Significance
Viruses express proteins that circumvent immune responses. In some cases, these viral proteins are homologous to cellular proteins. For example the poxviral MC159 and MC160 proteins are homologous to cellular FLICE-like inhibitory protein (FLIP). Each protein inhibited IFN-β activation, identifying a unique function for FLIPs. Surprisingly, the viral proteins possessed different biological mechanisms for inhibition. MC159 bound to upstream activators of this pathway (namely TBK1 and IKKε) to inhibit IRF3 but MC160 did not. Moreover, the MC159 and MC160 regions responsible for inhibition did not overlap. Scientists can uncover new pathways of immune system regulation and new means for manipulating immune responses by comparing the molecular functions of these viral and cellular FLIPs.
Keywords: host–pathogen interactions, immune evasion
Abstract
Apoptosis, NF-κB activation, and IRF3 activation are a triad of intrinsic immune responses that play crucial roles in the pathogenesis of infectious diseases, cancer, and autoimmunity. FLIPs are a family of viral and cellular proteins initially found to inhibit apoptosis and more recently to either up- or down-regulate NF-κB. As such, a broad role for FLIPs in disease regulation is postulated, but exactly how a FLIP performs such multifunctional roles remains to be established. Here we examine FLIPs (MC159 and MC160) encoded by the molluscum contagiosum virus, a dermatotropic poxvirus causing skin infections common in children and immunocompromised individuals, to better understand their roles in viral pathogenesis. While studying their molecular mechanisms responsible for NF-κB inhibition, we discovered that each protein inhibited IRF3-controlled luciferase activity, identifying a unique function for FLIPs. MC159 and MC160 each inhibited TBK1 phosphorylation, confirming this unique function. Surprisingly, MC159 coimmunoprecipitated with TBK1 and IKKε but MC160 did not, suggesting that these homologs use distinct molecular mechanisms to inhibit IRF3 activation. Equally surprising was the finding that the FLIP regions necessary for TBK1 inhibition were distinct from those MC159 or MC160 regions previously defined to inhibit NF-κB or apoptosis. These data reveal previously unappreciated complexities of FLIPs, and that subtle differences within the conserved regions of FLIPs possess distinct molecular and structural fingerprints that define crucial differences in biological activities. A future comparison of mechanistic differences between viral FLIP proteins can provide new means of precisely manipulating distinct aspects of intrinsic immune responses.
IFN-β provides an important defense against viral infections (1, 2). The pathway leading to IFN-β production is well characterized. By-products of viral infection, such as dsRNA, are detected by upstream cellular sensors, including retinoic acid-inducible gene 1 (RIG-I), melanoma differentiation-associated factor gene 5 (MDA5), and STING. RIG-I and MDA5 proteins then interact with mitochondrial antiviral signaling (MAVS) adaptor protein to trigger MAVS activation (3–8). The TNF receptor-associated factor 3 (TRAF3) adaptor protein is recruited to this complex, resulting in the activation of the kinase complex TANK-binding kinase 1 (TBK1)–IκB kinase ε (IKKε) (9–11). Alternatively, the STING molecule activates TBK1–IKKε (12, 13). In either case, TBK1–IKKε phosphorylates and activates IFN regulatory factor (IRF) transcription factor proteins (11), which migrate to the nucleus to bind to the IFN-β enhancer. IFN-β is secreted and binds to the IFN receptor (IFNR). This initiates a second signaling cascade in infected and neighboring cells, which promotes expression of IFN-α and IFN-stimulated genes whose products contribute to an antiviral state.
Identification of the above members of the IFN-β signal transduction pathway also resulted in the discovery of viral molecules that inhibit events in this pathway (14). Given that viral immunoevasion proteins often interact with host proteins in such a way as to provide maximal efficiency of replication, the study of these viral proteins has greatly increased our understanding of IFN-β production and regulation. One such virus that serves as an important model for viral effects on host immune function, molluscum contagiosum virus (MCV), does not encode any obvious IFN-β antagonists (15, 16). This was surprising given that related poxviruses, such as the vaccinia virus, express several intracellular proteins (E3, C6, and N2) that inhibit IFN-β (17–23). To date, only the MCV MC54 protein, a secreted interleukin binding protein (IL-18BP), is predicted to inhibit IFN expression indirectly by preventing IL-18–mediated IFN-β activation (24, 25). Unlike other poxviruses that have broad host and/or tissue tropism, MCV infections are limited to infection of the human skin and are often long lasting (26), suggesting that MCV may possess heretofore unappreciated and novel mechanisms to inhibit IFN-β. We examined this possibility in this study by characterizing MCV proteins that are known immune evasion molecules.
The MCV MC159 and MC160 immune evasion proteins are members of the FLIP family of proteins, which includes other viral and host cellular proteins, notably Kaposi sarcoma-associated herpesvirus (KSHV) K13 and the cellular cFLIP proteins, of which there are three variants: cFLIPL, cFLIPS, and cFLIPR (15, 27–29). These proteins possess characteristic tandem death-effector domains (DEDs). Initial reports show that FLIPs are immunomodulatory and inhibit death receptor-mediated apoptosis (27, 29–31). Now, it is known that the K13 and cFLIPL proteins also activate NF-κB (32–37). Surprisingly, the MCV MC159 and MC160 proteins have properties that are distinct from these earlier examples, in that they are known to inhibit TNF-induced NF-κB activation (38–42). Together, these publications highlight that although they are evolutionarily related and similar in amino acid sequence, each FLIP is distinct enough to possess unique biological properties, and that these properties may have evolved out of the different requirements a virus has to survive and replicate in its host cell environment.
The starting point for this study was a recent report showing that MC159 prevents IFN-β enhancer activation (43), which suggested a third function for FLIPs. However, because the IFN-β enhancer contains NF-κB–binding sites (44, 45), it remained unclear as to whether the IFN-β inhibitory effect of MC159 was indirect. Moreover, given the history of FLIP proteins exerting opposing effects on NF-κB, it was also unknown whether other members of the FLIP family possessed functions that were similar to or opposite of MC159. To address these specific questions and expand the general understanding of the functional repertoire of FLIPs, we assessed the functions of wild-type and mutated FLIPs under conditions in which NF-κB activation would not confound results.
Results
The MC159 and Other FLIPs Inhibit IRF3 Activation in an NF-κB–Independent Manner.
Recently, it was reported that MC159 inhibits dsRNA-induced activation of the IFN-β enhancer (43), suggesting that FLIPs may control IRF3 activation. However, the subcellular mechanism for this inhibition is ill defined, and it is not known if other FLIPs share a similar function. Interestingly, IFN-β protein levels are increased in cFLIPL−/− MEFs, suggesting cFLIPL inhibits IFN-β expression (46). To fill the gap in this knowledge, a model system was used in which the MCV FLIPs were expressed transiently. This strategy was taken because (i) there is no tissue culture-based propagation system for MCV (26) and (ii) expression of FLIPs independent of a parental or surrogate virus infection avoids confounding results due to other viral proteins that could potentially inhibit IFN-β activation directly or indirectly.
