Abstract
Background
Reliably abstracting outcomes from free-text electronic medical records remains a challenge. While automated classification of free text has been a popular medical informatics topic, performance validation using real-world clinical data has been limited. The two main approaches are linguistic (natural language processing [NLP]) and statistical (machine learning). The authors have developed a hybrid system for abstracting computed tomography (CT) reports for specified outcomes.
Objectives
The objective was to measure performance of a hybrid NLP and machine learning system for automated outcome classification of emergency department (ED) CT imaging reports. The hypothesis was that such a system is comparable to medical personnel doing the data abstraction.
Methods
A secondary analysis was performed on a prior diagnostic imaging study on 3,710 blunt facial trauma victims. Staff radiologists dictated CT reports as free text, which were then deidentified. A trained data abstractor manually coded the reference standard outcome of acute orbital fracture, with a random subset double-coded for reliability. The data set was randomly split evenly into training and testing sets. Training patient reports were used as input to the Medical Language Extraction and Encoding (MedLEE) NLP tool to create structured output containing standardized medical terms and modifiers for certainty and temporal status. Findings were filtered for low certainty and past/future modifiers and then combined with the manual reference standard to generate decision tree classifiers using data mining tools Waikato Environment for Knowledge Analysis (WEKA) 3.7.5 and Salford Predictive Miner 6.6. Performance of decision tree classifiers was evaluated on the testing set with or without NLP processing.
Results
The performance of machine learning alone was comparable to prior NLP studies (sensitivity = 0.92, specificity = 0.93, precision = 0.95, recall = 0.93, f-score = 0.94), and the combined use of NLP and machine learning shows further improvement (sensitivity = 0.93, specificity = 0.97, precision = 0.97, recall = 0.96, f-score = 0.97). This performance is similar to, or better than, that of medical personnel in previous studies.
Conclusions
A hybrid NLP and machine learning automated classification system shows promise in coding free-text electronic clinical data.
A well-known barrier to the use of electronic health records (EHRs) for clinical research is the prevalence of free-text data. Extraction of outcomes of interest requires trained data abstractors to manually process each report. This can consume significant time and resources, and extracting another outcome later may require repeating the whole process again.
Automated classification of free text has been an active area of informatics research.1 This task is often broken into two steps, namely, the structuring of text using standardized medical language and identification of outcomes of interest. Linguistics-based natural language processing (NLP) software for the medical domain has been shown to successfully perform the first step, although many NLP tools are developed for narrow medical domains.2–4 Furthermore, NLP traditionally needs to be paired with hand-crafted if–then rules (expert rules) for the second step of outcome identification.5 This approach is not easily generalizable because of the narrow scope of some NLP tools and the need to craft a new set of expert rules for each outcome of interest.
More recently, statistical machine learning techniques have shown promise for outcome identification, especially when dealing with large volumes of data. However, a number of machine learning classification techniques are not transparent, making them less likely to be adopted by clinicians.6 Regardless, more generalizable automated outcome classification pairing NLP software and machine learning techniques are now possible. This approach has shown to have the potential to code EHR data,7,8 although most prior studies have been performed on documents mocked up for NLP testing and never validated on real-world data.
Automated classification of outcomes, such as radiologic findings, could have a substantial effect on clinical research. A good example would be the project whose data were used for this study.9 To derive a clinical risk score to predict traumatic orbital fracture, a lengthy multicenter study was conducted that required physicians to fill out prospective surveys and research assistants to code the clinical outcomes from orbital computed tomography (CT) reports retrospectively. Data analysis was delayed by 1 year trying to secure a research assistant to work full time for 4 months to abstract the necessary information. While templated EHRs could obviate the need for prospective surveys to collect predictor variables, automated classification of the radiology reports would still prove crucial to generate the outcomes data. Furthermore, once clinical decision support tools are implemented in EHRs, auditing physician performance of CT would need to go beyond simple numbers of CTs ordered, but examine the positive yield of the CTs. The goal is to develop a translatable, accurate, and efficient computer system that structures free-text EHR data stored in clinical data warehouses and extracts outcomes suitable for clinical research and performance improvement.
