Abstract
The field of Membrane Protein Structural Biology has grown significantly since its first landmark in 1985 with the first three-dimensional atomic resolution structure of a membrane protein. Nearly twenty-six years later, the crystal structure of the beta2 adrenergic receptor in complex with G protein has contributed to another landmark in the field leading to the 2012 Nobel Prize in Chemistry. At present, more than 350 unique membrane protein structures solved by X-ray crystallography (http://blanco.biomol.uci.edu/mpstruc/exp/list, Stephen White Lab at UC Irvine) are available in the Protein Data Bank. The advent of genomics and proteomics initiatives combined with high-throughput technologies, such as automation, miniaturization, integration and third-generation synchrotrons, has enhanced membrane protein structure determination rate. X-ray crystallography is still the only method capable of providing detailed information on how ligands, cofactors, and ions interact with proteins, and is therefore a powerful tool in biochemistry and drug discovery. Yet the growth of membrane protein crystals suitable for X-ray diffraction studies amazingly remains a fine art and a major bottleneck in the field. It is often necessary to apply as many innovative approaches as possible. In this review we draw attention to the latest methods and strategies for the production of suitable crystals for membrane protein structure determination. In addition we also highlight the impact that third-generation synchrotron radiation has made in the field, summarizing the latest strategies used at synchrotron beamlines for screening and data collection from such demanding crystals. This article is part of a Special Issue entitled: Structural and biophysical characterisation of membrane protein-ligand binding.
Keywords: Membrane protein, Crystal dehydration, Crystal seeding, Macromolecular crystallography, In situ data collection, XFEL
Graphical abstract
Highlights
-
•
Overview of the most recent advances regarding the growth of membrane protein crystals
-
•
Rational design of new crystallization screens for membrane proteins
-
•
New automated method for dehydration of membrane proteins
-
•
High-throughput approach in seeding of membrane protein crystals
-
•
Recent developments in membrane protein structure determination
1. Introduction
Membrane proteins play a vital role in many critical biological processes. Nearly 30% of proteins in eukaryotic cells are known to be membrane proteins [1]. Mutations or improper folding of these proteins is associated with many known diseases such heart disease, cystic fibrosis, depression, obesity, cancer and many others. Currently approximately 60% of available drugs target membrane proteins of which G protein-coupled receptors (GPCRs) and ion channels constitute the largest groups [2,3]. Although most of the drugs commercially available have emerged through conventional drug discovery methods such as high-throughput screening (HTS), computational methods and functional assays, it is the structural information provided by the three-dimensional (3D) atomic structures that discloses details regarding the binding mode of such proteins. This information is critical in the rational design of better drugs with improved selectivity and pharmaceutical properties [4–7]. Since X-ray crystallography has been the only tool capable of delivering detailed empirical information on protein structures at atomic level, its use in drug discovery programs became popular and well established.
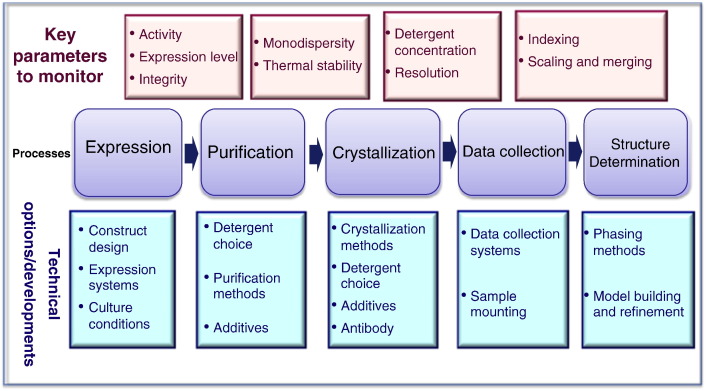
The first membrane protein structure solved by X-ray crystallography was reported in 1985 [8]. Since then more than 300 unique membrane protein structures have been solved using the same method (http://blanco.biomol.uci.edu/mpstruc/exp/list, Stephen White Lab at UC Irvine). Many high-resolution 3D structures of integral membrane proteins have proven to be fundamental for a better understanding of many biological processes [9–11]. Most recently the crystal structure of the beta2 adrenergic receptor in complex with the G protein was solved by Kobilka's group [12]. This structure has made an enormous contribution not only to biology but also to drug discovery by revealing the mechanism of action of GPCRs at the molecular level. However in spite of recent successes, the path to a high-resolution structure of a membrane protein still involves several bottlenecks including poor expression, limited extraction success, low purification yields and paucity of well-ordered 3D crystals (Fig. 1). Yet, the field of membrane protein structural biology is in a “log” phase. In recent years much effort has been put toward innovative developments to overcome the numerous obstacles associated with X-ray structure determination of membrane proteins. For instance much progress has been made regarding: (i) overexpression of recombinant membrane proteins in different expression hosts [13–18]; (ii) development of new detergents and lipids for more efficient solubilization and crystallization [19–22]; (iii) improvement in protein stability through mutations, deletions, engineering of fusion partners and monoclonal antibodies, to promote diffraction quality crystals [23–27]; (iv) developments in automation, miniaturization and integration which have contributed to the increasing number of initial crystallization conditions and crystal optimization strategies [28]; and (v) in synchrotron radiation and beamline developments [29]. This article provides an overview of the most recent advances regarding the growth of membrane protein crystals and how to best assess crystal quality-diffraction in a high-throughput fashion using synchrotron radiation.
Fig. 1.
Bottlenecks in membrane protein structure determination. Picture courtesy of Prof. So Iwata.
2. Overview of detergents
Detergents play a vital role in membrane protein structure determination. They are essential during the processes of solubilization, purification and crystallization. Once the protein of interest has been expressed at the required levels, it is usually necessary to extract it from its membrane environment. The biological membrane is a complex mosaic lipid bilayer in which membrane proteins interact closely with the nearby lipids and surrounding proteins. Structure and dynamics of integral membrane proteins (IMP) are intimately related to the properties of the surrounding phospholipids in the membrane [30]. Due to their amphipathic nature (hydrophobic tail and hydrophilic head) detergents used in membrane protein research are able to extract IMPs by disrupting the phospholipid bilayer without irreversibly disrupting the protein structure [31]. Detergents mimic the lipid membrane by surrounding the hydrophobic region of the IMP, generating a water-soluble protein–detergent complex (PDC). This prevents protein aggregation after its removal from the natural environment. The ability of a particular detergent to extract and solubilize a membrane protein is closely related to its aptitude to form micelles. In aqueous solution, the polar groups of the detergent form hydrogen bonds with water molecules, whereas the hydrophobic groups (tail) aggregate due to hydrophobic interactions. This results in organized spherical structures called micelles. Micelles in general are a few nanometers in diameter and have molecular weights of less than 100 kDa [19]. The minimal detergent concentration at which micelles are observed is called critical micelle concentration (CMC). At detergent concentrations below the CMC, only detergent monomers exist in solution whereas at detergent concentrations above the CMC, equilibrium between detergent monomers and micelles is observed. The CMC varies with physical factors such as pH, temperature and ionic strength. Another important physical property of micelles is the aggregation number. This is defined as the average number of detergent monomers in a micelle. While during protein extraction, the detergent concentration should be in excess of the CMC, during purification detergent concentration should be kept just above the CMC value [19,32]. Although it is important to keep in mind that lack of detergent can lead to protein aggregation, too much detergent can also lead to protein inactivation due to excess removal of essential lipids. Excess detergent can also interfere with the crystallization process leading to phase separation [33]. At high concentrations and low temperature, most detergents remain in their crystalline form. However, when in solution it is common for them to separate into two distinct phases (a detergent-poor and a detergent-rich phase). The temperature at which this occurs is called the cloud point. Cloud point is a specific characteristic of each detergent and should be taken into account when selecting a detergent for solubilization [34].
To date, a large number of detergents are commercially available. Yet many membrane proteins are still difficult to study due to the limited availability of a suitable detergent, not only for the solubilization and purification processes but also for the crystallization. Therefore, the development of new detergents is greatly welcomed by the membrane protein structural biology community [21].
2.1. Classification of detergents
In biology, detergents are conveniently classified according to the nature of their hydrophilic groups.
Ionic detergents possess a head group with a net charge that can either be positive (cationic) or negative (anionic). These detergents are well known for being harsh, mainly because of their ability to disrupt the hydrophobic interactions of the protein core leading to the unfolding or denaturing of the same. The most well known ionic detergents are sodium dodecyl sulfate (SDS), N-laurylsarcosine and sodium cholate. Bile acids are examples of anionic detergents with the cholic acid being one of the most widely used. They are unique for their “kidney-shaped” structure and also for being milder than other linear ionic detergents [35]. In general, the use of ionic detergents has been restricted to membrane proteins in which protein solubilization has proven potentially difficult or when the target protein is required in its denatured state.
Nonionic detergents contain uncharged hydrophilic head groups and are the most common detergents used in membrane protein research for solubilization, purification, stabilization, crystallization and functional assays. They are considered mild as they break the lipid–lipid and protein–lipid interactions rather than protein–protein interactions. Among the most popular nonionic detergents are the sugar based maltosides and glucosides. However it is important to note that detergents such as n-octyl-β-d-glucoside (β-OG) with short chains (C7–C10) may occasionally deactivate the target protein. Hence they are most commonly restricted to use in the solubilization process. On the other hand, detergents such as n-dodecyl-β-d-maltoside (DDM) and n-decyl-β-d-maltoside (DM) are frequently used in membrane protein research because of their mild and nondenaturing properties. A general rule has emerged in which shorter length detergents are considered particularly suitable for solubilization and crystallography while the longer chain detergents are used in the reconstitution procedures. Another advantage of sugar-based detergents is that they do not interfere with UV measurements, as is the case for Triton X-100. Although Triton and polyoxyethylene (e.g. C12E8) detergents are considered mild and nondenaturing, in general, they are extremely non-homogeneous detergents with large amounts of impurities.