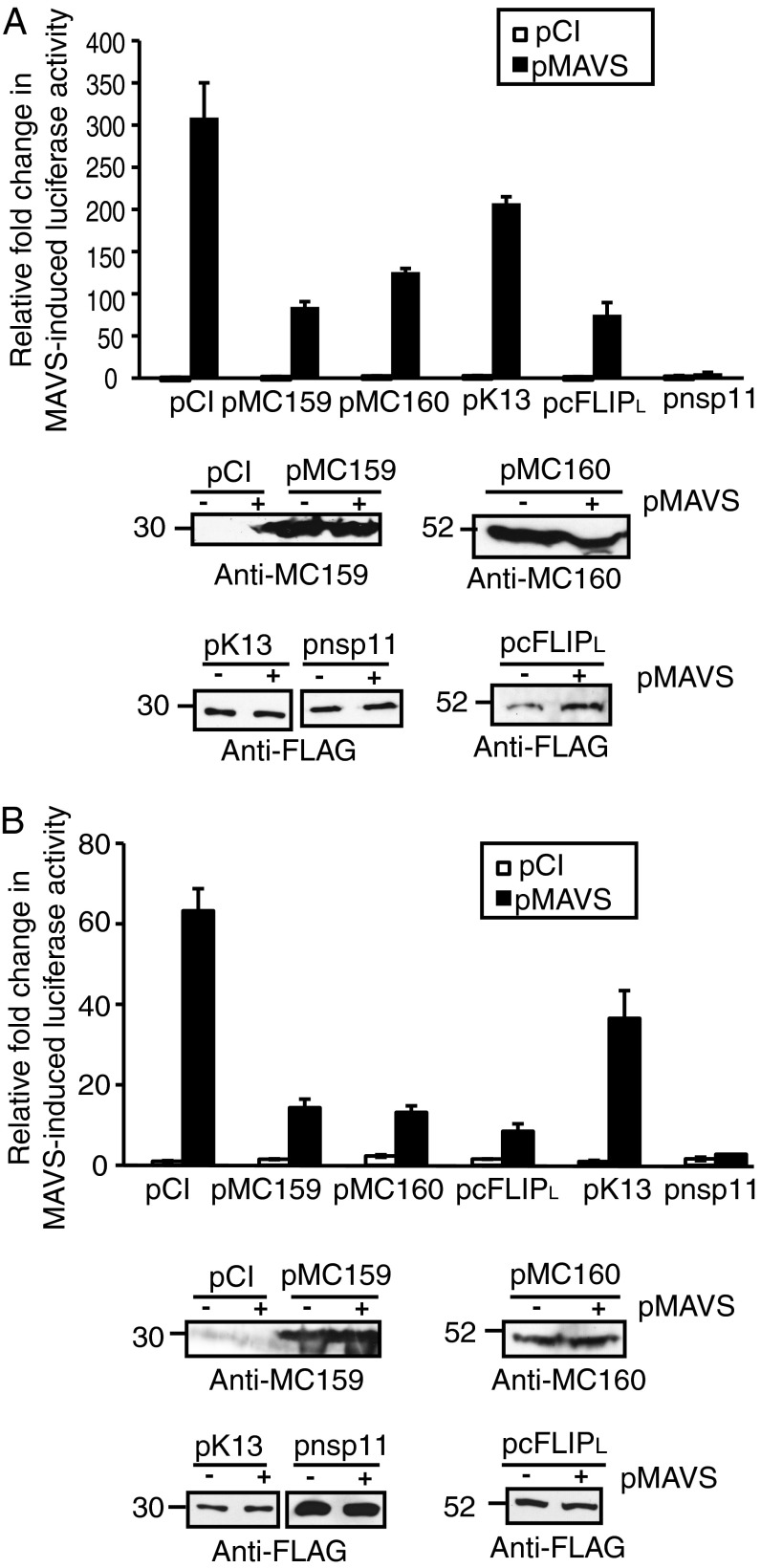
As an initial means to evaluate IFN-β gene activation, a reporter assay was used in which the firefly luciferase gene was under the transcriptional control of the natural IFN-β enhancer. The IFN-β enhancer is regulated by NF-κB, IRF3, IRF7, and AP-1 transcription factors (47). To study the effect of FLIP regulation of the IFN-β promoter independent of NF-κB, the luciferase reporter assay was performed using wild-type mouse embryo fibroblast (MEF) cells or MEFs lacking p65 (p65−/−). In both cases, cells were transfected with pMAVS to induce IFN-β enhancer-controlled gene transcription (Fig. 1). It should be noted that the p65−/− cells (Fig. 1B) were less responsive to MAVS-induced IFN-β enhancer activation than wild-type MEFs (Fig. 1A). Nevertheless, MC159, MC160, or cFLIPL expression greatly decreased IFN-β enhancer-controlled luciferase activity in comparison with cells transfected with empty vector, regardless of the presence or absence of the p65 subunit of NF-κB in MEFs. It should also be mentioned that a recent report shows that MC159 activates NF-κB (48). However, the overexpression of MC159, or any other FLIP, did not induce IFN-β enhancer activity in both cell types tested here. The porcine reproductive and respiratory syndrome virus (PRRSV) nsp11 protein is a known inhibitor of IFN-β activation via its ability to inhibit IRF3 and NF-κB (49, 50). As would be expected, luciferase activity was low in nsp11-expressing cells and was used as a positive control. Whereas KSHV K13 is reported to activate NF-κB (32), luciferase activity was decreased in cells here, albeit not to the levels observed with the other FLIPs or nsp11. When comparing the IRF3 inhibitory phenotypes to the reported effects of FLIPs on NF-κB, it was interesting that there was no correlation between FLIP modulation of NF-κB and its modulation of the IFN-β enhancer. Notably, cFLIPL activates NF-κB (32) while it inhibited activation of the IFN-β enhancer here. Expression of viral proteins was confirmed by immunoblotting lysates of both wild-type and p65−/− MEFs, showing that FLIP proteins were stably expressed under all conditions, and that differences in IFN-β activation among the FLIP proteins was not due to level of protein expression. It was difficult to reliably detect the FLAG-tagged MAVS protein in MEF or MEF p65−/−cells at the times when cells were harvested for luciferase activity assays. However, a profound induction of IFN-β–controlled luciferase activity was reliably observed in mouse cells transfected with the MAVS plasmid (Fig. 1 A and B), showing that the plasmid is indeed coding for a protein that has biological activity.
Fig. 1.
FLIP proteins inhibit IFN-β enhancer activation independent of NF-κB. Subconfluent (A) wild-type or (B) p65−/− MEF cellular monolayers were cotransfected with pIFN-β–luc (225 ng), pRL-null (25 ng), 500 ng pMAVS or pCI, and 750 ng of pCI, pMC159, pMC160, pcFLIPL, pK13, or pnsp11. Values are shown as mean ± SD (n = 3). Twenty-four hours later, cells were lysed and firefly and sea pansy luciferase activities were measured. A portion of each lysate was analyzed for FLIP expression by immunoblotting, using the antiserum indicated. These are representative results from experiments that were performed at least three times independently.
The MC159 and MC160 Proteins Inhibit TBK1 Activation.
Poxviruses code for a large number of proteins that inhibit innate immune responses. Because FLIPs are not expressed by any other poxvirus, it was of interest to continue study of the MC159 and MC160 molecules to identify new viral mechanisms to inhibit IFN-β. Next, it was of interest to determine the portion of the IRF3 activation pathway that each MCV FLIP targeted. For these experiments, a different luciferase reporter was used in which the firefly luciferase gene was controlled by the PRDIII (IRF3)-binding site of the IFN-β promoter as a means of exclusively detecting IRF3 activation. Both the STING- and MAVS-induced IRF3 activation pathway converge at activation of the TBK1–IKKε complex (11–13). As such, overexpression of either TBK1 or IKKε molecules activated the IRF3-controlled luciferase gene (Fig. 2). In conditions in which overexpression of TBK1 or IKKε activated luciferase expression, luciferase activity was significantly decreased when MC159 was present (Fig. 2 A and B, respectively). Expression of the MC160 protein also inhibited TBK1- and IKKε-induced IRF3 activation, and inhibition was consistently observed to be greater than that observed for MC159. When a constitutively active form of IRF3 was instead used to induce IRF3 activation, luciferase activity remained high in MC159- and MC160-expressing cells (Fig. 2C), suggesting that each MCV protein inhibited an event upstream of IRF3 activation. Nsp11 overexpression also did not inhibit constitutively active IRF3, which was expected given that nsp11 is thought to act as an endoribonuclease to target the degradation of MAVS transcripts (49, 50). Viral protein level expression was verified by immunoblotting to ensure that the lack of inhibition was not due to a lack of protein expression.
Fig. 2.
The effect of MC159 and MC160 proteins on IRF3 and IRF7 activation. Subconfluent 293T cells were transfected with pIRF3-luc (225 ng); pRL-null (25 ng); 1,000 ng of either pCI, pMC159, pMC160, or pnsp11; and 500 ng of (A) pTBK1, (B) pIKKε, or (C) IRF3CA. Forty-eight hours later, cells were lysed and firefly and sea pansy luciferase activities were measured. Values are shown as mean ± SD (n = 3). For C, a portion of each lysate was analyzed for protein expression by immunoblotting, using the antiserum indicated. These are representative results from experiments that were performed at least three times independently. (D) The 293T cells were cotransfected with 250 ng each of IRF7 and IFN-α plasmids, 500 ng TBK1 or pCI, and 750 ng pMC159, pMC160, or pnsp11. Twenty-four hours later, cells were lysed and probed for the presence of phospho-IRF7 by immunoblotting.
TBK1 activates IRF7 in addition to IRF3 (51, 52). It was shown previously that MC159 does not inhibit IRF7-induced IFN-β enhancer activation (43) when a constitutively active IRF7 is expressed, suggesting that MC159 does not directly interact with IRF7. However, the effect of MC160 on this process is unknown, and such knowledge may yield insights into the IRF3 inhibitory function of each MCV FLIP. To this end, IRF7 activation was induced by overexpressing TBK1 and IFN-α in 293T cells. As shown in Fig. 2D, the MC160 protein prevented IRF7 activation because phospho-IRF7 levels were greatly diminished in MC160-expressing cells versus vector-transfected cells. In contrast, the MC159 protein did not inhibit IRF7 activation under these conditions: phospho-IRF7 levels were similar in pMC159- versus pCI-transfected cells. These data were quite interesting because they demonstrated how these homologous MCV proteins have distinct phenotypes, and suggest that each MCV FLIP provides a unique role in the pathogenesis of molluscum contagiosum.
