Abstract
The ability to design proteins with high affinity and selectivity for any given small molecule would have numerous applications in biosensing, diagnostics, and therapeutics, and is a rigorous test of our understanding of the physiochemical principles that govern molecular recognition phenomena. Attempts to design ligand binding proteins have met with little success, however, and the computational design of precise molecular recognition between proteins and small molecules remains an “unsolved problem”1. We describe a general method for the computational design of small molecule binding sites with pre-organized hydrogen bonding and hydrophobic interfaces and high overall shape complementary to the ligand, and use it to design protein binding sites for the steroid digoxigenin (DIG). Of 17 designs that were experimentally characterized, two bind DIG; the highest affinity design has the lowest predicted interaction energy and the most pre-organized binding site in the set. A comprehensive binding-fitness landscape of this design generated by library selection and deep sequencing was used to guide optimization of binding affinity to a picomolar level, and two X-ray co-crystal structures of optimized complexes show atomic level agreement with the design models. The designed binder has a high selectivity for DIG over the related steroids digitoxigenin, progesterone, and β-estradiol, which can be reprogrammed through the designed hydrogen-bonding interactions. Taken together, the binding fitness landscape, co-crystal structures, and thermodynamic binding parameters illustrate how increases in binding affinity can result from distal sequence changes that limit the protein ensemble to conformers making the most energetically favorable interactions with the ligand. The computational design method presented here should enable the development of a new generation of biosensors, therapeutics, and diagnostics.
Current approaches for designing ligand binding proteins for medical2 and biotechnological uses rely upon raising antibodies against a target antigen in immunized animals3,4 and/or performing laboratory directed evolution of proteins with an existing low affinity for the desired ligand5-7, both of which offer incomplete control over molecular details. Computational design has the potential to provide a general, complementary approach for small molecule recognition in which design features and selectivity can be rationally programmed. Structural and biophysical characterization of previously designed ligand binding proteins has revealed numerous discrepancies with the design models, however, and it was concluded that protein-ligand interaction design is an unsolved problem1,8. The lack of accuracy in programming protein-small molecule interactions also contributes to low catalytic efficiencies of computationally designed enzymes relative to their natural counterparts9-14. The development of robust computational methods for the design of small molecule-binding proteins with high affinity and selectivity would have wide-ranging applications.
The goal of existing methods for computational enzyme design is to promote catalysis by creating energetically favorable hydrogen bonding, van der Waals, and electrostatic interactions to a high-energy reaction transition state(s) and/or intermediate(s). Although these interactions are also important for stabilizing the bound ground-state conformations of protein-small molecule complexes, they are not the sole determinant of small molecule binding. We developed a computational method for designing ligand binding proteins with two properties characteristic of naturally occurring binding sites in addition to specific energetically favorable interactions with the ligand: (1) high overall shape complementarity to the ligand, and (2) structural pre-organization in the unbound protein state, which minimizes entropy loss upon ligand binding15,16. To program in specific interactions with the small molecule, disembodied binding sites are created by positioning amino acid side chains around the ligand in orientations optimal for hydrogen bonding and other energetically favorable interactions and then placed at geometrically compatible binding sites in a set of scaffold protein structures using RosettaMatch17. The surrounding side chain identities and conformations are then optimized to generate additional protein-ligand interactions and buttressing protein-protein interactions (Fig 1a). Designs with protein-small molecule shape complementarity below those typical of native protein complexes18 or having interface side chain conformations with low Boltzmann-weighted probabilities in the unbound state16 are then discarded.
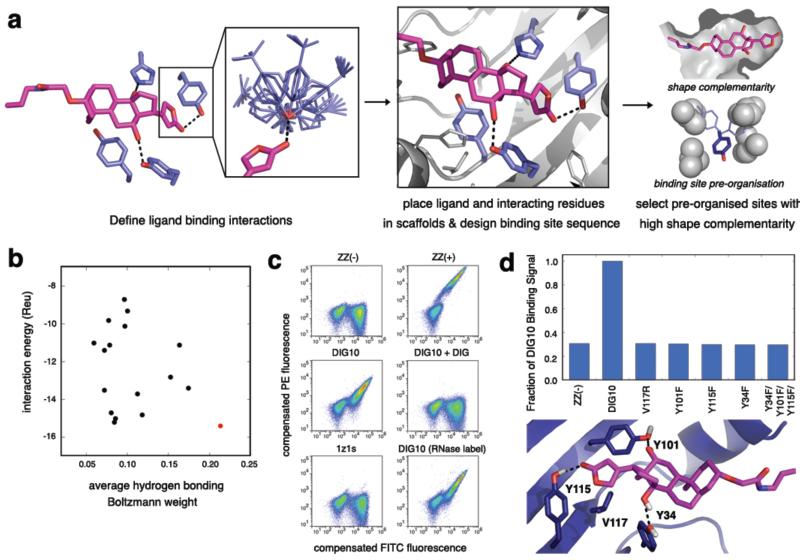
Figure 1. Computational Design Methodology and Experimental Validation.
