Abstract
Streptomyces species produce many clinically relevant secondary metabolites and exhibit a complex development that includes hyphal differentiation and sporulation in solid cultures. Industrial fermentations are usually performed in liquid cultures, conditions in which Streptomyces strains generally do not sporulate, and it was traditionally assumed that no differentiation took place. The aim of this work was to compare the transcriptomes of S. coelicolor growing in liquid and solid cultures, deepening the knowledge of Streptomyces differentiation. Microarrays demonstrated that gene expression in liquid and solid cultures were comparable and data indicated that physiological differentiation was similar for both conditions. Eighty-six percent of all transcripts showed similar abundances in liquid and solid cultures, such as those involved in the biosynthesis of actinorhodin (actVA, actII-4) and undecylprodigiosin (redF); activation of secondary metabolism (absR1, ndsA); genes regulating hydrophobic cover formation (aerial mycelium) (bldB, bldC, bldM, bldN, sapA, chpC, chpD, chpE, chpH, ramA, ramC, ramS); and even some genes regulating early stages of sporulation (wblA, whiG, whiH, whiJ). The two most important differences between transcriptomes from liquid and solid cultures were: first, genes related to secondary metabolite biosynthesis (CDA, CPK, coelichelin, desferrioxamine clusters) were highly up-regulated in liquid but not in solid cultures; and second, genes involved in the final stages of hydrophobic cover/spore maturation (chpF, rdlA, whiE, sfr) were up-regulated in solid but not in liquid cultures. New information was also provided for several non-characterized genes differentially expressed in liquid and solid cultures which might be regulating, at least in part, the metabolic and developmental differences observed between liquid and solid cultures.
Introduction
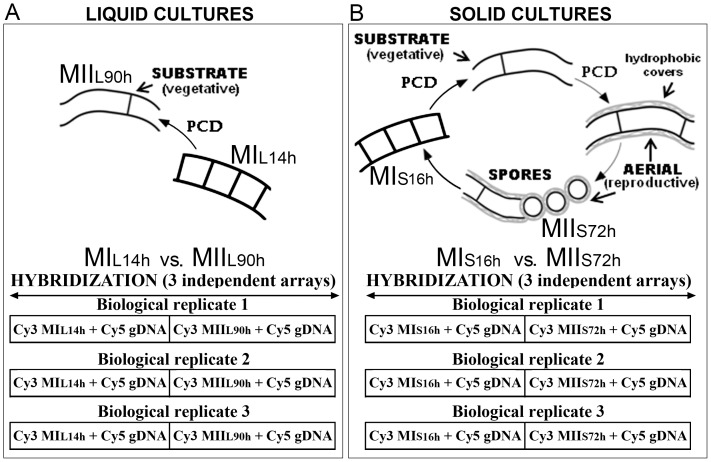
Approximately two-thirds of the clinical antibiotics, as well as a large number of eukaryotic cell differentiation inducers and inhibitors are synthesized by members of the Streptomyces genus [1]–[4]. Streptomycetes undergo a complex developmental cycle, which includes sporulation in solid cultures. Industrial processes for secondary metabolite production are performed in liquid cultures (large bioreactors), conditions in which most streptomycetes do not sporulate and it was generally assumed that differentiation processes were absent under these conditions [5]–[9]. During the last few years, new insights concerning Streptomyces differentiation during pre-sporulation stages were discovered in solid and liquid cultures. After spore germination, a compartmentalized mycelium (MI) initiates development and the MI compartments are separated by membranous septa which generally do not display thick cell walls (reviewed in [10]). A fraction of MI cells undergo a highly-ordered programmed cell death (PCD) [11] and the remaining viable cells differentiate into a multinucleated mycelium with sporadic septa (MII). It is the MII stage which produces antibiotics and sporulates on solid culture medium ([8], outlined in Fig. 1).
Figure 1. Streptomyces coelicolor development stages and sample preparation.
(A) Liquid non-sporulating cultures. (B) Solid sporulating cultures. Mycelial structures (MI, first compartmentalized mycelium; MII, second multinucleated mycelium). The classical nomenclature of substrate, aerial mycelium, and hydrophobic layers are indicated. Three independent biological replicates and two developmental stages (MI and MII) were processed and cDNA was labeled with Cy3 while chromosomal S. coelicolor DNA was used as reference and labeled with Cy5. The scheme was adapted from Manteca et al. [12].
Proteomic analyses demonstrate that differentiation in liquid non-sporulating cultures was more similar to sporulating cultures on solid medium than expected in the context of the classical Streptomyces developmental model focusing on hydrophobic cover formation and sporulation stages [12]. This work further extends upon studies analyzing gene expression during development.
There are several Streptomyces transcriptomic studies describing genetic expression at different developmental time points in liquid cultures [13], [14], at particular time points in solid cultures [15], [16], and comparing Streptomyces mutants with the wild type strain in liquid [17] or solid [18], [19] cultures. However, in our knowledge this study is the first to specifically compare differences between S. coelicolor liquid and solid cultures. Knowledge about the genes differentially expressed in liquid and solid cultures will contribute to understanding the biochemical pathways regulating pre-sporulation developmental stages in Streptomyces and the activation of secondary metabolism.
Materials and Methods
2.1. Bacterial Strains and Media
Streptomyces coelicolor M145 was used in this study. In order to facilitate data comparison with previous morphological [8] and proteomic [12] studies, the same culture conditions from those works were used: liquid cultures were performed in sucrose free R5A liquid media where 20 ml of culture medium were inoculated directly with spores (1×107 spores per ml) into flasks of 100 ml and incubated at 200 rpm and 30°C.
2.2. Sampling of Streptomyces coelicolor Cells throughout the Differentiation Cycle
S. coelicolor grown in liquid culture were processed at different developmental time points (14 and 90 hours) (outlined in Fig. 1). The 14-hour time point corresponded to the first compartmentalized mycelium (MIL) and 90 hours to the second multinucleated mycelium (MIIL). Three independent cultures were prepared and processed (biological replicates). Samples (20 ml from 14 h culture, 2 ml from 90 h culture) were centrifuged at 1000 g (5 minutes at 2°C) and the cells preserved in RNA Protect (Qiagen®) at −80°C.
2.3. RNA Isolation and Microarray Hybridization
Total RNA was isolated from 3 biological replicates using phenol extraction and the RNeasy Midi Kit (Qiagen). RNA integrity was verified using the 2100 Bioanalyzer (Agilent). cDNA samples were synthesized and labeled using random hexamers, SuperScript III reverse transcriptase (Invitrogen), and Cy3-dCTP (GE Healthcare Life Sciences). Remnant RNA was hydrolyzed with NaOH and retrotranscription products were purified using the MinElute PCR Purification Kit (Qiagen). Genomic DNA from S. coelicolor M145 was used as the common reference. gDNA was labeled with Cy5-dCTP (GE Healthcare Life Sciences) using the BioPrime Array CGH Genomic Labeling Module (Invitrogen), purified with the MinElute kit, and labeling efficiencies quantified with a NanoDrop ND-1000 spectrophotometer. Mixtures of Cy3-cDNA (825 ng)/Cy5-gDNA (20 pmol of Cy5) were prepared in 110 µl of hybridization buffer (1 M NaCl, 100 mM MES, pH 6.5, 20% formamide, 20 mM EDTA, 1% Triton X-100). The microarrays used for gene expression analysis were obtained from Oxford Gene Technology in the 4×44k format (Agilent ink-jet technology) comprised of 4 identical matrices of 43,798 experimentally-validated probes (60-mer oligonucleotides) covering ORFs and intergenic regions of the S. coelicolor genome [18]. The hybridization mixes (100 µl) were applied to the microarray surface following the manufacturer’s instructions, and hybridized at 55°C for 67 hours. The slide was washed in 50 ml of Agilent Gene Expression Wash Buffer 1 for 5 min at room temperature and then in 50 ml of Wash Buffer 2 preheated to 37°C for 1 min, both with horizontal agitation (85 rpm). The slide was then briefly immersed in the Agilent Stabilization and Drying Solution prior to measuring fluorescence with an Agilent DNA Microarray Scanner G2565BA using the extended dynamic mode. Quantification of fluorescence intensities was performed using the FeatureExtraction software (v9.5.1, Agilent).
2.4. Transcriptome Data Analysis
Fluorescence intensities were processed using the R environment (R Development Core Team 2011, version 2.12.2) and the limma package [20]. For each spot on the microarray, net fluorescence intensities were calculated subtracting the median of background pixels values from the mean of foreground pixels. When the intensity value was negative or lower than the background standard deviation, the standard deviation of the background pixels was used as the surrogate intensity, while S. coelicolor genomic DNA labeled with Cy5 was used as the common reference. The Mg values were calculated as the log2 of the Cy3-cDNA intensity divided by the Cy5-gDNA intensity and were normalized using probe weights by first cyclic loess (window of 0.3, 3 iterations) and then global median.
BLAST comparisons indicated that 943 of the array probes had potential cross-hybridization with more than one gene and that 7234 of the probes corresponded to intergenic regions. Weight values of 10−6 were assigned to these non-valid probes and a weight value of 1 assigned to the valid probes (35621). Values from valid probes of the same gene were averaged and limma linear models were used to obtain the log2 abundance values of comparisons between two conditions and the associated p-values, both FDR-corrected and uncorrected for multiple testing. The complete array data (Mg and p-values) for the 7721 transcripts quantified in this work are included in Table S1.