The MC159, but Not MC160 Viral FLIP, Associates with TBK1 and IKKε.
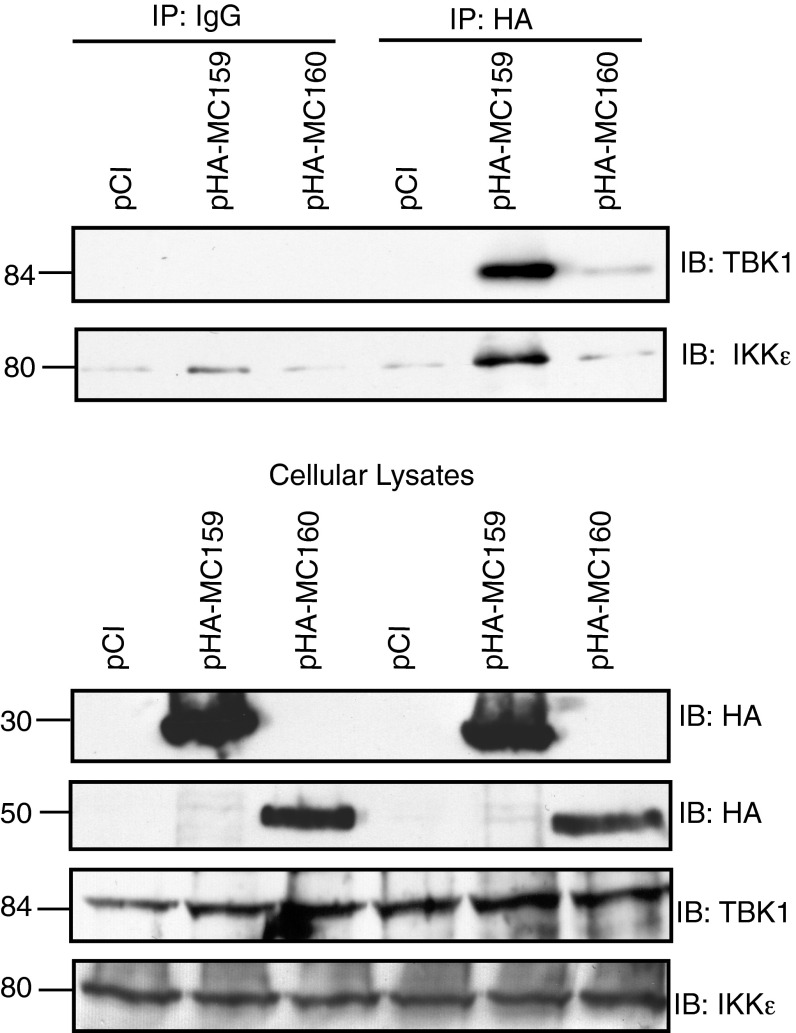
An attractive hypothesis was that the MC159 and MC160 proteins functioned at the level of the TBK1–IKKε complex, perhaps by each FLIP interacting with one or both of these molecules to prevent subsequent IRF activation. In support of this hypothesis were data showing that both the TBK1 and the IKKε molecules coimmunoprecipitated with MC159 (Fig. 3, Upper). TBK1 was absent in coimmunoprecipitations in which mouse IgG was used instead of anti-TBK1 antiserum. TBK1 was also absent from immunoprecipitates collected from vector-transfected cells. This showed specific interactions of MC159 with TBK1. Very small amounts of IKKε were visible in all immunoprecipitated samples and were considered background. Surprisingly, in MC160-expressing cells, only very low levels of TBK1 were present in immunoprecipitates and IKKε levels in immunoprecipitates were not above background levels. These data suggest that MC160 uses a molecular mechanism distinct from that of MC159 to inhibit IRF3 activation. Importantly, both TBK1 and IKKε proteins were present in preimmunoprecipitated lysates in equal amounts, regardless of transfection conditions, indicating that neither MCV FLIP affected the stability of these two cellular proteins as a mechanism of function (Fig. 3, Lower). As would be expected, both the MCV FLIP proteins were present in similar amounts in preimmunoprecipitated cellular lysates.
Fig. 3.
The MC159 protein coimmunoprecipitates with TBK1 and IKKε. Subconfluent 293T cells were transfected with 500 ng TBK1, 250 ng pIKKε, and 1,000 ng of pCI, pHA–MC159, or pHA–MC160. At 24 h posttransfection, cells were lysed. Immunoprecipitations (IPs) were performed by incubating clarified cellular lysates with protein G-Sepharose beads and either anti-HA or IgG. A portion of preimmunoprecipitated cellular lysates was used for detection of TBK1, MC159, MC160, or IKKε proteins by immunoblotting. IPs (Upper) or corresponding clarified cellular lysates (Lower) were analyzed for proteins by immunoblotting.
The MC159 and MC160 Proteins Each Inhibit TBK1 Activation Using Domains Distinct from Those Involved in Inhibiting Apoptosis or NF-κB.
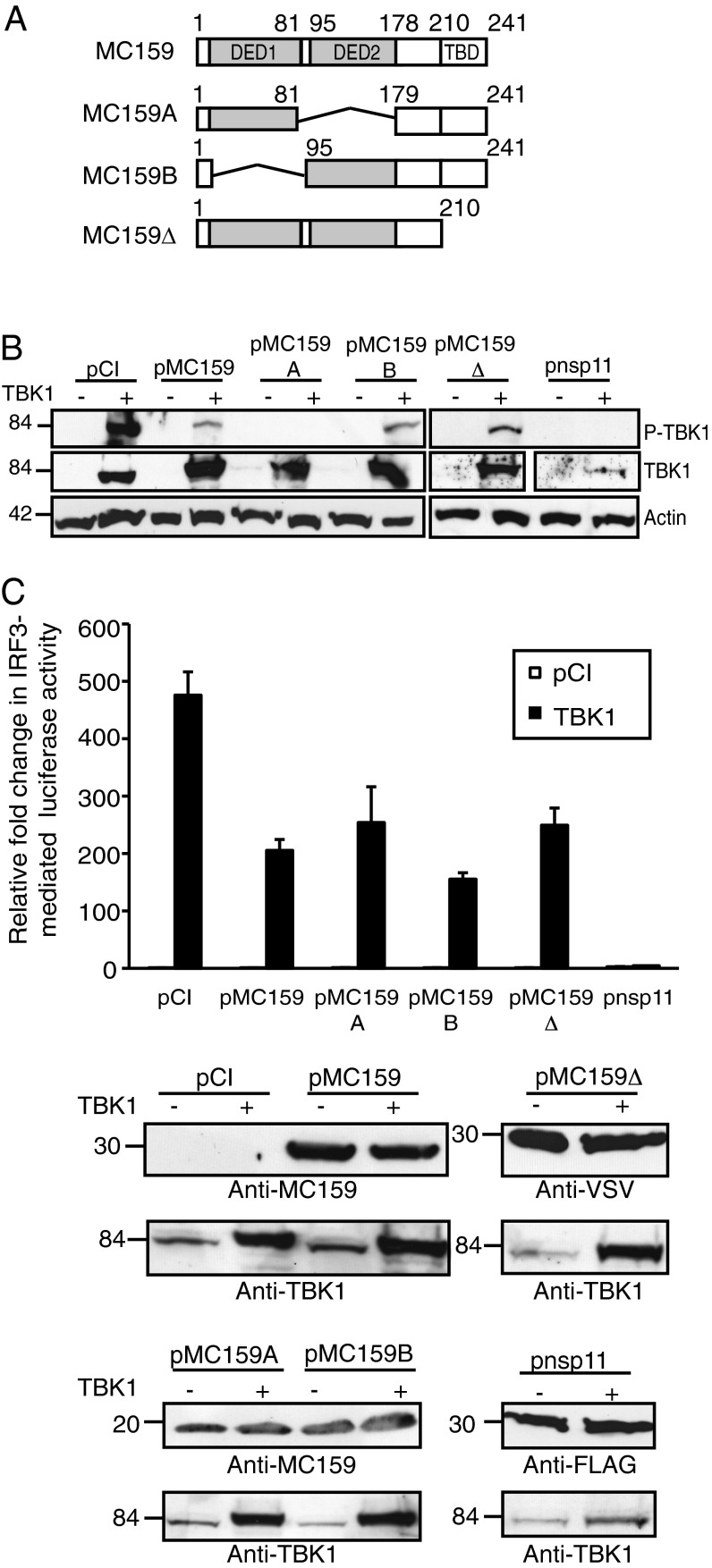
MC159 protein expression also prevented TBK1 activation as determined by the observation that TBK1 phosphorylation was greatly reduced when cells expressed MC159 (Fig. 4B). The MC159 protein has two tandem DEDs at residues 7–81 and 95–178 and then TRAF-binding motifs at residues 210–241 (Fig. 4A) (53–55). For the antiapoptosis function of MC159, all three motifs must be present to prevent cell death (53–55). Surprisingly, mutant MC159 proteins deleted for the DED1 region, DED2 region, or the TRAF-binding domains retained their TBK1 inhibitory functions (Fig. 4B). It was repeatedly observed that phospho-TBK1 levels were no longer visible in MC159A-expressing cells, and that MC159B and MC159Δ proteins inhibited TBK1 phosphorylation to the same extent as the wild-type MC159. TRAF3 is an adaptor molecule that acts on the TBK1–IKKε complex (9). Because a mutant MC159 protein deficient in TRAF3 binding (MC159Δ) retained its ability to inhibit TBK1 phosphorylation (Fig. 4B), it is likely that MC159 inhibits IFN-β expression independently of TRAF3. Actin levels were similar among all conditions, indicating equal protein loading. TBK1 levels remained similar in cells co-overexpressing TBK1 and wild-type or mutant MC159 protein, indicating that MC159 did not induce TBK1 protein degradation. TBK1 protein levels were diminished in cells coexpressing nsp11. Nsp11 possesses endoribonuclease activity and is known to target MAVS (49, 50). While nsp11 has not been reported to degrade TBK1 mRNA, it would be reasonable to predict that nsp11 could target other cellular mRNAs.
