Abstract
The epigenetic regulation of imprinted genes via monoallelic DNA methylation of either maternal or paternal alleles is critical for embryonic growth and development1. Imprinted genes were recently shown to be expressed in mammalian adult stem cells to support self-renewal of neural and lung stem cells2, 3,4; however, a role for imprinting per se in adult stem cells remains elusive. Here we show up-regulation of growth-restricting imprinted genes, including within the H19-Igf2 locus5, in long-term hematopoietic stem cells (LT-HSCs) and their down-regulation upon HSC activation and proliferation. A differentially methylated region (DMR) upstream of H19 (H19-DMR), serving as the imprinting control region, determines the reciprocal expression of H19 from the maternal allele and Igf2 from the paternal allele1. In addition, H19 also serves as a source of miR-675, which restricts Igf1r expression6. We demonstrated that conditional deletion of the maternal but not the paternal H19-DMR reduced adult HSC quiescence, a state required for long-term maintenance of HSCs, and compromised HSC function. Maternal-specific H19-DMR deletion resulted in activation of the Igf2-Igfr1 pathway as revealed by the translocation of phosphorylated Foxo3 (an inactive form) from nucleus to cytoplasm and the release of Foxo3-mediated cell-cycle arrest, thus leading to increased activation, proliferation, and eventual exhaustion of HSCs. Mechanistically, maternal-specific H19-DMR deletion led to Igf2 up-regulation and increased translation of Igf1r, which is normally suppressed by H19-derived miR-675. Similarly, genetic inactivation of Igf1r partially rescued the H19-DMR deletion phenotype. Our work establishes a novel role for this unique form of epigenetic control at the H19-Igf2 locus in maintaining adult stem cells.
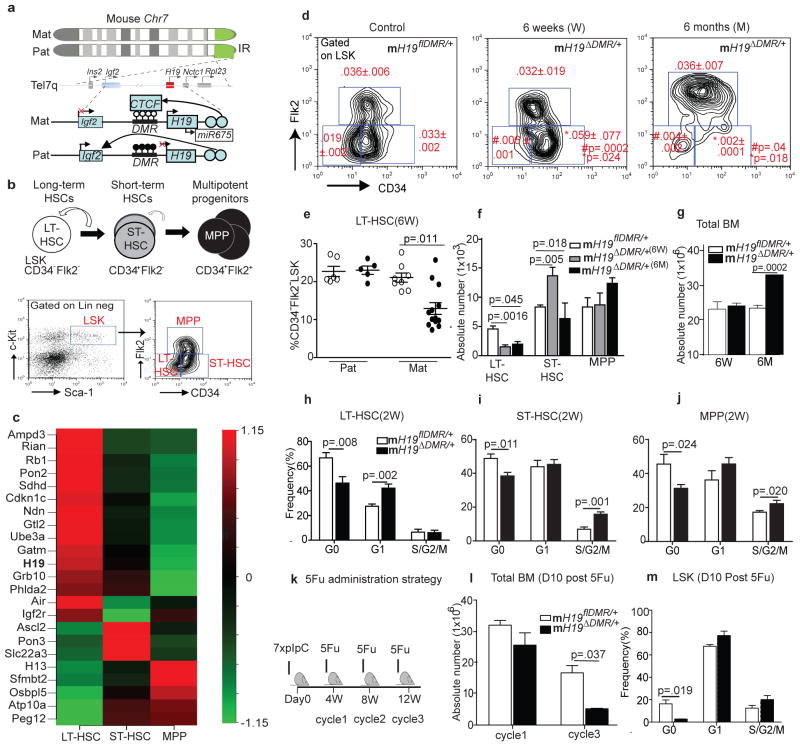
Our earlier studies had revealed that imprinted genes, including those within the H19-Igf2 locus (Fig. 1a), are differentially expressed in hematopoietic stem and progenitor cells (HSPCs)7. To explore this further, we systematically analyzed imprinted gene expression in quiescent-enriched long-term (LT) HSC, more active short-term (ST) HSC, and multipotent progenitor (MPP) populations (Fig. 1b)8. Out of 88 imprinted genes, 23 were differentially expressed in these populations. Of these 23, 15 were preferentially expressed in LT-HSCs, while the others were predominantly expressed in ST-HSCs and MPPs (Fig. 1c). Intriguingly, 80% of the imprinted genes with predominant expression in LT-HSCs were associated with growth restriction, including H19, Cdkn1c/p57, Ndn, Rb, Gtl2 and Grb109. In contrast, imprinted genes expressed preferentially in ST-HSCs and MPPs, including Ascl2, Peg12, Sfmbt2, Pon3, Atp10a, and Osbp15, were associated with growth-promotion and increased metabolism10. The expression of several representative genes was confirmed by qRTPCR (Supp.Fig. 1a).
Figure 1. Defective LT-HSC maintenance in mH19ΔDMR/+ mice.
a, H19-Igf2 cluster. Top image- red box, maternally-expressed; blue box, paternally-expressed; gray boxes, genes in the cluster. Bottom image- ○-unmethylated and ●-methylated CpG dinucleotides. X, no expression; arrow, active transcription. b, Upper cartoon- Hierarchical organization. Curved arrow, self-renewal. Lower image: FACS plot. Left panel: gated on Lineage−Sca-1+cKit+(LSK) or right panel: CD34 and FLk2. c, Heatmap of imprinted genes(n=3). d, FACS plot of mH19ΔDMR/+ (n=9) and mH19flDMRl/+(n=8. e, %LT-HSCs of LSK. Absolute numbers of f, HSCs and g, total BM cells. h–j, Cell cycle analysis 2W post-pIpC induction (n=5). k, 5FU cycles. l, Total BM cells(n=3). m, Cell cycle analysis. Error bars, s.e.m.