We performed this study to adapt an established broad-coverage medical NLP system and hybridize it with transparent modern machine learning techniques to enhance acceptability. We measured the diagnostic accuracy of a hybrid system using NLP and machine learning tools for automated classification of emergency department (ED) CT imaging reports by comparing to classification by traditional manual data abstraction.
METHODS
Study Design
This was a secondary analysis of data from a prior diagnostic imaging study on blunt facial trauma victims.9 Institutional review board (IRB) approval was obtained for this secondary analysis, which was a retrospective cohort study comparing automated classification of CT imaging reports against the reference standard of manual coding by trained data abstractors.
Study Setting and Population
The study setting and population of the original study are discussed in detail elsewhere.9 Briefly, the traumatic orbital fracture project was a prospective cohort study of ED patients presenting acutely with blunt orbital trauma who underwent CT imaging. The study derived a clinical risk score to help guide more efficient use of radiologic imaging, to improve the specificity for identifying trauma patients with orbital fracture using CT imaging, a high-radiation, time-consuming, and expensive test. Using conventional methods previously described,9 the investigators prospectively collected clinical data and outcomes on 3,710 consecutive patients, including CT imaging reports. Staff radiologists dictated each CT report.
The reference standard outcome of acute orbital fracture was extracted manually by a trained data abstractor, who determined whether an orbital fracture was present acutely, likely present, not likely present/chronic, or not present. To confirm reliability, a random subset of approximately 500 reports was checked by a study physician (??) blinded to the outcome. The binary reference standard outcome of acute fracture (present/not present) was created by grouping present acutely and likely present into “present” and the remaining two categories into “not present.”
Study Protocol
System Overview
For the secondary analysis reported here, patient reports were preprocessed for deidentification and processed by NLP (Figure 1). The NLP output was filtered to exclude findings with low certainty or negation, as well as findings linked with patients’ histories. The NLP-filtered findings were combined with the reference standard outcomes and then randomly divided into 50% training and 50% test sets to evaluate performance of machine learning classification. We optionally bypassed the NLP processing step after deidentification to evaluate machine learning classification performance directly on raw text.
Figure 1.
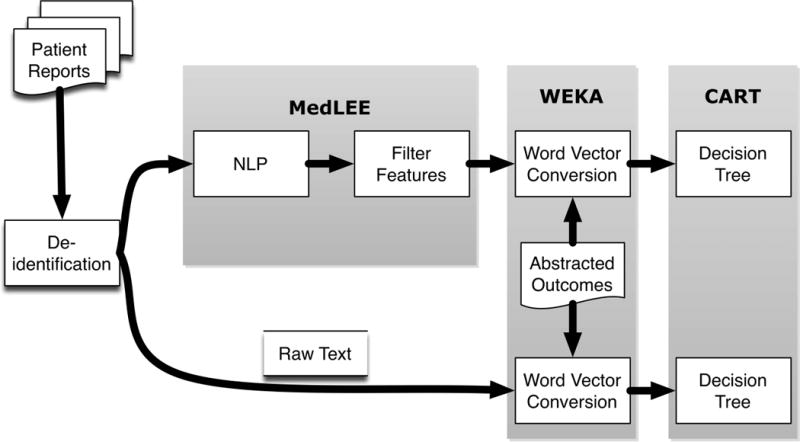
Overview of hybrid automated classification system approach. CART = Classification and Regression Trees; MedLEE = Medical Language Extraction and Encoding; NLP = natural language processing; WEKA = Waikato Environment for Knowledge Analysis.
Preprocessing
To secure IRB approval for waiver of consent, we manually removed all protected health information. This was performed after linking CT reports and the abstracted outcomes database using a script to replace medical record numbers with a matching unique sequenced study number.
Medical Language Extraction and Encoding Overview
Medical Language Extraction and Encoding (MedLEE; Columbia University, New York, NY; and Health Fidelity, Menlo Park, CA) was chosen as the NLP module because it is one of the most widely used NLP software packages in the medical research community7 and has previously successfully interpreted findings from free-text radiology procedure reports, including head CT imaging for stroke and chest radiography for respiratory diseases.10,11 It is available under both commercial and academic licenses. Figure 2 depicts MedLEE’s main components.12,13
Figure 2.
