Abstract
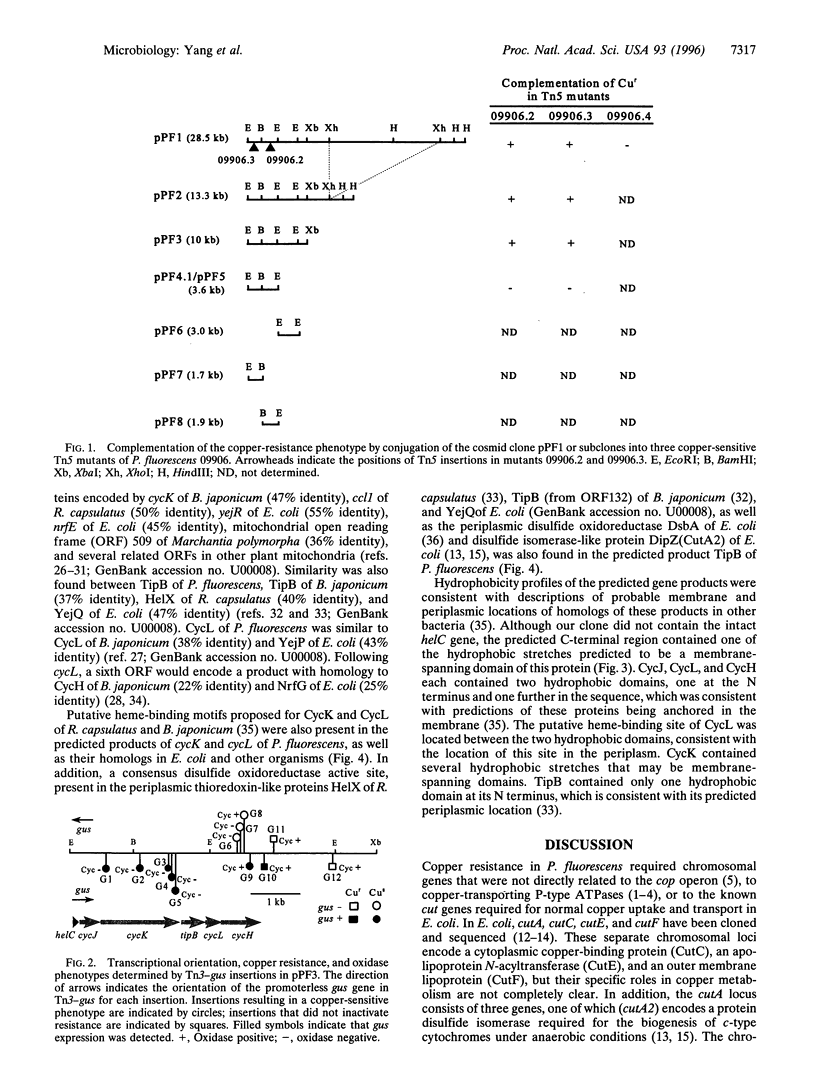
A chromosomal locus required for copper resistance and competitive fitness was cloned from a strain of Pseudomonas fluorescens isolated from copper-contaminated agricultural soil. Sequence analysis of this locus revealed six open reading frames with homology to genes involved in cytochrome c biogenesis in other bacteria, helC, cycJ, cycK, tipB, cycL, and cycH, with the closest similarity being to the aeg-46.5(yej) region of the Escherichia coli chromosome. The proposed functions of these genes in other bacteria include the binding, transport, and coupling of heme to apocytochrome c in the periplasm of these Gram-negative bacteria. Putative heme-binding motifs were present in the predicted products of cycK and cycL, and TipB contained a putative disulfide oxidoreductase active site proposed to maintain the heme-binding site of the apocytochrome in a reduced state for ligation of heme. Tn3-gus mutagenesis showed that expression of the genes was constitutive but enhanced by copper, and confirmed that the genes function both in copper resistance and production of active cytochrome c. However, two mutants in cycH were copper-sensitive and oxidase-positive, suggesting that the functions of these genes, rather than cytochrome c oxidase itself, were required for resistance to copper.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bardwell J. C., McGovern K., Beckwith J. Identification of a protein required for disulfide bond formation in vivo. Cell. 1991 Nov 1;67(3):581–589. doi: 10.1016/0092-8674(91)90532-4. [DOI] [PubMed] [Google Scholar]
- Beckman D. L., Kranz R. G. Cytochromes c biogenesis in a photosynthetic bacterium requires a periplasmic thioredoxin-like protein. Proc Natl Acad Sci U S A. 1993 Mar 15;90(6):2179–2183. doi: 10.1073/pnas.90.6.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckman D. L., Trawick D. R., Kranz R. G. Bacterial cytochromes c biogenesis. Genes Dev. 1992 Feb;6(2):268–283. doi: 10.1101/gad.6.2.268. [DOI] [PubMed] [Google Scholar]
- Blattner F. R., Burland V., Plunkett G., 3rd, Sofia H. J., Daniels D. L. Analysis of the Escherichia coli genome. IV. DNA sequence of the region from 89.2 to 92.8 minutes. Nucleic Acids Res. 1993 Nov 25;21(23):5408–5417. doi: 10.1093/nar/21.23.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bott M., Bolliger M., Hennecke H. Genetic analysis of the cytochrome c-aa3 branch of the Bradyrhizobium japonicum respiratory chain. Mol Microbiol. 1990 Dec;4(12):2147–2157. doi: 10.1111/j.1365-2958.1990.tb00576.x. [DOI] [PubMed] [Google Scholar]
- Bull P. C., Cox D. W. Wilson disease and Menkes disease: new handles on heavy-metal transport. Trends Genet. 1994 Jul;10(7):246–252. doi: 10.1016/0168-9525(94)90172-4. [DOI] [PubMed] [Google Scholar]
- Cha J. S., Cooksey D. A. Copper resistance in Pseudomonas syringae mediated by periplasmic and outer membrane proteins. Proc Natl Acad Sci U S A. 1991 Oct 15;88(20):8915–8919. doi: 10.1073/pnas.88.20.8915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe M., Reznikoff W. S. Anaerobically expressed Escherichia coli genes identified by operon fusion techniques. J Bacteriol. 1991 Oct;173(19):6139–6146. doi: 10.1128/jb.173.19.6139-6146.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooksey D. A., Azad H. R., Cha J. S., Lim C. K. Copper resistance gene homologs in pathogenic and saprophytic bacterial species from tomato. Appl Environ Microbiol. 1990 Feb;56(2):431–435. doi: 10.1128/aem.56.2.431-435.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooksey D. A. Copper uptake and resistance in bacteria. Mol Microbiol. 1993 Jan;7(1):1–5. doi: 10.1111/j.1365-2958.1993.tb01091.x. [DOI] [PubMed] [Google Scholar]