Fig. 4.
Mutational analysis of the MC159 protein. (A) A schematic of mutant MC159 constructs used in these experiments, in which the wild-type MC159 shows its two DEDs (DED1 and DED2) and the TRAF-binding domains (TBDs). (B) Subconfluent monolayers of 293T cells were transfected with 500 ng TBK1 or pCI and 1,000 ng pCI, pMC159, pMC159A, pMC159B, pMC159Δ, or pnsp11. At 24 h posttransfection, cells were lysed and clarified cellular lysates were analyzed by immunoblot. Following SDS-10% PAGE, proteins were transferred to PVDF membranes, and were incubated with anti–phospho-TBK1 (P-TBK1), TBK1, or anti-actin antiserum. (C) Subconfluent 293T cellular monolayers were transfected with pIRF3-luc (225 ng), pRL-null (25 ng), 500 ng of either pCI or TBK1, and (A) 1,000 ng of pCI, pMC159, pMC159A, pMC159B, pMC159Δ, or pnsp11. Values are shown as mean ± SD (n = 3). After 48 h incubation, cells were lysed and firefly and sea pansy luciferase activities were measured. A portion of each lysate was analyzed by immunoblotting for viral protein expression. These are representative results from experiments that were performed at least three times independently.
When assessing the TBK1 inhibitory function of the same mutant MC159 proteins by a luciferase reporter assay, wild-type MC159 protein reduced TBK1-induced luciferase activity by 57% compared with pCI-transfected cells (Fig. 4C). Whereas MC159B diminished luciferase activity to a slightly greater extent than MC159A or MC159Δ, the reduction in luciferase activities was statistically significant for each viral protein. Viral proteins were expressed at similar levels, indicating that TBK1 overexpression does not destabilize MC159 protein expression. As would be expected, nsp11 inhibited TBK1-induced luciferase activity the greatest.
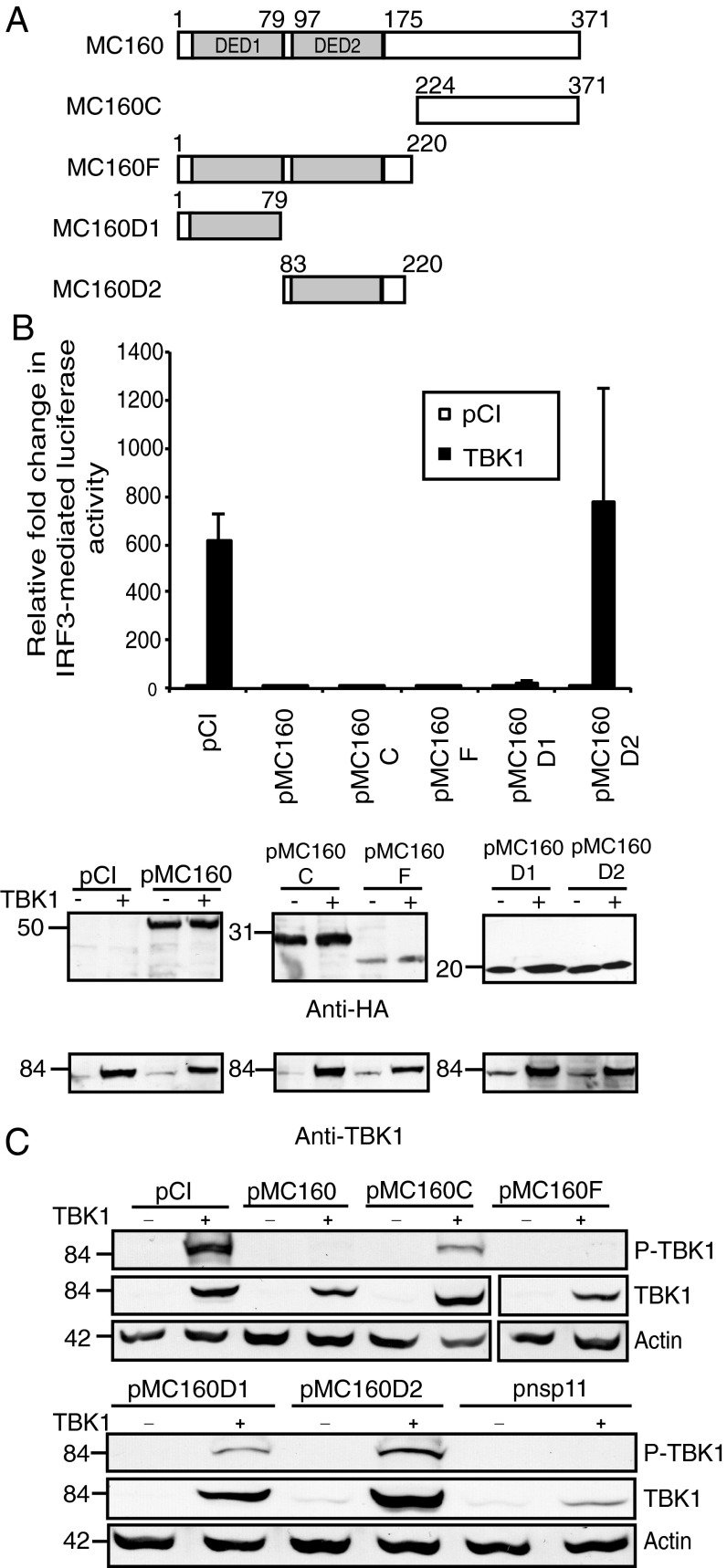
Based on the data using MC159 mutant proteins above, one concern was that DEDs themselves did not possess an IRF3 inhibitory function. Moreover, it was unclear how MC160 inhibited IRF3 activation. As a means of exploring both questions, we used a set of mutant MC160 proteins in which DED motifs were expressed independent of other portions of MC160 (Fig. 5A) and assessed their effect on TBK1 activation. As shown in Fig. 5A, the wild-type MC160 protein possesses two tandem DEDs; DED1 at residues 1–79 and DED2 at residues 97–175. The remainder of MC160, residues 175–371, lacks any known motifs. Two mutant MC160 constructs, MC160D1 and MC160D2, contained only the MC160 DED1 and DED2, respectively (Fig. 5A), and would afford us the opportunity to assess the inhibitory effect of DEDs to inhibit TBK1-induced luciferase activity (Fig. 5B) or TBK1 phosphorylation (Fig. 5C).
Fig. 5.