a, Overview of the computational design procedure. First, the geometric positions of a set of pre-chosen interaction side chains are defined with respect to the ligand (left panel), and rotamers for each interaction side chain are enumerated (left panel, inset). Second, a set of scaffolds is searched for backbones that can accommodate all of the desired interactions. For cases in which all chosen interaction residues can be placed in the scaffold protein and orient the ligand in the native binding cavity with no backbone clashes, the binding site sequence is optimized for binding affinity (center panel). Designs having native-like properties, such as high shape complementarity and binding site pre-organization, are chosen for experimental characterization (right panel). b, Ranking of the 17 experimentally characterized DIG designs by ligand interaction energy (Rosetta energy units, Reu) and the average (geometric mean) Boltzmann weight of the conformations of the side chains that hydrogen bond to the ligand. DIG10, depicted in red, scores the best by both metrics. c, Flow cytometric analysis of yeast cells expressing computationally designed proteins as part of a surface-targeted fusion protein with a C-terminal c-Myc tag. Yeast surface expression and DIG binding were confirmed by labeling the cells with a fluorescein (FITC)-conjugated anti-c-Myc antibody and a pre-incubated mixture of 2.7 μM biotinylated DIG-functionalized BSA (~10 DIG/BSA) and phycoerythrin (PE)-conjugated streptavidin, respectively. Cell populations shown are a negative control for binding (ZZ(-)), an anti-DIG antibody serving as a positive control for binding (ZZ(+)), DIG10, DIG10 in the presence of excess (730 μM) unlabeled DIG competitor, and scaffold 1z1s. DIG10 labeled with 2.7 μM biotinylated DIG-functionalized RNase (~6 DIG/RNase) is also shown. d, On-yeast substitutions of DIG10 designed hydrogen-bonding residues Tyr34, Tyr101, and Tyr115 to Phe and binding cavity residue Val117 to Arg reduces expressing-population compensated mean binding (PE) signals to background nonbinding (ZZ(-)) levels.
We used the method to design proteins that bind the steroid digoxigenin (DIG; Supplementary Fig. 1), the aglycone of digoxin, a cardiac glycoside used to treat heart disease19, and a commonly used non-radioactive biomolecular labeling reagent20. Anti-DIG antibodies are routinely administered to treat overdoses of digoxin, which has a narrow therapeutic window21, and are widely used to detect biomolecules in applications such as fluorescence in situ hybridization20. We created idealized DIG binding sites with hydrogen bonds from Tyr or His to the lactone carbonyl oxygen and both hydroxyl groups of DIG and hydrophobic packing interactions between Tyr, Phe, or Trp and the steroid ring system (Fig. 1a). These interactions were embedded in designed binding sites with high shape complementarity to DIG as outlined above, and 17 designs were selected for experimental characterization based on computed binding affinity, shape complementarity, and the extent of binding site pre-organization in the unbound state (Fig. 1b and Supplementary Tables 1 and 2).
Binding of the designed proteins to DIG was probed by yeast surface display22 and flow cytometry using DIG-functionalized bovine serum albumin (DIG-BSA) or ribonuclease (DIG-RNase). DIG5 and DIG10 bound to both labels (Fig. 1c and Supplementary Fig. 2), and binding was reduced to background levels when ~1 mM of unlabeled DIG was added as a competitor (Fig. 1c and Supplementary Fig. 3).
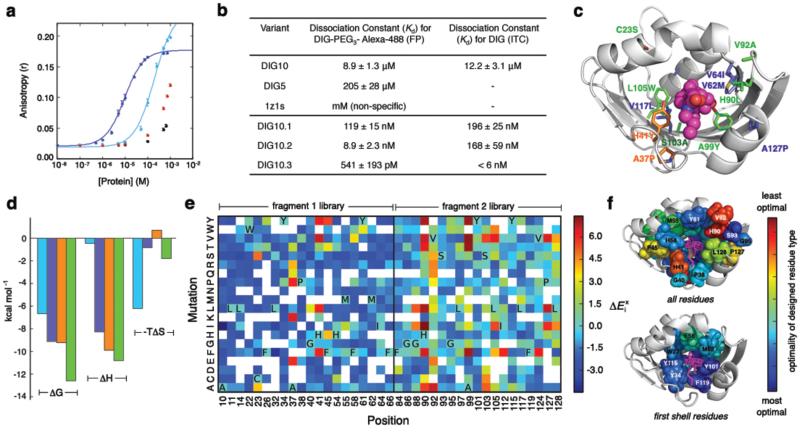
Fluorescence polarization (FP) measurements with purified proteins and Alexa488 fluorophore-conjugated DIG (DIG-PEG3-Alexa488) indicated affinities in the low to mid micromolar range, with DIG10 binding more tightly (Fig. 2a,b). Isothermal titration calorimetry (ITC) measurements confirmed that the affinity of DIG10 for unlabeled DIG is the same as that for the labeled DIG conjugate (Fig. 2b, Supplementary Fig. 4, and Supplementary Table 3). The scaffold from which both DIG5 and DIG10 derive, PDB ID 1z1s, a protein of unknown function from Pseudomonas aeruginosa, does not bind to either label (Fig. 1c and Supplementary Fig. 3a) when expressed on the yeast surface or to DIG-PEG3-Alexa488 in solution (Fig. 2a), suggesting that the binding activities of both proteins are mediated by the computationally designed interfaces. Indeed, substitution of small nonpolar residues in the central binding pockets of DIG5 and DIG10 with arginines resulted in complete loss of binding, and mutation of the designed hydrogen-bonding tyrosine and histidine residues to the nearly isosteric phenylalanine reduced binding; for DIG10, substitution of any of the three interacting tyrosines abolished binding completely (Fig. 1d and Supplementary Fig. 5). Optimization of DIG10 by a single round of mutagenesis and selections using yeast surface display and fluorescence-activated cell sorting (FACS) identified small-to-large hydrophobic amino acid changes that increase binding affinity 75-fold by enhancing the enthalpic contribution to binding, yielding DIG10.1 (Fig. 2b,c,d, Supplementary Figs 4,6-8, and Supplementary Table 3).