Zwitterionic detergents combine properties of both ionic and nonionic detergents and in general, are milder than ionic detergents. Although they are not the most suitable detergents for membrane protein purification and functional assays, they have been successfully used in crystallization and NMR studies. The most extensively used zwitterionic detergents are the 3-[(3-Cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS), 3-[(3-Cholamidopropyl) dimethylammonio]-2-hydroxy-1-propanesulfonate (CHAPSO), lauryldimethylamine-N-oxide (LDAO) and fos-choline 12.
2.2. Detergent purity
In membrane protein research, the use of high purity detergents is always recommended. Although today most of the detergents commercially available for membrane proteins research are of high purity, researchers are advised to pay particular attention to the purity value (> 99%). It is also advisable to know by which analytical method the purity has been assessed, e.g. HPLC or TLC. Detergent impurities not only interact and modify proteins during extraction/purification, they are also able to interfere with the crystallization process itself. The most common impurities in detergents are hydrophobic alcohols, peroxides and α-isomers. The majority of these impurities are insoluble in water. Therefore, if a detergent solution exhibits cloudiness at a concentration at which it is known to be soluble, it may be an indication of contaminants. Because glucoside and maltoside detergents are prepared from their corresponding alcohols, traces of hydrophobic alcohols may appear in the final product. In general, the presence of these alcohols is not a problem during the protein extraction and solubilization. But the same does not hold true regarding the existence of peroxides. Peroxides are a result of oxidation and hydrolysis of polyoxyethylene detergents (e.g. Triton-100 and Tween) [36]. The presence of peroxides in detergent solutions can be very damaging to biological materials since they react with protein sulfhydryl (SH) groups and other non-protein molecules. Also important is the fact that sugar-based detergents are synthesized in both isomeric forms α and β. The β isomer form is the most common anomer used in membrane protein research. The main reasons for this are its higher solubility when comparing with the α-isomer and the presence of α-isomer at certain levels interferes with crystallization. Therefore, when using β isomer detergents they should contain as little as possible of the α form [37]. Finally, it is advisable to always prepare detergent solutions fresh, filter them before use and keep protected from exposure to the light.
2.3. Detergent choice
Unfortunately, the choice of detergent for membrane protein investigation still remains largely a matter of trial-and-error approach. However, a rational approach may be taken based on (i) the properties of the detergents, (ii) on the downstream work planned (biochemical and biophysical assays, NMR, CD or crystallographic studies) and (iii) on the type of membrane protein being studied (α-helical, β-barrel, GPCR or others). Whereas detergent head groups have a strong influence on the way detergents interact with proteins, the presence of acyl chains exerts influence on detergent CMC and aggregation number [38]. For this reason, anionic and cationic detergents are considered harsher detergents, followed by the zwitterionic and nonionic detergents. For structure determination of membrane proteins, nonionic and zwitterionic detergents are most commonly used. Maltosides and glucosides are the most popular for their mild properties with the maltosides being milder than the glucosides.
Recently, the use of the Green Fluorescent Protein (GFP) fused to the membrane protein terminus has proven to be a useful tool for the rapid selection of the most effective detergent for the extraction/solubilization [39,40]. Upon ultraviolet light excitation at 395 nm or 498 nm the GFP fluorophore emits green fluorescence light at 509 nm (emission peak). Therefore, the membrane protein–GFP fusion is easily monitored and visualized at any stage during overexpression, solubilization and purification by a variety of instruments such as fluorometers and fluorescence microscopes. The use of the Fluorescence-detection Size Exclusion Chromatography (FSEC) provides a rapid and efficient method in detergent screening by assessing monodispersity and stability of the target protein. The great advantage of this approach is the requirement of only nanogram quantities of unpurified protein. A full description of the method can be found in [39]. Finally, it is important to note that in many cases the best detergent for protein solubilization is not usually the most suitable detergent for the purification and crystallization of the target protein. Often detergent exchange is necessary before proceeding to the purification and crystallization stages.
2.4. New detergents
New detergents are emerging as additional tools for membrane protein research. In common with traditional detergents, these can be used during the processes of protein extraction, purification and crystallization. However, they have significantly improved protein stability and endorsement of crystallization. The most successful new class of detergents is the neopentyl glycol (NG) amphiphilic class. These are sugar-based detergents that have proved very effective in the crystallization process because of their innovative architecture and low CMC values [21,41,42]. Architecture of the NG amphiphiles features two maltose units in the hydrophilic head and two n-decyl chains in the lipophilic tail connected to a central quaternary carbon. The central quaternary carbon is derived from neopentyl glycol molecules and thus the name of neopentyl glycol. Another new class is the cholate-based detergents in which the three parallel α-hydoxy groups in the cholate skeleton have been replaced by uncharged polar groups such glycosides [43,44]. These detergent molecules present high facial amphiphilicity that is distinct from the end polarity present in the conventional head-to-tail detergents. Their unique facial amphiphilic structure provides specific properties advantageous in the processes of protein solubilization, stabilization and crystallization [43,44].
3. Crystallization of membrane proteins
Every crystal structure must first start with a crystal. This inconvenient truth presents a significant problem in the field of membrane protein structural biology. Crystallization remains one of the most challenging hurdles that every structural group must tackle and unfortunately the one about which least is known or certain. Membrane proteins differ significantly from their water-soluble counterparts; they must be gently cosseted by the protective coat of detergent that shields the hydrophobic surfaces and allows the crystallographer to extract, purify and handle the proteins away from their membrane environment. Therein lies the challenge on how to promote such dynamic ensembles of protein and detergent molecules, which are in rapid equilibrium with the surrounding solution, into crystal formation.
The traditional approach to membrane protein crystallization, whatever the crystallization method chosen, is the screening of numerous potential crystallization conditions. Crystallization is multiparametric process where a great variety of biochemical, chemical and physical parameters needs to be explored. Examples of these are purity and aggregation state of the protein, buffers and their pH values, temperature, precipitants and additives. This is an exceptionally challenging and time-consuming process that is often limited by the amount of protein available. Automation and miniaturization of the protein crystallization processes have greatly contributed to the efficiency and effectiveness of the experimental technique. At present, integrated crystallization systems can perform more than 100,000 crystallization trials per day combined with robust automated visualization and powerful interface for data management [45–48].
3.1. Crystal lattice organization
Membrane protein crystals are classified according to how crystals are formed: 2D crystals, type I 3D crystals or type II 3D crystals [49]. 2D crystals are mainly considered to be reconstituted biomembranes formed by hydrophobic interactions between detergents and lipids. These crystals are generally used in electron microscopy and not suitable for X-ray crystallography studies. At present, all membrane protein X-ray structures have been solved from 3D crystals (type I or II) only. Within type I 3D crystals, proteins are organized in planar sheets through protein–detergent–lipid hydrophobic interactions (2D crystals) stacked on top of one another by polar interactions. Crystals grown from bicelles or lipidic cubic phase methods are known to be type I 3D crystals [50–52]. Type II 3D crystals are commonly observed when grown by in surfo methods. In this case, crystal packing is mostly due to the interactions between hydrophilic regions of protein molecules. The presence of detergent micelles shelters the protein hydrophobic region reducing the number of protein–protein contacts essential for crystal formation, resulting in extremely fragile crystals with large solvent content.
3.2. Crystallization in surfo
Until now crystallization in surfo has been the most common and successful method for crystallizing membrane proteins. Techniques such as vapor diffusion (sitting- or hanging-drop), microdialysis or microbatch (crystallization under oil) are normally used as the protein–detergent complex (PDCs) that is manipulated as a soluble protein. As mentioned above, crystallization in surfo generates type II 3D crystals. Often, the diffraction quality of these crystals is poor. The main reason for this is reduced essential protein–protein contacts due to the presence of the detergent micelles that cover most of the protein hydrophobic surface. For this reason when crystallizing in surfo, the choice of detergent is particularly important as the shape and size of the detergent micelle play a vital role in crystal formation. While detergents with small micelles such as octyl-β-d-maltoside (8M) or nonyl-β-d-maltoside (9M) hardly cover the hydrophobic surface of the protein leading to protein aggregation, detergents with large micelles such as tridecyl-β-d-maltoside (13 M) usually tend to engulf the entire protein [53]. The addition of small amphiphiles such as heptane-1,2,3-triol and benzamidine is frequently able to improve crystallography because they reduce detergent micelles thereby enhancing crystal contacts [54–56]. Alternative strategies to induce crystallization are the use of additives such as small micelle detergents, heavy metals, salts and organic solvents [57].
3.3. Crystallization in meso
Landau and Rosenbush were the first to report that integral membrane proteins could be crystallized from bilayers [58]. This concept was initially validated by the successful crystallization of bacteriorhodopsin [59,60] and halorhodopsin [61]. Since then, more than 114 membrane protein structures have been solved using the in meso method (http://cherezov.scripps.edu/structures.htm). Lipidic cubic phases (LCP) are mainly formed by energetically mixing the chosen lipid with an aqueous buffer (the protein–detergent complex) at certain ratio and temperature. The cubic phase is formed when the matrix becomes a transparent and non-birefringent gel-like material. Structurally, lipidic cubic phases are complex three-dimensional networks of a bicontinuous lipid bilayer and two separated water channels [58]. The most common lipidic cubic phases are Im3m, Ia3d and Pn3m [62]. For the monoolein/water system, the Pn3m phase has proven to be the most suitable phase for the crystallization of membrane proteins [63–68]. The addition of salts, high molecular weight PEGs, lipids and kosmotrope agents dehydrates the Pn3m cubic phase promoting protein–protein interactions that may result in crystal formation [69,70]. On the other hand, the addition of low molecular PEGs, MPD, detergents and chaotrope agents causes the cubic phase to swell forming a sponge phase. The sponge phase is most beneficial in the case of larger sized proteins or protein complexes [71,72]. A full collection of assays and methods relating to the behavior of membrane proteins and their interaction with lipids in LCP can be found in http://cherezov.scripps.edu/resources.htm.