Given the critical role of H19 during embryonic development and its preferential expression in LT-HSCs, we hypothesized that H19 plays a role in restricting LT-HSC activation. To test this idea, we conditionally deleted H19-DMR (an epigenetic regulator that controls expression of H19) by breeding H19flDMR/flDMR mice with Mx1-Cre mice to generate maternal (mH19ΔDMR/+) and paternal (pH19ΔDMR/+) allele specific mutants (Supp.Fig. 1b). The DMR region was deleted with 100% efficiency in LT-HSC (Supp.Fig. 1c,e–g,)11. As early as 6 weeks, flow cytometric analysis revealed a substantial decrease in frequency and absolute number of LT-HSCs in mH19ΔDMR/+ mice (Fig. 1 d–f, Supp.Fig. 1d), but not in pH19ΔDMR/+ mice (Fig. 1e, Supp.Fig. 2a,b). Concurrently, we observed a significant increase in frequency and absolute number of ST-HSCs (Fig. 1d–f); however, the total number of BM cells remained unchanged (Fig. 1g). By 6 months, both LT- and ST-HSCs were significantly decreased in frequency and absolute number while BM cellularity increased only in mH19ΔDMR/+ mice (Fig. 1d–g and Supp.Fig. 2c,d).
Cell cycle analysis of LT-HSCs at 6 weeks post pIpC induction revealed a decreased in the G0 phase fraction and a concomitant increased in the G1 phase fraction in mH19ΔDMR/+ relative to control (Fig. 1h–j). We then tested the response of mH19ΔDMR/+ mice to BM damage by administering 5-fluorouracil (5FU), which eliminates active HSPCs while sparing quiescent HSCs. Surviving quiescent HSCs later replenish lost HSPCs12 (Fig. 1k). In this context, a significant reduction in quiescent HSCs after 3 cycles of 5FU treatment led to deficient BM recovery in the mH19ΔDMR/+ mutant compared to control (Fig. 1l, m and Supp.Fig. 2f). Altogether, maternal but not paternal deletion of H19-DMR resulted in loss of HSC quiescence, leading to progressive loss of LT-HSCs and then ST-HSCs, acommpanied by increasing progenior cell proliferation and differentiation, thus ultimately increasing total BM cellularity (Fig. 1g, Supp.Fig. 2e and Supp.Fig. 3a–d).
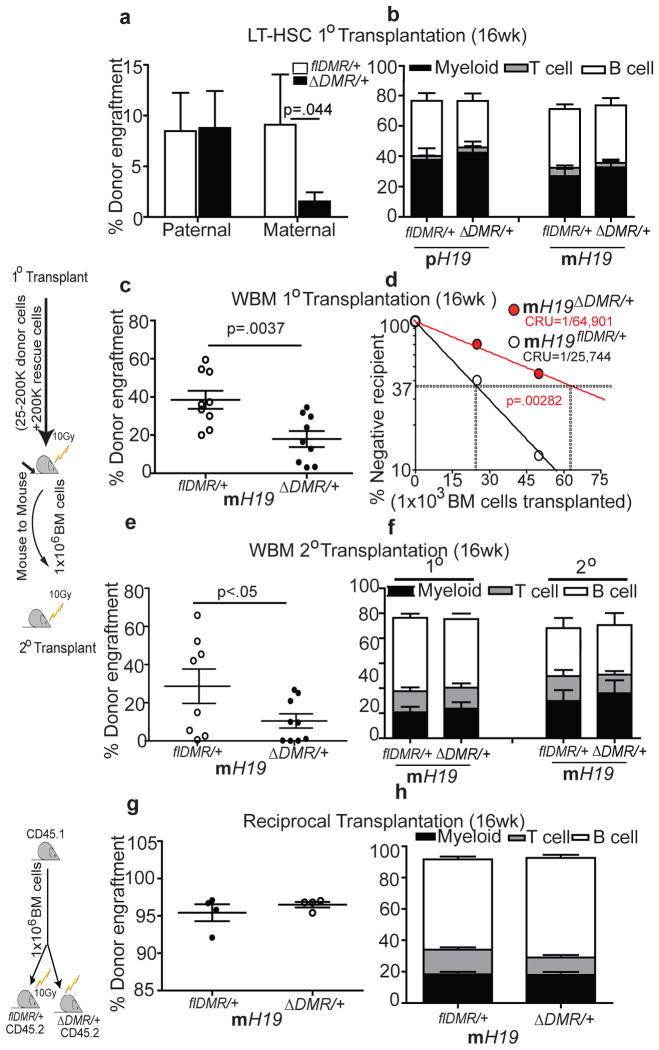
To functionally characterize the phenotype, we transplanted equal numbers of sorted LT-HSCs from mutants and their control littermates. We observed a significant reduction in reconstitution ability for LT-HSCs derived from mH19ΔDMR/+ but not pH19ΔDMR/+ mutants compared to controls. While overall engraftment was reduced in primary and secondary recipients, no mature lineage bias was apparent (Fig. 2a–f). Limiting dilution analysis to quantify functional HSCs revealed a 2.5-fold decrease in mH19ΔDMR/+ mutant HSCs relative to control (Fig. 2d). Reciprocal transplantation of Wt donor cells into either mH19ΔDMR/+ or control recipients did not result in alterations in hematopoiesis (Fig. 2g, h), suggesting that an intrinsic change in the mH19ΔDMR/+ mutant HSCs was the primary cause for the phenotype, although an environmental influence (such as an overall increase in Igf2 expression) may have enhanced the phenotype.
Figure 2. Compromised HSC function in mH19ΔDMR/+ mice.
Competitive repopulation assay 16W post transplantation a, sorted 100 LT-HSCs, n=10 c, primary transplant (test dosage of 2×105 BM cells), n=10 e, secondary transplant, n=10) g, reciprocal transplant, n=4. d, Competitive repopulation unit (CRU) content within each group of mice transplanted at each dose; (n = 60 mice total). Horizontal dashed line, 37% of recipient mice failed to engraft; vertical dashed lines, various CRU frequencies for each condition. Donor –derived lineage analysis after b, primary transplant f, secondary transplant h, reciprocal transplant. Error bars, s.e.m.