Figure 2. Affinity Maturation.
a, Equilibrium fluorescence polarization measurements of DIG-PEG3-Alexa488 treated with increasing amounts of DIG10 (blue), DIG5 (cyan), scaffold 1z1s (black), and negative control bovine serum albumin (red). Solid lines represent fits to the data to obtain dissociation constants (Kd values). Error bars represent standard deviations for at least three independent measurements. b, Kd values of relevant designs and affinity matured DIG10 variants. FP and ITC were used to measure the affinities for DIG-PEG3-Alexa488 and unlabeled DIG, respectively. The high affinity of DIG10.3 for DIG precluded measurement of a reliable Kd value by ITC. The differences in the Kd values observed by the two methods may be due to interactions with the PEG linker present in the DIG-PEG3-Alexa488 conjugate; DIG10.2, which shows the largest discrepancy, has two mutations at the cavity entrance. c, Mutations identified during affinity maturation to generate DIG10.1 (blue), DIG10.2 (orange), and DIG10.3 (green) mapped on to the computational model of DIG10.3. d, ITC thermodynamic parameters of DIG10 (cyan), DIG10.1 (blue), DIG10.2 (orange), and DIG10.3 (green) variants binding to DIG. The ΔG used to determine TΔS for DIG10.3 was calculated from the FP Kd value. e, Binding fitness landscape of DIG10.1 probed by deep sequencing. The effect of each amino acid substitution at 39 binding site residues on binding () was assessed by determining the log2 ratios of the frequencies of substitutions to each amino acid at each position after selection for DIG binding to the frequencies of the substitutions in the unselected population. Colored grids represent single point mutations having ≥7 counts in the unselected N-terminal (fragment 1) and C-terminal (fragment 2) libraries, where red and blue indicate high enrichment and depletion, respectively. White grids show mutations for which there were not enough sequences in the unselected library to make a statistically significant conclusion about function. The initial DIG10.1 amino acid at each position is indicated in bold using its one-letter amino acid code. f, The optimality of each DIG10.1 input residue type mapped onto the computational model of DIG10.1. Optimality is defined as the positional Z-score: where x is the sum of enrichment values at position i, μ is the mean sum of enrichment values for all interrogated positions within the fragment library, and, σ is the standard deviation of the sums of enrichment values for all interrogated positions within the fragment library. Blue is very optimal (mutations to all other amino acids are disfavored) and red is suboptimal (mutations are preferred).
To provide feedback for improving the overall design methodology and to evaluate the contribution of each residue in the DIG10.1 binding site, we used next generation sequencing to generate a comprehensive binding fitness map23-25. A library of variants with ~1-3 substitutions at 39 designed interface positions in DIG10.1 was generated using doped oligonucleotide mutagenesis, displayed on yeast, and subjected to selections using a monovalent DIG-PEG3-biotin conjugate (Supplementary Fig. 9). Variants with increased affinity for DIG were isolated by FACS, and next generation sequencing was used to quantify the frequency of every single point mutation in the unselected and selected populations. A large majority of the interrogated variants were depleted in the selected population relative to the unselected input library, suggesting that most of the designed residues are close to optimal for binding (Fig. 2e,f and Supplementary Fig. 10). In particular, mutation of the three designed hydrogen bonding residues, Tyr34, Tyr101, and Tyr115, to any other amino acid was disfavored. Several large hydrophobic residues that pack against the ligand in the computational model are also optimal for binding (e.g. Phe66 and Phe119). Besides A99, which contacts DIG directly, most of the observed mutations that improve binding are located in the second coordination shell of the ligand and fall into two functional categories: (1) core substitutions tolerating mutation to chemically-similar amino acids (e.g. Leu105 and Cys23), and (2) solvent-exposed loop amino acids having high sequence entropy (e.g. His90, Val92). The single best clone in the libraries, DIG10.2, features two of the most highly enriched mutations, Ala37Pro and His41Tyr (Fig. 2b,c and Supplementary Figs 6, 8, and 11).
Combination of the most highly enriched substitutions in a library followed by selections led to DIG10.3 (Supplementary Figs 6 and 12), which binds both DIG and its cardiac glycoside derivative, digoxin, with picomolar affinity (Fig. 2b, Supplementary Fig. 13, and Supplementary Table 4), rivaling the affinities of anti-digoxin antibody therapeutics21 and an evolved single-chain variable anti-DIG antibody fragment7. FP-based affinity measurements of DIG10.3 and its Tyr knockouts suggest that the designed hydrogen bonds each contribute ~2 kcal/mol to binding energy (Supplementary Table 5 and Supplementary Fig. 8).
