Abstract
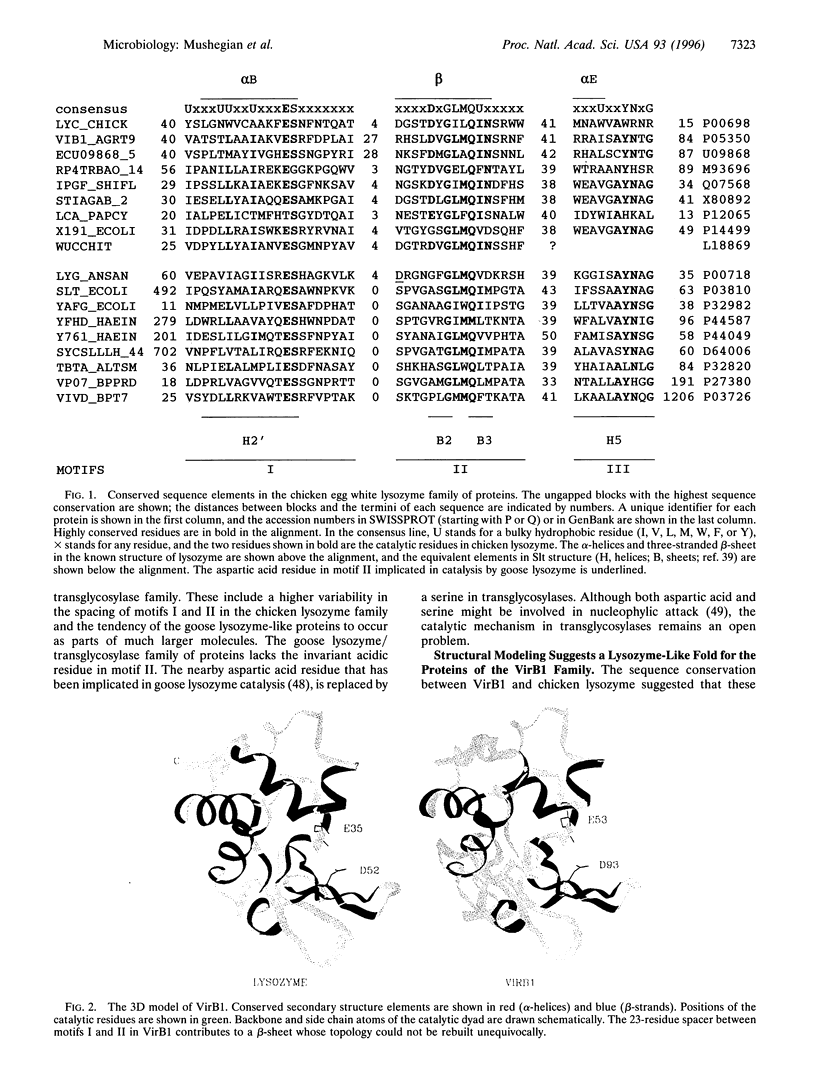
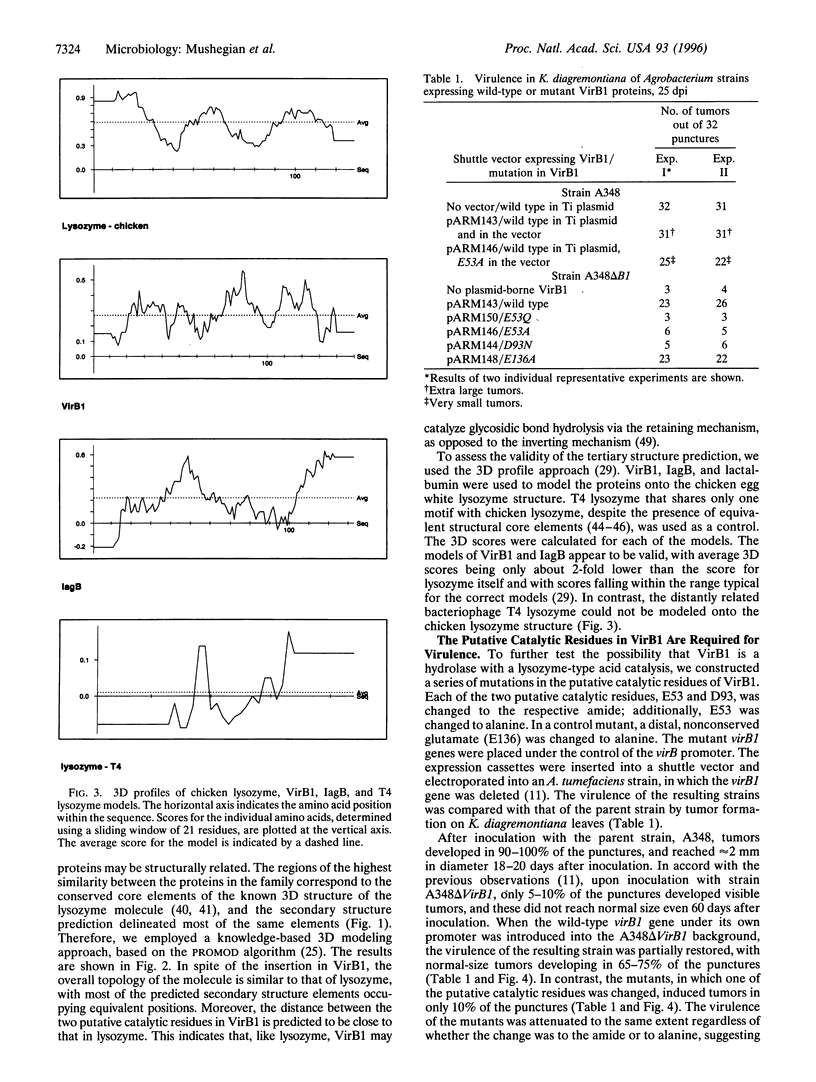
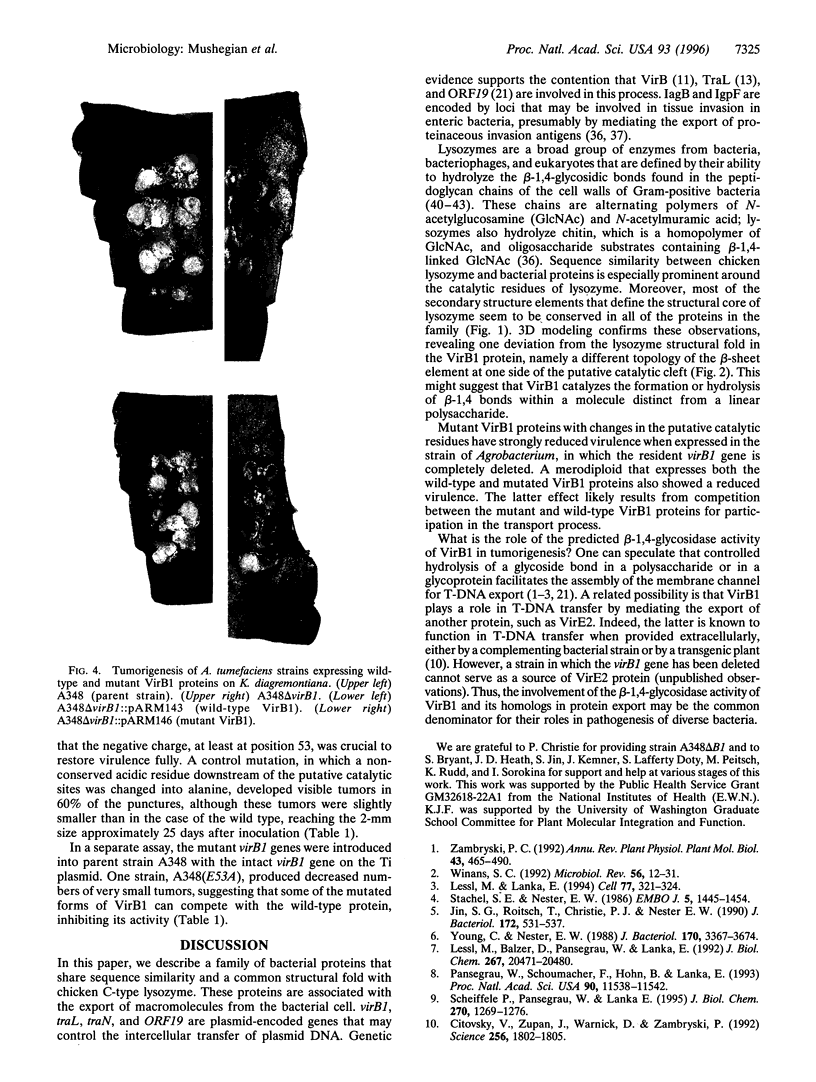
We describe a conserved family of bacterial gene products that includes the VirB1 virulence factor encoded by tumor-inducing plasmids of Agrobacterium spp., proteins involved in conjugative DNA transfer of broad-host-range bacterial plasmids, and gene products that may be involved in invasion by Shigella spp. and Salmonella enterica. Sequence analysis and structural modeling show that the proteins in this group are related to chicken egg white lysozyme and are likely to adopt a lysozyme-like structural fold. Based on their similarity to lysozyme, we predict that these proteins have glycosidase activity. Iterative data base searches with three conserved sequence motifs from this protein family detect a more distant relationship to bacterial and bacteriophage lytic transglycosylases, and goose egg white lysozyme. Two acidic residues in the VirB1 protein of Agrobacterium tumefaciens form a putative catalytic dyad, Each of these residues was changed into the corresponding amide by site-directed mutagenesis. Strains of A. tumefaciens that express mutated VirB1 proteins have a significantly reduced virulence. We hypothesize that many bacterial proteins involved in export of macromolecules belong to a widespread class of hydrolases and cleave beta-1,4-glycosidic bonds as part of their function.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allaoui A., Ménard R., Sansonetti P. J., Parsot C. Characterization of the Shigella flexneri ipgD and ipgF genes, which are located in the proximal part of the mxi locus. Infect Immun. 1993 May;61(5):1707–1714. doi: 10.1128/iai.61.5.1707-1714.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Boguski M. S., Gish W., Wootton J. C. Issues in searching molecular sequence databases. Nat Genet. 1994 Feb;6(2):119–129. doi: 10.1038/ng0294-119. [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bayer M., Eferl R., Zellnig G., Teferle K., Dijkstra A., Koraimann G., Högenauer G. Gene 19 of plasmid R1 is required for both efficient conjugative DNA transfer and bacteriophage R17 infection. J Bacteriol. 1995 Aug;177(15):4279–4288. doi: 10.1128/jb.177.15.4279-4288.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beijersbergen A., Smith S. J., Hooykaas P. J. Localization and topology of VirB proteins of Agrobacterium tumefaciens. Plasmid. 1994 Sep;32(2):212–218. doi: 10.1006/plas.1994.1057. [DOI] [PubMed] [Google Scholar]
