant Physiol. 2013;161(3):1066–1075. doi: 10.1104/pp.112.211763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.To JP, et al. Type-A ARRs are partially redundant negative regulators of cytokinin signaling in Arabidopsis. Plant Cell. 2004;16:658–671. doi: 10.1105/tpc.018978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Werner T, et al. Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell. 2003;15(11):2532–2550. doi: 10.1105/tpc.014928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swarup R, et al. Structure-function analysis of the presumptive Arabidopsis auxin permease AUX1. Plant Cell. 2004;16(11):3069–3083. doi: 10.1105/tpc.104.024737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu J, Mehdi S, Topping J, Tarkowski P, Lindsey K. Modelling and experimental analysis of hormonal crosstalk in Arabidopsis. Mol Syst Biol. 2010;6:373. doi: 10.1038/msb.2010.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rausenberger J, et al. Photoconversion and nuclear trafficking cycles determine phytochrome A’s response profile to far-red light. Cell. 2011;146(5):813–825. doi: 10.1016/j.cell.2011.07.023. [DOI] [PubMed] [Google Scholar]
- 29.Helariutta Y, et al. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell. 2000;101(5):555–567. doi: 10.1016/s0092-8674(00)80865-x. [DOI] [PubMed] [Google Scholar]
- 30.Nakajima K, Sena G, Nawy T, Benfey PN. Intercellular movement of the putative transcription factor SHR in root patterning. Nature. 2001;413(6853):307–311. doi: 10.1038/35095061. [DOI] [PubMed] [Google Scholar]
- 31.Cruz-Ramírez A, et al. A bistable circuit involving SCARECROW-RETINOBLASTOMA integrates cues to inform asymmetric stem cell division. Cell. 2012;150(5):1002–1015. doi: 10.1016/j.cell.2012.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lucas M, et al. Short-Root regulates primary, lateral, and adventitious root development in Arabidopsis. Plant Physiol. 2011;155(1):384–398. doi: 10.1104/pp.110.165126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mironova VV, et al. A plausible mechanism for auxin patterning along the developing root. BMC Syst Biol. 2010;4:98. doi: 10.1186/1752-0509-4-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dello Ioio R, et al. A PHABULOSA/cytokinin feedback loop controls root growth in Arabidopsis. Curr Biol. 2012;22(18):1699–1704. doi: 10.1016/j.cub.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 35.Savage NS, et al. A mutual support mechanism through intercellular movement of CAPRICE and GLABRA3 can pattern the Arabidopsis root epidermis. PLoS Biol. 2008;6(9):e235. doi: 10.1371/journal.pbio.0060235. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.