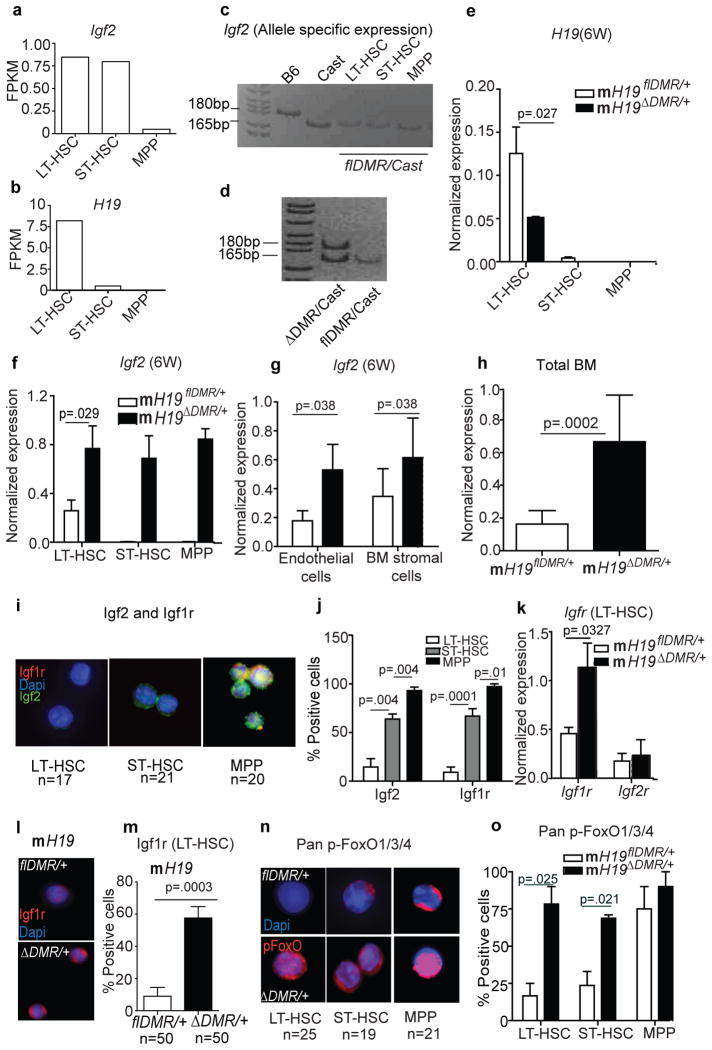
Next, we investigated whether H19-DMR controls the imprinted expression of H19 and Igf2 from the maternal and paternal alleles, respectively, in adult HSCs, as is observed in embryos11. Our RNA-seq analysis revealed differential expression of H19 as well as Igf2 in HSCs (Fig. 3a,b). By crossing H19flDMR/+ females with Mus castaneus (Cast) males, which enables parental allele-discrimination by SNP analysis, we further detected exclusive expression of Igf2 from the paternal allele in HSCs (Fig. 3c). However, after deletion of the maternal H19-DMR, we detected H19 down-regulation and Igf2 up-regulation, which resulted from biallelic Igf2 expression in HSCs (Fig. 3d–f). Igf2 was similarly up-regulated in BM, including surrounding stromal cells, after maternal deletion of H19-DMR (Fig. 3g,h). However, as revealed by reciprocal transplantation, an extrinsic increase of Igf2 expression alone is not sufficient to cause the mH19ΔDMR/+ HSC phenotype. We next investigated whether the Igf2 signaling13 pathway is activated in mH19ΔDMR/+ LT-HSCs. Binding of Igf2 to Igf1r activates signaling, whereas Igf2 binding to Igf2r attenuates signaling14. Igf2 and Igf1r levels were gradually increased from LT-HSCs to MPPs (Fig. 3i,j). However, mRNA, protein levels, and number of Igf1r+ cells was significantly increased in mH19ΔDMR/+ LT-HSCs compared to controls (Fig. 3k,l,m), with no change in Igf2r expression (Fig. 3k). Igf2-Igf1r signaling is known to activate PI3K-Akt, which phosphorylates and inactivates FoxO3, a transcription factor that arrests the cell cycle15,16. Inactive pFoxO3 was detected in only 15% of normal LT-HSCs but was substantially increased in ST-HSCs and MPPs (Fig. 3n,o); however in mH19ΔDMR/+ pFoxO3 was detected in 75% of LT-HSCs. Our data indicate that H19-DMR deletion increased Igf2 signaling, which released FoxO3-mediated suppression of HSC activation and proliferation.
Figure 3. Activation of Igf2-FoxO3 signaling in mH19ΔDMR/+ mice.
a, b, RNA-seq analysis of Igf2 and H19 transcripts. c,d, Allele-specific expression of the Igf2 transcript. qRT-PCR 6W post pIpC induction (n=3) for e, H19 f, and Igf2 g, stromal cells and h, total BM (n=5). i, j, Single-cell Igf1r and Igf2 staining from Wt BM cells and its quantitation (n=4). k, Igf1r and Igf2r expression in sorted LT-HSCs. l,m, Igf1r immune-staining and its quantitation (n=4). n,o, Single-cell phospho-FoxO1/3/4 staining and its quantitation (n=4). FPKM, fragments per kilobase exon per million fragments mapped. Scale bar 10μm. Error bars, s.e.m.