- Berger B. R., Christie P. J. Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J Bacteriol. 1994 Jun;176(12):3646–3660. doi: 10.1128/jb.176.12.3646-3660.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger B. R., Christie P. J. The Agrobacterium tumefaciens virB4 gene product is an essential virulence protein requiring an intact nucleoside triphosphate-binding domain. J Bacteriol. 1993 Mar;175(6):1723–1734. doi: 10.1128/jb.175.6.1723-1734.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky V., Zupan J., Warnick D., Zambryski P. Nuclear localization of Agrobacterium VirE2 protein in plant cells. Science. 1992 Jun 26;256(5065):1802–1805. doi: 10.1126/science.1615325. [DOI] [PubMed] [Google Scholar]
- Davies G., Henrissat B. Structures and mechanisms of glycosyl hydrolases. Structure. 1995 Sep 15;3(9):853–859. doi: 10.1016/S0969-2126(01)00220-9. [DOI] [PubMed] [Google Scholar]
- Dijkstra B. W., Thunnissen A. M. 'Holy' proteins. II: The soluble lytic transglycosylase. Curr Opin Struct Biol. 1994 Dec;4(6):810–813. doi: 10.1016/0959-440x(94)90261-5. [DOI] [PubMed] [Google Scholar]
- Fullner K. J., Stephens K. M., Nester E. W. An essential virulence protein of Agrobacterium tumefaciens, VirB4, requires an intact mononucleotide binding domain to function in transfer of T-DNA. Mol Gen Genet. 1994 Dec 15;245(6):704–715. doi: 10.1007/BF00297277. [DOI] [PubMed] [Google Scholar]
- Graus H., Hödl A., Wallner P., Högenauer G. The sequence of the leading region of the resistance plasmid R1. Nucleic Acids Res. 1990 Feb 25;18(4):1046–1046. doi: 10.1093/nar/18.4.1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L., Sander C. Structural similarity of plant chitinase and lysozymes from animals and phage. An evolutionary connection. FEBS Lett. 1994 Feb 28;340(1-2):129–132. doi: 10.1016/0014-5793(94)80187-8. [DOI] [PubMed] [Google Scholar]
- Jin S. G., Roitsch T., Christie P. J., Nester E. W. The regulatory VirG protein specifically binds to a cis-acting regulatory sequence involved in transcriptional activation of Agrobacterium tumefaciens virulence genes. J Bacteriol. 1990 Feb;172(2):531–537. doi: 10.1128/jb.172.2.531-537.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L. N., Cheetham J., McLaughlin P. J., Acharya K. R., Barford D., Phillips D. C. Protein-oligosaccharide interactions: lysozyme, phosphorylase, amylases. Curr Top Microbiol Immunol. 1988;139:81–134. doi: 10.1007/978-3-642-46641-0_4. [DOI] [PubMed] [Google Scholar]
- Kirby A. J. Illuminating the ancient retainer. Nat Struct Biol. 1996 Feb;3(2):107–108. doi: 10.1038/nsb0296-107. [DOI] [PubMed] [Google Scholar]