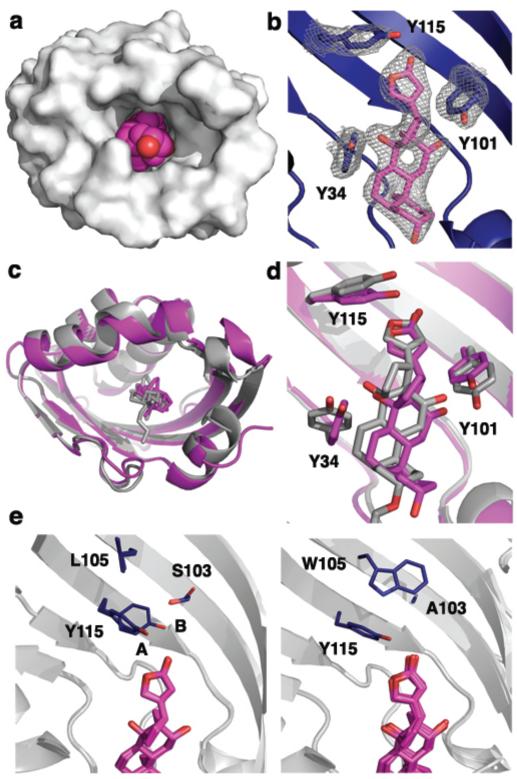
The crystal structures of DIG10.2 and DIG10.3 in complex with DIG were solved to 2.05 Å and 3.2 Å resolution, respectively (Fig. 3a,b and Supplementary Figs 14-16). The structure of DIG10.2 bound to DIG shows atomic-level agreement (average all atom root mean squared deviation (RMSD) of 0.54 Å) with the design model (Fig. 3c). The ligand-protein interface has high shape complementarity (Sc = 0.66) and there are no observable water molecules within the binding pocket. The DIG binding mode is nearly identical in the structure and the model, with an average RMSD of 0.99 Å for all 28 ligand heavy atoms (Fig. 3d). As anticipated, Tyr34, Tyr101, and Tyr115 make the designed hydrogen bonds with O3, O2, and O1 of DIG, respectively. Tyr41, a residue identified during affinity maturation, engages in an additional weak hydrogen bond with the terminal hydroxyl group of DIG (O5) (Supplementary Fig. 16). Of 27 non-glycine and non-alanine non-surface protein residues within ~10 Å of the ligand, 21 adopt rotamer conformations in the design model (Supplementary Fig. 17), including Tyr101 and Tyr115 (in chain B) as well as the first-shell packing residues Trp22, Phe58, and Phe119. The structure of DIG10.3 bound to DIG also agrees with the design model (average all-atom RMSD of 0.68 Å) (Supplementary Fig. 18).
Figure 3. Crystal Structures of the DIG10.2- and DIG10.3-DIG Complexes.
a, Surface representation of the DIG10.2-DIG complex showing the high overall shape complementarity of the interface. DIG is depicted in magenta spheres. DIG10.2 is a dimer and crystallized with four copies in the asymmetric unit. b, 2Fo - Fc omit map electron density of DIG interacting with Tyr34, Tyr101, and Tyr115 contoured at 1.0 sigma. c, Backbone superposition of the crystal structure of the DIG10.2-DIG complex (magenta) with the computational model (gray) shows close agreement between the two. d, Binding site backbone superposition shows that the ligand and the three programmed Tyr hydroxyl groups are in their designed conformations. e, Configurational side chain entropy between the four crystallographic copies of the DIG10.2-DIG (left panel) complex and chains A, B, C, H, and I of the DIG10.3-DIG (right panel) complex. The side chains of DIG10.3 at positions 103, 105, and 115 each adopt only a single rotamer. DIG10.2 Tyr115 conformation A features a more canonical hydrogen-bonding geometry than that of conformation B.
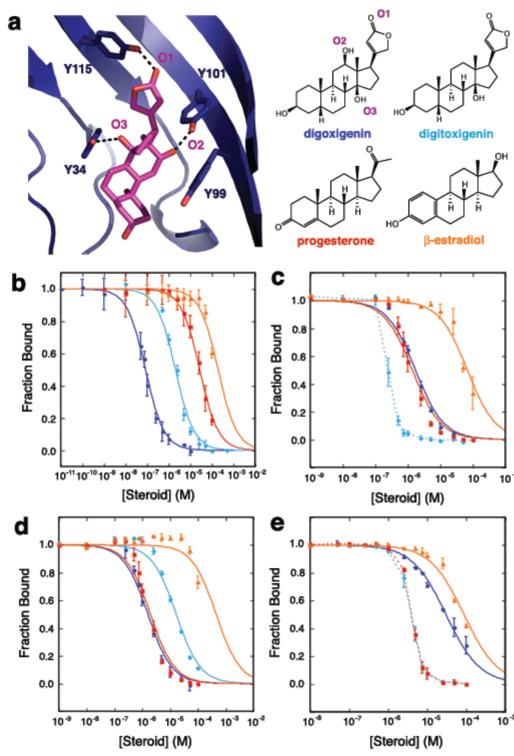
We assessed the binding specificity of DIG10.3 by determining affinities for a series of related steroids by equilibrium competition fluorescence polarization assays. Experiments with DIG, digitoxigenin, progesterone, and β-estradiol showed a decrease in affinity corresponding to the loss of one, two, and three hydrogen bonds respectively (assuming ~1.8 kcal/mol per hydrogen bond26), as expected from the structure if these compounds bind in the same orientation as DIG (Fig. 4a,b and Supplementary Table 4). We next investigated whether the observed steroid selectivity could be reprogrammed by mutagenesis of the key hydrogen-bonding tyrosine residues. The variants Tyr101Phe, Tyr34Phe, and Tyr34Phe/Tyr99Phe/Tyr101Phe show clear preferences for more hydrophobic steroids in a predictable manner that depends on the hydrogen bonding capabilities of both the protein and the steroid. Mutation of Tyr101 to Phe eliminates the DIG-specific hydrogen bond with O2 of DIG and provides better hydrophobic packing for the other three steroids lacking a hydroxyl group at that position (Fig. 4c). Substitution of Tyr34 with Phe removes a hydrogen bond to the C14 hydroxyl groups of both DIG (O3) and digitoxigenin, enhancing the preference for progesterone and maintaining the relative binding order of DIG and digitoxigenin due to the intact DIG-specific Tyr101-DIG O2 bond (Fig. 4d). Mutation of Tyr101, Tyr34, and binding site residue Tyr99 to Phe leads to decreased binding affinity for DIG and increased affinity for the more hydrophobic steroids (Fig. 4e). These results confirm that the selectivity of DIG10.3 for DIG is conferred largely through the designed hydrogen-bonding interactions and demonstrate how selectivity can be programmed through positive design alone by control of designed protein-ligand hydrogen bonding and non-polar interactions.