- Crooke H., Cole J. The biogenesis of c-type cytochromes in Escherichia coli requires a membrane-bound protein, DipZ, with a protein disulphide isomerase-like domain. Mol Microbiol. 1995 Mar;15(6):1139–1150. doi: 10.1111/j.1365-2958.1995.tb02287.x. [DOI] [PubMed] [Google Scholar]
- Dancis A., Yuan D. S., Haile D., Askwith C., Eide D., Moehle C., Kaplan J., Klausner R. D. Molecular characterization of a copper transport protein in S. cerevisiae: an unexpected role for copper in iron transport. Cell. 1994 Jan 28;76(2):393–402. doi: 10.1016/0092-8674(94)90345-x. [DOI] [PubMed] [Google Scholar]
- Fong S. T., Camakaris J., Lee B. T. Molecular genetics of a chromosomal locus involved in copper tolerance in Escherichia coli K-12. Mol Microbiol. 1995 Mar;15(6):1127–1137. doi: 10.1111/j.1365-2958.1995.tb02286.x. [DOI] [PubMed] [Google Scholar]
- García-Horsman J. A., Barquera B., Rumbley J., Ma J., Gennis R. B. The superfamily of heme-copper respiratory oxidases. J Bacteriol. 1994 Sep;176(18):5587–5600. doi: 10.1128/jb.176.18.5587-5600.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez D. H., Bonnard G., Grienenberger J. M. A gene involved in the biogenesis of c-type cytochromes is co-transcribed with a ribosomal protein gene in wheat mitochondria [corrected]. Curr Genet. 1993 Sep;24(3):248–255. doi: 10.1007/BF00351799. [DOI] [PubMed] [Google Scholar]
- Gupta S. D., Lee B. T., Camakaris J., Wu H. C. Identification of cutC and cutF (nlpE) genes involved in copper tolerance in Escherichia coli. J Bacteriol. 1995 Aug;177(15):4207–4215. doi: 10.1128/jb.177.15.4207-4215.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K. L., Merchant S. Coordinate expression of coproporphyrinogen oxidase and cytochrome c6 in the green alga Chlamydomonas reinhardtii in response to changes in copper availability. EMBO J. 1995 Mar 1;14(5):857–865. doi: 10.1002/j.1460-2075.1995.tb07067.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ish-Horowicz D., Burke J. F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981 Jul 10;9(13):2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. A., Hendson M., Panopoulos N. J., Schroth M. N. Molecular cloning, chromosomal mapping, and sequence analysis of copper resistance genes from Xanthomonas campestris pv. juglandis: homology with small blue copper proteins and multicopper oxidase. J Bacteriol. 1994 Jan;176(1):173–188. doi: 10.1128/jb.176.1.173-188.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellano M. A., Cooksey D. A. Nucleotide sequence and organization of copper resistance genes from Pseudomonas syringae pv. tomato. J Bacteriol. 1988 Jun;170(6):2879–2883. doi: 10.1128/jb.170.6.2879-2883.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills S. D., Jasalavich C. A., Cooksey D. A. A two-component regulatory system required for copper-inducible expression of the copper resistance operon of Pseudomonas syringae. J Bacteriol. 1993 Mar;175(6):1656–1664. doi: 10.1128/jb.175.6.1656-1664.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills S. D., Lim C. K., Cooksey D. A. Purification and characterization of CopR, a transcriptional activator protein that binds to a conserved domain (cop box) in copper-inducible promoters of Pseudomonas syringae. Mol Gen Genet. 1994 Aug 15;244(4):341–351. doi: 10.1007/BF00286685. [DOI] [PubMed] [Google Scholar]
- Oda K., Yamato K., Ohta E., Nakamura Y., Takemura M., Nozato N., Akashi K., Kanegae T., Ogura Y., Kohchi T. Gene organization deduced from the complete sequence of liverwort Marchantia polymorpha mitochondrial DNA. A primitive form of plant mitochondrial genome. J Mol Biol. 1992 Jan 5;223(1):1–7. doi: 10.1016/0022-2836(92)90708-r. [DOI] [PubMed] [Google Scholar]