2.5. Computational and Bioinformatics Analyses
For comparison of relative transcript abundance values between MI and MII, the MI stage was used as the reference, and abundance values were shown as the log2 ratio of MII/MI. Positive abundances corresponded to transcripts up-regulated in MII, negative abundance values to transcripts up-regulated in MI and abundance values were considered significant when they were higher than 1 (2-fold up-regulation in MII) or lower than −1 (0.5-fold up-regulation in MII). Differences between liquid and solid cultures were considered significant when their coefficients of variation were higher than 0.7 (Table 1 and Table 2). ProteinCenter 2.0 (Proxeon, Denmark) was used to analyze and process genes. Genes were classified into functional categories according to their annotated functions in the Gene Bank database, and by homology/functions according to the Gene Ontology, Conserved Domain, KEGG pathway and StrepDB databases.
Table 1. Transcripts showing opposite (positive values in solid and negative in liquid or vice versa) and significant (coefficient of variation between solid and liquid cultures higher than 0.7) abundances (Fig. 4C, F, I, L, O, R, U; Fig. 5C, F, I, L).
| Function | SCO no. | Description | Log 2 (MII/MI) | SCO no. | Description | Log 2 (MII/MI) | ||
| S | L | S | L | |||||
| PRIMARY METABOLISM | SCO0992 | Cysteine synthase | −1.1 | 1.2 | SCO3909 | 50S ribosomal protein L9, rplI | −1.2 | 0.3 |
| SCO1132 | Oxidoreductase | −0.4 | 1.8 | SCO4152 | 5′-nucleotidase | 0.7 | −1.1 | |
| SCO1134 | Oxidoreductase | −1.2 | 1.9 | SCO4284 | Deacetylase | −0.5 | 1.1 | |
| SCO1181 | Putative plasmid partition protein | −2.2 | 1.2 | SCO4577 | Helicase DEAD-like | −1.2 | 1.1 | |
| SCO1335 | Oxidoreductase | −1.7 | 0.2 | SCO4701 | 30S ribosomal protein S10 | 1.2 | −1.5 | |
| SCO1343 | Uracil-DNA glycosylase | −1.2 | 0.3 | SCO4702 | 50S ribosomal protein L3 | 0.5 | −2 | |
| SCO1600 | Translation initiation factor IF-3 | 1.2 | −0.9 | SCO4703 | 50S ribosomal protein L4 | 0.4 | −2.1 | |
| SCO1631 | Helicase | −1.2 | 0.8 | SCO4712 | 50S ribosomal protein L14 | 0.7 | −1.4 | |
| SCO1966 | Excinuclease ABC subunit B | −1.9 | 0.1 | SCO4734 | 50S ribosomal protein L13 | 0.6 | −1.8 | |
| SCO2003 | DNA polymerase I | −1.8 | 0.2 | SCO5494 | NAD-dependent DNA ligase ligA | −2.2 | 0.9 | |
| SCO2036 | Tryptophan synthase subunit alpha | −0.4 | 1.4 | SCO5566 | ATP-dependent DNA helicase recG | −1.2 | 0.1 | |
| SCO2597 | 50S ribosomal protein L21 | 0.9 | −1.4 | SCO5815 | ATP-dependent DNA helicase | −1.4 | 0.7 | |
| SCO2770 | Agmatinase. Urea cycle and metabolism of amino groups | 0.4 | −1.1 | SCO5920 | DEAD-box RNA helicase | −1.7 | 0 | |
| SCO3351 | DNA repair protein RadA | −1.6 | 0.2 | SCO6084 | DNA polymerase III subunit epsilon | −2.6 | 0.8 | |
| SCO3434 | DNA polymerase I | −1 | 0.8 | SCO6262 | Helicase | −1.6 | 0 | |
| SCO3543 | Putative DNA topoisomerase I | 1.2 | −0.2 | SCO6662 | Transaldolase | −3.8 | 0 | |
| SECONDARY METABOLISM | SCO0490 | Esterase | −1.1 | 1.4 | SCO3243 | Myo-inositol phosphate synthase | −0,6 | 1.6 |
| SCO0491 | ABC transporter | −0.8 | 1 | SCO6273 | Type I polyketide synthase cpkC | −1.3 | 2.5 | |
| SCO0492 | Peptide synthetase | −1.1 | 1.2 | SCO6274 | Type I polyketide synthase cpkB | −1.9 | 3.2 | |
| SCO0493 | ABC-transporter | −1 | 1.7 | SCO6275 | Type I polyketide synthase cpkA | −1.9 | 3.2 | |
| SCO0497 | Iron-siderophore permease | −1.9 | 0.9 | SCO6276 | Secreted monooxygenase | −2.3 | 7.9 | |
| SCO0498 | Peptide monooxygenase | −2.3 | 2.7 | SCO6277 | Epoxide hydrolase | −2.3 | 6 | |
| SCO0499 | Methionyl-tRNA formyltransferase | −1.6 | 1.9 | SCO6278 | Membrane transport protein | −2 | 5.2 | |
| SCO2782 | Pyridoxal-dependent decarboxylase | −3.2 | 0.4 | SCO6279 | Aminotransferase | −1.9 | 6.4 | |
| SCO3229 | 4-hydroxyphenylpyruvic dioxygenase | −1 | 1.6 | SCO6280 | Putative transcriptional regulator | −0.7 | 4.8 | |
| SCO3235 | ABC transporter | −1 | 1.8 | SCO6281 | FAD-binding protein | −0.6 | 4.3 | |
| DIFFERENTIA-TION | SCO2705 | ChpF | 1.4 | −0.4 | SCO5112 | BldKA | 1 | −2.2 |
| SCO2607 | Sfr protein; sporulation protein | 0.6 | −1.3 | SCO5114 | BldKC | 0.6 | −2.4 | |
| SCO4069 | SarA | −0.3 | 1.6 | SCO5190 | DNA-binding protein, wblC | 3.2 | −1.6 | |
| SCO4823 | Possible target for bldA regulation | 1 | −2.8 | |||||
| REGULATORY | SCO0646 | TetR family transcriptional regulator | −1.2 | 0.1 | SCO4303 | Transcriptional regulator | −2.2 | 0.3 |
| SCO1568 | TetR family transcriptional regulator | −1.8 | 2.4 | SCO4336 | MarR-family protein | −2.6 | 0.2 | |
| SCO2489 | TetR family transcriptional regulator | −2 | 1 | SCO4375 | MarR family regulatory protein | −1.6 | 0.1 | |
| SCO2845 | GntR family transcriptional regulator | −1.3 | 0.1 | SCO4413 | AraC transcription regulator similar to S griseus AdpA | −1 | 1.9 | |
| SCO2958 | Transcriptional regulator, nnaR | −1 | 0.2 | SCO5607 | Transcriptional regulator | −0.5 | 1.8 | |
| SCO3066 | Putative regulator of Sig15 | −0.5 | 1 | SCO6267 | Putative transcriptional regulator | −1.3 | 1.8 | |
| SCO3167 | TetR family transcriptional regulator | −3.1 | 0.6 | SCO6268 | Histidine kinase | −2.4 | 2.5 | |
| SCO3275 | merR family transcriptional regulator | 0.4 | −1.2 | SCO6565 | Transcriptional regulator | −1.4 | 0 | |
| SCO3390 | Two component sensor kinase | −1.7 | 1 | SCO6694 | Transcriptional regulator | −0.9 | 2.5 | |
| SCO3696 | Transcriptional regulator | −1.1 | 0.6 | SCO6743 | Transcriptional accessory protein | −0.9 | 1.2 | |
| SCO3848 | Serine/threonine protein kinase | 1 | −0.3 | SCO7364 | TetR transcriptional regulator | −2.3 | 0.4 | |
| SCO3879 | DnaA replication initiation protein | 0.6 | −1.4 | SCO7585 | MerR transcriptional regulator | −0.6 | 2 | |
| SCO4005 | RNA polymerase sigma factor | −2.1 | 3.5 | SCO7694 | TetR transcriptional regulator | −0.8 | 1 | |
| SCO4145 | Polyphosphate kinase, ppk | −1.3 | 0.5 | |||||
| TRASPOSONS -IS | SCO7074 | Transposase | 1.4 | −0.2 | ||||
| CONJUGATION-RECOMBINATION-MUTAGENESIS | SCO5102 | MutT-like protein | −0.6 | 1.6 | SCO5770 | Recombination regulator, recX | −1.3 | 3.3 |
| SCO5769 | Recombinase A, rec A | −1.3 | 2.2 | |||||
| STRESSS - DEFENSE | SCO0774 | Cytochrome P450 | −2.9 | 1.7 | SCO3890 | Thioredoxin reductase | −1.3 | 0.1 |
| SCO0180 | Usp, universal stress protein family | −1.1 | 1.5 | SCO4761 | Co-chaperonin GroES | −1.8 | 1.5 | |
| SCO3669 | Chaperone protein DnaJ | −0.5 | 1.6 | SCO4762 | GroEL1 | −1.4 | 0.5 | |
| SCO3670 | Heat shock protein GrpE | −0.8 | 1.6 | SCO5254 | SodN, superoxide dismutase | −1.2 | 1.3 | |
| SCO3701 | Putative thioredoxin reductase | −1.7 | 0.6 | |||||
| CATABOLISM -DEGRADATION | SCO3000 | Phosphatase | −1.2 | 1.2 | SCO4798 | Peptidase | 1.1 | −0.5 |
| SCO3487 | Agarase | 3.8 | −1 | SCO5285 | ATP-dependent protease | −1.3 | 3.6 | |
