Abstract
Increasing evidence demonstrates the importance of long coiled-coil proteins for the spatial organization of cellular processes. Although several protein classes with long coiled-coil domains have been studied in animals and yeast, our knowledge about plant long coiled-coil proteins is very limited. The repeat nature of the coiled-coil sequence motif often prevents the simple identification of homologs of animal coiled-coil proteins by generic sequence similarity searches. As a consequence, counterparts of many animal proteins with long coiled-coil domains, like lamins, golgins, or microtubule organization center components, have not been identified yet in plants. Here, all Arabidopsis proteins predicted to contain long stretches of coiled-coil domains were identified by applying the algorithm MultiCoil to a genome-wide screen. A searchable protein database, ARABI-COIL (http://www.coiled-coil.org/arabidopsis), was established that integrates information on number, size, and position of predicted coiled-coil domains with subcellular localization signals, transmembrane domains, and available functional annotations. ARABI-COIL serves as a tool to sort and browse Arabidopsis long coiled-coil proteins to facilitate the identification and selection of candidate proteins of potential interest for specific research areas. Using the database, candidate proteins were identified for Arabidopsis membrane-bound, nuclear, and organellar long coiled-coil proteins.
The coiled-coil protein oligomerization motif consists of two or more amphipathic alpha helices that twist around each other in a supercoil (Burkhard et al., 2001). It was one of the earliest protein structures discovered, first described for the hair protein alpha keratin (Crick, 1952). Sequences with the capacity to form coiled-coils are characterized by a heptad repeat pattern in which residues in the first and fourth positions are hydrophobic, and residues in the fifth and seventh position are predominantly charged or polar. The stability of the coiled-coil is derived from a characteristic packing of the hydrophobic side chains into a hydrophobic core (“knobs in holes”; Crick, 1952).
It has been estimated that approximately 10% of all proteins of an organism contain a coiled-coil motif (Liu and Rost, 2001). Roughly, coiled-coil proteins can be grouped into two classes: Short coiled-coil domains of six or seven heptad repeats, also called Leucine zippers, are frequently found as homo- and heterodimerization motifs in transcription factors (Jakoby et al., 2002; Vinson et al., 2002). In contrast, long coiled-coil domains of several hundred amino acids are found in a number of functionally distinct proteins, which are often involved in attaching functional protein complexes to larger cellular structures, such as the Golgi, centrosomes, centromers, or the nuclear envelope.
Some large coiled-coil proteins oligomerize into filaments or networks and have themselves a structural role. One of the three main classes of cytoskeletal proteins, the intermediate filament proteins, represents a well-characterized group of coiled-coil proteins (Strelkov et al., 2003). In addition, the cytoskeletal motor proteins myosin, dynein, and kinesin contain coiled-coil motifs (Schliwa and Woehlke, 2003).
In the past few years, the number of investigated long coiled-coil proteins from animals and yeast has rapidly grown. They include proteins involved in nuclear organization, such as lamins (Goldman et al., 2002; Holaska et al., 2002), NuMA (nuclear mitotic apparatus protein; Compton et al., 1992; Yang et al., 1992), or the SMC (structural maintenance of chromosomes) proteins (Hirano, 2000; Jessberger, 2002). A number of coiled-coil proteins have been characterized that associate with the kinetochore/centromere regions of chromosomes in vertebrates and are involved in assembling other proteins on the kinetochore (Liao et al., 1995; Sugata et al., 1999, 2000; Starr et al., 2000; Fukagawa et al., 2001).
Long coiled-coil proteins play a role in microtubule nucleation and spindle organization during cell division. For example, coiled-coil proteins are involved in the architecture of the spindle pole body, the nuclear envelope-embedded microtubule organization center in yeast (Saccharomyces cerevisiae). They are required for insertion of the spindle pole body into the nuclear envelope (Schramm et al., 2000; Le Masson et al., 2002) and for the precise spatial positioning of the outer plaque, central plaque, and inner plaque (Kilmartin et al., 1993; Chen et al., 1998; Souès and Adams, 1998; Schaerer et al., 2001). The vertebrate microtubule organization center, the centrosome, also contains a number of long coiled-coil proteins. They are involved in microtubule nucleation, scaffolding/bridging of other proteins, and the anchoring of signaling components such as calmodulin, protein kinase C, and protein kinase A (Fava et al., 1999; Takahashi et al., 1999; Witczak et al., 1999; Flory et al., 2000; Li et al., 2000; Takahashi et al., 2000; Moisoi et al., 2002; Sillibourne et al., 2002; Takahashi et al., 2002). In nematodes, the coiled-coil proteins PUMA1 (Esteban et al., 1998) and LIN-5 (Lorson et al., 2000) have been found to localize to the spindle apparatus in a cell cycle- and microtubule-dependent manner. PUMA1 might be part of a “centromeric matrix,” whereas LIN-5 is thought to play a role in localizing or regulating a motor-protein complex and/or connecting the spindle apparatus with the cell cortex.