Figure 4. Steroid Binding Selectivity.
a, The x-ray crystal structure of the DIG10.3-DIG complex (left panel) and the chemical structures of steroids interrogated in equilibrium competition fluorescence polarization assays (right panel). b, Steroid selectivity profile of DIG10.3. Solid lines represent fits to the data to obtain half-maximal inhibitory concentrations (IC50 values) and error bars indicate standard deviations for at least three independent measurements. c, Steroid selectivity profile of DIG10.3 Tyr101Phe. Dashed lines show qualitative assessments of the inhibitory effects for cases in which the data could not be fit due to experimental limitations (see Supplementary Methods). d, Steroid selectivity profile of DIG10.3 Tyr34Phe. e, Steroid selectivity profile of DIG10.3 Tyr34Phe/Tyr99Phe/Tyr101F.
Comparison of the properties of successful and unsuccessful designs provides a test of the hypotheses underlying the design methodology. Although all 17 designed proteins had high computed shape complementarity to DIG by construction, the DIG10 design, which had the highest affinity for DIG, had the most favorable computed ligand interaction energies and was predicted to have the most pre-organized binding site (Fig. 1b and Supplementary Table 6), suggesting that these attributes should continue to be the focus of future design methodology development. One potential avenue for obtaining more favorable interaction energies would be the incorporation of additional binding site backbone flexibility to achieve more tightly packed binding sites: the observation that substitution of small hydrophobic interface residues to larger amino acids increased binding affinity indicates that the original DIG10 design was under-packed.
The binding fitness landscape in combination with the x-ray co-crystal structures highlight the importance of second shell interactions in stabilizing binding competent conformations. The fitness landscape favors substitution of Leu105, adjacent to the key hydrogen-bonding residue Tyr115, to Trp or other large hydrophobic residues (Fig. 2e). Tyr115 exhibits obvious conformational side chain heterogeneity in the four independent protein subunits of the 2.05 Å resolution DIG10.2 crystal structure. Mutation of Leu105 to Trp results in a single uniform set of side chain conformations for Tyr115 in the lower resolution DIG10.3 design (which contains nine independently visualized subunits) that features a more canonical hydrogen bond geometry between Tyr115 and DIG than the alternative DIG10.2 Tyr115 conformation (Fig. 3e and Supplementary Fig. 19). The higher affinity of DIG10.3 might result from a higher population of the pre-organized, higher affinity conformation of the protein15,27, and ITC studies confirmed that entropic as well as enthalpic factors contribute to its enhanced binding affinity (Fig. 2d and Supplementary Table 3). Indeed, all key hydrogen-bonding tyrosines, particularly Tyr115, have higher computed Boltzmann weighted side chain probabilities in apo-DIG10.3 than in apo-DIG10.2 and apo-DIG10 (Supplementary Table 7). Similarly, reduced backbone conformational entropy is likely responsible for the increased fitness of substitutions increasing β-sheet propensity at positions 90 and 92 which likely stabilize a more ordered extended strand backbone conformation (Fig. 2e). That conformational flexibility is selected against during affinity maturation suggests that accounting for free energy gaps between binding-competent and alternative states of the binding site28, possibly by better assessing side chain entropy or explicitly designing second shell buttressing interactions for key contacts, should aid in achieving high affinity in the next generation of computationally designed ligand binding proteins.
The binding affinity of DIG10.3 is within a factor of two of that of the widely used anti-DIG antibodies,21 and because it is stable for extended periods at ambient temperatures (Supplementary Fig. 20) and can be expressed at high levels in bacteria, it could provide more cost-effective alternative for biotechnological applications and for therapeutic purposes if it can be engineered for compatibility with the human immune response. With continued improvement in the methodology and feedback from experimental results, computational protein design should provide an increasingly powerful approach to creating a new generation of small molecule receptors for synthetic biology, therapeutic scavengers for toxic compounds, and robust binding domains for diagnostic devices.
Methods
Computational methods
Full details for all computational methods are given in Supplementary Methods. Example command lines and RosettaScripts31 design protocols are provided in Supplementary Data. Source code is freely available to academic users through the Rosetta Commons agreement (http://www.rosettacommons.org/). Design models, the scaffold library, and scripts for running design calculations are provided on the Baker lab website.
Matching
A set of 401 scaffolds was searched for backbones that can accommodate five pre-defined side chain interactions with DIG using RosettaMatch17 (Supplementary Table 8). This set contained scaffolds previously used for design projects within our lab10,13,32 as well as structural homologs of a subset of these scaffolds that are known to tolerate mutations. Full details are given in Supplementary Methods.
Rosetta sequence design
Two successive rounds of sequence design were employed. The purpose of the first was to maximize binding affinity for the ligand33. The goal of the second was to minimize protein destabilization due to aggressive scaffold mutagenesis while maintaining the binding interface designed during the first round. During the latter round, ligand-protein interactions were up-weighted by a factor of 1.5 relative to intra-protein interactions to ensure that binding energy was preserved. Two different criteria were used to minimize protein destabilization: (1) native scaffold residues identities were favored by 1.5 Rosetta energy units (Reu), and (2) no more than five residues were allowed to change from residue types observed in a multiple sequence alignment (MSA) of the scaffold if (a) these residues were present in the MSA with a frequency greater than 0.6 and, (b) if the calculated ∆∆G for mutation of the scaffold residue to alanine34 was greater than 1.5 Reu in the context of the scaffold sequence. In some design calculations, identities of the matched hydrogen bonding residues were allowed to vary according to the MSA and ∆∆G criteria described above. Designs having fewer than three hydrogen bonds between the protein and the ligand were rejected.