| SCO3661 | ATP-dependent protease, clpB | −2.1 | 0.7 | SCO5446 | Probable zinc metalloprotease | 1.6 | 0 | |
| SCO3712 | Hydrolase | −1.9 | 1.1 | SCO6109 | Probable secreted hydrolase | −1.4 | 0.1 | |
| SCO4241 | Proteinase | 1.2 | −0.3 | SCO7263 | Chitinase | 1.2 | −0.2 | |
| TRANSPORTERS - SECRETED | SCO0119 | Possible small secreted protein | 1 | −0.3 | SCO3607 | Secreted protein | −4.1 | 0.1 |
| SCO0994 | Putative transport permease protein | −0.7 | 2 | SCO4243 | Putative secreted protein | 0.7 | −1.5 | |
| SCO0286 | Putative peptidoglycan binding protein | 0.5 | −2.8 | SCO4289 | Possible secreted protein | −1.9 | 0.1 | |
| SCO1292 | Putative secreted protein | −1.6 | 1.3 | SCO4585 | ABC transporter protein | −0.3 | 2.4 | |
| SCO1515 | Preprotein translocase subunit SecF | 0.4 | −1.2 | SCO4722 | Preprotein translocase SecY | 0.5 | −1.5 | |
| SCO1516 | Preprotein translocase subunit SecD | 0.5 | −1.5 | SCO4993 | Metal ion transport protein | 0.8 | −1.1 | |
| SCO1567 | Transmembrane-transport protein pqrB | −1.4 | 2.3 | SCO5016 | Possible secreted protein | 1.1 | −0.3 | |
| SCO1785 | Iron-siderophore | −1.3 | 0.1 | SCO5130 | ABC transporter | 0.6 | −1.3 | |
| SCO2074 | Lipoprotein signal peptidase | 0.7 | −1.3 | SCO6199 | Possible secreted esterase | −1.3 | 2.1 | |
| SCO2949 | Carboxyvinyltransferase, murA | 0.5 | −1.2 | SCO6272 | Possible secreted protein | −1.9 | 1.9 | |
| SCO3024 | Transport protein | 0.5 | −1.3 | SCO6320 | Transport membrane protein | −1.1 | 0.1 | |
| SCO3166 | Membrane transport protein | −2.3 | 0.6 | SCO6665 | Probable secreted glucosidase | 0.4 | −1 | |
| SCO3286 | Putative secreted protin | −3.1 | 0.5 | SCO6980 | ABC transporter membrane protein | 1.4 | 0 | |
| SCO3366 | Exporter | −1.8 | 0.4 | SCO7453 | Putative secreted protein | 3.4 | −0.6 | |
Average log2 abundance values from three biological replicates of the MII with respect to MI in solid (S) and liquid (L) cultures. Only transcripts with significant abundances are shown (log2 abundance greater than ±1 in liquid and/or solid cultures). Functions (according to Gene bank, Gene Ontology, Conserved Domain, and KEGG and StrepDB databases): Primary metabolism (DNA/RNA replication, aerobic and anaerobic energy production, glycolysis and glyconeogenesis, pentose phosphate pathway, amino acid metabolism, nucleotide metabolism, translation, protein folding, RNA/protein processing, nucleases/RM methylases); secondary metabolism (secondary metabolites synthesis); differentiation (TTA bldA targets, Bld and Whi proteins); regulatory genes (transcriptional regulators, kinases, other regulatory genes); transposons - insertion sequences; conjugation – recombination - mutagenesis; stress and defense proteins; catabolism - degradation; lipid metabolism; transporters and secreted (ABC transporters, transporters and secreted proteins). Genes with “unknown” functions (Table S1) were not included.
Table 2. Transcripts up- or down- regulated in MI or MII liquid and solid cultures, showing differences in their abundances (coefficient of variation between liquid and solid higher than 0.7).
| Function | SCO no. | Description | Log 2 (MII/MI) | SCO no. | Description | Log 2 (MII/MI) | ||
| S | L | S | L | |||||
| PRIMARY METABOLISM | SCO0922 | Succinate dehydrogenase | −2.3 | −0.5 | SCO4631 | Type IV restriction endonuclease | −3 | −0.7 |
| SCO1321 | Elongation factor Tu | 1.4 | 0 | SCO4685 | DEAD-box RNA helicase | −1.1 | 0 | |
| SCO1522 | Glutamine amidotransferase | −1.8 | −0.6 | SCO5059 | Polyphosphate glucokinase, ppgK | −2 | −0.4 | |
| SCO1679 | Gluconokinase | −2.8 | −0.5 | SCO5657 | Aldehyde dehydrogenase | −1.4 | 0 | |
| SCO2470 | Deoxyguanosinetriphosphate Triphosphohydrolase-like protein | −1.7 | −0.2 | SCO6211 | Uricase | 2.1 | 0.4 | |
| SCO2655 | Putative nuclease | 0.9 | 3.9 | SCO6341 | Exonuclease | −2 | −0.3 | |
| SCO3023 | S-adenosyl-L-homocysteine hydrolase | 1.7 | 0 | SCO6415 | Probable dihydropyrimidinase | 1.1 | 0.3 | |
| SCO3303 | Lysyl-tRNA synthetase | −0.4 | −1.4 | SCO6661 | Glucose-6-phosphate dehydrogenase | −2.7 | −0.4 | |
| SCO3542 | Putative thymidine kinase | −1 | −0.3 | SCO6769 | Aminotransferase | 0.3 | 1.6 | |
| SCO4606 | NADH dehydrogenase subunit NuoL2 | −0.3 | −1.3 | SCO6962 | Glutamine synthetase | 0.9 | 3.2 | |
| SCO4608 | NADH dehydrogenase subunit NuoN2 | −0.5 | −2.1 | SCO7511 | Glyceraldehyde 3-phosphate dehydrogenase, gap2 | −2.7 | −0.7 | |
| SECONDARY METABOLISM | SCO0188 | Methylesterase | −1.3 | −0.4 | SCO5693 | Acyl CoA dehydrogenase | 1.4 | 0.5 |
| SCO0190 | Methyltransferase | −2.4 | −0.7 | SCO6286 | ScbR2 | 1.8 | 6.6 | |
| SCO1267 | Acyl carrier protein | −3.9 | 0 | SCO6750 | Isopentenyl-diphosphate isomerase | 0.6 | 2.8 | |
| SCO2478 | Reductase activated by actinorhodin | 0.2 | 4.2 | SCO6760 | Phytoene synthase | 0.4 | 1.2 | |
| DIFFERENTIA-TION | SCO2718 | RdlA | 7.4 | 0.9 | SCO5316 | WhiE | 4 | 0.1 |
| SCO4346 | Possible target for bldA regulation | −1.5 | −0.1 | |||||
| REGULATORY | SCO0193 | Putative transcriptional regulator | −2.5 | −0.4 | SCO5006 | Septum site-determining protein | 0.4 | 1.3 |
| SCO0447 | MarR family regulatory protein | −1.5 | −0.5 | SCO5008 | Putative septum site-determining protein | 0.6 | 2 | |
| SCO0767 | Putative transcriptional regulator | 0.3 | 1.3 | SCO5264 | Putative transcriptional regulator | −1.7 | −0.2 | |
| SCO1034 | TetR family transcriptional regulator | 0.2 | 1.9 | SCO5540 | Histidine kinase-like ATPases | 1.1 | 0.3 | |
| SCO1801 | Two component response regulator | 0.5 | 2.6 | SCO5616 | Putative transcriptional regulator | −1.6 | −0.5 | |
| SCO2223 | TetR family transcriptional regulator | −0.5 | −1.7 | SCO5656 | Transcriptional regulatory protein | −3.8 | −0.5 | |
| SCO2253 | Putative transcriptional regulator | 1.2 | 0.1 | SCO5785 | Transcriptional regulator | 2 | 0.5 | |
| SCO2730 | Putative TetR- transcriptional regulator | −1.9 | −0.4 | SCO6154 | Putative transcriptional regulator | 0.6 | 2 | |
| SCO2879 | Putative MinC septum formation inhibitor | 0.4 | 1.7 | SCO6219 | Ser/Thr protein kinase | −2.2 | −0.5 | |
| SCO3367 | TetR family transcriptional regulator | −1.2 | −0.2 | SCO6424 | Putative histidine kinase | 2.3 | 0.7 | |
| SCO4019 | GntR family regulatory protein | −1.4 | −0.3 | SCO6566 | ROK family protein | 1.8 | 0.2 | |
| SCO4021 | Two component system histidine kinase | −0.5 | −1.9 | SCO6696 | Regulatory protein | 0.2 | 1.6 | |
| SCO4032 | MarR regulatory protein | −5.4 | −0.2 | SCO6778 | Transcriptional regulator | −1.2 | −0.3 | |
| SCO4223 | AraC family transcription regulator | −1.4 | −0.1 | SCO7014 | LacI transcriptional regulator | −4.5 | −0.8 | |
| SCO4261 | LuxR regulatory protein | −3.6 | −0.3 | SCO7016 | LacI transcriptional regulator | −2.1 | −0.6 | |
| SCO4308 | Transcriptional regulator | −1.4 | −0.2 | SCO7061 | MarR transcriptional regulator | −1.3 | −0.2 | |