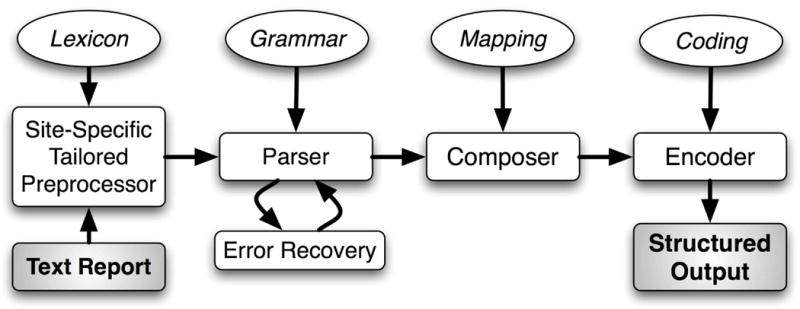
Main components of MedLEE. MedLEE = Medical Language Extraction and Encoding.
MedLEE parses text using a grammar to recognize syntactic and semantic patterns, generating structured text with contextual modifiers that are organized in tables and assigned to codes.13 To adapt MedLEE for new clinical applications, its lexicon, abbreviations, and section names can be extended dynamically to reflect the terms and organization seen in the documents to be interpreted. This is necessary because of the need for disambiguation, where terms have different meanings in different contexts (e.g. “ventricle” in an echocardiogram report is anatomically different from “ventricle” in a CT head report). For this study, we randomly sampled a set of 200 reports that had been processed by MedLEE to identify any problematic terms that would need MedLEE adaptation.
Unified Medical Language System
Unified Medical Language System (UMLS) is a repository of many controlled vocabularies developed by the U.S. National Library of Medicine for use in the biomedical sciences.14,15 It is a comprehensive thesaurus and ontology of biomedical concepts consisting of 6.4 million unique terms for 1.3 million concepts from more than 119 families of biomedical vocabularies. MedLEE matches its findings to Concept Unique Identifiers (CUIs) from UMLS, which increases interoperability of the system (Figure 3).
Figure 3.

Example of MedLEE structured output. MedLEE = Medical Language Extraction and Encoding.
Feature Selection Filtering
MedLEE output includes problems, findings, and procedures with associated modifiers that report specific body locations, certainty, and temporal status (Figure 3). We used the certainty and temporal status modifiers to include only likely acute findings, filtering out findings associated with negated and low-probability certainty modifiers, as well as those associated with historical or chronic temporal status modifiers. We did not plan on limiting the structured output to UMLS codes with body locations specific to orbital anatomy as we expected the machine learning algorithms to detect these on their own, and we wanted to minimize human supervision of the study approach.
Postprocessing Using Waikato Environment for Knowledge Analysis
Waikato Environment for Knowledge Analysis (WEKA; Waikato University, Hamilton, New Zealand) is an open-source collection of machine learning algorithms for data mining tasks written in Java.16 WEKA 3.7.5 contains tools for data preprocessing, classification, regression, clustering, association rules, and visualization, although we used it solely for postprocessing. It is capable of importing multiple forms of data, including the deidentified raw-text reports and the filtered feature sets from NLP output. These were separately compiled with the reference standard outcomes of acute orbital fracture (Figure 1) into individual files in attribute relation file format (arff), where each line represents one report with its associated outcome. These arff files underwent conversion into word vector representations.17 The word vector representations for the raw text were unigram words. The word vector representations for the NLP-processed reports combined unigram words and UMLS CUI phrases. We justified this approach to allow the machine learning classification to utilize the NLP-processed UMLS CUIs, yet also allow it to identify simple key words that could potentially perform highly as well.