Design evaluation
Designs were evaluated on interface energy, ligand solvent exposed surface area, ligand orientation, shape complementarity, and apo-protein binding site pre-organization. The latter was enforced by two metrics: (1) explicitly introducing second-shell amino acids that hold the pre-selected residues in place using Foldit35, and (2) selecting designs having rotamer Boltzmann probabilities16 > 0.1 for at least one hydrogen bonding residue (Supplementary Table 6). High shape complementary was enforced using by rejecting designs having Sc < 0.5. Shape complimentary was computed using the CCP4 package v.6.0.236 using the Sc program18 and the Rosetta radii library. All designs were evaluated for local sequence secondary structure compatibility, and those predicted to have backbone conformations that varied by > 0.8 Å from their native scaffold were rejected (see Supplementary Methods).
General experimental methods
Detailed procedures for the syntheses of DIG-BSA-biotin, DIG-RNase-biotin, DIG-PEG3-biotin, and DIG-PEG3-Alexa488, as well as protein expression, purification, and crystallization, cloning, and mutagenesis methods are given in Supplementary Methods. Details about fluorescence polarization binding assays, isothermal titration calorimetry, gel filtration analysis, analytical ultracentrifugation experiments, and circular dichroism protein stability measurements are also provided in Supplementary Methods.
Yeast surface display
Designed proteins were tested for binding using yeast-surface display22. Yeast surface protein expression was monitored by binding of anti-cmyc FITC to the C-terminal myc epitope tag of the displayed protein. DIG binding was assessed by quantifying the phycoerythrin (PE) fluorescence of the displaying yeast population following incubation with DIG-BSA-biotin, DIG-RNase-biotin, or DIG-PEG3-biotin, and streptavidin-phycoerythrin (SAPE). In a typical experiment using DIG-BSA-biotin or DIG-RNase-biotin, cells were resuspended in a premixed solution of PBSF (PBS + 1 g/L of BSA) containing a 1:100 dilution of anti-cmyc FITC, 2.66 μM DIG-BSA-biotin or DIG-RNase biotin, and 664 nM SAPE for 2-4 hr at 4 °C. Cellular fluorescence was monitored on an Accuri C6 flow cytometer using a 488 nm laser for excitation and a 575 nm band pass filter for emission. Phycoerythrin fluorescence was compensated to minimize bleed-over contributions from the FITC channel. Competition assays with free digoxigenin were performed as above except that between 750 μM and 1.5 mM of digoxigenin was added to each labeling reaction mixture. Full details are given in Supplementary Methods.
Affinity maturation
Detailed procedures for constructing and selecting all libraries, including those for deep sequencing, are provided in Supplementary Methods. Yeast surface display library selections were conducted on a Cytopeia inFlux cell sorter using increasingly stringent fluorescence gates. In all labeling reactions for selections, care was taken to maintain at least a 10-fold molar excess of label to cell surface protein. Cell surface protein molarity was estimated by assuming that an O.D.600 of 1.0 = 1e7 cells/mL and that each cell displays 50,000 copies of protein22. For each round of sorting, we sorted at least 10 times the theoretical library size. FlowJo software v. 7.6 was used to analyze all data. Cell sorting parameters and statistics for all selections are given in Supplementary Table 9.
Next-generation sequencing
Two sequencing libraries based on DIG10.1 were assembled by recursive PCR: an N-terminal library (fragment 1 library) and a C-terminal library (fragment 2 library). To introduce mutations, we used degenerate PAGE-purified oligos in which 39 selected positions within the binding site were doped with a small amount of each non-native base at a level expected to yield 1-2 mutations per gene (TriLink BioTechnologies) (Supplementary Table 10). Yeast cells were transformed with DNA insert and restriction-digested pETCON37. Surface protein expression was induced22 and cells were labeled with anti-cymc-FITC and sorted for protein expression. Expressing cells were recovered, induced, labeled with 100 nM of DIG-PEG3-biotin for > 3 hrs at 4 °C and then SAPE and anti-cymc-FITC for 8 min at 4 °C, and then sorted. For each library, clones having binding signals higher than that of DIG10.1 were collected (Supplementary Fig. 9). To reduce noise from the first round of cell sorting, the sorted libraries were recovered, induced, and subjected to a second round of sorting using the same conditions (Supplementary Methods).