| SCO4640 | TetR transcriptional regulator | −4.3 | −0.2 | |||||
| TRASPOSONS -IS | SCO7740 | Insertion element | 1.5 | 0 | ||||
| CONJUGATION-RECOMBINATION-MUTAGENESIS | SCO7442 | Putative conjugal transfer protein TrbL | −2 | 0 | ||||
| STRESSS - DEFENSE | SCO0885 | Thioredoxin | 0.5 | 1.8 | SCO4609 | Putative heat shock peptidase htpX | −1.5 | −0.4 |
| CATABOLISM -DEGRADATION | SCO0591 | Putative lysozyme | 2.7 | 0.2 | SCO6078 | Probable alpha amylase | 0.3 | 1.4 |
| SCO0740 | Probable hydrolase | −0.9 | −2.8 | SCO6324 | Putative hydrolase | 2.2 | 0.4 | |
| SCO1509 | Possible hydrolase | −0.5 | −1.6 | SCO6414 | Possible hydrolase | 1.2 | 0.4 | |
| SCO1643 | 20S proteasome alpha-subunit | 0.5 | 2 | SCO7205 | Putative hydrolase | −1.7 | 0 | |
| SCO3779 | Probable nucleoside hydrolase | −0.3 | −1.4 | SCO7473 | Phenylacetic acid degradation protein PaaC | 0.7 | 3.3 | |
| SCO4108 | Probable peptidase | 1.8 | 0.5 | SCO7474 | Phenylacetic acid degradation protein PaaD | 0.4 | 1.5 | |
| SCO4288 | Possible phosphatase | −1.4 | −0.5 | SCO7590 | Catalase | 1.6 | 0.2 | |
| SCO5456 | Putative glycosyl hydrolase | 0.3 | 1.7 | |||||
| TRANSPORTERS - SECRETED | SCO0796 | Putative membrane transporter | −0.3 | −1.4 | SCO3507 | Integral membrane efflux protein | −0.3 | −1.3 |
| SCO1044 | Possible secreted protein | −2 | −0.2 | SCO3513 | Possible secreted protein | −1.5 | −0.2 | |
| SCO1144 | ABC transporter ATP-binding protein | −1.9 | −0.5 | SCO3718 | Potassium-transporting ATPase | −0.3 | −2.1 | |
| SCO1147 | ABC transporter | −0.5 | −2 | SCO4031 | Membrane transport protein | −3 | −0.5 | |
| SCO1822 | Transmembrane transport protein | 1.6 | 0.4 | SCO4424 | Possible secreted protein | −1 | −0.2 | |
| SCO2010 | ABC transporter Branched chain amino acid transport permease | 1.1 | 0.3 | SCO4471 | Possible secreted protein | −1.9 | −0.6 | |
| SCO2011 | Putative amino acid transport | 0.5 | 2.2 | SCO4641 | Transmembrane efflux protein | −2.5 | −0.5 | |
| SCO2383 | Possible secreted protein | −1.1 | −0.3 | SCO6086 | Transport system integral membrane protein | −1.9 | −0.1 | |
| SCO2756 | Predicted permease | −0.5 | −1.4 | SCO6104 | Possible secreted protein | 0.7 | 3.2 | |
| SCO2780 | Secreted protein | −2 | −0.3 | SCO6258 | Sugar ABC transporter permease | −0.4 | −1.4 | |
| SCO2905 | Putative sugar permease | −1.5 | −0.2 | SCO6417 | Integral membrane transporter | 1.5 | 0.3 | |
| SCO3402 | Possible secreted protein | −1.1 | −0.1 | |||||
(Fig. 4B, E, H, K, N, Q, T; Fig. 5B, E, H, K). Average log2 abundance values from three biological replicates of MII with respect to MI in solid (S) and liquid (L) cultures. Only transcripts with significant abundances are shown (log2 abundance greater than ±1 in liquid and/or solid cultures). Functions as in Table 1. Genes with “unknown” functions (Table S1) were not included.
Results and Discussion
3.1. Global Quantification of Gene Transcriptions in Liquid Cultures
In order to facilitate comparison with previous work [16], MI and MII transcriptomes were analyzed in liquid non-sporulating cultures using a workflow similar to that used for solid sporulating cultures [16]: three independent biological replicates and two developmental stages (MI and MII) were processed (outlined in Fig. 1). MI from liquid cultures (MIL) was obtained at early time points (14 hours), long before the onset of the PCD and differentiation of MII, and MII (MIIL) was obtained at 90 hours long after the disappearance of the MI [8] (Fig. 1A). The transcriptomes from both developmental stages were compared with those of solid cultures (MIS and MIIS) [16] (Fig. 1B).
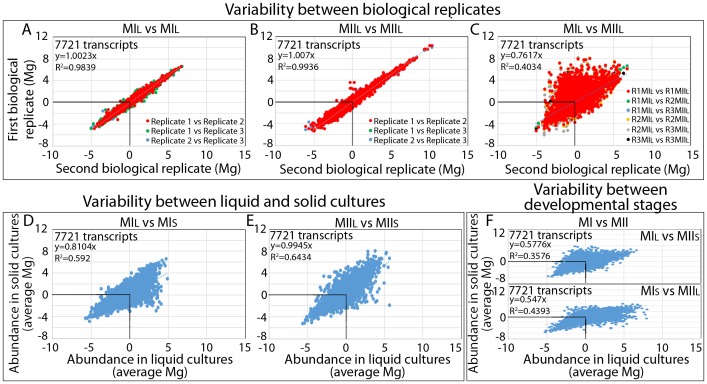
The relative abundances of 7721 transcripts were estimated and normalized to chromosomal DNA (see full Mg data in Table S1). Reproducibility between biological replicates of MIL and MIIL (Fig. 2A, B) was high (regression coefficients of 0.98 and 0.99 respectively) and much higher than correlation between abundance values from different developmental stages (MIL vs. MIIL; regression coefficient of 0.4) (Fig. 2C). Correlation between the Mg values from liquid cultures obtained in this work (average from the three biological replicates) and the Mg values reported for sporulating solid cultures [16] was also high for MIL vs. MIS (Fig. 2D) and MIIL vs. MIIS (Fig. 2E) (regression coefficient of 0.6 in both cases). Correlation between different developmental stages (MI vs. MII) in solid and liquid cultures (Fig. 2F) was comparable to the correlation observed between different developmental stages in liquid cultures (regression coefficients of 0.4 in all cases) (compare Fig. 2C with Fig. 2F).
Figure 2. Quantitative transcriptomic data analysis.
Correlation of transcription abundance values (log2 ratio against chromosomal DNA). Upper panels - biological replicates: (A) MIL vs. MIL, (B) MIIL vs. MIIL, (C) MIL vs. MIIL (three biological replicates compared in pairs). Lower panels - developmental stages (average abundance values from three biological replicates: (D) MIL vs. MIS, (E) MIIL vs. MIIS, (F) MIL vs. MIIS and MIS vs. MIIL.
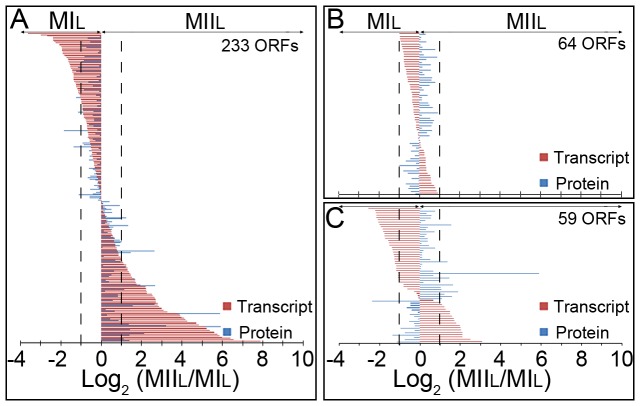
The abundances of MI and MII transcripts (this work) were compared with the MI and MII protein abundances previously reported for liquid cultures [12] using the MI stage as reference, and both, transcript/protein abundances were shown as the log2 ratio of MII/MI. Three hundred fifty-six of the proteins encoded by the 7721 transcripts identified in this work have been previously quantified by proteomics [12]. Two hundred thirty-three proteins and transcripts were significantly up-regulated in MII (positive abundance values higher than 1) or MI (negative abundance values lower than −1) (Fig. 3A); 64 did not differ significantly (log 2 abundances within ±1 interval) (Fig. 3B); and 59 ORFs showed divergent abundances (positive protein abundance and negative transcript abundance or vice versa) (Fig. 3C). Overall, correlation between protein and transcript abundance was reasonable, considering they are different biomolecules with different kinetics and turnover rates.
Figure 3. Comparison between transcriptomics (this work) and proteomics [12].
Transcription and protein abundance values correspond to log2 MIIL/MIL and are the average of two (in the case of proteomics) or three (in the case of transcriptomics) biological replicates. Dashed lines indicate the limit for considering abundance variations as significant (log2 abundances greater than ±1). (A) Proteins and transcripts significantly up-regulated in MIIL (positive abundance values higher than 1) or in MIL (negative abundance values lower than −1). (B) Proteins and transcripts without significant variations (log 2 abundances within ±1 interval). (C) Proteins and transcripts showing divergent abundance values (positive values in MIL and negative in MIIL or vice versa).