Decision Tree Classification
We used decision trees for classification because of their explicit rule-based output, which can be easily evaluated for content validity. We used the Classification and Regression Trees (CART) module of Salford Predictive Miner 6.6 (Salford Systems, San Diego, CA) to generate decision trees using the word vector attributes as predictors, without explicit constraints, minimum performance cutoffs, or maximum number of nodes. The goal was to generate a parsimonious tree that was robust to varying misclassification costs. We opted to use training and testing sets to evaluate performance instead of cross-validation because we wanted to see how decision tree classifiers would work in a real-world scenario of training an automated classifier using a subset of data to then be applied to the remaining testing subset.
RESULTS
Of the 3,710 CT imaging reports, 460 (12.4%) were coded positive for orbital fractures by the trained data abstractor. A random subset of 507 CT reports were double coded, and interrater analysis revealed excellent agreement between the data abstractor and study physician, with Cohen’s kappa of 0.97 (95% = confidence interval [CI] = 0.94 to 0.99). Random screening of initial MedLEE output identified only minor lexical modifications (Data Supplement S1, available as supporting information in the online version of this paper) that were incorporated into the final MedLEE command line to extend the lexicon (Data Supplement S2, available as supporting information in the online version of this paper). After association with the reference standard outcome files, the raw text and NLP outputs were converted to word vector representations using WEKA using the StringtoWordVector filter with OutputWordCounts option set to true. The word vector representations consisted of 1,296 raw-text attributes and 1,371 NLP attributes.
Decision tree modeling using CART was successfully performed using either the attributes from the raw-text word vector representations or the attributes from the NLP word vector representations. Using the binary outcome of fracture/no fracture, both test sets had high classification accuracies, exceeding 90% on almost all measures (Table 1). Decision trees were qualitatively selected to balance improving classification performance (i.e. maximizing the area under the receiver operating characteristic curve) and minimizing relative cost and tree complexity. Final decision trees were parsimonious at eight nodes and nine nodes, respectively, and were robust to varying misclassification costs increasingly favoring sensitivity (Data Supplements S3 and S4, available as supporting information in the online version of this paper). The training and testing data sets were confirmed to include similar proportions of positive and negative fracture CT reports.
Table 1.
Classification Performance of Decision Trees on Test Sets
| Test | Raw Text | NLP |
|---|---|---|
| Sensitivity (95% CI) | 0.925 (0.883–0.954) | 0.933 (0.897–0.959) |
| Specificity (95% CI) | 0.933 (0.928–0.937) | 0.969 (0.964–0.973) |
| PPV (95% CI) | 0.650 (0.621–0.671) | 0.816 (0.785–0.839) |
| NPV (95% CI) | 0.989 (0.983–0.993) | 0.990 (0.985–0.994) |
| Weighted precision | 0.949 | 0.968 |
| Weighted recall | 0.932 | 0.964 |
| Weighted F-score | 0.940 | 0.966 |
Weighted performance scores are commonly reported in the information retrieval literature. Weighted precision is the weighted average of PPV and NPV, weighted recall is the weighted average of sensitivity and specificity, and the weighted F-score is the weighted average of precision and recall for positive cases and negative cases.
NLP = natural language processing; NPV = negative predictive value; PPV = positive predictive value.
Misclassified CT reports, both false positives and false negatives, were reviewed in both the training and the test sets and the causes of error were categorized (Table 2). Of 102 total misclassified reports (2.7%), the main sources of classification error were nonorbital fractures being classified as orbital fractures and disagreement between radiologists. The latter classification error stems from the fact that the next-day radiology overread is appended to the preliminary reading to create the final CT report of record.
Table 2.
Classification Errors (Combination of Training and Test Sets)*
| Cause | Frequency (%) |
|---|---|
| Nonorbital fracture | 32 (31.4) |
| Final reading disagrees with preliminary reading | 19 (18.6) |
| Vague certainty | 9 (8.8) |
| Fracture acuity | 9 (8.8) |
| Recent facial fracture surgery | 6 (5.9) |
| MedLEE miscoding | 5 (4.9) |
| Other† | 22 (21.6) |
Total sample of 3,710. Errors total 102 instances (2.7%).
Includes dictation error, filtering error, fracture implied but not stated, and miscellaneous poor wording.