The MC160 protein inhibits TBK1 activation independent of DED1. (A) A schematic of mutant MC160 constructs used in these experiments, in which the two tandem DEDs of MC160 (DED1 and DED2) are shown. (B) Subconfluent 293T cellular monolayers were transfected with pIRF3-luc (225 ng), pRL-null (25 ng), 500 ng of either pCI or TBK1, and (A) 1,000 ng of pCI, pMC160, pMC160C, pMC160F, pMC160D1, or pMC160D2. Values are shown as mean ± SD (n = 3). After 48 h incubation, cells were lysed and firefly and sea pansy luciferase activities were measured. A portion of each lysate was analyzed by immunoblotting for viral protein expression. These are representative results from experiments that were performed at least three times independently. (C) Subconfluent monolayers of 293T cells were transfected with 500 ng TBK1 or pCI and 1,000 ng pCI, pMC160, pMC160C, pMC160F, pMC160D1, pMC160D2, or pnsp11. At 24 h posttransfection, cells were lysed and clarified cellular lysates were analyzed by immunoblotting for phospho-TBK1 (P-TBK1), TBK1, or actin proteins.
When mapping the MC160 region responsible for inhibition of TBK1-induced luciferase activity (Fig. 5B), it was observed that MC160F, which expressed both DEDs, inhibited IRF3-mediated luciferase activity. The MC160C construct also inhibited IRF3 activation. Interestingly, when each MC160 DED was expressed independently, only the DED1 region (protein MC160D1) retained its ability to inhibit luciferase activity. Using the same set of mutant proteins, a second method (detecting TBK1 phosphorylation) was used to evaluate TBK1 activation (Fig. 5C). The MC160F protein inhibited TBK1 phosphorylation to the same degree at wild-type MC160. In contrast, phospho-TBK1 levels were now detected in cells expressing the MC160D2 proteins, albeit not to the levels observed in vector-transfected cells. Phospho-TBK1 levels were also slightly elevated in cells expressing MC160C and MC160D1, despite that fact that each construct inhibited IRF3-induced luciferase activity quite strongly. As would be expected, all mutant and wild-type MC160 proteins were expressed in similar amounts. In addition, ectopic expression of TBK1 was not compromised by coexpression by any of the MC160 constructs. Similar to Fig. 4, TBK1 protein levels were reduced when cells coexpressed nsp11.
Discussion
MCV is a dermatotropic poxvirus that causes benign skin neoplasms in humans (26). MCV is a common and worldwide infection, and is an opportunistic infection of immunocompromised patients (56–59). MCV is understudied due to the lack of an animal model for disease or a tissue culture-based propagation system. Despite these technical difficulties, great insights into viral immune evasion have occurred by studying these proteins independent of MCV infection (16). Because there is no cure for MCV infections, continued study of these immune evasion proteins on a subcellular level will provide information that may allow for the reconstitution of the immune response to resolve MCV infections.
The TBK1–IKKε complex is an essential complex for activation of IFN-β expression in response to viral infection (11). Therefore, it is not surprising that viral immune evasion proteins target TBK1–IKKε. For example, herpes simplex virus, another virus with a DNA genome, expresses the γ1 34.5 protein, which also prevents TBK1 activation to block IFN-β expression (60). Vaccinia virus, a well-studied poxvirus, expresses the C6 protein to interact with TBK1–IKKε complex scaffold proteins to block IRF3- and IRF7-controlled IFN-β production (22). With respect to C6, it would be expected that MC159 would possess a different molecular mechanism to inhibit TBK1 activation because MC159 inhibits TBK1-induced IRF3 activation but not TBK1-induced IRF7 phosphorylation. Despite the fact that both C6 and MC160 inhibited IRF3 and IRF7, it is presumed that MC160 and C6 inhibitory mechanisms are distinct because MC160 did not coimmunoprecipitate with TBK1 or IKKε (22).
For MC159, the current model is that MC159 binds simultaneously to TBK1 and IKKε to prevent subsequent TBK1 activation through either TRAF3 or STING (9, 12, 13). Another possibility is that MC159 prevents TBK1–IKKε complex formation by binding to TBK1 or IKKε. It is highly speculative at this point to predict which DED of MC159 would specifically interact with TBK1 versus IKKε. Recently it was reported that the cellular TRAF-interacting protein (TRIP) inhibits TBK1 by targeting it for ubiquitin-mediated degradation (61). Here, TBK1 protein levels were unaffected by MC159, suggesting that MC159 is not acting as a functional TRIP homolog. Although TRAF3–MC159 interactions are known (55), it is unlikely that MC159 competitively binds to TRAF3 to prevent TBK1 activation because we observed that a mutant MC159 protein that lacks TRAF-binding motifs still inhibited TBK1 activation. A previous study shows that MC159 inhibits IFN-β expression indirectly, by interacting with FADD (43). Both MC159A and MC159B bind to FADD (53) and retain IRF3 inhibitory function. As such, we cannot rule out MC159–FADD interactions playing a role in the IRF3 inhibitory function of MC159.
How MC160 prevents TBK1 activation remains unknown. There are some potential mechanisms that are less likely. For example, coimmunoprecipitation data shown here would suggest that MC160 does physically interact with this complex as a means of inhibition. Second, it is doubtful that MC160 interferes with TRAF3, a protein that triggers TBK1–IKKε complex formation, because MC160 lacks consensus motifs for TRAF3 binding (38). MC160 is known to interact with procaspase-8 (39). It was published recently that procapase-8 cleaves IRF3 to induce IRF3 degradation as a means to down-regulate expression of genes induced by IRF3 (62). However, the IRF3 protein is neither decreased nor cleaved in MC160-expressing cells under conditions in which IRF3 is inactive or triggered (S1), suggesting that MC160 did not directly or indirectly target the IRF3 protein as its mechanism of inhibition.
An analysis of mutant MC160 proteins was performed here as one approach to elucidate IRF3 inhibitory function. What is clear from these data are that more than one region of MC160 inhibits IRF3, namely DED1 and the C-terminal region of MC160. Interestingly, the C terminus of MC160 binds to Hsp90 (39). During Sendai virus infection, Hsp90 binds to and stabilizes both IRF3 and TBK1, forming a complex to facilitate signal transduction from TBK1 to IRF3 (63, 64). Thus, one future area of investigation would be to evaluate whether the C terminus of MC160 prevents Hsp90-mediated stabilization of TBK1 and IRF3. It was also observed that the MC160F protein, which expresses both MC160 DEDs, inhibited TBK1 phosphorylation to a greater extent that either DED. One interpretation of this data is that MC160F may be superior for inhibiting TBK1 activation because it has a larger surface area that could allow for more stable interactions with its upstream cellular-binding partners than the smaller MC160D1 protein, and thus this is an area for future investigation.
Whereas IRF3 and IRF7 proteins are both activated by TBK1, each protein has distinct functions (65). IRF3 is constitutively expressed in most cells. In contrast, lymphoid cells express IRF7 constitutively and other cells express IRF7 after prolonged IRF3 activation. Although IRF7 is dispensable for IFN-β gene expression, this transcription factor is critical for IFN-α gene activation. It is unclear how MC159 would inhibit IRF3 but not IRF7, and future studies that map the region of TBK1–MC159 interactions versus residues important for TBK1 activation may shed light on this problem (52). In comparison, MC160 inhibited both IRF3 and IRF7 despite its lack of interactions with the TBK1–IKKε complex. Such a phenotype would be expected if MC160 targeted more than one cellular protein. To address this possibility, future directions will include comparing the MC160 regions required for inhibition of TBK-1 versus IRF3 versus IRF7. A remaining question is what benefit IRF7 inhibition would afford MCV, given that MCV is thought to not replicate in lymphoid cells (26).
It was observed that the cFLIPL protein inhibits IFN-β enhancer activation in an NF-κB–independent manner. cFLIPL is up-regulated in several cancers (66–68). At first glance, it would seem that this unique inhibitory property of cFLIPL would aid in tumor cell survival. As such, small molecules that target cFLIPL may eliminate cancer cells. Because cFLIPS and cFLIPR variants also possess tandem DEDs, it is of great interest to determine the effect of these two forms of cFLIP on IFN-β gene activation.
Work shown here highlights the diverse effects of the FLIPs on intrinsic immune responses. Notably, MC159 is the only FLIP to date that inhibits apoptosis, NF-κB, and IRF3 activation (27, 29–31, 40, 41). Although MC160 inhibits NF-κB and IRF3 activation (38, 39), its function as an antiapoptosis protein is less convincing, with only one of several publications showing MC160 inhibition of apoptosis, whereas other reports show no such function (29, 31, 69). The K13 protein of KSHV is a robust inducer of NF-κB in addition to its ability to inhibit apoptosis (70). Here, K13 gave an intermediate phenotype for its ability to inhibit IRF3-induced IFN-β activation. Interestingly, KSHV and MCV both infect the skin. However, only KSHV has a latent life cycle. One tantalizing possibility is that the KSHV and MCV FLIPs each possess different patterns for inhibiting apoptosis, NF-κB, or IRF3 to overcome different host barriers for virus replication. For example, KSHV activation of NF-κB is required for the transformation of cells (33, 35), whereas MCV inhibition of NF-κB may be necessary for dampening localized antiviral immune responses triggered by an acute infection.