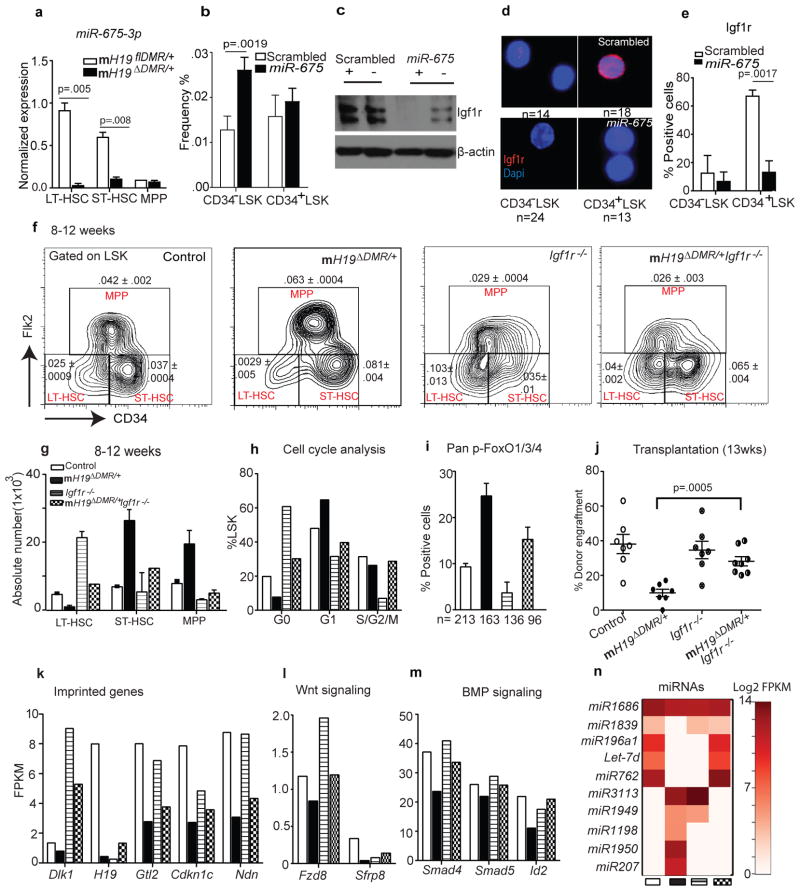
In the placenta, H19 functions as a precursor of miR-675, which in turn suppresses Igf1r6. We next investigated whether this regulation exists in adult HSCs. Expression of miR-675 was highest in LT-HSCs in the control mice but was substantially reduced in mH19ΔDMR/+ LT-HSCs (Fig. 4a). To explore the potential role of miR-675 in Igf1r regulation, we transplanted BM cells over-expressing miR-675 into Wt mice. Over-expression of miR-675 increased the percentage of quiescent CD34−LSK cells but did not significantly affect active CD34+ LSK cells (Fig. 4b). Western blot analysis showed a significant reduction of Igf1r by miR-675 compared to control (Fig. 4c). Furthermore, Igf1r levels were lower in CD34−LSK compared to CD34+LSK cells in the control. However, miR-675 overexpression significantly decreased Igf1r (Fig. 4c–e). These data demonstrate that H19-derived miR-675 regulates Igf1r and the corresponding quiescent state in HSCs.
Figure 4. Igf1r regulation and rescue by genetic blockage of Igf2-Igf1r signaling.
a, miR-675 analysis by qRT-PCR (n=3). b, Frequency of CD34− vs. CD34+ LSK cells 8W post-lentiviral infection. c, Immunoblot analysis of Igf1r in sorted BM cells (positive and negative for GFP). d–e, Single-cell immunostaining of Igf1r 8W post-lentiviral infection and its quantitation (n=4). f, Representative FACS plot with frequency (n=4) g, absolute number and h, cell-cycle analysis. i, Pan-FoxO staining (n=3). j, Donor engraftment 12W post transplantation. RNA sequencing analysis in sorted stem cells. k, imprinted genes, l–m, Wnt and BMP signaling n, heatmap of miRNA expression. Error bars s.e.m.
To further confirm that H19-DMR controls Igf2-Igf1r signaling, we crossed female H19flDMR/+ mice with male Mx1-Cre:Igf1rfl/fl mice (Supp.Fig. 4a)17. While mH19 DMR/+ mutants (Fig. 3f,g) showed a decrease in LT-HSCs and an increase in ST-HSCs and MPPs, Igf1r−/− mutants showed an increase in LT-HSCs and a decrease in MPPs. This suggests that Igf1r regulates the transitions from LT-HSCs to ST-HSCs and further to MPPs. Interestingly, mH19 DMR/+Igf1r−/− double mutants showed a partial restoration of LT-HSC frequency (Fig. 4f), while the transition from ST-HSCs to MPPs was still blocked (Fig. 4f–g). This suggests that Igf2-Igf1r signaling is partially responsible for the mH19 DMR/+ phenotype. An increase in ST-HSC frequency in double mutant mice was likely due to blocked transition from ST-HSC to MPP by down-regulation of Igf1r. Furthermore, cell cycle analysis and pFoxO3 staining in double mutants revealed partial rescue of the loss of quiescence phenotype (Fig. 4h,i).