MedLEE = Medical Language Extraction and Encoding.
DISCUSSION
The results of this study support our hypothesis that a hybrid automated classification system can perform comparably to medical personnel. The performance of our system is similar to that of physician raters in previous studies (Table 3).2,18–21 Furthermore, the classification performance was comparable to the high interrater reliability between the trained data abstractor and study physician in this study. Even though this study was performed using data from real-world settings, the automated classification performance was similar to the performance in simulated conditions in previous studies.1
Table 3.
Performance of Automated Classification Compared to Physician Raters
| Study and Coding Method | Sensitivity | Specificity |
|---|---|---|
| This study, hybrid automated (95% CI) | 0.933 (0.897–0.959) | 0.969 (0.964–0.973) |
| Hripcsak 1995,18 7 internists*† | 0.839 | 0.983 |
| Hripcsak 1995,18 7 radiologists*† | 0.854 | 0.986 |
| Hripcsak 1998,19 12 physicians* (95% CI) | 0.87 (0.84–0.90) | 0.98 (0.98–0.99) |
| Chapman 1999,2 1 physician (95% CI) | 0.900 (0.812–0.956) | 0.811 (0.753–0.848) |
| Fiszman 2000,20 4 physicians*† | 0.94 | 0.91 |
| Solti 2009,21 11 physicians*† | 0.85 | 0.95 |
Average.
95% CI not reported or not calculable from published results.
A few prior studies have examined the use of classification algorithms to interpret radiology report free text data from real-world sources. Two early studies used MedLEE for chest radiograph reports, but used expert rules for the classification step.18,19 Although less generalizable in approach, these early studies suggested that a hybrid approach to automated classification was comparable to medical personnel. A more recent study by the same authors used MedLEE and expert rules on a random selection of 150 chest radiographs to identify 24 different pathologic conditions and demonstrated an average sensitivity of 0.81 and an average specificity of 0.99.22 The largest real-world study examined 7,928 chest radiographs on a consecutive cohort of 1,277 neonates to detect pneumonia.12 Although this meant each unique patient was the source of multiple reports and there was a low prevalence of positive cases (seven cases, 0.5%), MedLEE and expert rules identified pneumonia with a sensitivity of 0.71 and specificity of 0.99. The best performance was found in a study comparing the precision and accuracy of NegEx (negative identification for clinical conditions, https://code.google.com/p/negex/) and SQL Server 2008 Free Text Search (Microsoft Corp., Redmond, WA) to identify acute fractures in 400 randomly selected extremity and spine radiograph reports.23 Although the expert rules were constructed to broadly identify any acute fractures and there was a low prevalence of positive cases (13 cases, 3.25%), NegEx performance was perfect, while modified SQLServer also did well (precision = 1.00, recall = 0.92; F-score = 0.96).
This study improves on previous work in two ways. First, we achieved similar performance using data sourced from real-world clinical settings despite being the first to use machine learning for outcome identification. Second, we analyzed the largest number of unique patients to date.
In selecting MedLEE for our hybrid approach, we did consider other available NLP tools. Alternative medical NLP tools, such as the open-source Clinical Text Analysis and Knowledge Extraction System (cTAKES; Mayo Clinic, Rochester, MN), are still being developed to understand temporality, among other features.24 More general NLP tools, such as Stanford NLP,25 are not customized for medical terms and often lack contextual modifiers. MedLEE, on the other hand, produces modifiers to determine the validity of the findings and also codes them to UMLS medical terms, which provides semantic standardization.