- Kirby A. J. Mechanism and stereoelectronic effects in the lysozyme reaction. CRC Crit Rev Biochem. 1987;22(4):283–315. doi: 10.3109/10409238709086959. [DOI] [PubMed] [Google Scholar]
- Koonin E. V., Rudd K. E. A conserved domain in putative bacterial and bacteriophage transglycosylases. Trends Biochem Sci. 1994 Mar;19(3):106–107. doi: 10.1016/0968-0004(94)90201-1. [DOI] [PubMed] [Google Scholar]
- Koonin E. V., Tatusov R. L. Computer analysis of bacterial haloacid dehalogenases defines a large superfamily of hydrolases with diverse specificity. Application of an iterative approach to database search. J Mol Biol. 1994 Nov 18;244(1):125–132. doi: 10.1006/jmbi.1994.1711. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lessl M., Balzer D., Pansegrau W., Lanka E. Sequence similarities between the RP4 Tra2 and the Ti VirB region strongly support the conjugation model for T-DNA transfer. J Biol Chem. 1992 Oct 5;267(28):20471–20480. [PubMed] [Google Scholar]
- Lessl M., Lanka E. Common mechanisms in bacterial conjugation and Ti-mediated T-DNA transfer to plant cells. Cell. 1994 May 6;77(3):321–324. doi: 10.1016/0092-8674(94)90146-5. [DOI] [PubMed] [Google Scholar]
- Lüthy R., Bowie J. U., Eisenberg D. Assessment of protein models with three-dimensional profiles. Nature. 1992 Mar 5;356(6364):83–85. doi: 10.1038/356083a0. [DOI] [PubMed] [Google Scholar]
- Malcolm B. A., Rosenberg S., Corey M. J., Allen J. S., de Baetselier A., Kirsch J. F. Site-directed mutagenesis of the catalytic residues Asp-52 and Glu-35 of chicken egg white lysozyme. Proc Natl Acad Sci U S A. 1989 Jan;86(1):133–137. doi: 10.1073/pnas.86.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miras I., Hermant D., Arricau N., Popoff M. Y. Nucleotide sequence of iagA and iagB genes involved in invasion of HeLa cells by Salmonella enterica subsp. enterica ser. Typhi. Res Microbiol. 1995 Jan;146(1):17–20. doi: 10.1016/0923-2508(96)80267-1. [DOI] [PubMed] [Google Scholar]
- Monzingo A. F., Marcotte E. M., Hart P. J., Robertus J. D. Chitinases, chitosanases, and lysozymes can be divided into procaryotic and eucaryotic families sharing a conserved core. Nat Struct Biol. 1996 Feb;3(2):133–140. doi: 10.1038/nsb0296-133. [DOI] [PubMed] [Google Scholar]
- Pansegrau W., Schoumacher F., Hohn B., Lanka E. Site-specific cleavage and joining of single-stranded DNA by VirD2 protein of Agrobacterium tumefaciens Ti plasmids: analogy to bacterial conjugation. Proc Natl Acad Sci U S A. 1993 Dec 15;90(24):11538–11542. doi: 10.1073/pnas.90.24.11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peitsch M. C., Jongeneel C. V. A 3-D model for the CD40 ligand predicts that it is a compact trimer similar to the tumor necrosis factors. Int Immunol. 1993 Feb;5(2):233–238. doi: 10.1093/intimm/5.2.233. [DOI] [PubMed] [Google Scholar]