Crystallization using bicelles is another successful in meso method that was firstly introduced by Bowie and co-workers during the crystallization of bacteriorhodopsin [73]. Since then, several membrane protein structures have been solved using the bicelle method [74–79]. Bicelles are small bilayer disks formed from a number of lipid:amphiphile combinations when mixed at low temperature [73,80,81]. They offer a native-like bilayer environment to the membrane proteins enhancing the growth of type I crystals. Crystallization in bicelles is very simple to set up [81,82]. And, because of the protein–bicelle mixture presents a viscosity similar to the detergent-based drop, crystallization can be carried out in a traditional set up including the use of standard crystallization robotics and commercial crystallization screens [81,82].
3.4. Heading toward to rational design of membrane protein crystallization screens
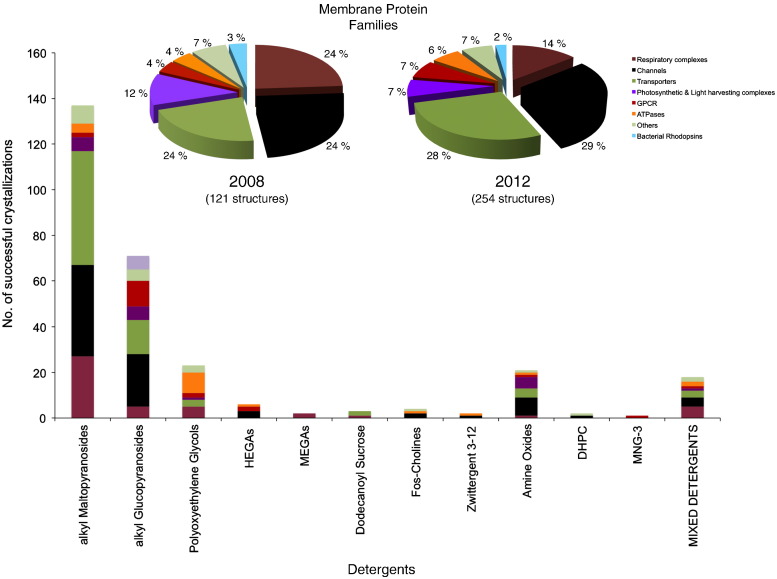
When crystallizing membrane proteins there are many physicochemical parameters to be considered (e.g. different buffers and pH, precipitants, salts, additives and many others). At the present moment a number of membrane crystallization screens are readily available from various companies (Molecular Dimensions, Hampton Research, Jena Biosciences, Emerald Biostructures and Qiagen). However, with the exponential increase in the number of membrane protein structures deposited in the Protein Data Bank (PDB) there exists considerable potential for specific membrane protein crystallization screens to be developed. Using the information present in the Protein Data Bank and respective journal articles, a database of crystallization conditions was generated for all alpha helical membrane proteins up to and including 2012 [83] (Fig. 2 inset). This database has provided unique insights into many aspects surrounding successful membrane protein crystallization. Perhaps unsurprisingly it was discovered that membrane proteins do indeed crystallize in conditions that are very different from their water-soluble counterparts [57]. Although PEG has been the most effective precipitant, as shown for water-soluble proteins [84], it was the small molecular weight (MW) PEGs (400, 600) rather than their larger MW counterparts (3350, 6000), which proved most successful for crystallography of hydrophobic channel and transporter proteins. The effective concentration of the small MW PEGs was also different, being ~ 20% higher than traditional screening kits. The database enabled a more rational approach to the design of tailored screening kits, MemGold and MemGold2 to facilitate crystallization of alpha helical MPs. The approach was simple; conditions previously reported to be successful were recreated and arranged for the 96 well crystallization formats popular with structural biology groups. Additionally, data mining could now be undertaken on successful detergents and these sub-divided by membrane protein family (Fig. 2). Trends were clearly visible, with ion channels being more successfully crystallized using shorter chain detergents such as octyl glucoside, while crystals of transporters and respiratory complexes were more successful obtained using dodecyl maltoside and ATPases using polyoxyethylene glycols. Recently the emergence of mixed detergents has occurred in the reporting of crystallization conditions. Although too early for any conclusions to be drawn or trends suggested, the use of mixed detergent micelles for crystallization is likely to grow in the coming years.
Fig. 2.
Analyzing current trends in alpha helical membrane protein crystallization. Inset, pie charts showing the change in the proportion of structures between 2008 and 2012 and used in the analysis of their crystallization conditions. Respiratory complexes (brown), Channels (black), Transporters (green), Photosynthetic and Light Harvesting Complexes (purple), GPCRs (red), ATPases (orange), Bacterial Rhodopsins (blue) and the Others category (olive), those not fitting the seven main groupings. Stacked bar chart showing the breakdown of successful detergents used for crystallization is shown, subdivided into the eight MP families. Analyses such as these can help formulate successful strategies for crystallizing new MP targets.
Reproduced with permission from [83].
Crystal optimization is a common requirement following initial success in the broad screens; indeed analysis of the recent literature strongly suggests that additive screening is likely to be required for optimal diffraction [57,71]. Using this information a table was constructed based on successful additives reported in the PDB for membrane proteins and formatted into a recent additive screen, MemAdvantage [83], for the purpose of providing an ‘off the shelf’ option for crystal optimization. This analysis has shown that all types of alpha helical membrane proteins have benefited from the presence of additional additives, suggesting additive screening should always be attempted to increase resolution. Multivalent salts and poly alcohols appear particularly advantageous in our analysis, accounting for 10 and 15% of all structures analyzed. Of note is the rapid increase since 2008 in the use of secondary detergents, which account for 19% of all structures reported in 2012 [83], making secondary detergent screens a promising avenue for optimization strategies.
3.5. Dehydration (automated) of membrane protein crystals
Since the early days of crystallography crystal dehydration has been noticed, studied and often put in good use [85–95]. Along with other post-crystallization techniques such as soaking, cross-linking and annealing it is used to try to improve the diffraction properties of crystals in order to permit structural resolution [93–101]. Dehydration has been successful in many cases when applied to membrane proteins [102,103]. Given the cost and time spent in obtaining membrane protein crystals it is worth exploring every option available for optimizing diffracting conditions. As membrane protein crystals have, in general, very high solvent content, dehydration success is dependent on being able to extract part of the available water, inducing reorganization in the diffracting molecules. This may lead to new contacts between the hydrophilic areas of the proteins potentially leading to better general order. There are currently a number of classic protocols to achieve dehydration based on either simple air drying, vapor diffusion or soaking with dehydrating salts and/or precipitants [98,99,101]. The advantage of classic methods is that salt standards are well characterized [104], the experiment can be performed with very small humidity steps, over long periods of time and that a large number of crystals can be treated at the same time. The limitation is that the outcomes of the dehydration experiments are generally unknown until cryo data are collected. Furthermore, at practical level there are a number of complex protocols involved that are quite difficult to establish and assess. This problem was addressed by developing a system that despite using a different methodology (dew point control) correlates well with the classic approaches [105] and easy to use. The Humidity Control device (HC) [106,107] system allows diffraction monitoring while dehydration is being induced on the samples either within the lab environment (home source) or within the constraints of modern MX beamline environments.
The HC device generates an airstream around the samples at controlled relative humidity based on dew point. Vapor saturated water is passed through a cooled condenser where excess water is removed prior reaching the sample. By monitoring the airflow temperature as it reaches the sample the temperature of the condenser (dew point) is altered such that the resulting airflow will be at the correct relative humidity when it reaches the sample. The airflow is delivered over the sample via a nozzle, with a very similar footprint to most gas nitrogen cryo-stats, so it requires no alteration to the sample environment or the experimental considerations. Sample relative humidity is determined using the mother liquor of the grown crystals by direct observation of droplet upon different relative humidity values. Users later mount their samples by hand on Kapton meshes at the predetermined initial relative humidity, where crystals will be stable. The meshes allow wicking the excess mother liquor around the crystal preventing crystal slippage and improving the dehydration process. From this point onwards data collections are carried out in a standard manner. Diffraction images are collected while the HC device alters the hydration status of the samples and users follow the progress of the experiment by analyzing the subsequent diffraction patterns.
The HC device is currently operational in several laboratories and synchrotrons across the world including the European Synchrotron Radiation Facility (ESRF), MaxLab, Helmholtz-Zentrum Berlin (HZB), Diamond Light Source (DLS), the Advanced Light Source (ALS), the Advanced Photon Source (APS) and the Canadian Light Source (CLS).
3.6. High-throughput seeding of membrane protein crystals
Seeding techniques have been successfully applied to membrane proteins [108,109]. Random Microseed Matrix-Screening (rMMS), where seed crystals are added automatically to random crystallization screens, is a significant recent breakthrough in protein crystallization [110]. The rMMS allows more crystals to grow in the metastable zone [111,112] and is therefore a useful method of finding new crystallization conditions. We have applied the rMMS to the crystallization of membrane proteins following the adapted protocol: (i) seed stocks were made as soon as the crystals stopped growing; (ii) crystals were thoroughly crushed in the wells with a rounded glass probe without adding any extra solution; (iii) seed-stock was transferred to an Eppendorf tube and used as quickly as possible, with the remainder being frozen for future use; (iv) sitting drops were set up automatically with the Oryx4 robot (Douglas Instruments, Hungerford, Berkshire). Drops comprised 0.3 μl (protein sample in detergent) + 0.29 μl (reservoir solution) + 10 nl (seed stock). Only 1.5 μl of seed-stock was used per 96-well crystallization plate. Initially, five different test membrane proteins were used (Mhp1 transporter, antibiotic transporter A, antibiotic transporter B, enzyme protein and a bacterial cytochrome). While the transporter A gave no apparent improvement with rMMS, the method yielded significant increased crystallization successes for the cytochrome (16 extra new crystallization conditions), the enzyme protein (12 extra new crystallization conditions) and Mhp1 (8 extra new crystallization conditions).