To functionally demonstrate phenotypic rescue in mH19 DMR/+Igf1r−/− mice, we performed BM transplantation assays. While mH19 DMR/+ mutants had significantly reduced engraftment due to LT-HSC loss, engraftment of mH19 DMR/+Igf1r−/− BM cells increased to a level between that of the mH19 DMR/+ and the Igf1r−/− single mutants, indicating a partial functional rescue (Fig. 4j, Supp.Fig. 4b). These results indicate that the maternal H19-DMR controls Igf2-Igf1r signaling which regulates HSC state; however, the partial rescue suggests that deletion of H19-DMR also affects other pathways required for LT-HSC maintenance. To investigate this possibility, we performed RNA-seq analysis of HSCs isolated from control, mH19ΔDMR single, Igf1r−/− single, and mH19ΔDMR/+:Igf1r−/− double mutants. mH19ΔDMR/+ HSPCs showed widespread alterations in expression of imprinted genes in all three populations (Supp.Fig. 4c, and Supp.Fig. 5). Genes involved in cell cycle arrest (Cdkn1c)18,19, tumor suppression and stem cell maintenance (Ndn, Gtl2)20,21 were down-regulated in LT-HSCs. However, Igf1r−/− LT-HSCs largely maintained expression patterns similar to control, with high expression levels of Cdkn1c, Ndn, Gtl2 and Dlk1. The double mutants generally showed partial rescue of the alterations observed in single mutants, suggesting either compensation in gene expression and/or existence of a proposed imprinted gene network22,23 (Fig. 4k, Supp.Fig. 4c). Gene expression profiling of non-imprinted genes showed many overlapping downstream genes and miRNAs that were abnormally expressed in single mutants but partially rescued in double mutants (Fig. 4l–n, Supp.Fig. 6a–c and Supp.Fig. 7e). These included components of the Wnt and Tgfβ/BMP pathways such as Smad4, Id2, and Fzd824–27 as well as Let-7, which is known to repress cell proliferation28 and Igf signaling29 (Fig. 4l–n). Interestingly, H19-DMR potentially controls other miRNAs, SnoRNAs, and genes, such as Dusp26, p2rx2, and Gpr63, independent of the Igf2-Igf1r pathway (Supp.Fig. 7a–f). Dysregulation of some of these genes is known to cause cellular hyperproliferation30,31. Taken together, our data reveal that maternal H19-DMR primarily restricts Igf2-Igf1r signaling but also influences other imprinted and non-imprinted genes as well as miRNAs involved in maintaining HSC quiescence.
By studying the H19-DMR locus in an allele-specific manner, we demonstrate that a specialized form of epigenetic control—genomic imprinting—is critical to the maintenance of adult stem cells. This is accomplished by maintaining LT-HSC quiescence, which can be attributed largely to Igf2-Igf1r dependent signaling, but also to additional Igf2-Igf1r independent effects on the regulation of cell cycle, proliferation, and growth.
Full Methods
Animals
All mice used in this study were housed in the animal facility at the Stowers Institute for Medical Research (SIMR) and handled according to SIMR and National Institutes of Health (NIH) guidelines. All procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of SIMR. H19-DMR flDMR/flDMR mice on B6 background were kindly provided by Dr. Marisa S. Bartolomei (University of Pennsylvania Perelman School of Medicine)31. Conditional mutant Igf1rfl/+ was kindly provided by Thomas L Clemens (Center for Musculoskeletal Research, Johns Hopkins Medicine, 601 North Caroline Street, Baltimore, MD 21287)32. Interferon inducible Mx1-Cre or tamoxifen inducible Scl-Cre mouse strains were used to delete the floxed H19-DMR and Igf1r. For Mx1-Cre activation, 250μg of pIpC was injected intraperitoneal every other day for 14 days at 5 weeks of age. For Scl-CreER activation, 2mg tamoxifen dissolved in 0.1 ml of corn oil was injected intraperitoneal every day for 5 days.
Single-cell HSC Genotyping
Single CD34− Flk2− LSK cells were sorted into 96 well plates (1 cell/well) containing 50μl MethoCult® complete media (M3434; Stem Cell Technologies, Vancouver, Canada) and incubated (37°C, 5% CO2) for 12 days. Individual colonies were harvested separately, and DNA was purified using QIAGENamp DNA Blood Kit (QIAGEN, Valencia, CA). PCR genotyping of H19 locus was performed using G1, G5 and G7 primers described elsewhere31.
Flow cytometry
Phenotypic analysis of hematopoietic cells harvested from BM (femur and tibia) and peripheral blood were performed as described previously33,34. Cell sorting and analysis were performed with a MoFlo (Dako) and/or CyAn ADP (Dako). Data analysis was performed with FlowJo software (Ashland).
5 Fluorouracil treatment
Cohorts of mH19ΔDMR/+ and mH19flDMR/+ were injected with 5FU (Sigma-Aldrich) at 150 μ/g body weight (BW)34 4 weeks post pIpC induction. For one cycle, 5FU was injected once intravenously, and for 3 cycles, 5FU was injected 3 times at 4-week intervals. BM cells were analyzed 10 days after 5FU injections.
Cell cycle analysis
Cell cycle analysis of BM Lineage− Sca-1+cKit+ (LSKs) was performed. BM cells (5 × 106) were stained for LSKs, fixed overnight at 4°C in 4% para-formaldehyde, and permeabilized with 0.2% triton x-100. Cells were further stained with BD PharmingenTM FITC conjugated-Mouse Anti-Human Ki67 Set (BD Pharmingen) according to manufacturer’s instruction and 0.1ug/uL DAPI. This was followed by flow cytometric analysis with InFlux Cell Sorter (BD Biosciences).
Transplantation studies
For competitive repopulation assays, 2 × 105 BM cells congenic with the host (CD45.1+) were included per mouse. One-hundred sorted LT-HSCs from mH19ΔDMR/+ or mH19flDMR/+were transplanted intravenously into lethally irradiated (10 Gy) Ptprc (CD45.1) recipient mice. Mice were placed on Baytril water 3 days prior to irradiation, which continued for 2 weeks post irradiation. Each transplanted group consisted of 8–10 recipients. Donor-derived engraftment was assayed every 4 weeks post transplant by collection of peripheral blood, RBC lysis and staining of CD45.1 (recipient) vs. CD45.2 (donor). Multi-lineage reconstitution was determined by CD3, B220 (for T and B lymphoid, respectively) and Gr1, Mac-1 (for myeloid) gating on donor (CD45.2+) cells. Limiting dilution experiments were performed by transplanting 3 doses (200K, 100K and 25K) of test samples (n=2) from mH19ΔDMR/+ or mH19flDMR/+(CD45.2) along with fixed number of 2×105 rescue cells (CD45.2) into groups of 10 lethally irradiated (10gy) recipient mice (CD45.1). CRU frequency was determined with L-Calc software (Stem Cell Technologies, Inc.) based on Poisson statistics35. The plot was made based on percentage of recipient mice containing 1% CD45.2+ cells in the peripheral blood at 16 weeks post transplant versus the number of cells injected per mouse. For secondary transplantation, the original, primary transplant recipients were sacrificed; BM was harvested from the femur and then transplanted mouse-to-mouse at a dosage of 1×106 cells per mouse. For reciprocal transplantation, wild-type congenic B6.SJL (CD45.1+) BM cells (1 × 106 cells per recipient) were transplanted into lethally irradiated Mx-1 Cre induced mH19flDMR/+ and mH19ΔDMR/+ (CD45.2+) recipients. Complete donor cell engraftment by wild-type CD45.1+ cells was confirmed by flow cytometric analysis. For rescue transplants 2 × 105 (CD45.2) BM cells from the mH19flDMR/+, Igf1r−/− and mH19ΔDMR/+Igf1r−/− mutants and the controls along with 2 × 105 (CD45.1+) Ptprc BM cells were transplanted into CD45.1 recipient mice. Complete donor cell engraftment by wild-type CD45.1+ cells was confirmed by flow cytometric analysis.