3.2. Similarities and Differences between MI and MII Transcriptomes in Solid and Liquid Cultures
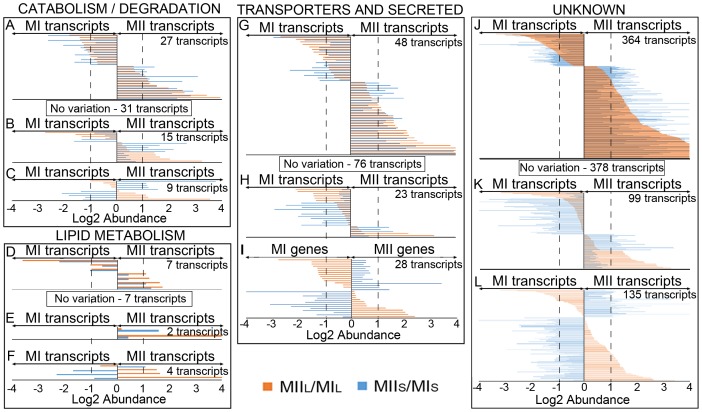
MI and MII transcriptomes from liquid (this work) and solid cultures [16] were compared, using the MI stage as the reference, and showing transcript abundances as the log2 ratio of MII/MI (Fig. 4, Fig. 5) (Table S1). When transcript abundances with very high confidence, described by Yagüe et al [16] in solid cultures (1901 transcripts), were compared to the abundances detected in liquid cultures (this work), it was evident that transcriptomes of MI and MII were very similar for both conditions (Fig. 4, Fig. 5): 1420 transcripts (75% of the total) were up-regulated (positive abundance values higher than 1), down-regulated (negative abundance values lower than −1) in MII with respect to MI, or were not significantly different (log 2 abundances within ±1 interval) (Fig. 4A, D, G, J, M, P, S; Fig. 5A, D, G, J); 204 transcripts (11% of the total) were up- or down- regulated in MI or MII in liquid and solid cultures but showed important differences in their abundance (coefficient of variations between liquid and solid abundances higher than 0.7) (Fig. 4B, E, H, K, N, Q, T; Fig. 5B, E, H, K) while 277 (14% of the total) showed significantly (coefficient of variation between liquid and solid cultures higher than 0.7) opposite (positive values in solid and negative in liquid or vice versa) abundances (Fig. 4C, F, I, L, O, R, U; Fig. 5C, F, I, L).
Figure 4. Abundance values of transcripts quantified in solid and liquid cultures (1901 in total) and grouped into functional categories.
Abundance values (average of log2 MII/MI from three biological replicates) and transcripts with significant variations in MIIL and/or MIIS (log2 abundance greater than ±1) are shown. Transcripts without significant (log2 abundance within the ±1interval) are indicated into squares and labeled as “No variation”. Functional categories: primary metabolism (DNA/RNA replication, aerobic and anaerobic energy production, glycolysis and glyconeogenesis, pentose phosphate pathway, amino acid metabolism, nucleotide metabolism, translation, protein folding, RNA/protein processing, nucleases/RM methylases); secondary metabolism (secondary metabolite synthesis); differentiation (TTA bldA targets, Bld and Whi proteins); regulatory genes (transcriptional regulators, kinases, other regulatory genes); transposons - insertion sequences; conjugation, recombination, mutagenesis; stress and defense proteins. Dashed lines indicate the limit for considering abundance variations significant (log2 abundance ±1).
Figure 5. Transcripts abundance values (log2 MII/MI) grouped in functional categories (continuation).
Functional categories: catabolism and degradation; lipid metabolism; transporters and secreted (ABC transporters, transporters and secreted proteins); genes with unknown function. Dashed lines indicate the limit for considering abundance variations significant (log2 abundance ±1).
3.2.1. Transcripts showing opposite abundances in solid and liquid cultures
The most important differences between liquid and solid cultures were for the 277 transcripts showing opposite (positive values in solid and negative in liquid or vice versa) and significant abundances (coefficient of variation between solid and liquid cultures higher than 0.7) (Table 1). These transcripts included 21 genes encoding for proteins involved in “primary metabolism” whose biological roles in development remain unknown. Interestingly, one third of these 21 genes encode for ribosomal proteins, indicating that translational machinery may play a relevant role in controlling parts of Streptomyces differentiation, as has been previously suggested [21].
The 20 genes involved in “secondary metabolism” which had opposite differences in liquid and solid cultures (Table 1), were up-regulated in MIIL (up to 239-fold), and down-regulated in MIIS (up-to 0.1-fold) with respect to MI. These included genes from calcium-dependent antibiotic (CDA), yellow antibacterial pigment (Cpk), and coelichelin clusters [22] as well as SCO2782, one of the genes putatively involved in desferrioxamine biosynthesis [22]. These results indicated production of these secondary metabolites in liquid but not in solid cultures, and illustrated that in addition to differentiation (development of MII), there were specific regulatory mechanisms for the production of different secondary metabolites under different culture conditions.
Six of the 7 genes related to hyphal “differentiation” differentially expressed in liquid and solid cultures were up-regulated in MII from solid sporulating cultures, and down-regulated in MII from non-sporulating liquid cultures (Table 1) and included genes related to aerial mycelium/spore hydrophobic cover formation (chpF, sfr, bldkA, bldkC) [23]. Also included were wblC, one of the whiB-like regulatory genes highly conserved in Streptomyces, whose biological function remains to be characterized [24] and SCO4823, encoding for a putative protein with a TTA leucine codon which converts it to a putative target for bldA [25]. SarA was the only “differentiation” transcript up-regulated in MIIL (3-fold) and down-regulated in MIIS (0.8-fold). Interestingly, sarA was conserved in the Streptomyces genus and it is known to repress sporulation and activate antibiotic production in solid cultures [26]. Repression of sporulation and increased antibiotic production in liquid cultures are the most important phenotypic differences between MIIL and MIIS, and sarA may be one of the master genes regulating this process.
Twenty-four of the 27 “regulatory” genes differentially expressed in liquid and solid cultures (Table 1) were up-regulated in MIIL (up to 11-fold) and down-regulated in MIIS (up to 0.18-fold). These genes included pqrA, a TetR-like transcriptional regulator involved in resistance to oxidative stress [27]; nnaR, a transcriptional regulator which appears to be involved in the regulation of nitrate/nitrite assimilation [28]; SCO4005, a sigma factor which is activated by the stringent factor ppGpp [29]; and ppk (SCO4145) which represses antibiotic production [30]. The biological functions of the other 21 “regulatory” genes up-regulated in MIIL have yet to be characterized. Only 3 “regulatory” genes were up-regulated in MIIS (up to 2-fold) and down-regulated in MIIL (up to 0.4-fold) (Table 1): SCO3275, a putative MerR transcriptional regulator of unknown function; SCO3848, a putative serine/threonine protein kinase of unknown function and dnaA a chromosomal replication initiator protein [31]. Up-regulation of dnaA in MIIS may be involved with the division of chromosomal DNA during sporulation.
Other genes differentially expressed in liquid and solid cultures were SCO7074, a putative transposase up-regulated in MIIS (2.6-fold), and genes related to “conjugation, recombination, or mutagenesis” such as recA, recX and SCO5102 (mutT-like), which were up-regulated in MIIL (up to 10-fold), and down-regulated in MIIS (up to 0.4-fold), which suggested activation of mechanisms of genetic variability in liquid but not in solid cultures.
All transcripts encoding for “stress and defense” proteins (9 genes) were up-regulated in MIIL (up to 3.2-fold), and down-regulated in MIIS (up to 0.1-fold), which suggested the existence of greater stress in liquid cultures than in solid cultures. These transcripts included well-characterized genes such as chaperones (dnaJ, grpE, groES, groEL1), superoxide dismutases (sodN), and thioredoxin reductases (SCO3890).
Ten genes encoding for proteins related to “catabolism and degradation” were also differentially expressed in liquid and solid, but their expression was not clearly biased toward MI or MII (Table 1). These transcripts included SCO3487, a gene encoding for an agarase [32], which was up regulated in MIIS (14-fold) but down-regulated in MIIL (0.5-fold), and could be involved in agar degradation in solid cultures. The biological significance of the remainder of these catabolic MI or MII genes in liquid and solid cultures, is yet to be characterized.
Twenty-eight transcripts encoding for “transporters and secreted proteins” were differentially expressed in liquid and solid cultures (Table 1) though the biological role of these genes in controlling development remains unknown. Interestingly pqrB, encoding for a transmembrane-transport protein, was co-expressed with SCO1568 (a TetR transcriptional regulator, discussed above), and both were up-regulated in MIIL (up to 5-fold) and are known to play a role in oxidative stress [27].
3.2.2. Transcripts up- or down- regulated in MI or MII in liquid and solid cultures showing significant differences in their abundances
Two hundred and nine transcripts were up- or down- regulated in MI or MII for liquid and solid cultures and showed important differences in abundance (coefficient of variation between liquid and solid higher than 0.7) (Fig. 4B, E, H, K, N, Q, T; Fig. 5B, E, H, K) (Table 2). These genes, together with the genes described in the previous paragraph (Table 1), were potentially involved, in the regulation of the developmental/metabolic differences between liquid and solid cultures. Of these, 24 transcripts were involved in “primary metabolism” (Table 2): genes for oxidative phosphorylation, TCA cycle, glycolysis, glyconeogenesis, biosynthesis of amino acids, RNA translation, etc. Eight genes putatively involved in “secondary metabolism” also showed significant differences between liquid and solid cultures (Table 2). SCO2478, a reductase induced by actinorhodin [33], SCO6750, an isopentenyl-diphosphate isomerase putatively involved in terpenoid biosynthesis (sco00900 KEGG pathway), SCO6286 or scbR2, a pseudo-γ-butyrolactone receptor that represses the SARP regulator kasO of the yellow antibacterial pigment (Cpk) cluster [34], and SCO6760, a putative phytoene synthase [22], were highly up-regulated in MII with respect to MI in liquid cultures (18-, 7-, 97- and 2.3-fold respectively) but not in solid (1.1-, 1.5-, 3.5- and 1.3-fold respectively). The opposite was seen for SCO5693, a putative acyl CoA dehydrogenase involved in the biosynthesis of secondary metabolites (the sco01110 KEGG pathway), which was overexpressed 2.6-fold in MII with respect to MI in solid cultures, but only 1.4-fold in liquid. The other 3 genes encoding for proteins putatively involved in “secondary metabolism” were down-regulated in MII liquid and solid cultures, and were the only exceptions to secondary metabolite genes up-regulated in MI: two genes putatively involved in isorenieratene biosynthesis (SCO0188 and SCO0190); and SCO1267, one of the genes encoding for an acyl carrier protein [22]. These genes may not be activated under the culture conditions used in this work.