Library DNA was prepared as described25. Illumina adapter sequences and unique library barcodes were appended to each library pool by PCR amplification using population-specific primers (Supplementary Table 11). DNA was sequenced in paired-end mode on an Illumina MiSeq using a 300-cycle reagent kit and custom primers (see Supplementary Methods). Of a total 5,630,105 paired-end reads, 2,531,653 reads were mapped to library barcodes (Supplementary Table 12). For each library, paired end reads were fused and filtered for quality (Phred ≥ 30). The resulting full-length reads were aligned against DIG10.1 using Enrich38. For single mutations having ≥ 7 counts in the original input library, a relative enrichment ratio between the input library and each selected library was calculated23-25. The effect of each amino acid substitution at 39 binding site residues on binding () is given as the log base 2 frequency of observing mutation x at position i in the selected versus the unselected population, relative to that of the DIG10.1 residue (orig) at position i:
Fluorescence polarization binding assays
Fluorescence polarization-based affinity measurements of designs and their evolved variants were performed as described30 using Alexa488-conjugated DIG (DIG-PEG3-Alexa488). Fluorescence anisotropy (r) was measured in 96-well plate format on a SpectraMax M5e microplate reader (Molecular Devices) with λex = 485 nM and λem = 538 nM using a 515 nm emission cutoff filter. Fluorescence polarization equilibrium competition binding assays were used to determine the binding affinities of DIG10.3 and its variants for unlabeled digoxigenin, digitoxigenin, progesterone, β-estradiol, and digoxin. The inhibition constant for each protein-ligand interaction, Ki, was calculated from the measured total unlabeled ligand producing 50% binding signal inhibition (I50) and the Kd of the protein-label interaction according to a model accounting for receptor-depletion conditions30. Full details are provided in Supplementary Methods.
Isothermal Titration Calorimetry
ITC studies were performed on an iTC200 microcalorimeter (MicroCal) at 25 °C in PBS, pH 7.4. Ligand solutions were prepared by diluting a stock solution of digoxigenin (100 mM in 100% DMSO) into the flow-through of the last buffer aliquot used to exchange the protein (final DMSO concentrations were 1-3%). ITC titration data were integrated and analyzed with Origin 7.0 (MicroCal). Full details are provided in Supplementary Methods.
Supplementary Material
Acknowledgements
We thank Dr. E.-M. Strauch and Dr. P.-S. Huang for providing the ZZ/pETCON and S2/pETCON plasmids, respectively, and Dr. B. Shen for assistance with data processing, modeling, and refinement of the x-ray crystal structures. We thank Dr. J. P. Sumida for assistance with analytical ultracentrifugation data collection, processing, and analysis. Analytical ultracentrifugation experiments were performed at the University of Washington Analytical Biopharmacy Core, which is supported by the Washington State Life Sciences Discovery Fund and the Center for the Intracellular Delivery of Biologics. We thank Dr. S. Fleishman, Dr. O. Khersonsky, and Dr. P.-S. Huang for comments on the manuscript. J.W.N. acknowledges a pre-doctoral fellowship from the National Human Genome Research Institute under the Interdisciplinary Training in Genome Sciences program (T32 HG00035). This work was supported by grants from the HHMI and DTRA to D.B., a grant from DARPA to D.B. and B.L.S., a grant from the Swiss National Science Foundation to K.J., and NSF grant MCB1121896 to C.G.K.
Footnotes
Author Contributions
C.E.T., S.D.K., and D.B. designed the research. S.D.K., C.E.T., and J.D. performed computations. S.D.K. wrote program code. C.E.T. experimentally characterized the designs, constructed libraries, performed affinity maturation and deep sequencing selections, and conducted binding and biochemical studies of DIG10. J.D. characterized DIG5. J.W.N. analyzed deep sequencing data. C.E.T. and J.D. prepared protein samples for crystallographic analysis. L.D. and B.S. crystallized the protein samples and analyzed crystallographic data. A.S. and K.J. synthesized DIG-PEG3-biotin and DIG-PEG3-Alexa488. C.E.T. prepared protein samples for ITC studies, and W.J. and C.G.K. performed ITC experiments and analyzed ITC data. C.E.T., S.D.K., and D.B. analyzed data and wrote the manuscript. All authors discussed the results and commented on the manuscript.
Author Information
The crystal structures of DIG10.2 and DIG10.3 bound to DIG have been deposited in the RCSB Protein Data Bank under the accession numbers 4J8T (DIG10.2) and 4J9A (DIG10.3). The authors declare no competing financial interest.
Supplementary Information is available in the online version of the paper.
Full Methods and any associated references are available in the online version of the paper.
References
- 1.Schreier B, Stumpp C, Wiesner S, Höcker B. Computational design of ligand binding is not a solved problem. Proc. Natl. Acad. Sci. USA. 2009;106:18491–18496. doi: 10.1073/pnas.0907950106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Wolf FA, Brett GM. Ligand-binding proteins: their potential for application in systems for controlled delivery and uptake of ligands. Pharmacol. Rev. 2000;52:207–236. [PubMed] [Google Scholar]
- 3.Hunter MM, Margolies MN, Ju A, Haber E. High-affinity monoclonal antibodies to the cardiac glycoside, digoxin. J. Immunol. 1982;129:1165–1172. [PubMed] [Google Scholar]
- 4.Shen XY, Orson FM, Kosten TR. Vaccines against drug abuse. Clin. Pharmacol. Ther. 2012;91:60–70. doi: 10.1038/clpt.2011.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradbury ARM, Sidhu S, Dübel S, McCafferty J. Beyond natural antibodies: the power of in vitro display technologies. Nat. Biotechnol. 2011;29:245–254. doi: 10.1038/nbt.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brustad EM, Arnold FH. Optimizing non-natural protein function with directed evolution. Curr. Opin. Chem. Biol. 2011;15:201–210. doi: 10.1016/j.cbpa.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen G, et al. Isolation of high-affinity ligand-binding proteins by periplasmic expression with cytometric screening (PECS) Nat. Biotechnol. 2001;19:537–542. doi: 10.1038/89281. [DOI] [PubMed] [Google Scholar]