Lentivirus infection
Mice were treated with 150 μg/g body weight of 5FU to activate and enrich for HSPCs35. 4 days later, BM was harvested and cultured overnight in ST media and transduced by Magnetofection™ using ViroMag R/L particles according to the manufacturer’s protocol (OZ Biosciences). Transplantation experiments conducted in the knockdown model were done with unsorted 300,000 infected BM cells (CD45.2). The cells were transplanted into each lethally irradiated (10Gy) Ptprc (CD45.1). 8 weeks post engraftment, BM cells double positive for GFP and CD45.2 were sorted for CD34−LSK and CD34+LSK.
Lentivirus construction
The pSicoR-EF1α promoter-IRES-EGFP lentiviral construct was kindly provided by T. Xie (SIMR).
mir-675_Fwd AGCGTGCGGCCCAGGGACTGGTGCGGAAAGGGCCCACAGTGGACTTGGTACACTGTATGCCCTAACCGCTCAGTCCCTGGGTCTGGCA
mir-675_Rev GGCATGCCAGACCCAGGGACTGAGCGGTTAGGGCATACAGTGTACCAAGTCCACTGTGGGCCC TTTCCGCACCAGTCCCTGGGCCGCA
IGF2 shRNA
FWD AGCGCGCCCAAATTTGATTGGCTCTAAATAGTGAAGCCACAGATGTATTTAG AGCCAATCAAATTTGGTCA
REV GGCATGACCAAATTTGATTGGCTCTAAATACATCTGTGGCTTCACTATTTAGA GCCAATCAAATTTGGGCG
Allele specific Igf2 expression
Male Castaneous mice (Cast) were crossed with female flDMR or ΔDMR. Heterozygous progeny at SNPs differ between the two strains. In mice that inherited Cast allele paternally and flDMR maternally, LT-HSCs, ST-HSCs and MPPs were sorted from total BM cells. RNA was extracted and DNase treated using RQ1 RNase-Free DNase per manufacturer’s instruction (Promega). This RNA was reversed transcribed in the presence of SuperScriptIII Reverse Transcriptase (SSIII) using Igf2 specific primer Igf2-20r (5′-gggttgtttagagccaatcaa-3′) per manufacturer’s instructions (Invitrogen); simultaneously, equal concentrations of RNA were identically treated in the absence of SSIII (for minus (-) RT). Equal volumes of RT and –RT were amplified using 0.5 μM of primers Igf2-18f (5′atctgtgacctcctcttgagcagg-3′) and Igf2-20r and Go-Taq Green Master Mix (Promega) using the following PCR conditions: 94°C 2min 1 cycle; 94°C 15 sec, 58°C 15 sec and 72°C 20 sec for 43 cycles. No product was detected in –RT samples. Amplified Igf2 was digested with MluCI (NEB) and the paternal Cast product (165 bp) and the maternal B6 product (180 bp) products were resolved on a 15% polyacrylamide gel similar to methods described earlier31.
Microarray
RNA was extracted by conventional trizol method from sorted LT-HSCs, ST-HSCs and MPPs (10,000 cells each)36. Samples were analyzed with Affymetrix MouseGenome430_2 arrays and scanned with a GeneChip Scanner 3000 7G using GeneChip Fluidics Station 450 and GeneChip Operating Software (GCOS 1.4). Heatmap data represent the fold change between 2 populations from at least 3 independent biological samples. Three samples were run on Affymetrix Mouse 430.2 arrays in triplicate, using the standard Affymetrix protocols. CEL files were read into the R software environment http://www.cran.r-project.org/ and normalized with RMA37–39. Normalized data were fit with a linear model using the limma package40 and three contrasts were fit: CD34p/CD34n, FLK2p/CD34p, FLK2p/CD34n. Probes that were significant for at least one contrast (BH adj p <= 0.05) went to further analysis.
A list of imprinted genes was taken from the catalogue of imprinting genes at http://igc.otago.ac.nz/1101Summary-table.pdf. Names were matched to MGI and Ensembl 63 genes and then converted to probeset ids. Of 125 input genes, 86 could be mapped to probesets, and of these, 23 were significant. Sample expression coefficients per probeset were averaged together by gene. Expression levels varied widely, from 4–14 in log2 scale, which obscured the regulatory trend across samples during clustering. We constructed a heatmap to show only the trending of expression, and not the magnitude, by subtracting the mean from each row and dividing by the standard deviation. Thus the heatmap scale shows expression z-scores. Row ordering reflects hierarchical clustering, average linkage, using Pearson dissimilarity for distance. Microarray data are submitted to ArrayExpress with accession number E-MTAB-1644.
qRT-PCR
Total RNA (2–50ng) was extracted from sorted LT-HSCs, ST-HSCs and MPPs directly into trizol. This was followed by DNAse I treatment (Ambion) and reverse transcription with a high capacity cDNA reverse transcription kit (Applied Biosystems). cDNA was pre-amplified by TaqMan® PreAmplification master mix (Applied Biosystems) according to the manufacturer’s instruction. TaqMan® gene expression assays (Applied Biosystems) were performed on triplicate samples with a 7500 real-time cycler (Applied Biosystems). Data were normalized relative to Gapdh and Hprt1. For mi-R-675-3p assay, extracted RNA were reverse transcribed using TaqMan miRNA reverse transcription kit (Applied Biosystems). TaqMan pre-amplification and TaqMan gene expression assay were performed per manufacturer’s instruction. All the qRT-PCR was performed using TaqMan probes.