Two key “differentiation” genes were also differentially expressed in liquid and solid cultures (Table 2). RdlA, one of the genes involved in the last stages of aerial mycelium/spore hydrophobic cover maturation [35] was up-regulated 168-fold for MII in solid cultures with respect to MI, but only 1.8-fold in liquid cultures. WhiE, a gene responsible for the biosynthesis of an aromatic polyketide precursor to the gray spore pigment [36] was up-regulated 16-fold for MII in solid cultures but expressed at the same level in MI and MII liquid cultures.
Two of the “regulatory” genes differentially expressed in liquid and solid cultures had already been characterized (Table 2): SCO4223, an AraC transcriptional regulator involved in resistance to oxidative stress [37] up-regulated in MI liquid cultures (2.6-fold vs. 1-fold in solid) and SCO5785, a transcriptional regulator which enhances antibiotic production [38] up-regulated in MII solid cultures (4-fold vs. 1.4-fold in solid). The biological functions of the other 31 “regulatory” transcripts differentially expressed in liquid and solid cultures remain unknown (Table 2).
One putative insertion element (SCO7740) was up-regulated in MII with respect to MI in solid cultures (2.8-fold), but not in liquid (1-fold, no variation) and a putative conjugal transfer protein (SCO7442) was up-regulated in MI liquid (4-fold) but not solid cultures (1-fold, no variation) (Table 2). Two genes related to “stress and defense” (SCO0885, SCO4609) were differentially expressed in liquid and solid cultures (Table 2). SCO0885 encoded for a thioredoxin which is induced under oxidative stress [29] and was especially up-regulated for MII in liquid (3.5-fold vs. 1.4-fold in solid), suggesting the existence of more stress in liquid than in solid cultures.
For genes involved in “catabolism and degradation”, a putative lysozyme (SCO0591) was up-regulated in MII solid cultures (6.5-fold vs. 1.1-fold in liquid) and may play a role during the excision of individual spores. Twenty-three genes putatively encoding for “transporters and secreted” proteins (Table 2) and 95 genes encoding for “unknown” proteins (Fig. 5K. Table S1) were also differentially expressed in liquid and solid cultures, though further work will be necessary to characterize their biological function.
3.2.3. Transcripts showing similar abundances in solid and liquid cultures
The 1420 transcripts with similar abundance in solid and liquid cultures (Table S1), included several well-characterized genes (summarized in Table 3). Genes involved in “primary metabolism” (Fig. 4A) were mostly up-regulated in MI: up to 18-fold in the case of oxidative phosphorylation and the TCA cycle genes, up to 4-fold in the case of genes encoding proteins involved in glycolysis and glyconeogenesis, and up to 3.7-fold in the case of genes encoding ribosomal proteins (Table 3).
Table 3. Summary of well characterized genes whose transcripts showed similar abundances in liquid and solid cultures (coefficient of variation between liquid and solid cultures lower than 0.7) (Fig. 4A, D, G, J, M, P, S; Fig. 5A, D, G, J).
| Function | SCO no. | Description | Log 2 (MII/MI) | SCO no. | Description | Log 2 (MII/MI) | ||
| S | L | S | L | |||||
| PRIMARY METABOLISM | SCO1947 | Glyceraldehyde-3-phosphate dehydrogenase | −2 | −2 | SCO3945 | Cytochrome oxidase CydA | −4.2 | −2.2 |
| SCO2972 | PrfB | −0.6 | −1 | SCO3946 | Cytochrome oxidase CydB | −3.4 | −1.7 | |
| SCO3425 | 30S ribosomal protein S18 | −0.8 | −1.9 | SCO4607 | NADH dehydrogenase NuoM2 | −0.7 | −1.8 | |
| SECONDARY METABOLISM | SCO5077 | ActVA | 1.3 | 2.7 | SCO5898 | RedF | 1.7 | 1.7 |
| SCO5085 | ActII-4 | 2.6 | 5.9 | SCO6992 | AbsR1 | 5.8 | 5.8 | |
| DIFFERENTIA-TION | SCO0409 | SapA | 2.7 | 3.4 | SCO4768 | BldM | 3.5 | 4.5 |
| SCO1674 | ChpC | 4.3 | 4.5 | SCO5582 | NdsA | 3.1 | 6.2 | |
| SCO1675 | ChpH | 4.7 | 4.9 | SCO5621 | WhiG | 0.7 | 1.7 | |
| SCO1800 | ChpE | 5.3 | 7.2 | SCO5723 | BldB | 1 | 2.6 | |
| SCO2717 | ChpD | 5.6 | 3.4 | SCO5819 | WhiH | 1.8 | 1.5 | |
| SCO3323 | BldN | 4.2 | 4.8 | SCO6681 | RamC | 1 | 0.9 | |
| SCO3579 | WblA | 3.5 | 6.7 | SCO6682 | RamS | 4.6 | 5.8 | |
| SCO4091 | BldC | 1.4 | 2.7 | SCO6683 | RamA | 0.9 | 0.7 | |
| SCO4543 | WhiJ | −1.4 | −0.9 | |||||
| REGULATORY | SCO0155 | TetR family transcriptional regulator | −2.1 | −0.8 | SCO2232 | Maltose operon repressor | −0.8 | −1 |
| SCO1193 | TetR family transcriptional regulator | −1.3 | −0.7 | SCO4034 | Sigma factor sigN | 1.3 | 2.8 | |
| SCO1626 | Cytochrome P450, rarE | 2.4 | 3.9 | SCO4180 | Iron uptake regulatory protein | −0.7 | −1.3 | |
| SCO1628 | RarC homologue | 2.7 | 4.3 | SCO4377 | Serine-threonine kinase, afsL | −1.2 | −1 | |
| SCO1629 | RarB homologue | 2.8 | 4.6 | SCO4850 | TetR transcriptional regulator | −1.1 | −0.6 | |
| SCO1630 | RarA homologue | 3.7 | 5.8 | SCO5820 | hrdB sigma factor | 1.7 | 1 | |
| SCO2077 | DivIVA | 0.9 | 2.2 | SCO7809 | TetR transcriptional regulator | −2.2 | −1.3 | |
| TRASPOSONS -IS | SCO2236 | Plasmid maintenance killer protein | 2.3 | 1.5 | SCO6208 | Putative Transposase | 1.2 | 2.2 |
| SCO2311 | Putative transposes | 2.5 | 1.7 | SCO6393 | Transposase | 1.6 | 1.7 | |
| SCO3714 | Transposase | 3.1 | 3.1 | SCO6394 | IS element ATP binding protein | 1.7 | 1.1 | |
| SCO4350 | Integrase | 1.7 | 2.8 | SCO6395 | Putative IS element transposase | 1 | 1.1 | |
| SCO4772 | Transposase | 2.9 | 4.7 | |||||
| CONJUGATION-RECOMBINATION-MUTAGENESIS | SCO1520 | Holliday junction resolvase ruvC | −0.7 | −1.5 | SCO3876 | Recombination protein F | −0.8 | −1.1 |
| STRESSS – DEFENSE | SCO0379 | Catalase, katA | 1.2 | 2.2 | SCO5803 | SOS regulatory protein LexA | 2.3 | 3.8 |
| SCO0560 | Catalase/peroxidase cpeB | 1.2 | 2.2 | |||||
| CATABOLISM -DEGRADATION | SCO5444 | Possible glycogen phosphorylase, glgP | 2.1 | 4.1 | ||||
| TRANSPORTERS – SECRETED | SCO2008 | Branched chain amino acid binding protein | 1.9 | 2.8 | ||||
Average log2 abundance values (from three biological replicates) for the MII with respect to MI in solid (S) and liquid (L) cultures. Only transcripts with significant abundances are shown (log2 abundance greater than ±1 in liquid and/or solid cultures). Functions as in Table 1. Genes with “unknown” functions (Table S1) were not included.
Most genes involved in “secondary metabolism” (28 of 32) were up-regulated in MII (Fig. 4D): up to 60-fold in the case of genes from the actinorhodin cluster, 4.3-fold in the case of redF, a gene belonging to the prodigiosin cluster, and 56-fold in the case of absR1, a well-known activator of secondary metabolism [39].
Most of the well-characterized genes that participated in hyphal “differentiation” (30 of 32) were up-regulated in MII (Fig. 4G): up to 28-fold for activators of aerial mycelium differentiation (bldB, bldC, bldN, bldM); up to 147-fold for transcripts involved in the formation of hydrophobic covers (sapA, chpC, chpD, chpE, chpH, ramA, ramC, ramS); up to 3.2-fold for sporulation regulatory genes (wblA, whiG, whiH); and 73-fold for ndsA, a gene affecting antibiotic production [40]. WhiJ, a repressor of sporulation [41], was the only exception, as it was up-regulated during the non-sporulating phase (MI).