More curious is that the DEDs of these FLIPs, although similar, are not functionally identical. For example, within the context of a single FLIP, both DED1 and DED2 must be present for antiapoptosis function of MC159 (53, 54, 71). In contrast, MC159 DED1 is sufficient to inhibit NF-κB activation (41). Here, either DED1 or DED2 of MC159 is sufficient to inhibit IRF3 activation. For the MC160 protein, neither DED inhibits apoptosis (31), but DED2 or the C terminus inhibits NF-κB (39), whereas DED1 or the MC160 C terminus is sufficient to inhibit IRF3 activation. These data suggest that these DEDs, which appear to be homologous at first glance, actually possess distinct molecular and structural fingerprints within their DEDs that are responsible for these specificities. Currently it is unclear what differences among these FLIPs and their DEDs allow them to modulate apoptosis, NF-κB activation, and IFN-β expression by such different mechanisms. A more careful and precise analysis of the sequences, structure, and molecular functions of FLIPs, and the comparison of FLIPs to each other, can give new information into protein–protein interactions that are critical for regulating intrinsic immune responses.
Materials and Methods
Cell Culture and Plasmids.
Wild-type MEF and p65−/− MEFs were obtained from Laurent Poliquin (University of Quebec, Montreal). Human embryonic kidney 293T cells were obtained from the American Type Culture Collection. Plasmid pCI was obtained from Promega. Plasmid pMC159 and pHA–MC159, and the construction of plasmids pMC159A and pMC159B are described previously (53). pMC159Δ (Margot Thome; University of Lausanne, Lausanne, Switzerland) encodes a vesicular stomatitis virus epitope-tagged MC159 protein in which the TRAF3-binding sites are deleted (55). Plasmids pMC160, pHA–MC160, pMC160C, pMC160F, pMC160D1, and pMC160D2 were described previously (38, 39). All mutant MC160 proteins express a hemagglutinin epitope tag at the N terminus of the protein (38, 39). pK13 encodes a FLAG-tagged K13 gene from KSHV and a the pcFLIPL plasmid encoding a FLAG-tagged human cFLIPL gene were provided by Jeffery Cohen [National Institutes of Health (NIH), Bethesda]. Dongwan Yoo (University of Illinois, Urbana-Champaign, IL) provided pnsp11, which encodes a FLAG-tagged nsp11 protein from the porcine respiratory virus (50), pMAVS, which encodes a FLAG-tagged MAVS protein, IRF3CA, which encodes a FLAG-tagged constitutively active mutant IRF3. Plasmids that encode either IRF7 or IFN-α were gifts from Fanxiu Zhu (Florida State University, Tallahassee, FL). Plasmid TBK1, which encodes a FLAG epitope-tagged TBK1 protein, was a kind gift from Siddharth Balachandran (Fox Chase Cancer Center, Philadelphia). A pcDNA3.1 vector encoding a FLAG-tagged IKKε plasmid was obtained from Addgene (11). The following plasmids were used for luciferase reporter assays: pRL-null (Promega), pIFN-β−luc (containing the natural IFN-β promoter), and pIRF3-luc (containing four copies of the PRDIII promoter sequence 5′-GAAANNGAAANN-3′). For all experiments involving plasmids, DNA was transfected into cells using TransIT-2020 transfection reagent (Mirus Bio), following the manufacturer’s protocol.
Luciferase Assays.
For assays in which MAVS overexpression triggered IFN-β–controlled luciferase activity, Subconfluent MEFs or p65−/− MEFs in 12-well plates were cotransfected with 225 ng pIFN-β–luc, 25 ng pRL-null, 500 ng pCI, or pMAVS and 750 ng of either pCI, pMC159, pMC159A, pMC159B, pMC160, pcFLIPL, pK13, or pnsp11. At 24 h posttransfection, cells were lysed in 1× passive lysis buffer (PLB) (Promega). Lysates were either used for evaluating luciferase activity or for evaluating the expression of viral or cellular proteins by immunoblotting. For all other luciferase assays, 293T cells were used, and the pIRF3-luc plasmid was used in place of pIFN-β–luc. In experiments using overexpression of TBK1, IKKε, or constitutively active IRF3 to stimulate IRF3-controlled luciferase activity, cells were cotransfected with 225 ng pIRF3-luc, 25 ng pRL-null, 500 ng TBK1, or pIKKε or IRF3CA or pCI, and 750 ng of either pCI, pMC159, pMC160, or pnsp11. Forty-eight hours later, cells were lysed in 1× PLB. For luciferase assays in which mutant MC159 or MC160 proteins were used, subconfluent 293T cells were cotransfected with 225 ng pIRF3-luc, 25 ng pRL-null, 500 ng TBK1, and either 1000 ng pCI, pMC159, pMC159A, pMC159B, pMC159Δ, pMC160, pMC160C, pMC160F, pMC160D1, or pMC160D2. At 48 h posttransfection, cells were harvested and lysed in 1× PLB. All lysates were analyzed in triplicate for luciferase activities. All luciferase activities were measured as relative light units using a BioTek luminometer and the Dual Luciferase Reporter Assay System (Promega). Relative luciferase activity ratios were calculated as described previously (41). In some instances, a portion of each cellular lysate from luciferase assays was analyzed for protein expression by immunoblotting, using the antiserum indicated under each immunoblot in the figures. For luciferase assays, the magnitude of the signal induced by overexpression of signaling proteins can vary from experiment to experiment. This is likely due to differences in cell passage numbers. However, despite these differences in magnitude, the extent of inhibition observed by each FLIP protein or nsp11 protein remains constant between experiments.
Coimmunoprecipitations.
The 293T cellular monolayers in six-well plates were transfected with 500 ng TBK1, 250 ng pIKKε, and 1,000 ng pCI, pHA–MC159, or pHA–MC160. At 24 h posttransfection, cells were detached from plates by scraping and collected by centrifugation (1,000 × g for 10 min). Cellular pellets were resuspended in 150 μL DED lysis buffer (41) for 30 min at 4 °C. Cellular lysates were centrifuged (14,000 × g for 10 min). Clarified supernatants were collected and 50 μL of the supernatants were removed to a new tube, and 30 μg of protein from this sample was used to assess expression levels of proteins. The remaining 100 μL of clarified lysates were incubated overnight with 1 μg monoclonal mouse anti-HA (Sigma-Aldrich) with constant rotation at 4 °C. Fifty microliters of protein G-Sepharose (Invitrogen) beads were added the next day to the sample and continued rotation at 4 °C for another 6 h. As a control, an identical set of clarified lysates was incubated instead with 1 μg mouse IgG (Sigma-Aldrich). For all samples, beads were collected by centrifugation (14,000 × g for 2 min), and washed three times with large volumes of DED lysis buffer. Pelleted bead–protein complexes were suspended in 30 μL of 2× Laemmli buffer containing 5% (vol/vol) 2-mercaptoethanol and boiled for 5 min. Samples were then analyzed by immunoblot for the presence of proteins. Primary antibodies used for these experiments were as follows: polyclonal rabbit anti-MC159 (31), polyclonal rabbit anti-MC160 (31), monoclonal mouse anti-FLAG (Sigma-Aldrich), monoclonal rabbit TBK1 (Cell Signaling), and polyclonal rabbit anti-actin (Calbiochem).
Immunoblotting.
The activation state of TBK1 was determined by detecting the phosphorylated (activated) form of the protein by using immunoblotting. The 293T cells were cotransfected with 500 ng TBK1 or pCI and 1,000 ng pMC159-based vectors, pMC160-based vectors, or pnsp11. The activation state of IRF7 was determined by detecting the phosphorylated form of IRF7. The 293T cells were cotransfected with 250 ng each of IRF7 and IFN-α plasmids, 500 ng TBK1 or pCI, and 750 ng pMC159, pMC160, or pnsp11. For both assays, 24 h later, cells were lysed in cytoplasmic extraction buffer. The protein concentration of each clarified cellular lysate was determined using bicinchoninic acid assay (Pierce), and 20 μg of protein from each lysate was prepared for immunoblotting as previously described (41). Anti-serum used to probe immunoblots for this assay was as follows: polyclonal rabbit anti–phospho-TBK1 (Ser-172; Millipore), monoclonal rabbit anti-TBK1 (Cell Signaling), and polyclonal rabbit anti-actin (Calbiochem), rabbit anti–phospho-IRF7 (Cell Signaling), monoclonal anti-IRF7 (Cell Signaling), polyclonal rabbit anti-MC159 (31), polyclonal rabbit anti-MC160 (31), and monoclonal mouse anti-FLAG (Sigma-Aldrich). Secondary antibodies that were used were either HRP-conjugated goat anti-mouse IgG (Thermo Scientific) or HRP-conjugated goat anti-rabbit IgG (Calbiochem).