Immunostaining
Immunostaining was performed as described previously33. For immunostaining of sorted cells, cells were sorted onto lysine-coated slides, fixed with chilled methanol for 10 minutes, followed by blocking and staining with primary antibody41. The following primary antibodies were used: Chicken anti Igf1r (Abcam, 1:100), Rabbit anti Igf2 (Abcam, 1:100), Rabbit anti FoxO3a (1:100), Rabbit anti Foxo1/3/4-Pan and phosphor_Thr24/32 (Assay biotech, 1:50).
RNA-seq
The RNA-sequencing library was prepared from approximately 200 ng of total RNA (mH19ΔDMR/+ Igf1r−/−, mH19ΔDMR/+Igf1rf−/− and mH19flDMR/+) for each sample using illumina TruSeq RNA Sample Prep Kit (Catalog #: FC-122-1001). The fragment size in the generated library ranged from 220 to 500 bps with a pick at 280 bps. A total of 10 fmol library fragments were loaded to cBot to generate clusters, followed by sequencing on an Illumina HiSeq 2000 to produce 10–30 million paired-end 100 bp reads per sample. Reads were trimmed to 70bp due to quality and aligned to mm9 with Tophat 1.3.1 42 / Bowtie 0.12.743, using the Ensembl 63 GTF file for gene models. Parameters were -g 1 --mate-inner-dist 200 --mate-std-dev 70 --segment-length 35 --segment-mismatches 2; this allows for 4 mismatches per read (two per read half) and unique alignments only.
Gene expression was quantitated using Cufflinks 1.0.344. We chose any imprinted genes with an absolute log-fold-change of 1.3. A total of 38 imprinted genes were selected this way, 32 having measurements on both samples and 6 having measurements in only one sample. We heatmapped the genes found in both samples using FPKMs only. For the genes found in both, the range of expression was skewed enough to make visualization by heatmap difficult, so we created a row-normalized heatmap as with the microarray data. RNA seq data set were submitted to Array Express with accession number E-MTAB-1628.
Supplementary Material
Acknowledgments
We thank M. Hembree, T. Johnson, H. Marshall, B. Lewis, D. Dukes, C. Semerad, J. Park, and A. Box for technical support; and members of the Li laboratory for scientific discussion. We thank J. Lu and Y. Huang for communications regarding H19, miR-675, and Let-7. We thank K. Tannen for editing and proofreading. This work is supported by Stowers Institute for Medical Research and by the Department of Biotechnology, Ministry of Science and Technology, Govt. of India in the form of an overseas associateship to A.Venkatraman. M. Bartolomei is supported by NIH (GM51279).
Footnotes
Author Contributions
A.V. designed and performed experiments, analyzed data, and wrote the manuscript. X.H. provided training, performed transplantations and RNA seq. F.T., J.T., M.C., L.P., X.Z., A.P., H.L, J.P. M.Z., and J.H. performed part of experiments. M.B. and T.C. contributed the mouse lines. L.L. directed the overall project and co-wrote the manuscript. All authors contributed to critical reading and editing the manuscript.
References
- 1.Bartolomei MS. Genomic imprinting: employing and avoiding epigenetic processes. Genes Dev. 2009;23:2124–2133. doi: 10.1101/gad.1841409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berg JS, et al. Imprinted genes that regulate early Mammalian growth are coexpressed in somatic stem cells. PLoS One. 2011;6:e26410. doi: 10.1371/journal.pone.0026410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferron SR, et al. Postnatal loss of Dlk1 imprinting in stem cells and niche astrocytes regulates neurogenesis. Nature. 2011;475:381–385. doi: 10.1038/nature10229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zacharek SJ, et al. Lung stem cell self-renewal relies on BMI1-dependent control of expression at imprinted loci. Cell Stem Cell. 2011;9:272–281. doi: 10.1016/j.stem.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeChiara TM, Robertson EJ, Efstratiadis A. Parental imprinting of the mouse insulin-like growth factor II gene. Cell. 1991;64:849–859. doi: 10.1016/0092-8674(91)90513-x. [DOI] [PubMed] [Google Scholar]
- 6.Keniry A, et al. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nature cell biology. 2012;14:659–665. doi: 10.1038/ncb2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haug JS, et al. N-cadherin expression level distinguishes reserved versus primed states of hematopoietic stem cells. Cell Stem Cell. 2008;2:367–379. doi: 10.1016/j.stem.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 8.Yang L, et al. Identification of Lin(-)Sca1(+)kit(+)CD34(+)Flt3- short-term hematopoietic stem cells capable of rapidly reconstituting and rescuing myeloablated transplant recipients. Blood. 2005;105:2717–2723. doi: 10.1182/blood-2004-06-2159. [DOI] [PubMed] [Google Scholar]
- 9.Frost JM, Moore GE. The importance of imprinting in the human placenta. PLoS Genet. 2010;6:e1001015. doi: 10.1371/journal.pgen.1001015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hudson QJ, Kulinski TM, Huetter SP, Barlow DP. Genomic imprinting mechanisms in embryonic and extraembryonic mouse tissues. Heredity (Edinb) 2010;105:45–56. doi: 10.1038/hdy.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thorvaldsen JL, Fedoriw AM, Nguyen S, Bartolomei MS. Developmental profile of H19 differentially methylated domain (DMD) deletion alleles reveals multiple roles of the DMD in regulating allelic expression and DNA methylation at the imprinted H19/Igf2 locus. Mol Cell Biol. 2006;26:1245–1258. doi: 10.1128/MCB.26.4.1245-1258.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lerner C, Harrison DE. 5-Fluorouracil spares hemopoietic stem cells responsible for long-term repopulation. Exp Hematol. 1990;18:114–118. [PubMed] [Google Scholar]
- 13.Smith FM, Garfield AS, Ward A. Regulation of growth and metabolism by imprinted genes. Cytogenet Genome Res. 2006;113:279–291. doi: 10.1159/000090843. [DOI] [PubMed] [Google Scholar]