Interestingly, the expression of most “regulatory” genes (65 of 87) (Fig. 4J) was up-regulated in MII: up to 55-fold for “restoration of aerial mycelium formation” genes rarA-C, rarE homologues [42]; up to 4.6-fold for DivIVA, a gene essential for polar growth and morphogenesis [43]; and up to 7-fold for sigma factors sigN and hrdB (Table 3). The other 22 “regulatory” genes were up-regulated in MI, which included TetR transcriptional regulators, afsL, a serine-threonine kinase, as well as regulators involved in repressing maltose utilization (malR) [44] or regulation of nickel homeostasis/antioxidant response (nur) [45]. Several TetR family transcriptional regulators have been described in Streptomyces which function as repressors of antibiotic biosynthesis and export [46]–[48], so a role for these putative TetR transcriptional regulators in repressing the onset of antibiotic production in MI may be feasible; AfsL could be one of the proteins inactivating secondary metabolism in MI, as it has been shown to phosphorylate and regulate different regulators such as AfsR, a transcriptional activator involved in the regulation of secondary metabolism [49]; and the two regulators involved in repressing maltose utilization and nickel homeostasis/antioxidant response may be regulating metabolism of these compounds in MI and MII.
The expressions of the 9 genes encoding for putative “transposons and insertion sequences” which had similar abundances in liquid and solid cultures were up-regulated in MII (Fig. 4M) (Table 3), which suggested the activation of mobile genetic elements in solid and liquid MII cultures. The 4 transcripts involved in “conjugation, recombination, or mutagenesis” were up-regulated by as much 2.8-fold in MI (Fig. 4P) (Table S1) which included the well-characterized recF [50] and holliday junction resolvase ruvC [22] genes (Table 3), and may indicate the activation of DNA recombination in MI (prior to MII).
Most genes encoding for “stress and defense” proteins (8 of 10) were up-regulated up to 15-fold in MII (Fig. 4S). These transcripts included catalases, or the SOS regulatory protein LexA. The expression of genes related to “catabolism/degradation” (Fig. 5A), “lipid metabolism” (Fig. 5D), and “transport and secretion” (Fig. 5G), was not clearly biased toward MI or MII (Table S1). Some of these genes are well characterized, including glgP, a glycogen phosphorylase up-regulated in MII (up to 16-fold) which participates in glycogen and trehalose metabolism during sequential stages of aerial mycelium development [51] and SCO2008, encoding for a branched chain amino acid binding protein up-regulated in MII (up to 7-fold), which may participate in regulation of morphological differentiation in S. coelicolor [52]. Interestingly, most genes encoding for proteins of “unknown” function were up-regulated in MII (Fig. 5J).
Conclusions and Future Perspectives
This work was the first specifically focused on comparing MI and MII transcriptomes from S. coelicolor liquid and solid cultures. Expression of most transcripts (86% of all the identified transcripts) was comparable between non-sporulating liquid cultures and sporulating solid cultures, including genes involved in the biosynthesis of actinorhodin (actVA, actII-4) and undecylprodigiosin (redF); certain activators of secondary metabolism (absR1, ndsA); genes regulating hydrophobic cover formation (aerial mycelium) (bldB, bldC, bldM, bldN, sapA, chpC, chpD, chpE, chpH, ramA, ramC, ramS) and a few genes regulating early stages of sporulation (wblA, whiG, whiH, whiJ). The important similarities between transcriptomes from liquid and solid cultures were especially relevant considering that MIIS was collected at the sporulation phase, and that different culture media were used for liquid and solid cultures. Consequently, the developmental stages (MI and MII) were comparable, independent of age (developmental time points) or culture conditions.
The two major important differences between transcriptomes from liquid and solid cultures were: first, the expression of genes related to secondary metabolite biosynthesis (CDA, CPK, coelichelin, desferrioxamine clusters) which were up-regulated in liquid but not solid cultures; and second, genes involved in the last stages of hydrophobic cover/spore maturation (chpF, rdlA, whiE, sfr), which were up-regulated in solid with respect to liquid cultures.
Overall, this work extended previous morphological and proteomic studies, demonstrating that differentiation in liquid non-sporulating cultures was more similar to solid sporulating cultures than expected based on hyphal morphology (aerial mycelium formation and sporulation), and concluded that physiological differentiation was similar under both culture conditions. New information was also provided for several un-characterized genes differentially expressed in liquid and solid cultures (Table 1 and Table S1), which may be regulating, at least in part, the metabolic and developmental differences observed in liquid and solid cultures. This study contributes to the knowledge needed to further understand the biochemical pathways controlling pre-sporulation developmental stages and the activation of secondary metabolism in Streptomyces.
Supporting Information
Quantitative data for the expression of Streptomyces coelicolor transcripts. The relative abundances of 7721 transcripts quantified in this work are shown as the log2 ratio of transcript abundance with respect to chromosomal DNA (Mg values) and to MI in both liquid (this work) and solid [16] cultures. Data are the average of three biological replicates and P-values are indicated. The 1901 transcripts quantified with very high confidence by Yagüe et al [16] in solid cultures (1901 transcripts) were compared with the transcripts obtained in this work for liquid cultures. Data were separated in four folders: three of them include data of Tables 1, 2 and 3; the other include data for transcripts without significant variations between solid and liquid cultures.
(XLSX)
Acknowledgments
Our thanks to Beatriz Gutiérrez Magán (Universidad de Oviedo, Dpto. Biología Funcional, Área de Microbiología) for laboratory assistance, and Proof-Reading-Service.com for proofreading the text, Yasuo Ohnishi.
Funding Statement
This research was funded by an ERC Starting Grant (Strp-differentiation 280304). Jesús Sánchez was funded by project BIO2010-16303. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Tamaoki T, Nakano H (1990) Potent and specific inhibitors of protein kinase C of microbial origin. Biotechnology (NY) 8: 732–735. [DOI] [PubMed] [Google Scholar]
- 2. Omura S (1992) The expanded horizon for microbial metabolites-a review. Gene 115: 141–149. [DOI] [PubMed] [Google Scholar]
- 3. Umezawa K (1997) Induction of cellular differentiation and apoptosis by signal transduction inhibitors. Adv Enzyme Regul 37: 393–401. [DOI] [PubMed] [Google Scholar]
- 4.Champness WC (2000) Prokaryotic Development. YV Brun and Skimkets LJ, editors. Actinomycete development, antibiotic production and phylogeny: questions and challenges. American Society for Microbiology Washington, DC 11–31.
- 5. Rueda B, Miguelez EM, Hardisson C, Manzanal MB (2001) Mycelial differentiation and spore formation by Streptomyces brasiliensis in liquid culture. Can J Microbiol 47: 1042–1047. [PubMed] [Google Scholar]
- 6. Stocks SM, Thomas CR (2001) Viability, strength, and fragmentation of Saccharopolyspora erythraea in submerged fermentation. Biotechnol Bioeng 75: 702–709. [DOI] [PubMed] [Google Scholar]
- 7. Pamboukian CRD, Guimaraes LM (2002) Candida, M. Applications of image analysis in the characterization of Streptomyces olindensis in submerged culture. Braz J Microbiol 33: 17–21. [Google Scholar]
- 8. Manteca A, Alvarez R, Salazar N, Yagüe P, Sánchez J (2008) Mycelium differentiation and antibiotic production in submerged cultures of Streptomyces coelicolor . Appl Environ Microbiol 74: 3877–3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yagüe P, Manteca A, Simon A, Diaz-Garcia ME, Sanchez J (2010) A new method for monitoring programmed cell death and differentiation in submerged cultures of Streptomyces . Appl Environ Microbiol 76: 3401–3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yagüe P, López-García MT, Rioseras B, Sánchez J, Manteca A (2013) Pre-sporulation stages of Streptomyces differentiation, state-of-the-art and future perspectives. FEMS Microbiol Lett 342: 79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Manteca A, Fernández M, Sánchez J (2006) Cytological and biochemical evidence for an early cell dismantling event in solid cultures of Streptomyces antibioticus . Res Microbiol 157: 143–152. [DOI] [PubMed] [Google Scholar]
- 12. Manteca A, Jung HR, Schwämmle V, Jensen ON, Sánchez J (2010) Quantitative proteome analysis of Streptomyces coelicolor nonsporulating liquid cultures demonstrates a complex differentiation process comparable to that occurring in sporulating solid cultures. J Proteome Res 9: 4801–4811. [DOI] [PubMed] [Google Scholar]