The main success of this method is to provide a larger number/selection of crystallization starting points that can be used for optimization. Combining the improved technique with in situ plate screening (see Section 4.2) we believe that rMMS has strong potential for crystallizing membrane proteins.
4. Synchrotron radiation in membrane protein structure determination
For more than 50 years, synchrotron radiation (SR) has been fundamental in many areas of science discovery including physics, material sciences, chemistry, biology and medicine. In the field of structural biology, SR has been crucial for the structure determination of numerous important biological macromolecules including the atomic structure of the ribosome [113,114], structure of large virus [115] and structures of membrane protein complexes [116,12]. The third-generation of X-ray sources, with their associated insertion devices (undulators and wigglers), high brilliance beamlines with tunable X-ray wavelength and state-of-the-art end-stations incorporating multi-axes goniometers, cryo-cooling devices, microbeams, pixel array detectors and automated sample exchange [29,117–119] has transformed macromolecular the technique of crystallography (MX). The arrival of high-throughput structural genomics and proteomics initiatives simultaneously with an increase in the awareness of the value of SR in biology has led to a high demand in the number of synchrotron MX beamlines [29,119]. Today, more than 120 MX beamlines from 22 different synchrotrons around the world are available to researchers (http://biosync.sbkb.org/) [29,119].
In the field of membrane protein structural biology, advances in SR have also been significant. Despite the advent of many innovative crystallization approaches (see Section 3) the growth of well-ordered membrane protein crystals is still a major problem. Crystals are often very small, extremely fragile, poorly ordered (high mosaicity) and very sensitive to radiation damage. Collecting data from such crystals is not a straightforward task and the resolution of the X-ray diffraction data not only depends on the crystal quality but also on the characteristics of the data collection apparatus. A major breakthrough in addressing these difficulties has been the arrival of dedicated microfocus beamlines and the use of in situ diffraction [120,132]. The challenges associated with the structure determination of membrane proteins are still numerous but the combination developments in crystallization automation and synchrotron instrumentation and software make the future brighter.
The Membrane Protein Laboratory (MPL) at Diamond Light Source was created to help researchers address the difficulties highlighted above. The MPL is a state-of-the-art research and training user facility open to scientists from laboratories anywhere in the world interested in determining 3D membrane protein structures by X-ray crystallography. The lab combines recently developed high throughput technologies for protein production and crystallization with the latest developments in X-ray diffraction data collection systems at Diamond MX beamlines (http://www.diamond.ac.uk/Home/MPL.html)
4.1. The use of microfocus beamlines
Microfocus beamlines combined with modern developments in sample handling [121], sample visualization [122], automatic crystal centering [123,124], cryo-cooling systems [125,126], fast readout detectors [127–129], new data collection strategies [130] and the appearance of new algorithms for merging data collected from different crystals [131] have recently yielded many novel high-resolution membrane protein structures (http://blanco.biomol.uci.edu/mpstruc/listAll/list). In fact, all GPCR structures in the past few years have been solved using microfocus beamlines. Microfocus beamlines are highly developed to target very small and/or weakly diffracting crystals [120]. While a small sized beam can reduce the background scattering resultant from the mother liquor surrounding a crystal and allow the measurement of data from more ordered regions of inhomogeneous crystals, the high flux increases the scattered reflection intensities and makes measurements from a crystal possible in seconds — an essential requirement when hundreds or thousands of crystals are required to obtain the 3D structure. The combination of these two features is essential for structure determination. However, radiation damage can be a limiting factor in data collection at microfocus beamlines. This is particularly true for membrane protein crystals because of their high solvent content. Nevertheless, the combination between cryogenic cooling of the protein crystals and the use of a micro or submicron beam to explore different unexposed zones of a crystal has overcome many of the challenges associated with the radiation damage. The use of raster scanning is also advantageous [120,127]. The raster scanning system takes a series of X-ray snapshots across the sample and determines the best diffraction region of a disordered crystal. It is also useful for locating crystals when they cannot be seen with visible light, as often happens in the case of crystals in lipid cubic phase. Furthermore, using fast read-out detectors (Pilatus 6M) large areas can be scanned, for example a grid scan of 225 images at 0.2 s per exposure takes only 99 s.
4.2. In situ data collection of membrane-protein crystals
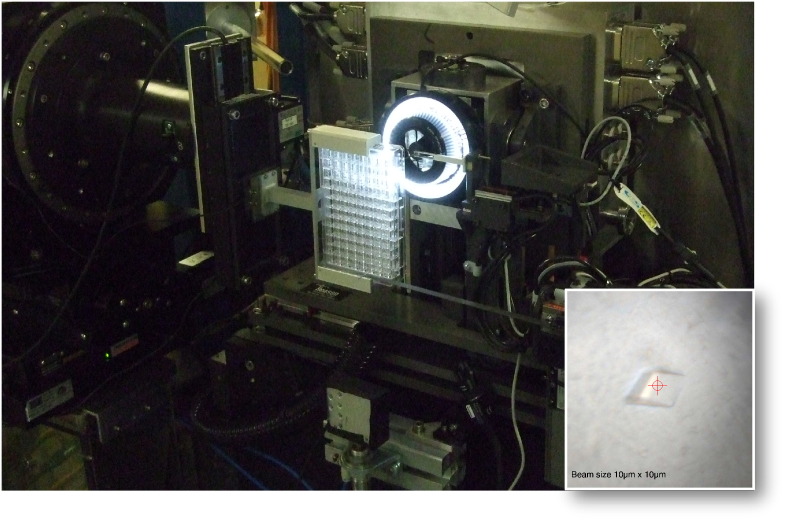
A critical aspect of crystal production is the optimization of crystal quality to achieve diffraction that will yield a structure. In order to improve crystal quality, a large number of crystallization conditions must be explored to increase the number and quality of protein crystals produced. However, dealing with large number of crystals that are small in size and extremely fragile to loop mounting (due to mechanical shocks) can be challenging. In situ screening of membrane protein crystals has proved to be valuable during early stage characterization of crystal diffraction properties [132]. This technique allows crystals to be tested directly in their crystallization plates at room temperature without any physical manipulation of crystals. The in situ screening approach consists of a gripper attached to a goniometer that supports most SBS-format plates including glass LCP plates. The plates are positioned perpendicular to the X-ray beam and are able to travel along a vertical and horizontal axis in order to accurately position each drop, and each crystal within a drop, onto the rotation axis (ω-axis) (Fig. 3). At the MPL in situ screening using the I24 microfocus beamline (Diamond Light Source) is routinely used to assess the diffraction quality of the membrane protein crystals prior to any handling or optimization.
Fig. 3.
Picture showing the environment setup used for the in situ data collection at I24 beamline, Diamond Light Source. Inset, on-axis microscope image of an in situ crystal hit is shown. The red circle and cross-hair represent the beam size and position.
4.3. The use of free electron lasers (FEL)
Although the use of conventional synchrotron radiation has revolutionized the field of membrane protein structure determination, the growth of well-diffracting crystals of sufficient size for X-ray studies is still a challenge. In addition, as described above radiation damage is another prominent barrier present during data collection [133]. New high brilliant X-ray sources such free-electron lasers (XFEL) have provided innovative new opportunities in the field of structure determination of membrane proteins. This new technology is able to obtain structural information from tiny crystals without interference from radiation damage [134]. XFELs produce pulses of light that are a million times more intense than those from standard synchrotron facilities. The combination of intense and ultra-short pulses (< 10 femtoseconds) by XFELs offers the great advantage of “diffraction before destruction” [134,135]. During the experiment, the incident pulse terminates before the destruction of the sample (femtosecond exposure time). This permits the acquisition of structural information before the disintegration of the sample [134,135]. At present, development of XFEL sources in US, Japan and Europe is moving fast to the beginning of a new era: “The femtosecond nanocrystallography” [136].
5. Conclusions
For many years membrane protein structure determination was seen as a “near-impossible mission” and therefore considered to be one of the “last frontiers” in the field of structural biology. Nevertheless, in recent years we have seen many new exciting technologies leading to an exponential rate increase in solved membrane protein structures. As a result, many more research groups are working in the field with great dedication and enthusiasm. Therefore, today membrane protein structural biology presents itself to the scientific community as an exciting field with a bright future.
“…The structure of proteins is the major unsolved problem on the boundary of chemistry and biology to-day. We have not yet found the key to the problem, but in recent years a mass of new evidence and new lines of attack have enabled us to see it in a far more concrete and precise form, and to have some hope that we are near to solving it…” J.D. Bernal, Nature 143 (1939).
Acknowledgments
The Membrane Protein Laboratory is kindly funded by the Wellcome Trust grant No. 099165/Z/12/Z.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
This article is part of a Special Issue entitled: Structural and biophysical characterisation of membrane protein-ligand binding.