- 8.Telmer PG, Shilton BH. Structural studies of an engineered zinc biosensor reveal an unanticipated mode of zinc binding. J. Mol. Biol. 2005;354:829–840. doi: 10.1016/j.jmb.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 9.Baker D. An exciting but challenging road ahead for computational enzyme design. Protein Sci. 2010;19:1817–1819. doi: 10.1002/pro.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang L, et al. De novo computational design of retro-Aldol enzymes. Science. 2008;319:1387–1391. doi: 10.1126/science.1152692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khare SD, Fleishman SJ. Emerging themes in the computational design of novel enzymes and protein-protein interfaces. FEBS Lett. 2013;587:1147–1154. doi: 10.1016/j.febslet.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 12.Khersonsky O, et al. Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59. Proc. Natl. Acad. Sci. USA. 2012;109:10358–10363. doi: 10.1073/pnas.1121063109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Röthlisberger D, et al. Kemp elimination catalysts by computational enzyme design. Nature. 2008;453:190–195. doi: 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]
- 14.Wang L, et al. Structural analyses of covalent enzyme-substrate analog complexes reveal strengths and limitations of de novo enzyme design. J. Mol. Biol. 2012;415:615–625. doi: 10.1016/j.jmb.2011.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boehr DD, Nussinov R, Wright PE. The role of dynamic conformational ensembles in biomolecular recognition. Nat. Chem. Biol. 2009;5:789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fleishman SJ, Khare SD, Koga N, Baker D. Restricted sidechain plasticity in the structures of native proteins and complexes. Protein Sci. 2011;20:753–757. doi: 10.1002/pro.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zanghellini A, et al. New algorithms and an in silico benchmark for computational enzyme design. Protein Sci. 2006;15:2785–2794. doi: 10.1110/ps.062353106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J. Mol. Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 19.The Digitalis Investigation Group The effect of digoxin on mortality and morbidity in patients with heart failure. N. Engl. J. Med. 1997;336:525–533. doi: 10.1056/NEJM199702203360801. [DOI] [PubMed] [Google Scholar]
- 20.Eisel D, Seth O, Grünewald-Janho, Kruchen B. DIG application manual for nonradioactive in situ hybridization. 4th ed. Roche Diagnostics, Penzberg: 2008. [Google Scholar]
- 21.Flanagan RJ, Jones AL. Fab antibody fragments: some applications in clinical toxicology. Drug Safety. 2004;27:1115–1133. doi: 10.2165/00002018-200427140-00004. [DOI] [PubMed] [Google Scholar]
- 22.Chao G, et al. Isolating and engineering human antibodies using yeast surface display. Nat. Protoc. 2006;1:755–768. doi: 10.1038/nprot.2006.94. [DOI] [PubMed] [Google Scholar]
- 23.Fowler DM, et al. High-resolution mapping of protein sequence-function relationships. Nat. Methods. 2010;7:741–746. doi: 10.1038/nmeth.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McLaughlin RN, Jr, Poelwijk FJ, Raman A, Gosal WS, Ranganathan R. The spatial architecture of protein function and adaptation. Nature. 2012;491:138–142. doi: 10.1038/nature11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whitehead TA, et al. Optimization of affinity, specificity and function of designed influenza inhibitors using deep sequencing. Nat. Biotechnol. 2012;30:543–548. doi: 10.1038/nbt.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fersht AR, et al. Hydrogen bonding and biological specificity analysed by protein engineering. Nature. 1985;314:235–238. doi: 10.1038/314235a0. [DOI] [PubMed] [Google Scholar]
- 27.Frederick KK, Marlow MS, Valentine KG, Wand AJ. Conformational entropy in molecular recognition by proteins. Nature. 2007;448:325–329. doi: 10.1038/nature05959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fleishman SJ, Baker D. Role of the biomolecular energy gap in protein design, structure, and evolution. Cell. 2012;149:262–273. doi: 10.1016/j.cell.2012.03.016. [DOI] [PubMed] [Google Scholar]
- 29.Kuhlman B, Baker D. Native protein sequences are close to optimal for their structures. Proc. Natl. Acad. Sci. USA. 2000;97:10383–10388. doi: 10.1073/pnas.97.19.10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rossi AM, Taylor CW. Analysis of protein-ligand interactions by fluorescence polarization. Nat. Protoc. 2011;6:365–387. doi: 10.1038/nprot.2011.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fleishman SJ, et al. RosettaScripts: A scripting language interface to the Rosetta macromolecular modeling suite. PLOS ONE. 2011;6:e20161. doi: 10.1371/journal.pone.0020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siegel JB, et al. Computational design of an enzyme catalyst for a stereoselective bimolecular Diels-Alder reaction. Science. 2010;329:309–313. doi: 10.1126/science.1190239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richter F, Leaver-Fay A, Khare SD, Bjelic S, Baker D. De novo enzyme design using Rosetta3. PLOS ONE. 2011;6:e19230. doi: 10.1371/journal.pone.0019230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kellogg EH, Leaver-Fay A, Baker D. Role of conformational sampling in computing mutation-induced changes in protein structure and stability. Proteins. 2010;79:830–838. doi: 10.1002/prot.22921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cooper S, et al. Predicting protein structures with a multiplayer online game. Nature. 2010;466:756–760. doi: 10.1038/nature09304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Collaborative Computational Project, N. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. Sect. D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 37.Benatuil L, Perez JM, Belk J, Hsieh C-M. An improved yeast transformation method for the generation of very large human antibody libraries. Protein Eng. Des. Sel. 2010;23:155–159. doi: 10.1093/protein/gzq002. [DOI] [PubMed] [Google Scholar]
- 38.Fowler DM, Araya CL, Gerard W, Fields S. Enrich: software for analysis of protein function by enrichment and depletion of variants. Bioinformatics. 2011;27:3430–3431. doi: 10.1093/bioinformatics/btr577. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.