- Odermatt A., Suter H., Krapf R., Solioz M. Primary structure of two P-type ATPases involved in copper homeostasis in Enterococcus hirae. J Biol Chem. 1993 Jun 15;268(17):12775–12779. [PubMed] [Google Scholar]
- Olsen G. J., Woese C. R., Overbeek R. The winds of (evolutionary) change: breathing new life into microbiology. J Bacteriol. 1994 Jan;176(1):1–6. doi: 10.1128/jb.176.1.1-6.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page M. D., Ferguson S. J. Cloning and sequence analysis of cycH gene from Paracoccus denitrificans: the cycH gene product is required for assembly of all c-type cytochromes, including cytochrome c1. Mol Microbiol. 1995 Jan;15(2):307–318. doi: 10.1111/j.1365-2958.1995.tb02245.x. [DOI] [PubMed] [Google Scholar]
- Phung L. T., Ajlani G., Haselkorn R. P-type ATPase from the cyanobacterium Synechococcus 7942 related to the human Menkes and Wilson disease gene products. Proc Natl Acad Sci U S A. 1994 Sep 27;91(20):9651–9654. doi: 10.1073/pnas.91.20.9651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramseier T. M., Winteler H. V., Hennecke H. Discovery and sequence analysis of bacterial genes involved in the biogenesis of c-type cytochromes. J Biol Chem. 1991 Apr 25;266(12):7793–7803. [PubMed] [Google Scholar]
- Ritz D., Bott M., Hennecke H. Formation of several bacterial c-type cytochromes requires a novel membrane-anchored protein that faces the periplasm. Mol Microbiol. 1993 Aug;9(4):729–740. doi: 10.1111/j.1365-2958.1993.tb01733.x. [DOI] [PubMed] [Google Scholar]
- Ritz D., Thöny-Meyer L., Hennecke H. The cycHJKL gene cluster plays an essential role in the biogenesis of c-type cytochromes in Bradyrhizobium japonicum. Mol Gen Genet. 1995 Apr 10;247(1):27–38. doi: 10.1007/BF00425818. [DOI] [PubMed] [Google Scholar]
- Rogers S. D., Bhave M. R., Mercer J. F., Camakaris J., Lee B. T. Cloning and characterization of cutE, a gene involved in copper transport in Escherichia coli. J Bacteriol. 1991 Nov;173(21):6742–6748. doi: 10.1128/jb.173.21.6742-6748.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster W., Combettes B., Flieger K., Brennicke A. A plant mitochondrial gene encodes a protein involved in cytochrome c biogenesis. Mol Gen Genet. 1993 May;239(1-2):49–57. doi: 10.1007/BF00281600. [DOI] [PubMed] [Google Scholar]
- Staskawicz B., Dahlbeck D., Keen N., Napoli C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1987 Dec;169(12):5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thöny-Meyer L., Ritz D., Hennecke H. Cytochrome c biogenesis in bacteria: a possible pathway begins to emerge. Mol Microbiol. 1994 Apr;12(1):1–9. doi: 10.1111/j.1365-2958.1994.tb00988.x. [DOI] [PubMed] [Google Scholar]
- Trenor C., 3rd, Lin W., Andrews N. C. Novel bacterial P-type ATPases with histidine-rich heavy-metal-associated sequences. Biochem Biophys Res Commun. 1994 Dec 30;205(3):1644–1650. doi: 10.1006/bbrc.1994.2856. [DOI] [PubMed] [Google Scholar]
- Voloudakis A. E., Bender C. L., Cooksey D. A. Similarity between Copper Resistance Genes from Xanthomonas campestris and Pseudomonas syringae. Appl Environ Microbiol. 1993 May;59(5):1627–1634. doi: 10.1128/aem.59.5.1627-1634.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C. H., Menge J. A., Cooksey D. A. Mutations Affecting Hyphal Colonization and Pyoverdine Production in Pseudomonads Antagonistic toward Phytophthora parasitica. Appl Environ Microbiol. 1994 Feb;60(2):473–481. doi: 10.1128/aem.60.2.473-481.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C. H., Menge J. A., Cooksey D. A. Role of copper resistance in competitive survival of Pseudomonas fluorescens in soil. Appl Environ Microbiol. 1993 Feb;59(2):580–584. doi: 10.1128/aem.59.2.580-584.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]