References
- 1.Wallin E., von Heijne G. Genome-wide analysis of integral membrane proteins from eubacterial, archaean and eukaryotic organisms. Protein Sci. 1998;7:1029–1038. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Terstappen G.C., Reggiani A. In silico research in drug discovery. Trends Pharmacol. Sci. 2001;22:23–26. doi: 10.1016/s0165-6147(00)01584-4. [DOI] [PubMed] [Google Scholar]
- 3.Davey J. G-protein-coupled receptors: new approaches to maximise the impact of GPCRs in drug discovery. Expert Opin. Ther. Targets. 2004;8:165–170. doi: 10.1517/14728222.8.2.165. [DOI] [PubMed] [Google Scholar]
- 4.Nam H.-J., Jouhyun J., Sanguk K. Bioinformatic approaches for the structure and function of membrane proteins. BMB Rep. 2009;42(11):697–704. doi: 10.5483/bmbrep.2009.42.11.697. [DOI] [PubMed] [Google Scholar]
- 5.Deschamps J.R. The role of crystallography in drug design. AAPS J. 2005;7(4):813–819. doi: 10.1208/aapsj070478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murray C.W., Blundell T.L. Structural biology in fragment-based drug design. Curr. Opin. Struct. Biol. 2010;20:497–507. doi: 10.1016/j.sbi.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 7.Blundell T.L., Jhoti H., Abell C. High-throughput crystallography for lead discovery in drug design. Nat. Rev. Drug Discov. 2002;1:45–54. doi: 10.1038/nrd706. [DOI] [PubMed] [Google Scholar]
- 8.Deisenhofer J., Epp O., Miki R.H., Huber R., Michel H. Structure of the protein subunits in the photosynthetic reaction centre of Rhodopseudomonas viridis at 3 Å resolution. Nature. 1985;318:618–624. doi: 10.1038/318618a0. [DOI] [PubMed] [Google Scholar]
- 9.Rosenbaum D.M., Rasmussen S.G.F., Kobilka B.K. The structure and function of G protein-coupled receptors. Nature. 2009;459:356–363. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fredriksson R., Lagerström M.C., Lundin L.-G., Schiöth H.B. The G-protein coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol. Pharmacol. 2003;63:1256–1272. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- 11.McCusker E.C., Bagnéris C., Naylor C.E., Cole A.R., D'Avanzo N., Nichols C.G., Wallace B.A. Structure of a bacterial voltage-gated sodium channel pore reveals mechanisms of opening and closing. Nat. Commun. 2012;3:1102. doi: 10.1038/ncomms2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rasmussen S.G.F., DeVree B.T., Zou Y., Kruse A.C., Chung K.Y., Kobilka T.S., Thian F.S., Chae P.S., Pardon E., Calinski D., Mathiesen J.M., Shah S.T.A., Lyons J.A., Cafferey M., Gellman S.H., Steyaert J., Skiniotis G., Weis W.I., Sunahara R.K., Kobilka B. Crystal structure of the beta 2 adrenergic receptor–Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wagner S., Klepsch M.M., Schlegel S., Appel A., Draheim R., Tarry M., Högbom M., van Wijk K.J., Slotboom D.J., Persson J.O., de Gier J.-W. Tuning Escherichia coli for membrane protein overexpression. Proc. Natl. Acad. Sci. U. S. A. 2008;105:14371–14376. doi: 10.1073/pnas.0804090105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schlegel S., Löfblom J., Lee C., Hjelm A., Klepsch M., Strous M., Drew D., Slotboom D.J., de Gier J.-W. Optimizing membrane protein overexpression in the Escherichia coli strain Lemo21 (DE3) J. Mol. Biol. 2012;423:648–659. doi: 10.1016/j.jmb.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 15.Hays F.A., Zygy R.-Z., Stroud R.M. Overexpression and purification of integral membrane proteins in yeast. Methods Enzymol. 2010;470:695–707. doi: 10.1016/S0076-6879(10)70029-X. [DOI] [PubMed] [Google Scholar]
- 16.Clark K.M., Fedoriw N., Robinson K., Connelly S.M., Randles J., Malkowski M.G., DeTitta G.T., Dumont M.E. Purification of transmembrane proteins from Saccharomyces cerevisiae for X-ray crystallography. Protein Expr. Purif. 2010;71:207–223. doi: 10.1016/j.pep.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drew D., Slotboom D.-J., Friso G., Reda T., Genevaux P., Rapp M., Meindl‐Beinker N.M., Lambert W., Lerch M., Daley D.O., Van Wijk K.-J., Hirst J., Kunji E., De Gier J.-W. A scalable, GFP‐based pipeline for membrane protein overexpression screening and purification. Protein Sci. 2005;14:2011–2017. doi: 10.1110/ps.051466205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tate C.G. Overexpression of mammalian integral membrane proteins for structural studies. FEBS Lett. 2001;504:94–98. doi: 10.1016/s0014-5793(01)02711-9. [DOI] [PubMed] [Google Scholar]
- 19.Seddon A.M., Curnow P., Booth P.J. Membrane proteins, lipids and detergents: not just a soap opera. Biochim. Biophys. Acta. 2004;1666:105–117. doi: 10.1016/j.bbamem.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 20.Privé G.G. Detergents for the stabilization and crystallization of membrane proteins. Methods. 2007;41:388–397. doi: 10.1016/j.ymeth.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 21.Chae P.S., Rasmussen S.G.F., Rana R.R., Gotfryd K., Chandra R., Goren M.A., Kruse A.C., Nurva S., Loland C.J., Pierre Y., Drew D., Popot J.-L., Picot D., Fox B.G., Guan L., Gether U., Byrne B., Kobilka B., Gellman S.H. Maltose-neopentyl glycol (MNG) amphiphiles for solubilization, stabilization and crystallization of membrane proteins. Nat. Methods. 2010;7:1003–1008. doi: 10.1038/nmeth.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Serebryany E., Zhu G.A., Yan E.C.Y. Artificial membrane-like environments for in vitro studies of purified G-protein coupled receptors. Biochim. Biophys. Acta. 2012;1818:225–233. doi: 10.1016/j.bbamem.2011.07.047. [DOI] [PubMed] [Google Scholar]
- 23.Serrano-Vega M.J., Magnani F., Shibata Y., Tate C.G. Conformational thermostabilization of the β1-adrenergic receptor in a detergent-resistant form. Proc. Natl. Acad. Sci. U. S. A. 2008;105:877–882. doi: 10.1073/pnas.0711253105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Serrano-Vega M.J., Tate C.G. Transferability of thermostabilizing mutations between β-adrenergic receptors. Mol. Membr. Biol. 2009;26:385–396. doi: 10.3109/09687680903208239. [DOI] [PubMed] [Google Scholar]
- 25.Tate C.G., Schertler G.F.X. Engineering G protein-coupled receptors to facilitate their structure determination. Curr. Opin. Struct. Biol. 2009;19:386–395. doi: 10.1016/j.sbi.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 26.Chun E., Thompson A.A., Liu W., Roth C.B., Griffith M.T., Katritch V., Kunken J., Xu F., Cherezov V., Hanson M.A., Stevens R.C. Fusion partner toolchest for the stabilization and crystallization of G protein-coupled receptors. Structure. 2012;20:967–976. doi: 10.1016/j.str.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steyaert J., Kobilka B. Nanobody stabilization of G protein-coupled receptor conformational states. Curr. Opin. Struct. 2011;21:567–572. doi: 10.1016/j.sbi.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stevens R.C., Yokoyama S., Wilson I.A. Global efforts in structural genomics. Sci. Signal. 2001;294:89. doi: 10.1126/science.1066011. [DOI] [PubMed] [Google Scholar]
- 29.Duke E.M.H., Johnson L.N. Macromolecular crystallography at synchrotron radiation sources: current status and future developments. Proc. R. Soc. A. 2010;466:3421–3452. [Google Scholar]
- 30.Phillips R., Ursell T., Wiggins P., Sens P. Emerging roles for lipids in shaping membrane-protein function. Nature. 2009;459:379–385. doi: 10.1038/nature08147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Linke D. Detergents: an overview. Methods Enzymol. 2009;463:603–617. doi: 10.1016/S0076-6879(09)63034-2. [DOI] [PubMed] [Google Scholar]
- 32.Arachea B.T., Sun Z., Potente N., Malik R., Isailovic D., Viola R.E. Detergent selection for enhanced extraction of membrane proteins. Protein Expr. Purif. 2012;86:12–20. doi: 10.1016/j.pep.2012.08.016. [DOI] [PubMed] [Google Scholar]
- 33.Guan L., Smirnova I.N., Verner G., Nagamori S., Kaback H.R. Manipulating phospholipids for crystallization of a membrane transport protein. Proc. Natl. Acad. Sci. U. S. A. 2006;103:1723–1726. doi: 10.1073/pnas.0510922103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arai H. The relationship between the cloud points and the properties of micelles of nonionic detergents. J. Colloid Interface Sci. 1967;23:348–351. [Google Scholar]
- 35.Lund S., Orlowski S., De Foresta B., Champeil P., Le Maire M., Møller J.V. Detergent structure and associated lipid as determinants in the stabilization of solubilized Ca2 +-ATPase from sarcoplasmic reticulum. J. Biol. Chem. 1989;264:4907–4915. [PubMed] [Google Scholar]
- 36.Catravas G.N. Highly reactive impurities in Triton X-100 and Brij 35: partial characterization and removal. Anal. Biochem. 1980;109:55–62. doi: 10.1016/0003-2697(80)90009-3. [DOI] [PubMed] [Google Scholar]