Supplementary Material
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1314569111/-/DCSupplemental.
References
- 1.Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8(7):559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Müller U, et al. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264(5167):1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 3.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122(5):669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 4.Kawai T, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6(10):981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 5.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5(7):730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 6.Hornung V, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314(5801):994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 7.Pichlmair A, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314(5801):997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 8.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441(7089):101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 9.Oganesyan G, et al. Critical role of TRAF3 in the Toll-like receptor-dependent and -independent antiviral response. Nature. 2006;439(7073):208–211. doi: 10.1038/nature04374. [DOI] [PubMed] [Google Scholar]
- 10.Sharma S, et al. Triggering the interferon antiviral response through an IKK-related pathway. Science. 2003;300(5622):1148–1151. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- 11.Fitzgerald KA, et al. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4(5):491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 12.Nakhaei P, Hiscott J, Lin R. STING-ing the antiviral pathway. J Mol Cell Biol. 2010;2(3):110–112. doi: 10.1093/jmcb/mjp048. [DOI] [PubMed] [Google Scholar]
- 13.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455(7213):674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Randall RE, Goodbourn S. Interferons and viruses: An interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol. 2008;89(Pt 1):1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- 15.Senkevich TG, et al. Genome sequence of a human tumorigenic poxvirus: prediction of specific host response-evasion genes. Science. 1996;273(5276):813–816. doi: 10.1126/science.273.5276.813. [DOI] [PubMed] [Google Scholar]
- 16.Randall CMH, Shisler J. Molluscum contagiosum virus: Persistence pays off. Future Virology. 2013;8(6):561–573. [Google Scholar]
- 17.Waibler Z, et al. Vaccinia virus-mediated inhibition of type I interferon responses is a multifactorial process involving the soluble type I interferon receptor B18 and intracellular components. J Virol. 2009;83(4):1563–1571. doi: 10.1128/JVI.01617-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Myskiw C, Arsenio J, van Bruggen R, Deschambault Y, Cao J. Vaccinia virus E3 suppresses expression of diverse cytokines through inhibition of the PKR, NF-kappaB, and IRF3 pathways. J Virol. 2009;83(13):6757–6768. doi: 10.1128/JVI.02570-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zurkova K, et al. The expression of the soluble isoform of hFlt3 ligand by recombinant vaccinia virus enhances immunogenicity of the vector. Oncol Rep. 2009;21(5):1335–1343. doi: 10.3892/or_00000359. [DOI] [PubMed] [Google Scholar]
- 20.Deng L, et al. Vaccinia virus subverts a mitochondrial antiviral signaling protein-dependent innate immune response in keratinocytes through its double-stranded RNA binding protein, E3. J Virol. 2008;82(21):10735–10746. doi: 10.1128/JVI.01305-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Domingo-Gil E, Pérez-Jiménez E, Ventoso I, Nájera JL, Esteban M. Expression of the E3L gene of vaccinia virus in transgenic mice decreases host resistance to vaccinia virus and Leishmania major infections. J Virol. 2008;82(1):254–267. doi: 10.1128/JVI.01384-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Unterholzner L, et al. Vaccinia virus protein C6 is a virulence factor that binds TBK-1 adaptor proteins and inhibits activation of IRF3 and IRF7. PLoS Pathog. 2011;7(9):e1002247. doi: 10.1371/journal.ppat.1002247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferguson BJ, et al. Vaccinia virus protein N2 is a nuclear IRF3 inhibitor that promotes virulence. J Gen Virol. 2013;94(Pt 9):2070–2081. doi: 10.1099/vir.0.054114-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiang Y, Moss B. IL-18 binding and inhibition of interferon gamma induction by human poxvirus-encoded proteins. Proc Natl Acad Sci USA. 1999;96(20):11537–11542. doi: 10.1073/pnas.96.20.11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiang Y, Moss B. Molluscum contagiosum virus interleukin-18 (IL-18) binding protein is secreted as a full-length form that binds cell surface glycosaminoglycans through the C-terminal tail and a furin-cleaved form with only the IL-18 binding domain. J Virol. 2003;77(4):2623–2630. doi: 10.1128/JVI.77.4.2623-2630.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Damon IK. Poxviruses. In: Knipe D, Howley P, editors. Fields Virology. Philadelphia: Lippincott Williams & Wilkins; 2007. 5th Ed, Vol 2, pp 2947–2976. [Google Scholar]
- 27.Bertin J, et al. Death effector domain-containing herpesvirus and poxvirus proteins inhibit both Fas- and TNFR1-induced apoptosis. Proc Natl Acad Sci USA. 1997;94(4):1172–1176. doi: 10.1073/pnas.94.4.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Irmler M, et al. Inhibition of death receptor signals by cellular FLIP. Nature. 1997;388(6638):190–195. doi: 10.1038/40657. [DOI] [PubMed] [Google Scholar]
- 29.Hu S, Vincenz C, Buller M, Dixit VM. A novel family of viral death effector domain-containing molecules that inhibit both CD-95- and tumor necrosis factor receptor-1-induced apoptosis. J Biol Chem. 1997;272(15):9621–9624. doi: 10.1074/jbc.272.15.9621. [DOI] [PubMed] [Google Scholar]
- 30.Thome M, et al. Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature. 1997;386(6624):517–521. doi: 10.1038/386517a0. [DOI] [PubMed] [Google Scholar]
- 31.Shisler JL, Moss B. Molluscum contagiosum virus inhibitors of apoptosis: The MC159 v-FLIP protein blocks Fas-induced activation of procaspases and degradation of the related MC160 protein. Virology. 2001;282(1):14–25. doi: 10.1006/viro.2001.0834. [DOI] [PubMed] [Google Scholar]
- 32.Chaudhary PM, et al. Activation of the NF-kappaB pathway by caspase 8 and its homologs. Oncogene. 2000;19(39):4451–4460. doi: 10.1038/sj.onc.1203812. [DOI] [PubMed] [Google Scholar]
- 33.Sun Q, Zachariah S, Chaudhary PM. The human herpes virus 8-encoded viral FLICE-inhibitory protein induces cellular transformation via NF-kappaB activation. J Biol Chem. 2003;278(52):52437–52445. doi: 10.1074/jbc.M304199200. [DOI] [PubMed] [Google Scholar]
- 34.Matta H, et al. Kaposi’s sarcoma-associated herpesvirus (KSHV) oncoprotein K13 bypasses TRAFs and directly interacts with the IkappaB kinase complex to selectively activate NF-kappaB without JNK activation. J Biol Chem. 2007;282(34):24858–24865. doi: 10.1074/jbc.M700118200. [DOI] [PubMed] [Google Scholar]
- 35.Matta H, Sun Q, Moses G, Chaudhary PM. Molecular genetic analysis of human herpes virus 8-encoded viral FLICE inhibitory protein-induced NF-kappaB activation. J Biol Chem. 2003;278(52):52406–52411. doi: 10.1074/jbc.M307308200. [DOI] [PubMed] [Google Scholar]
- 36.Kataoka T, et al. The caspase-8 inhibitor FLIP promotes activation of NF-kappaB and Erk signaling pathways. Curr Biol. 2000;10(11):640–648. doi: 10.1016/s0960-9822(00)00512-1. [DOI] [PubMed] [Google Scholar]
- 37.Field N, et al. KSHV vFLIP binds to IKK-gamma to activate IKK. J Cell Sci. 2003;116(Pt 18):3721–3728. doi: 10.1242/jcs.00691. [DOI] [PubMed] [Google Scholar]