Other techniques for machine learning can be faster than decision trees, but they do not provide readily interpretable output that would be important to clinicians. When reviewing the decision tree splitting criteria (Data Supplements S3 and S4), it should be noted that the classification algorithms used clinically sensible terms. Most criteria were words or UMLS CUIs that describe facial fractures or orbital anatomy. Support Vector Machines (SVM) have been previously shown to be very successful in text classification,26 including the medical domain,8 but provide no way of verifying the sensibility of the algorithm. Although we planned to perform only decision trees as a transparent form of machine learning, we did confirm that the results of classification using SVM (using the WEKA SMO classifier) showed similar performance.27
For this study, an unexpected finding was the high classification performance of machine learning techniques applied to raw text. The high performance of the decision tree classifiers without NLP processing may be due to the fact that certain anatomical terms were present in CT reports only when describing orbital fractures without modifiers (Data Supplement S3). The notable exception was the use of the modifier “associated” as the final splitting criteria after the term “orbital” was found. It was likely serving as a surrogate for complex concepts like fluid in a sinus or overlying soft tissue swelling. The persistence of simple key anatomic terms for a few of the splitting criteria in the NLP decision tree supports this observation (Data Supplement S4). Previous examples of machine learning classification performing well without the benefit of NLP exist in the literature, although the identified research all used simulated data sets.4,21,28 However, one of the goals of applying a hybrid system is to take advantage of the pattern-matching benefits of machine learning to identify key words, while leveraging the power of NLP to understand more complex prose. For a given outcome, one approach may be better than the other, but combining them will be better.8 We expect a hybrid approach to excel when associating complex findings with outcomes of clinical importance. For instance, a hybrid automated classification system should perform better for conditions in which the outcome itself may be described by multiple synonyms or findings of varying severity (e.g., degree of injury or volume of blood).
One of the challenges of developing an automated classification system using NLP is the need to adjust the system for a particular task. For explicit rule-based systems, this may involve actually changing the rules themselves. In the case of MedLEE, the adjustments required were simply lexical in nature, to address disambiguation and add missing anatomical terms (Data Supplement S2). Regardless, there are no methodologic standards for conducting this adjustment, and a large number of classification errors were related to anatomic terminology errors (Table 2). In this study, we conducted a random sampling of reports for the study physician to review and analyze. More comprehensive methods may be needed when conducting multicenter studies. Content analysis, a well-established qualitative research methodology, could be used to exert rigor to the process of identifying and categorizing important terms and concepts.29 Content analysis would involve multiple raters and require defining a structured, inductive approach to evaluation that would afford a measure of objectivity, reliability, and reproducibility.
While this study adds to a growing body of research demonstrating the utility of automated classification to support outcomes research, further studies are needed. It remains to be seen if this hybrid system will outperform simple machine learning in identification of clinical outcomes of interest from medical free text. We are currently testing this system on the more complex findings of traumatic brain injury in head CT imaging reports from several sites to see if the NLP processing aspect of the hybrid demonstrates superior performance over exclusive use of machine learning.
LIMITATIONS
As a representative corpus of radiology reports, this data set may not have enough challenging text to demonstrate NLP’s dexterity, such as lacking more of a variety of temporal or conditional modifiers. However, this was a real-world data set, consisting of 2 years of consecutive CT reports. It may not reflect a “typical” radiology corpus that is used to test automated classification systems by informatics researchers, but it does represent a typical data set that could have used automated classification for clinical application.
Another limitation was the nature of documentation at the sites that generated the CT reports, namely, combining preliminary and final findings into the final CT report of record. While this is likely not common practice, it created a challenge for automated classification as contradictory findings and impressions existed in a single CT report. We are not aware of established techniques to overcome this problem. In the meantime, careful consideration should be made in applying this approach in future studies if the reporting behavior is similar at a participating institution.
CONCLUSIONS
Combining natural language processing and machine learning techniques shows promise in automating outcome classification from free-text electronic clinical data. This hybrid approach should be broadly applicable to outcomes of clinical interest, whereas relying on machine learning alone likely is not. Future work will address creating robust approaches to refining natural language processing tools, as well as validating that a hybrid approach provides more consistent performance for complex outcomes. Validated performance, when tested on data from other real-world settings, could lead to potentially streamlining data collection for clinical research and performance improvement.
Supplementary Material
Data Supplement S1. MedLEE command line.
Data Supplement S2. Lexicon modification file.
Data Supplement S3. CART decision tree for automated classification of raw text.
Data Supplement S4. CART decision tree for automated classification of NLP-structured text.