- 37.Screpanti E., Padan E., Rimon A., Michel H., Hunte C. Crucial steps in the structure determination of the Na+/H+ antiporter NhaA in its native conformation. J. Mol. Biol. 2006;362:192–202. doi: 10.1016/j.jmb.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 38.le Maire M., Champeil P., Møller J.V. Interaction of membrane proteins and lipids with solubilizing detergents. Biochim. Biophys. Acta. 2000;1508:86–111. doi: 10.1016/s0304-4157(00)00010-1. [DOI] [PubMed] [Google Scholar]
- 39.Drew D., Lerch M., Kunji E., Slotboom D.-J., de Gier J.-W. Optimization of membrane protein overexpression and purification using GFP fusions. Nat. Methods. 2006;3:303–313. doi: 10.1038/nmeth0406-303. [DOI] [PubMed] [Google Scholar]
- 40.Drew D., Slotboom D.J., Friso G., Reda T., Genevaux P., Rapp M., Nadja M., Meindl‐Beinker W., Lambert M. Lerch, Daley D.O., van Wijk K.-J., Hirst J., Kunji E., de Gier J.-W., Scalable A. GFP‐based pipeline for membrane protein overexpression screening and purification. Protein Sci. 2009;14:2011–2017. doi: 10.1110/ps.051466205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Granier S., Manglik A., Kruse A.C., Kobilka T.S., Thian F.S., Weis W.I., Kobilka B.K. Structure of the δ-opioid receptor bound to naltrindole. Nature. 2012;485:400–404. doi: 10.1038/nature11111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kellosalo J., Kajander T., Kogan K., Pokharel K., Goldman A. The structure and catalytic cycle of a sodium-pumping pyrophosphatase. Science. 2012;337:473–476. doi: 10.1126/science.1222505. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Q., Ma X., Ward A., Hong W.X., Jaakola V.-P., Stevens R.C., Finn M.G., Chang G. Designing facial amphiphiles for the stabilization of integral membrane proteins. Angew. Chem. 2007;119:7153–7715. doi: 10.1002/anie.200701556. [DOI] [PubMed] [Google Scholar]
- 44.Chae P.S., Gotfryd K., Pacyna J., Miercke L.J.W., Rasmussen S.G.F., Robbins R.A., Rana R.R., Loland C.J., Kobilka B., Stroud R., Byrne B., Gether U., Gellman S.H. Tandem facial amphiphiles for membrane protein stabilization. J. Am. Chem. Soc. 2010;132:16750–16752. doi: 10.1021/ja1072959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abola E., Kuhn P., Earnest T., Stevens R.C. Automation of X-ray crystallography. Nat. Struct. Biol. 2000;7:973–977. doi: 10.1038/80754. [DOI] [PubMed] [Google Scholar]
- 46.Hosfield D., Palan J., Hilgers M., Scheibe D., McRee D.E., Stevens R.C. A fully integrated protein crystallization platform for small-molecule drug discovery. J. Struct. Biol. 2003;142:207–217. doi: 10.1016/s1047-8477(03)00051-0. [DOI] [PubMed] [Google Scholar]
- 47.Pusey M.L., Liu Z.-J., Tempel W., Praissman J., Lin D., Wang B.-C., Gavira J.A., Ng J.D. Life in the fast lane for protein crystallization and X-ray crystallography. Prog. Biophys. Mol. Biol. 2005;88:359. doi: 10.1016/j.pbiomolbio.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 48.Kissick D.J., Gualtieri E.J., Simpson G.J., Cherezov V. Nonlinear optical imaging of integral membrane protein crystals in lipidic mesophases. Anal. Chem. 2009;82:491–497. doi: 10.1021/ac902139w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Michel H. Crystallization of membrane proteins. Trends Biochem. Sci. 1983;8:56–59. [Google Scholar]
- 50.Qutub Y., Reviakine I., Maxwell C., Navarro J., Landau E.M., Vekilov P.G. Crystallization of Transmembrane Proteins in cubo. Mechanisms of Crystal Growth and Defect Formation. J. Mol. Biol. 2004;343:1243–1254. doi: 10.1016/j.jmb.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 51.Caffrey M., Li D., Dukkipati A. Membrane protein structure determination using crystallography and lipidic mesophases — recent advances and successes. Biochemistry. 2012;51:6266–6288. doi: 10.1021/bi300010w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Michel H. Crystallization of membrane proteins. Trends Biochem. Sci. 1983;8:56–59. [Google Scholar]
- 53.Bamber Lisa, Harding M., Monné M., Slotboom D.-J., Kunji E.R.S. The yeast mitochondrial ADP/ATP carrier functions as a monomer in mitochondrial membranes. Proc. Natl. Acad. Sci. U. S. A. 2007;104:10830–10834. doi: 10.1073/pnas.0703969104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Timmins P.A., Hauk J., Wacker T., Welte W. The influence of heptane-1,2,3-triol on the size and shape of LDAO micelles. Implications for the crystallisation of membrane proteins. FEBS Lett. 1991;280:115–120. doi: 10.1016/0014-5793(91)80217-q. [DOI] [PubMed] [Google Scholar]
- 55.Schertler G.F., Bartunik H.D., Michel H., Oesterhelt D. Orthorhombic crystal form of bacteriorhodopsin nucleated on benzamidine diffracting to 3.6 Å resolution. J. Mol. Biol. 1993;234:156. doi: 10.1006/jmbi.1993.1570. [DOI] [PubMed] [Google Scholar]
- 56.Timmins P.A., Pebay-Peyroula E., Welte W. Detergent organisation in solutions and in crystals of membrane proteins. Biophys. Chem. 1994;53:27–36. doi: 10.1016/0301-4622(94)00073-5. [DOI] [PubMed] [Google Scholar]
- 57.Newstead S., Ferrandon S., Iwata S. Rationalizing α‐helical membrane protein crystallization. Protein Sci. 2008;17:466–472. doi: 10.1110/ps.073263108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Landau E.M., Rosenbusc J.P. Lipidic cubic phases: a novel concept for the crystallization of membrane proteins. Proc. Natl. Acad. Sci. U. S. A. 1996;93:14532–14535. doi: 10.1073/pnas.93.25.14532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pebay-Peyroula E., Rummel G., Rosenbusch J.P., Landau E.M. X-ray structure of bacteriorhodopsin at 2.5 angstroms from microcrystals grown in lipidic cubic phases. Science. 1997;277:1676–1681. doi: 10.1126/science.277.5332.1676. [DOI] [PubMed] [Google Scholar]
- 60.Luecke H., Schobert B., Richter H.-T., Cartailler J.-P., Lanyi J.K. Structure of bacteriorhodopsin at 1.55 A resolution. J. Mol. Biol. 1999;291:899–911. doi: 10.1006/jmbi.1999.3027. [DOI] [PubMed] [Google Scholar]
- 61.Kolbe M., Besir H., Essen L.-O., Oesterhelt D. Structure of the light-driven chloride pump halorhodopsin at 1.8 Å resolution. Science. 2000;288:1390–1396. doi: 10.1126/science.288.5470.1390. [DOI] [PubMed] [Google Scholar]
- 62.Lindblom G., Rilfors L. Cubic phases and isotropic structures formed by membrane lipids — possible biological relevance. Biochim. Biophys. Acta Rev. Biomembr. 1989;988:221–256. [Google Scholar]
- 63.Chiu M.L., Nollert P., Loewen M.C., Belrhali H., Pebay-Peyroula E., Rosenbusch J.P., Landau E.M. Crystallization in cubo: general applicability to membrane proteins. Acta Crystallogr. D Biol. Crystallogr. 2000;56:781–784. doi: 10.1107/s0907444900004716. [DOI] [PubMed] [Google Scholar]
- 64.Nollert P., Qiu H., Caffrey M., Rosenbusch J.P., Landau E.M. Molecular mechanism for the crystallization of bacteriorhodopsin in lipidic cubic phases. FEBS Lett. 2001;504:179–186. doi: 10.1016/s0014-5793(01)02747-8. [DOI] [PubMed] [Google Scholar]
- 65.Chung H., Caffrey Martin. The curvature elastic-energy function of the lipid–water cubic mesophase. Nature. 1994;368:224–226. doi: 10.1038/368224a0. [DOI] [PubMed] [Google Scholar]
- 66.Chung H., Caffrey Martin. The neutral area surface of the cubic mesophase: location and properties. Biophys. J. 1994;66:377–381. doi: 10.1016/s0006-3495(94)80787-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Seddon J.M., Templer R.H., Warrender N.A., Huang Z., Cevc G., Marsh D. Phosphatidylcholine–fatty acid membranes: effects of headgroup hydration on the phase behaviour and structural parameters of the gel and inverse hexagonal (HII) phases. Biochim. Biophys. Acta Rev. Biomembr. 1997;1327:131–147. doi: 10.1016/s0005-2736(97)00047-3. [DOI] [PubMed] [Google Scholar]
- 68.Templer R.H. Thermodynamic and theoretical aspects of cubic mesophases in nature and biological amphiphiles. Curr. Opin. Colloid Interface Sci. 1998;3:255–263. [Google Scholar]
- 69.Cherezov V., Fersi H., Caffrey M. Crystallization screens: compatibility with the lipidic cubic phase for in meso crystallization of membrane proteins. Biophys. J. 2001;81:225–242. doi: 10.1016/S0006-3495(01)75694-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cherezov V., Clogston J., Misquitta Y., Abdel-Gawad W., Caffrey M. Membrane protein crystallization in meso: lipid type-tailoring of the cubic phase. Biophys. J. 2002;83:3393–3407. doi: 10.1016/S0006-3495(02)75339-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cherezov V., Clogston J., Papiz M.Z., Caffrey Martin. Room to move: crystallizing membrane proteins in swollen lipidic mesophases. J. Mol. Biol. 2006;357:1605–1618. doi: 10.1016/j.jmb.2006.01.049. [DOI] [PubMed] [Google Scholar]