- 14.Kang HM, Park S, Kim H. Insulin-like growth factor 2 enhances insulinogenic differentiation of human eyelid adipose stem cells via the insulin receptor. Cell Prolif. 2011;44:254–263. doi: 10.1111/j.1365-2184.2011.00755.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J, et al. PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature. 2006;441:518–522. doi: 10.1038/nature04747. [DOI] [PubMed] [Google Scholar]
- 16.Tothova Z, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128:325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 17.Klinakis A, et al. Igf1r as a therapeutic target in a mouse model of basal-like breast cancer. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:2359–2364. doi: 10.1073/pnas.0810221106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zou P, et al. p57(Kip2) and p27(Kip1) cooperate to maintain hematopoietic stem cell quiescence through interactions with Hsc70. Cell stem cell. 2011;9:247–261. doi: 10.1016/j.stem.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 19.Walkley CR, Shea JM, Sims NA, Purton LE, Orkin SH. Rb Regulates Interactions between Hematopoietic Stem Cells and Their Bone Marrow Microenvironment. Cell. 2007;129:1081–1095. doi: 10.1016/j.cell.2007.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kubota Y, Osawa M, Jakt LM, Yoshikawa K, Nishikawa S. Necdin restricts proliferation of hematopoietic stem cells during hematopoietic regeneration. Blood. 2009;114:4383–4392. doi: 10.1182/blood-2009-07-230292. [DOI] [PubMed] [Google Scholar]
- 21.Stadtfeld M, et al. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao Z, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- 23.Varrault A, et al. Zac1 regulates an imprinted gene network critically involved in the control of embryonic growth. Dev Cell. 2006;11:711–722. doi: 10.1016/j.devcel.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 24.Karlsson G, et al. Smad4 is critical for self-renewal of hematopoietic stem cells. The Journal of experimental medicine. 2007;204:467–474. doi: 10.1084/jem.20060465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sugimura R, et al. Noncanonical wnt signaling maintains hematopoietic stem cells in the niche. Cell. 2012;150:351–365. doi: 10.1016/j.cell.2012.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perry JM, et al. Cooperation between both Wnt/{beta}-catenin and PTEN/PI3K/Akt signaling promotes primitive hematopoietic stem cell self-renewal and expansion. Genes Dev. 2011;25:1928–1942. doi: 10.1101/gad.17421911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dijke P, Heldin CH. Smad Signal Transduction: Smads in Proliferation, Differentiation and Disease. Springer London, Limited; 2006. [Google Scholar]
- 28.Johnson CD, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer research. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 29.Toledano H, D’Alterio C, Czech B, Levine E, Jones DL. The let-7-Imp axis regulates ageing of the Drosophila testis stem-cell niche. Nature. 2012;485:605–610. doi: 10.1038/nature11061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu W, et al. A novel amplification target, DUSP26, promotes anaplastic thyroid cancer cell growth by inhibiting p38 MAPK activity. Oncogene. 2007;26:1178–1187. doi: 10.1038/sj.onc.1209899. [DOI] [PubMed] [Google Scholar]
- 31.Niedernberg A, Tunaru S, Blaukat A, Ardati A, Kostenis E. Sphingosine 1-phosphate and dioleoylphosphatidic acid are low affinity agonists for the orphan receptor GPR63. Cell Signal. 2003;15:435–446. doi: 10.1016/s0898-6568(02)00119-5. [DOI] [PubMed] [Google Scholar]
- 31.Thorvaldsen JL, Fedoriw AM, Nguyen S, Bartolomei MS. Developmental profile of H19 differentially methylated domain (DMD) deletion alleles reveals multiple roles of the DMD in regulating allelic expression and DNA methylation at the imprinted H19/Igf2 locus. Molecular and cellular biology. 2006;26:1245–1258. doi: 10.1128/MCB.26.4.1245-1258.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dietrich P, Dragatsis I, Xuan S, Zeitlin S, Efstratiadis A. Conditional mutagenesis in mice with heat shock promoter-driven cre transgenes. Mamm Genome. 2000;11:196–205. doi: 10.1007/s003350010037. [DOI] [PubMed] [Google Scholar]
- 33.Zhang J, et al. Identification of the haematopoietic stem cell niche and control of the niche size. Nature. 2003;425:836–841. doi: 10.1038/nature02041. [DOI] [PubMed] [Google Scholar]
- 34.Haug JS, et al. N-cadherin expression level distinguishes reserved versus primed states of hematopoietic stem cells. Cell stem cell. 2008;2:367–379. doi: 10.1016/j.stem.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 35.Miller CL, Eaves CJ. Expansion in vitro of adult murine hematopoietic stem cells with transplantable lympho-myeloid reconstituting ability. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:13648–13653. doi: 10.1073/pnas.94.25.13648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akashi K, et al. Transcriptional accessibility for genes of multiple tissues and hematopoietic lineages is hierarchically controlled during early hematopoiesis. Blood. 2003;101:383–389. doi: 10.1182/blood-2002-06-1780. [DOI] [PubMed] [Google Scholar]
- 37.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 38.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 39.Irizarry RA, et al. Summaries of Affymetrix GeneChip probe level data. Nucleic acids research. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article 3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 41.Ema H, et al. Adult mouse hematopoietic stem cells: purification and single-cell assays. Nature protocols. 2006;1:2979–2987. doi: 10.1038/nprot.2006.447. [DOI] [PubMed] [Google Scholar]
- 42.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.