- 38.Nichols DB, Shisler JL. The MC160 protein expressed by the dermatotropic poxvirus molluscum contagiosum virus prevents tumor necrosis factor alpha-induced NF-kappaB activation via inhibition of I kappa kinase complex formation. J Virol. 2006;80(2):578–586. doi: 10.1128/JVI.80.2.578-586.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nichols DB, Shisler JL. Poxvirus MC160 protein utilizes multiple mechanisms to inhibit NF-kappaB activation mediated via components of the tumor necrosis factor receptor 1 signal transduction pathway. J Virol. 2009;83(7):3162–3174. doi: 10.1128/JVI.02009-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murao LE, Shisler JL. The MCV MC159 protein inhibits late, but not early, events of TNF-α-induced NF-κB activation. Virology. 2005;340(2):255–264. doi: 10.1016/j.virol.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 41.Randall CM, Jokela JA, Shisler JL. The MC159 protein from the molluscum contagiosum poxvirus inhibits NF-κB activation by interacting with the IκB kinase complex. J Immunol. 2012;188(5):2371–2379. doi: 10.4049/jimmunol.1100136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gil J, Rullas J, Alcamí J, Esteban M. MC159L protein from the poxvirus molluscum contagiosum virus inhibits NF-kappaB activation and apoptosis induced by PKR. J Gen Virol. 2001;82(Pt 12):3027–3034. doi: 10.1099/0022-1317-82-12-3027. [DOI] [PubMed] [Google Scholar]
- 43.Balachandran S, Venkataraman T, Fisher PB, Barber GN. Fas-associated death domain-containing protein-mediated antiviral innate immune signaling involves the regulation of Irf7. J Immunol. 2007;178(4):2429–2439. doi: 10.4049/jimmunol.178.4.2429. [DOI] [PubMed] [Google Scholar]
- 44.Maniatis T, et al. Structure and function of the interferon-beta enhanceosome. Cold Spring Harb Symp Quant Biol. 1998;63:609–620. doi: 10.1101/sqb.1998.63.609. [DOI] [PubMed] [Google Scholar]
- 45.Panne D. The enhanceosome. Curr Opin Struct Biol. 2008;18(2):236–242. doi: 10.1016/j.sbi.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 46.Handa P, Tupper JC, Jordan KC, Harlan JM. FLIP (Flice-like inhibitory protein) suppresses cytoplasmic double-stranded-RNA-induced apoptosis and NF-κB and IRF3-mediated signaling. Cell Commun Signal. 2011;9:16. doi: 10.1186/1478-811X-9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sato M, et al. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity. 2000;13(4):539–548. doi: 10.1016/s1074-7613(00)00053-4. [DOI] [PubMed] [Google Scholar]
- 48.Challa S, Woelfel M, Guildford M, Moquin D, Chan FK. Viral cell death inhibitor MC159 enhances innate immunity against vaccinia virus infection. J Virol. 2010;84(20):10467–10476. doi: 10.1128/JVI.00983-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shi X, et al. Endoribonuclease activities of porcine reproductive and respiratory syndrome virus nsp11 was essential for nsp11 to inhibit IFN-β induction. Mol Immunol. 2011;48(12-13):1568–1572. doi: 10.1016/j.molimm.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun Y, Han M, Kim C, Calvert JG, Yoo D. Interplay between interferon-mediated innate immunity and porcine reproductive and respiratory syndrome virus. Viruses. 2012;4(4):424–446. doi: 10.3390/v4040424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ning S, Pagano JS, Barber GN. IRF7: Activation, regulation, modification and function. Genes Immun. 2011;12(6):399–414. doi: 10.1038/gene.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Helgason E, Phung QT, Dueber EC. Recent insights into the complexity of Tank-binding kinase 1 signaling networks: The emerging role of cellular localization in the activation and substrate specificity of TBK1. FEBS Lett. 2013;587(8):1230–1237. doi: 10.1016/j.febslet.2013.01.059. [DOI] [PubMed] [Google Scholar]
- 53.Garvey T, Bertin J, Siegel R, Lenardo M, Cohen J. The death effector domains (DEDs) of the molluscum contagiosum virus MC159 v-FLIP protein are not functionally interchangeable with each other or with the DEDs of caspase-8. Virology. 2002;300(2):217–225. doi: 10.1006/viro.2002.1518. [DOI] [PubMed] [Google Scholar]
- 54.Garvey TL, et al. Binding of FADD and caspase-8 to molluscum contagiosum virus MC159 v-FLIP is not sufficient for its antiapoptotic function. J Virol. 2002;76(2):697–706. doi: 10.1128/JVI.76.2.697-706.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thurau M, Everett H, Tapernoux M, Tschopp J, Thome M. The TRAF3-binding site of human molluscipox virus FLIP molecule MC159 is critical for its capacity to inhibit Fas-induced apoptosis. Cell Death Differ. 2006;13(9):1577–1585. doi: 10.1038/sj.cdd.4401847. [DOI] [PubMed] [Google Scholar]
- 56.Tyring SK. Molluscum contagiosum: The importance of early diagnosis and treatment. Am J Obstet Gynecol. 2003;189(3) Suppl:S12–S16. doi: 10.1067/s0002-9378(03)00793-2. [DOI] [PubMed] [Google Scholar]
- 57.Konya J, Thompson CH. Molluscum contagiosum virus: Antibody responses in persons with clinical lesions and seroepidemiology in a representative Australian population. J Infect Dis. 1999;179(3):701–704. doi: 10.1086/314620. [DOI] [PubMed] [Google Scholar]
- 58.Birthistle K, Carrington D. Molluscum contagiosum virus. J Infect. 1997;34(1):21–28. doi: 10.1016/s0163-4453(97)80005-9. [DOI] [PubMed] [Google Scholar]
- 59.Watanabe T, et al. Antibodies to molluscum contagiosum virus in the general population and susceptible patients. Arch Dermatol. 2000;136(12):1518–1522. doi: 10.1001/archderm.136.12.1518. [DOI] [PubMed] [Google Scholar]
- 60.Ma Y, et al. Inhibition of TANK binding kinase 1 by herpes simplex virus 1 facilitates productive infection. J Virol. 2012;86(4):2188–2196. doi: 10.1128/JVI.05376-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang M, et al. TRAF-interacting protein (TRIP) negatively regulates IFN-β production and antiviral response by promoting proteasomal degradation of TANK-binding kinase 1. J Exp Med. 2012;209(10):1703–1711. doi: 10.1084/jem.20120024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sears N, Sen GC, Stark GR, Chattopadhyay S. Caspase-8-mediated cleavage inhibits IRF-3 protein by facilitating its proteasome-mediated degradation. J Biol Chem. 2011;286(38):33037–33044. doi: 10.1074/jbc.M111.257022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu XY, Wei B, Shi HX, Shan YF, Wang C. Tom70 mediates activation of interferon regulatory factor 3 on mitochondria. Cell Res. 2010;20(9):994–1011. doi: 10.1038/cr.2010.103. [DOI] [PubMed] [Google Scholar]
- 64.Yang K, et al. Hsp90 regulates activation of interferon regulatory factor 3 and TBK-1 stabilization in Sendai virus-infected cells. Mol Biol Cell. 2006;17(3):1461–1471. doi: 10.1091/mbc.E05-09-0853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol. 2008;26:535–584. doi: 10.1146/annurev.immunol.26.021607.090400. [DOI] [PubMed] [Google Scholar]
- 66.Murtaza I, Saleem M, Adhami VM, Hafeez BB, Mukhtar H. Suppression of cFLIP by lupeol, a dietary triterpene, is sufficient to overcome resistance to TRAIL-mediated apoptosis in chemoresistant human pancreatic cancer cells. Cancer Res. 2009;69(3):1156–1165. doi: 10.1158/0008-5472.CAN-08-2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Medema JP, de Jong J, van Hall T, Melief CJ, Offringa R. Immune escape of tumors in vivo by expression of cellular FLICE-inhibitory protein. J Exp Med. 1999;190(7):1033–1038. doi: 10.1084/jem.190.7.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee TJ, Lee JT, Park JW, Kwon TK. Acquired TRAIL resistance in human breast cancer cells are caused by the sustained cFLIP(L) and XIAP protein levels and ERK activation. Biochem Biophys Res Commun. 2006;351(4):1024–1030. doi: 10.1016/j.bbrc.2006.10.163. [DOI] [PubMed] [Google Scholar]
- 69.Shisler JL, Senkevich TG, Berry MJ, Moss B. Ultraviolet-induced cell death blocked by a selenoprotein from a human dermatotropic poxvirus. Science. 1998;279(5347):102–105. doi: 10.1126/science.279.5347.102. [DOI] [PubMed] [Google Scholar]
- 70.Chaudhary PM, Jasmin A, Eby MT, Hood L. Modulation of the NF-kappa B pathway by virally encoded death effector domains-containing proteins. Oncogene. 1999;18(42):5738–5746. doi: 10.1038/sj.onc.1202976. [DOI] [PubMed] [Google Scholar]
- 71.Yang JK, et al. Crystal structure of MC159 reveals molecular mechanism of DISC assembly and FLIP inhibition. Mol Cell. 2005;20(6):939–949. doi: 10.1016/j.molcel.2005.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.