- 72.Wadsten P., Wöhri A.B., Snijder A., Katona G., Gardiner A.T., Cogdell R.J., Neutze R., Engström S. Lipidic sponge phase crystallization of membrane proteins. J. Mol. Biol. 2006;364:44–53. doi: 10.1016/j.jmb.2006.06.043. [DOI] [PubMed] [Google Scholar]
- 73.Faham S., Bowie J.U. Bicelle crystallization: a new method for crystallizing membrane proteins yields a monomeric bacteriorhodopsin structure. J. Mol. Biol. 2002;316:1–6. doi: 10.1006/jmbi.2001.5295. [DOI] [PubMed] [Google Scholar]
- 74.Faham S., Boulting G.L., Massey E.A., Yohannan S., Yang D., Bowie J.U. Crystallization of bacteriorhodopsin from bicelle formulations at room temperature. Protein Sci. 2005;14:836–840. doi: 10.1110/ps.041167605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rasmussen S.G.F., Choi H.-J., Rosenbaum D.M., Kobilka T.S., Thian F.S., Edwards P.C., Burghammer M., Ratnala V.R.P., Sanishvili R., Fischetti R.F., Schertler G.F.X., Weis W.I., Kobilka B.K. Crystal structure of the human of the β2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 76.Luecke H., Schobert B., Stagno J., Imasheva E.S., Wang J.M., Balashov S.P., Lanyi J.K. Crystallographic structure of xanthorhodopsin, the light-driven proton pump with a dual chromophore. Proc. Natl. Acad. Sci. U. S. A. 2008;105:16561–16565. doi: 10.1073/pnas.0807162105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ujwal R., Cascio D., Colletier J.-P., Faham S., Zhang J., Toro L., Ping P., Abramson J. The crystal structure of mouse VDAC1 at 2.3 Å resolution reveals mechanistic insights into metabolite gating. Proc. Natl. Acad. Sci. U. S. A. 2008;105:17742–17747. doi: 10.1073/pnas.0809634105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vinothkumar K.R. Structure of rhomboid protease in a lipid environment. J. Mol. Biol. 2011;407:232–247. doi: 10.1016/j.jmb.2011.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Payandeh J., Scheuer T., Zheng N., Catterall W.A. The crystal structure of a voltage-gated sodium channel. Nature. 2011;475:353–358. doi: 10.1038/nature10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sanders C.R., Prosser R.S. Bicelles: a model membrane system for all seasons? Structure. 1998;6:1227–1234. doi: 10.1016/s0969-2126(98)00123-3. [DOI] [PubMed] [Google Scholar]
- 81.Ujwal R., Bowie J.U. Crystallizing membrane proteins using lipidic bicelles. Methods. 2011;55:337–341. doi: 10.1016/j.ymeth.2011.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ujwal R., Abramson J. High-throughput crystallization of membrane proteins using the lipidic bicelle method. J. Vis. Exp. 2012;59:e3383. doi: 10.3791/3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Parker J.L., Newstead Simon. Current trends in α‐helical membrane protein crystallization: an update. Protein Sci. 2012;21:1358–1365. doi: 10.1002/pro.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Page R., Stevens R.C. Crystallization data mining in structural genomics: using positive and negative results to optimize protein crystallization screens. Methods. 2004;34:373–389. doi: 10.1016/j.ymeth.2004.03.026. [DOI] [PubMed] [Google Scholar]
- 85.Perutz M.F. The composition and swelling properties of haemoglobin crystals. Trans. Faraday Soc. 1946;42:B187–B195. [Google Scholar]
- 86.Huxley H.E., Kendrew J.C. Discontinuous lattice changes in haemoglobin crystals. Acta Crystallogr. 1953;6:76–80. [Google Scholar]
- 87.Einstein J.R. Humidity control device for the Buerger precession camera. J. Sci. Instrum. 2002;38:449. [Google Scholar]
- 88.Einstein J.R., Low B.W. Insulin. Some shrinkage stages of sulfate and citrate crystals. Acta Crystallogr. 1962;15:32–34. [Google Scholar]
- 89.McGavin A.S., Einstein J.R., Low B.W. Insulin — gross molecular structure: trial-and-error studies using transform and Patterson function techniques. Proc. Natl. Acad. Sci. U. S. A. 1962;48:2150. doi: 10.1073/pnas.48.12.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Einstein J.R., McGavin A.S., Low B.W. Insulin. Some shrinkage stages of sulfate and citrate crystals. 1963;49:74. [Google Scholar]
- 91.Stammers D.K., Somers D.O., Ross C.K., Kirby I., Ray P.H., Wilson J.E., Norman M., Ren J.S., Esnouf R.M., Garman E.F., Jones E.Y., Stuart D.I. Crystals of HIV-1 reverse transcriptase diffracting to 2.2 A resolution. J. Mol. Biol. 1994;242:586–588. doi: 10.1006/jmbi.1994.1604. [DOI] [PubMed] [Google Scholar]
- 92.Ren J., Esnouf R., Garman E.F., Somers D.O., Ross C., Kirby I., Keeling J., Darby G., Jones E.Y., Stuart D.I., Stammers D.K. High resolution structures of HIV-1 RT from four RT-inhibitor complexes. Nat. Struct. Mol. Biol. 1995;2:293–302. doi: 10.1038/nsb0495-293. [DOI] [PubMed] [Google Scholar]
- 93.Esnouf R.M., Ren J., Garman E.F., Somers D.O., Ross C.K., Jones E.Y., Stammers D.K., Stuart D.I. Continuous and discontinuous changes in the unit cell of HIV-1 reverse transcriptase crystals on dehydration. Acta Crystallogr. D Biol. Crystallogr. 1998;54:938–953. doi: 10.1107/s0907444998004284. [DOI] [PubMed] [Google Scholar]
- 94.Yeh J.I., Hol W.G. A flash-annealing technique to improve diffraction limits and lower mosaicity in crystals of glycerol kinase. Acta Crystallogr. D Biol. Crystallogr. 1998;54:479–480. doi: 10.1107/s0907444998004697. [DOI] [PubMed] [Google Scholar]
- 95.Harp J.M., Hanson B.L., Timm D.E., Bunick G.J. Macromolecular crystal annealing: evaluation of techniques and variables. Acta Crystallogr. D Biol. Crystallogr. 1999;55:1329–1334. doi: 10.1107/s0907444999005442. [DOI] [PubMed] [Google Scholar]
- 96.Abergel C. Spectacular improvement of X-ray diffraction through fast desiccation of protein crystals. Acta Crystallogr. D Biol. Crystallogr. 2004;60:1413–1416. doi: 10.1107/S0907444904013678. [DOI] [PubMed] [Google Scholar]
- 97.Juers D.H., Matthews B.W. The role of solvent transport in cryo-annealing of macromolecular crystals, Acta Crystallogr. D Biol. Crystallogr. 2004;60:412–421. doi: 10.1107/S0907444903027938. [DOI] [PubMed] [Google Scholar]
- 98.Heras B., Martin J.L. Post-crystallization treatments for improving diffraction quality of protein crystals. Acta Crystallogr. D Biol. Crystallogr. 2005;61:1173–1180. doi: 10.1107/S0907444905019451. [DOI] [PubMed] [Google Scholar]
- 99.Newman J. A review of techniques for maximizing diffraction from a protein crystal in stilla. Acta Crystallogr. D Biol. 2006;62:27–31. doi: 10.1107/S0907444905032130. [DOI] [PubMed] [Google Scholar]
- 100.Nakamura A., Wada C., Miki K. Expression and purification of F-plasmid RepE and preliminary X-ray crystallographic study of its complex with operator DNA. Acta Crystallogr. F Struct. Biol. Cryst. Commun. 2007;63:346–349. doi: 10.1107/S1744309107012894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Krauss I.R., Sica F., Mattia C.A., Merlino A. Increasing the X-ray diffraction power of protein crystals by dehydration: the case of bovine serum albumin and a survey of literature data. Int. J. Mol. Sci. 2012;13:3782–3800. doi: 10.3390/ijms13033782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hu N.-J., Iwata S., Cameron A.D., Drew D. Crystal structure of a bacterial homologue of the bile acid sodium symporter ASBT. Nature. 2011;478:408–411. doi: 10.1038/nature10450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.McCusker E.C., Bagnéris C., Naylor C.E., Cole A.R., D'Avanzo N., Nichols C.G., Wallace B.A. Structure of a bacterial voltage-gated sodium channel pore reveals mechanisms of opening and closing. Nat. Commun. 2012;3:1102. doi: 10.1038/ncomms2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Greenspan L. Humidity fixed points of binary saturated aqueous solutions. J. Res. Natl. Bur. Stand. A. 1977;81:89–96. [Google Scholar]
- 105.Sjogren T., Carlsson G., Larsson G., Hajdu A., Andersson C., Pettersson H., Hajdu J. Protein crystallography in a vapour stream: data collection, reaction initiation and intermediate trapping in naked hydrated protein crystals. J. Appl. Crystallogr. 2002;35:113–116. [Google Scholar]
- 106.Sanchez-Weatherby J., Bowler M.W., Huet J., Gobbo A., Felisaz F., Lavault B., Moya R., Kadlec J., Ravelli R.B.G., Cipriani F. Improving diffraction by humidity control: a novel device compatible with X-ray beamlines. Acta Crystallogr. D Biol. Crystallogr. 2009;65:1237–1246. doi: 10.1107/S0907444909037822. [DOI] [PubMed] [Google Scholar]