Acknowledgments
The authors acknowledge the assistance of Dr. Carol Friedman, Dr. Ethan Cowan, Lauren Winter, Lyudmila Ena, and Dr. James Chamberlain in the execution and completion of this project.
This publication was supported through the National Institutes of Health (NIH) Clinical and Translational Science Award (CTSA) program, grants UL1TR000075 and KL2TR000076. The CTSA program is led by the NIH’s National Center for Advancing Translational Sciences (NCATS). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. Medical Language Extraction and Encoding (MedLEE) was developed with support from the National Library of Medicine (R01LM010016 and R01LM008635).
Footnotes
Presented at The SHARPn Summit on Secondary Use, Rochester, MN, June 2012; The NLM/NIBIB Natural Language Processing: State of the Art, Future Directions and Applications for Enhancing Clinical Decision-Making Workshop, Bethesda, MD, April 2012; The ACRT/AFMR/SCTS Translational Science Meeting, Washington, DC, April 2012; The 2012 AMIA Clinical Research Informatics Summit, San Francisco, CA, March 2012; and The Workshop for Women in Machine Learning, Granada, Spain, December 2011.
The following supporting information is available in the online version of this paper:
Please note: Wiley Periodicals Inc. is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
The authors have no conflicts of interest to declare.
References
- 1.Stanfill MH, Williams M, Fenton SH, Jenders RA, Hersh WR. A systematic literature review of automated clinical coding and classification systems. J Am Med Inform Assoc. 2010;17:646–51. doi: 10.1136/jamia.2009.001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chapman WW, Haug PJ. Comparing expert systems for identifying chest x-ray reports that support pneumonia. Proc AMIA Symp. 1999:216–20. [PMC free article] [PubMed] [Google Scholar]
- 3.Dreyer KJ, Kalra MK, Maher MM, et al. Application of recently developed computer algorithm for automatic classification of unstructured radiology reports: validation study. Radiology. 2005;234:323–9. doi: 10.1148/radiol.2341040049. [DOI] [PubMed] [Google Scholar]
- 4.Percha B, Nassif H, Lipson J, Burnside E, Rubin D. Automatic classification of mammography reports by BI-RADS breast tissue composition class. J Am Med Inform Assoc. 2012;19:913–6. doi: 10.1136/amiajnl-2011-000607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turchin A, Kolatkar NS, Grant RW, Makhni EC, Pendergrass ML, Einbinder JS. Using regular expressions to abstract blood pressure and treatment intensification information from the text of physician notes. J Am Med Inform Assoc. 2006;13:691–5. doi: 10.1197/jamia.M2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greenes RA. Clinical Decision Support. 1. Philadelphia, PA: Elsevier; 2007. [Google Scholar]
- 7.Meystre SM, Savova GK, Kipper-Schuler KC, Hurdle JF. Extracting information from textual documents in the electronic health record: a review of recent research. IMIA Yearb Med Inform. 2008;47(Suppl 1):138–54. [PubMed] [Google Scholar]
- 8.Minard AL, Ligozat AL, Ben Abacha A, et al. Hybrid methods for improving information access in clinical documents: concept, assertion, and relation identification. J Am Med Inform Assoc. 2011;18:588–93. doi: 10.1136/amiajnl-2011-000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yadav K, Cowan E, Haukoos JS, et al. Derivation of a clinical risk score for traumatic orbital fracture. J Trauma Acute Care Surg. 2012;73:1313–8. doi: 10.1097/TA.0b013e318265cf61. [DOI] [PubMed] [Google Scholar]
- 10.Elkins JS, Friedman C, Boden-Albala B, Sacco RL, Hripcsak G. Coding neuroradiology reports for the Northern Manhattan Stroke study: a comparison of natural language processing and manual review. Comput Biomed Res. 2000;33:1–10. doi: 10.1006/cbmr.1999.1535. [DOI] [PubMed] [Google Scholar]