- 13. Jayapal KP, Philp RJ, Kok YJ, Yap MG, Sherman DH, et al. (2008) Uncovering genes with divergent mRNAprotein dynamics in Streptomyces coelicolor . PLoS One 3: e2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nieselt K, Battke F, Herbig A, Bruheim P, Wentzel A, et al. (2010) The dynamic architecture of the metabolic switch in Streptomyces coelicolor. BMC genomics. 11: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gatewood M, Bralley P, Weil MR, Jones GH (2012) RNA-Seq and RNA immunoprecipitation analyses of the transcriptome of Streptomyces coelicolor identify substrates for RNase III. J Bacteriol 194: 2228–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yagüe P, Rodríguez-García A, López-García MT, Martín JF, Rioseras B, et al. (2013) Transcriptomic analysis of Streptomyces coelicolor differentiation in solid sporulating cultures: first compartmentalized and second multinucleated mycelia have different and distinctive transcriptomes. PLoS One 8: e60665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hesketh A, Bucca G, Laing E, Flett F, Hotchkiss G, et al. (2007) New pleiotropic effects of eliminating a rare tRNA from Streptomyces coelicolor, revealed by combined proteomic and transcriptomic analysis of liquid cultures. BMC Genomics 8: 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. de Jong W, Manteca A, Sánchez J, Bucca G, Smith CP, et al. (2009) NepA is a structural cell wall protein involved in maintenance of spore dormancy in Streptomyces coelicolor . Mol Microbiol 71: 1591–1603. [DOI] [PubMed] [Google Scholar]
- 19. Hesketh A, Chen WJ, Ryding J, Chang S, Bibb M (2007) The global role of ppGpp synthesis in morphological differentiation and antibiotic production in Streptomyces coelicolor A3(2). Genome Biol 8: R161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smyth GK (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl Genet Mol Biol 3: Article 3. [DOI] [PubMed]
- 21. Shima J, Hesketh A, Okamoto S, Kawamoto S, Ochi K (1996) Induction of actinorhodin production by rpsL (encoding ribosomal protein S12) mutations that confer streptomycin resistance in Streptomyces lividans and Streptomyces coelicolor A3(2). J Bacteriol 178: 7276–7284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bentley SD, Chater KF, Cerdeño-Tárraga AM, Challis GL, Thomson NR, et al. (2002) Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417: 141–147. [DOI] [PubMed] [Google Scholar]
- 23. Claessen D, de Jong W, Dijkhuizen L, Wösten HA (2006) Regulation of Streptomyces development, reach for the sky! Trends Microbiol. 14: 313–319. [DOI] [PubMed] [Google Scholar]
- 24. Fowler-Goldsworthy K, Gust B, Mouz S, Chandra G, Findlay KC, et al. (2011) The actinobacteria-specific gene wblA controls major developmental transitions in Streptomyces coelicolor A3(2). Microbiology 157: 1312–1328. [DOI] [PubMed] [Google Scholar]
- 25. Chandra G, Chater KF (2008) Evolutionary flux of potentially bldA-dependent Streptomyces genes containing the rare leucine codon TTA. Antonie Van Leeuwenhoek 94: 111–126. [DOI] [PubMed] [Google Scholar]
- 26. Ou X, Zhang B, Zhang L, Dong K, Liu C, et al. (2008) SarA influences the sporulation and secondary metabolism in Streptomyces coelicolor M145. Acta Biochim Biophys Sin (Shanghai) 40: 877–882. [PubMed] [Google Scholar]
- 27. Cho YH, Kim EJ, Chung HJ, Choi JH, Chater KF, et al. (2003) The pqrAB operon is responsible for paraquat resistance in Streptomyces coelicolor . J Bacteriol 185: 6756–6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Amin R, Reuther J, Bera A, Wohlleben W, Mast Y (2012) A novel GlnR target gene, nnaR, is involved in nitrate/nitrite assimilation in Streptomyces coelicolor . Microbiology 158: 1172–82. [DOI] [PubMed] [Google Scholar]
- 29. Paget MS, Molle V, Cohen G, Aharonowitz Y, Buttner MJ (2001) Defining the disulphide stress response in Streptomyces coelicolor A3(2): identification of the sigmaR regulon. Mol Microbiol 42: 1007–1020. [DOI] [PubMed] [Google Scholar]
- 30. Chouayekh H, Virolle MJ (2002) The polyphosphate kinase plays a negative role in the control of antibiotic production in Streptomyces lividans . Mol Microbiol 43: 919–930. [DOI] [PubMed] [Google Scholar]
- 31. Majka J, Zakrzewska-Czerwiñska J, Messer W (2001) Sequence recognition, cooperative interaction, and dimerization of the initiator protein DnaA of Streptomyces . J Biol Chem 276: 6243–6252. [DOI] [PubMed] [Google Scholar]
- 32. Temuujin U, Chi WJ, Chang YK, Hong SK (2012) Identification and biochemical characterization of SCO3487 from Streptomyces coelicolor A3(2), an exo- and endo-type β-agarase-producing neoagarobiose. J Bacteriol 194: 142–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shin JH, Singh AK, Cheon DJ, Roe JH (2011) Activation of the SoxR regulon in Streptomyces coelicolor by the extracellular form of the pigmented antibiotic actinorhodin. J Bacteriol 193: 75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xu G, Wang J, Wang L, Tian X, Yang H, et al. (2010) “Pseudo” gamma-butyrolactone receptors respond to antibiotic signals to coordinate antibiotic biosynthesis. J Biol Chem 285: 27440–27448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Claessen D, Wösten HA, van Keulen G, Faber OG, Alves AM, et al. (2002) Two novel homologous proteins of Streptomyces coelicolor and Streptomyces lividans are involved in the formation of the rodlet layer and mediate attachment to a hydrophobic surface. Molecular microbiology 44: 1483–1492. [DOI] [PubMed] [Google Scholar]
- 36. Lee MY, Ames BD, Tsai SC (2012) Insight into the molecular basis of aromatic polyketide cyclization: crystal structure and in vitro characterization of WhiE-ORFVI. Biochemistry 51: 3079–3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Darbon E, Martel C, Nowacka A, Pegot S, Moreau PL, et al. (2012) Transcriptional and preliminary functional analysis of the six genes located in divergence of phoR/phoP in Streptomyces lividans . Appl Microbiol Biotechnol 95: 1553–1566. [DOI] [PubMed] [Google Scholar]
- 38. Rozas D, Gullón S, Mellado RP (2012) A novel two-component system involved in the transition to secondary metabolism in Streptomyces coelicolor . PLoS One 7: e31760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park UM, Suh JW, Hong SK. (2000) Genetic Analysis of absR, a new abs locus of Streptomyces coelicolor. J Microbiol Biotechnol 10: 169∼175.
- 40. Wang XJ, Guo SL, Guo WQ, Xi D, Xiang WS (2009) Role of nsdA in negative regulation of antibiotic production and morphological differentiation in Streptomyces bingchengensis . J Antibiot (Tokyo) 62: 309–313. [DOI] [PubMed] [Google Scholar]
- 41. Aínsa JA, Bird N, Ryding NJ, Findlay KC, Chater KF (2010) The complex whiJ locus mediates environmentally sensitive repression of development of Streptomyces coelicolor A3(2). Antonie Van Leeuwenhoek 98: 225–236. [DOI] [PubMed] [Google Scholar]
- 42. Komatsu M, Takano H, Hiratsuka T, Ishigaki Y, Shimada K, et al. (2006) Proteins encoded by the conservon of Streptomyces coelicolor A3(2) comprise a membrane-associated heterocomplex that resembles eukaryotic G protein-coupled regulatory system. Mol Microbiol 62: 1534–1546. [DOI] [PubMed] [Google Scholar]
- 43. Flärdh K (2003) Essential role of DivIVA in polar growth and morphogenesis in Streptomyces coelicolor A3(2). Mol Microbiol 49: 1523–1536. [DOI] [PubMed] [Google Scholar]
- 44. van Wezel GP, White J, Young P, Postma PW, Bibb MJ (1997) Substrate induction and glucose repression of maltose utilization by Streptomyces coelicolor A3(2) is controlled by malR, a member of the lacl-galR family of regulatory genes. Mol Microbiol 23: 537–549. [DOI] [PubMed] [Google Scholar]
- 45. Ahn BE, Cha J, Lee EJ, Han AR, Thompson CJ, et al. (2006) Nur, a nickel-responsive regulator of the Fur family, regulates superoxide dismutases and nickel transport in Streptomyces coelicolor. Mol Microbiol 59: 1848–1858. [DOI] [PubMed] [Google Scholar]
- 46. Tahlan K, Yu Z, Xu Y, Davidson AR, Nodwell JR (2008) Ligand recognition by ActR, a TetR-like regulator of actinorhodin export. J Mol Biol 383: 753–761. [DOI] [PubMed] [Google Scholar]
- 47. Ou X, Zhang B, Zhang L, Zhao G, Ding X (2009) Characterization of rrdA, a TetR family protein gene involved in the regulation of secondary metabolism in Streptomyces coelicolor . Appl Environ Microbiol 75: 2158–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Duong CT, Lee HN, Choi SS, Lee SY, Kim ES (2009) Functional expression of SAV3818, a putative TetR-family transcriptional regulatory gene from Streptomyces avermitilis, stimulates antibiotic production in Streptomyces species. J Microbiol Biotechnol 19: 136–139. [DOI] [PubMed] [Google Scholar]
- 49. Sawai R, Suzuki A, Takano Y, Lee PC, Horinouchi S (2004) Phosphorylation of AfsR by multiple serine/threonine kinases in Streptomyces coelicolor A3(2). Gene 334: 53–61. [DOI] [PubMed] [Google Scholar]
- 50. Kieser HM, Henderson DJ, Chen CW, Hopwood DA (1989) A mutation of Streptomyces lividans which prevents intraplasmid recombination has no effect on chromosomal recombination. Mol Gen Genet 220: 60–64. [DOI] [PubMed] [Google Scholar]
- 51. Schneider D, Bruton CJ, Chater KF (2000) Duplicated gene clusters suggest an interplay of glycogen and trehalose metabolism during sequential stages of aerial mycelium development in Streptomyces coelicolor A3(2). Mol Gen Genet 263: 543–553. [DOI] [PubMed] [Google Scholar]
- 52. Penyige A, Keseru J, Fazakas F, Schmelczer I, Szirák K, et al. (2009) Analysis and identification of ADP-ribosylated proteins of Streptomyces coelicolor M145. J Microbiol 47: 549–556. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Quantitative data for the expression of Streptomyces coelicolor transcripts. The relative abundances of 7721 transcripts quantified in this work are shown as the log2 ratio of transcript abundance with respect to chromosomal DNA (Mg values) and to MI in both liquid (this work) and solid [16] cultures. Data are the average of three biological replicates and P-values are indicated. The 1901 transcripts quantified with very high confidence by Yagüe et al [16] in solid cultures (1901 transcripts) were compared with the transcripts obtained in this work for liquid cultures. Data were separated in four folders: three of them include data of Tables 1, 2 and 3; the other include data for transcripts without significant variations between solid and liquid cultures.
(XLSX)