- Pohlman R. F., Genetti H. D., Winans S. C. Common ancestry between IncN conjugal transfer genes and macromolecular export systems of plant and animal pathogens. Mol Microbiol. 1994 Nov;14(4):655–668. doi: 10.1111/j.1365-2958.1994.tb01304.x. [DOI] [PubMed] [Google Scholar]
- Raghavan N., Freedman D. O., Fitzgerald P. C., Unnasch T. R., Ottesen E. A., Nutman T. B. Cloning and characterization of a potentially protective chitinase-like recombinant antigen from Wuchereria bancrofti. Infect Immun. 1994 May;62(5):1901–1908. doi: 10.1128/iai.62.5.1901-1908.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B., Sander C. Prediction of protein secondary structure at better than 70% accuracy. J Mol Biol. 1993 Jul 20;232(2):584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- Scheiffele P., Pansegrau W., Lanka E. Initiation of Agrobacterium tumefaciens T-DNA processing. Purified proteins VirD1 and VirD2 catalyze site- and strand-specific cleavage of superhelical T-border DNA in vitro. J Biol Chem. 1995 Jan 20;270(3):1269–1276. doi: 10.1074/jbc.270.3.1269. [DOI] [PubMed] [Google Scholar]
- Schuler G. D., Altschul S. F., Lipman D. J. A workbench for multiple alignment construction and analysis. Proteins. 1991;9(3):180–190. doi: 10.1002/prot.340090304. [DOI] [PubMed] [Google Scholar]
- Shirasu K., Kado C. I. Membrane location of the Ti plasmid VirB proteins involved in the biosynthesis of a pilin-like conjugative structure on Agrobacterium tumefaciens. FEMS Microbiol Lett. 1993 Aug 1;111(2-3):287–294. doi: 10.1111/j.1574-6968.1993.tb06400.x. [DOI] [PubMed] [Google Scholar]
- Stachel S. E., Nester E. W. The genetic and transcriptional organization of the vir region of the A6 Ti plasmid of Agrobacterium tumefaciens. EMBO J. 1986 Jul;5(7):1445–1454. doi: 10.1002/j.1460-2075.1986.tb04381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens K. M., Roush C., Nester E. Agrobacterium tumefaciens VirB11 protein requires a consensus nucleotide-binding site for function in virulence. J Bacteriol. 1995 Jan;177(1):27–36. doi: 10.1128/jb.177.1.27-36.