- 11.Friedman C, Alderson PO, Austin JH, Cimino JJ, Johnson SB. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc. 1994;1:161–74. doi: 10.1136/jamia.1994.95236146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mendonça EA, Haas J, Shagina L, Larson E, Friedman C. Extracting information on pneumonia in infants using natural language processing of radiology reports. J Biomed Inform. 2005;38:314–21. doi: 10.1016/j.jbi.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 13.Friedman C. A broad-coverage natural language processing system. Proc AMIA Symp. 2000:270–4. [PMC free article] [PubMed] [Google Scholar]
- 14.Federal Coordinating Council for Comparative Effectiveness Research. Report to the President and the Congress. Washington, DC: US Department of Health and Human Services; Available at: http://www.hhs.gov/recovery/programs/cer/cerannualrpt.pdf. Accessed May 29, 2013. [Google Scholar]
- 15.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004;32:D267–70. doi: 10.1093/nar/gkh061. database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall M, Frank E, Holmes G, Pfahringer B. The WEKA data mining software: an update. SIGKDD Explorations. 2009;11:10–18. [Google Scholar]
- 17.Salton G. Automatic Text Processing. Boston, MA: Addison-Wesley; 1989. [Google Scholar]
- 18.Hripcsak G, Friedman C, Alderson PO, DuMouchel W, Johnson SB, Clayton PD. Unlocking clinical data from narrative reports: a study of natural language processing. Ann Intern Med. 1995;122:681–8. doi: 10.7326/0003-4819-122-9-199505010-00007. [DOI] [PubMed] [Google Scholar]
- 19.Hripcsak G, Kuperman GJ, Friedman C. Extracting findings from narrative reports: software transferability and sources of physician disagreement. Methods Inform Med. 1998;37:1–7. [PubMed] [Google Scholar]
- 20.Fiszman M, Chapman WW, Aronsky D, Evans RS, Haug PJ. Automatic detection of acute bacterial pneumonia from chest X-ray reports. J Am Med Inform Assoc. 2000;7:593–604. doi: 10.1136/jamia.2000.0070593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Solti I, Cooke CR, Xia F, Wurfel MM. Automated classification of radiology reports for acute lung injury: comparison of keyword and machine learning based natural language processing approaches. Proc IEEE Int Conf Bioinformat Biomed. 2009:314–9. doi: 10.1109/BIBMW.2009.5332081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hripcsak G, Austin JH, Alderson PO, Friedman C. Use of natural language processing to translate clinical information from a database of 889,921 chest radiographic reports. Radiology. 2002;224:157–63. doi: 10.1148/radiol.2241011118. [DOI] [PubMed] [Google Scholar]
- 23.Womack JA, Scotch M, Gibert C, et al. A comparison of two approaches to text processing: facilitating chart reviews of radiology reports in electronic medical records. Perspect Health Inform Manag. 2010;7:1a. [PMC free article] [PubMed] [Google Scholar]
- 24.Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical text analysis and knowledge extraction system (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc. 2010;17:507–13. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Marneffe MC, MacCartney B, Manning CD. Generating typed dependency parses from phrase structure parses. Proc LREC. 2006;6:449–54. [Google Scholar]
- 26.Joachims T. Text categorization with support vector machines: learning with many relevant features. Mach Learn: ECML-98. 1998:137–42. [Google Scholar]
- 27.Sarioglu E, Yadav K, Choi HA. Efficient classification of clinical reports utilizing natural language processing. 2012 AAAI Fall Symp Series. 2012;FS-12-05:88–9. [Google Scholar]
- 28.Chapman WW, Fizman M, Chapman BE, Haug PJ. A comparison of classification algorithms to automatically identify chest X-ray reports that support pneumonia. J Biomed Inform. 2001;34:4–14. doi: 10.1006/jbin.2001.1000. [DOI] [PubMed] [Google Scholar]
- 29.Krippendorff KH. Content Analysis: An Introduction to Its Methodology. 2. Thousand Oaks, CA: Sage Publications Inc.; 2003. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Supplement S1. MedLEE command line.
Data Supplement S2. Lexicon modification file.
Data Supplement S3. CART decision tree for automated classification of raw text.
Data Supplement S4. CART decision tree for automated classification of NLP-structured text.