- 107.Russi S., Juers D.H., Sanchez-Weatherby J., Pellegrini E., Mossou E., Forsyth V.T., Huet J., Gobbo A., Felisaz F., Moya R., McSweeney S.M., Cusack S., Cipiani F., Bowler M.W. Inducing phase changes in crystals of macromolecules: status and perspectives for controlled crystal dehydration. J. Struct. Biol. 2011;175:236–243. doi: 10.1016/j.jsb.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 108.Fromme P., Witt H.T. Improved isolation and crystallization of photosystem I for structural analysis. Biochim. Biophys. Acta Bioenerg. 1998;1365:175–184. [Google Scholar]
- 109.Lancaster C.R.D., Kröger A., Auer M., Michel H. Structure of fumarate reductase from Wolinella succinogenes at 2.2 A resolution. Nature. 1999;402:377–385. doi: 10.1038/46483. [DOI] [PubMed] [Google Scholar]
- 110.D'Arcy A., Villard F., Marsh M. An automated microseed matrix-screening method for protein crystallization. Acta Crystallogr. D Biol. Crystallogr. 2007;63:550–554. doi: 10.1107/S0907444907007652. [DOI] [PubMed] [Google Scholar]
- 111.Stewart P.D.S., Kolek S.A., Briggs R.A., Chayen N.E., Baldock P.F.M. Random microseeding: a theoretical and practical exploration of seed stability and seeding techniques for successful protein crystallization. Cryst. Growth Des. 2011;11:3432–3441. [Google Scholar]
- 112.Villasenor A.G., Wong A., Shao A., Garg A., Kuglstatter A., Harris S.F. Acoustic matrix microseeding: improving protein crystal growth with minimal chemical bias. Acta Crystallogr. D Biol. Crystallogr. 2010;66:568–576. doi: 10.1107/S0907444910005512. [DOI] [PubMed] [Google Scholar]
- 113.Von Böhlen K., Makowski I., Hansen H.A.S., Bartels H., Berkovitch-Yellin Z., Zaytzev-Bashan A., Meyer S., Paulke C., Franceschi F., Yonath A. Characterization and preliminary attempts for derivatization of crystals of large ribosomal subunits from Haloarcula marismortui diffracting to 3 Å resolution. J. Mol. Biol. 1991;222:11–15. doi: 10.1016/0022-2836(91)90730-t. [DOI] [PubMed] [Google Scholar]
- 114.Ben-Shem A., Loubresse N.G., Melnikov S., Jenner L., Yusupova G., Yusupov M. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 115.Kuhn R.J., Zhang W., Rossmann M.G., Pletnev S.V., Corver J., Lenches E., Jones C.T., Mukhopadhyay S., Chipman P.R., Strauss E.G., Baker T.S., Strauss J.H. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell. 2002;108:717–725. doi: 10.1016/s0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yankovskaya V., Horsefield R., Törnroth S., Luna-Chavez C., Miyoshi H., Léger C., Byrne B., Cecchini G., Iwata S. Architecture of succinate dehydrogenase and reactive oxygen species generation. Science. 2003;299:700–704. doi: 10.1126/science.1079605. [DOI] [PubMed] [Google Scholar]
- 117.Smith J.L., Fischetti R.F., Yamamoto M. Microcrystallography comes of age. Curr. Opin. Struct. Biol. 2012;22:602–612. doi: 10.1016/j.sbi.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Stojanoff V., Northrup P., Pietri R., Zhong Z. Synchrotron radiation in life sciences. Protein Pept. Lett. 2012;19:761–769. doi: 10.2174/092986612800793163. [DOI] [PubMed] [Google Scholar]
- 119.Dauter Z., Jaskolski M., Wlodawer A. Impact of synchrotron radiation on macromolecular crystallography: a personal view. J. Synchrotron Radiat. 2010;17:433–444. doi: 10.1107/S0909049510011611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Evans G., Axford D., Waterman D., Owen R.L. Macromolecular microcrystallography. Crystallogr. Rev. 2011;17:105–142. [Google Scholar]
- 121.Cipriani F., Felisaz F., Launer L., Aksoy J.-S., Caserotto H., Cusack S., Dallery M., di-Chiaro F., Guijarro M., Huet J., Larsen S., Lentini M., McCarthy J., McSweeney S., Ravelli R., Renier M., Taffut C., Thompson A., Leonard G.A., Walsh M.A. Automation of sample mounting for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1251–1259. doi: 10.1107/S0907444906030587. [DOI] [PubMed] [Google Scholar]
- 122.Perrakis A., Cipriani F., Castagna J.-C., Claustre L., Burghammer M., Riekel C., Cusack S. Protein microcrystals and the design of a microdiffractometer: current experience and plans at EMBL and ESRF/ID13. Acta Crystallogr. D Biol. Crystallogr. 1999;55:1765–1770. doi: 10.1107/s0907444999009348. [DOI] [PubMed] [Google Scholar]
- 123.Song J., Mathew D., Jacob S.A., Corbett L., Moorhead P., Soltis S.M. Diffraction-based automated crystal centering. J. Synchrotron Radiat. 2007;14:191–195. doi: 10.1107/S0909049507004803. [DOI] [PubMed] [Google Scholar]
- 124.Lavault B., Ravelli R.B.G., Cipriani F. C3D: a program for the automated centring of cryocooled crystals. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1348–1357. doi: 10.1107/S0907444906031234. [DOI] [PubMed] [Google Scholar]
- 125.Hope H. Cryocrystallography of biological macromolecules: a generally applicable method. Acta Crystallogr. B Struct. Sci. 1988;44:22–26. doi: 10.1107/s0108768187008632. [DOI] [PubMed] [Google Scholar]
- 126.Garman E.F., Mcsweeney S.M. Progress in research into radiation damage in cryo-cooled macromolecular crystals. J. Synchrotron Radiat. 2006;14:1–3. doi: 10.1107/S0909049506053015. [DOI] [PubMed] [Google Scholar]
- 127.Aishima J., Owen R.L., Axford D., Shepherd E., Winter G., Levik K., Gibbons P., Ashton A., Evans Gwyndaf. High-speed crystal detection and characterization using a fast-readout detector. Acta Crystallogr. D Biol. Crystallogr. 2010;66:1032–1035. doi: 10.1107/S0907444910028192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Broennimann C., Eikenberry E.F., Henrich B., Horisberger R., Huelsen G., Pohl E., Schmitt B., Schulze-Briese C., Suzuki M., Tomizaki T., Toyokawa H., Wagner A. The PILATUS 1M detector. J. Synchrotron Radiat. 2006;13:120–130. doi: 10.1107/S0909049505038665. [DOI] [PubMed] [Google Scholar]
- 129.Kraft P., Bergamaschi A., Broennimann C., Dinapoli R., Eikenberry E.F., Henrich B., Johnson I., Mozzanica A., Schleputz C.M., Willmott P.R., Schmitt B. Performance of single-photon-counting PILATUS detector modules. J. Synchrotron Radiat. 2009;16:368–375. doi: 10.1107/S0909049509009911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Winter G., McAuley K.E. Automated data collection for macromolecular crystallography. Methods. 2011;55:81–93. doi: 10.1016/j.ymeth.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 131.Foadi J., Alguel Y., Armour W., Axford D., Cameron A., Owen R., Waterman D., Evans G. On the systematic scaling and merging of multiple datasets in macromolecular crystallography. Acta Crystallogr. 2011;A67:C162. [Google Scholar]
- 132.Axford D., Owen R.L., Foadi J., Morgan A.W., Robinson James I., Nettleship J., Owens R., Moraes I., Stuart D.I., Ren J., Fry E.E., Harlos K., Kotecha A., Evans G. In situ macromolecular crystallography using microbeams. Acta Crystallogr. D Biol. Crystallogr. 2012;68:592–600. doi: 10.1107/S0907444912006749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Owen R.L., Rudino-Pinera E., Garman E.F. Experimental determination of the radiation dose limit for cryocooled protein crystals. Proc. Natl. Acad. Sci. U. S. A. 2006;103:4912–4917. doi: 10.1073/pnas.0600973103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Neutze R., Wouts R., van der Spoel D., Weckert E., Hajdu J. Potential for biomolecular imaging with femtosecond X-ray pulses. Nature. 2000;406:752–757. doi: 10.1038/35021099. [DOI] [PubMed] [Google Scholar]
- 135.Borgan M.J. X‑ray free electron lasers motivate bioanalytical characterization of protein nanocrystals: serial femtosecond crystallography. Anal. Chem. 2013;85:3464–3471. doi: 10.1021/ac303716r. [DOI] [PubMed] [Google Scholar]
- 136.Chapman H.N., Fromme P., Barty A., White T.A., Kirian R.A., Aquila A., Hunter M.S., Schulz J., DePonte D.P., Weiersta U., Doak R.B., Maia F.R.N.C., Martin A.V., Schlichting I., Lomb L., Coppola N., Shoeman R.L., Epp S.W., Hartmann R., Rolles D., Rudenko A., Foucar L., Kimmel N., Weidenspointner G., Holl P., Liang M., Barthelmess M., Caleman C., Boutet S., Bogan M.J., Krzywinski J., Bostedt C., Bajt S., Gumprecht L., Rudek B., Erk B., Schmidt C., Homke A., Reich C., Pietschner D., Struder L., Hauser G., Gorke H., Ullrich J., Herrmann S., Schaller G., Schopper F., Soltau H., Kuhnel K.-U., Messerschmidt M., Bozek J.D., Hau-Riege S.P., Frank M., Hampton C.Y., Sierra R.G., Starodub D., Williams G.J., Hajdu J., Timneanu N., Seibert M.M., Andreasson J., Rocker A., Jonsson O., Svenda M., Stern S., Nass K., Andritschke R., Schroter C.-D., Krasniqi F., Bott M., Schmidt K.E., Wang X., Grotjohann I., Holton J.M., Barends T.R.M., Neutze R., Marchesini S., Fromme R., Schorb S., Rupp D., Adolph M., Gorkhover T., Andersson I., Hirsemann H., Potdevin G., Graafsma H., Nilsson B., Spence J.C.H. Nature. 2011;470:73–77. [Google Scholar]