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov R. L., Altschul S. F., Koonin E. V. Detection of conserved segments in proteins: iterative scanning of sequence databases with alignment blocks. Proc Natl Acad Sci U S A. 1994 Dec 6;91(25):12091–12095. doi: 10.1073/pnas.91.25.12091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorstenson Y. R., Kuldau G. A., Zambryski P. C. Subcellular localization of seven VirB proteins of Agrobacterium tumefaciens: implications for the formation of a T-DNA transport structure. J Bacteriol. 1993 Aug;175(16):5233–5241. doi: 10.1128/jb.175.16.5233-5241.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thunnissen A. M., Dijkstra A. J., Kalk K. H., Rozeboom H. J., Engel H., Keck W., Dijkstra B. W. Doughnut-shaped structure of a bacterial muramidase revealed by X-ray crystallography. Nature. 1994 Feb 24;367(6465):750–753. doi: 10.1038/367750a0. [DOI] [PubMed] [Google Scholar]
- Ward J. E., Akiyoshi D. E., Regier D., Datta A., Gordon M. P., Nester E. W. Characterization of the virB operon from an Agrobacterium tumefaciens Ti plasmid. J Biol Chem. 1988 Apr 25;263(12):5804–5814. [PubMed] [Google Scholar]
- Ward J. E., Akiyoshi D. E., Regier D., Datta A., Gordon M. P., Nester E. W. Correction: characterization of the virB operon from Agrobacterium tumefaciens Ti plasmid. J Biol Chem. 1990 Mar 15;265(8):4768–4768. [PubMed] [Google Scholar]
- Ward J. E., Jr, Dale E. M., Christie P. J., Nester E. W., Binns A. N. Complementation analysis of Agrobacterium tumefaciens Ti plasmid virB genes by use of a vir promoter expression vector: virB9, virB10, and virB11 are essential virulence genes. J Bacteriol. 1990 Sep;172(9):5187–5199. doi: 10.1128/jb.172.9.5187-5199.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver L. H., Grütter M. G., Matthews B. W. The refined structures of goose lysozyme and its complex with a bound trisaccharide show that the "goose-type" lysozymes lack a catalytic aspartate residue. J Mol Biol. 1995 Jan 6;245(1):54–68. doi: 10.1016/s0022-2836(95)80038-7. [DOI] [PubMed] [Google Scholar]
- Weiss A. A., Johnson F. D., Burns D. L. Molecular characterization of an operon required for pertussis toxin secretion. Proc Natl Acad Sci U S A. 1993 Apr 1;90(7):2970–2974. doi: 10.1073/pnas.90.7.2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winans S. C. Two-way chemical signaling in Agrobacterium-plant interactions. Microbiol Rev. 1992 Mar;56(1):12–31. doi: 10.1128/mr.56.1.12-31.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young C., Nester E. W. Association of the virD2 protein with the 5' end of T strands in Agrobacterium tumefaciens. J Bacteriol. 1988 Aug;170(8):3367–3374. doi: 10.1128/jb.170.8.3367-3374.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]