In the cytoplasm, long coiled-coil proteins are involved in the organization of and targeting to membrane systems. The golgin family comprises a group of coiled-coil peripheral or integral membrane proteins associated with the Golgi apparatus. They have been shown to function in a variety of membrane-membrane and membrane-cytoskeleton tethering events at the Golgi and are regulated by small GTPases of the Rab and Arl families (Barr and Short, 2003). It has been suggested that golgins and the related fruitfly (Drosophila melanogaster) protein Lva (Lava Lamp; Sisson et al., 2000) are forming a Golgi matrix that serves as the structural scaffold for the enzyme-containing membranes of the Golgi apparatus and may provide the means of partitioning the Golgi during mitosis (Seemann et al., 2000, 2002). A group of long coiled-coil proteins associated with both the centrosome and the Golgi are involved in anchoring both cyclic nucleotide phosphodiesterase and cAMP-dependent protein kinase A to the centrosome/Golgi, suggesting a role of these coiled-coil proteins in cAMP signal compartmentalization (Witczak et al., 1999; Diviani and Scott, 2001; Verde et al., 2001).
These examples serve to illustrate the emerging function of long coiled-coil proteins as anchors for the regulation of protein positioning in the cell, thus both separating and coordinating signaling pathways in a temporal and spatial manner and organizing cellular processes like cell division. In contrast to animals and yeast, only a handful of long coiled-coil proteins have been studied in plants. Besides the large families of myosins and kinesins (Reddy and Day, 2001a, 2001b; Smith, 2002), the homologs of the mammalian SMC proteins have been characterized in Arabidopsis (Mengiste et al., 1999; Hanin et al., 2000; Liu et al., 2002). In addition, a small number of apparently plant-specific coiled-coil proteins have been identified. The carrot (Daucus carota) coiled-coil protein NMCP1 (Nuclear Matrix Constituent Protein 1) is located at the nuclear rim during interphase and at the spindle poles in mitotic cells (Masuda et al., 1997). CIP1 (COP1-interactive protein 1), a cytoskeleton-associated coiled-coil protein, binds to the photomorphogenesis suppressor COP1 (Matsui et al., 1995). MFP1 is a DNA-binding protein and associated with the thylakoids in plant chloroplasts (Jeong et al., 2003). PF2 is a large coiled-coil protein found in a screen for motility mutants in the algae Chlamydomonas reinhardtii (Rupp and Porter, 2003), where it is required for the assembly of the dynein regulatory complex. Besides these few examples, nothing is presently known about plant long coiled-coil proteins and their potential functions in the anchoring and structuring of cellular events.
In BLAST searches of the whole Arabidopsis genome for all animal and yeast proteins discussed above, significant homologies can only be found for the protein families of the SMC proteins and myosins, with E values typically below e-100, kinesins with E values in the e-50 to e-100 range, and for the nuclear pore complex protein Tpr (5e-78). In all other cases, the best hits for functionally very different proteins are the same three proteins from the Arabidopsis genome, indicating the difficulty in using sequence similarity algorithms to identify functional homologs of long coiled-coil proteins. The multiple heptad repeats in long coiled-coil domains cause a low and promiscuous sequence similarity between long coiled-coil proteins, which leads to meaningless results. This clearly demonstrates the need to use other methods than sequence comparison for the identification of plant long coiled-coil proteins potentially involved in the diverse cellular functions discussed above.
Although the heptad repeat pattern causes false hits in sequence similarity searches, it can be easily exploited by computational methods to predict coiled-coil domains in amino acid sequences (Parry, 1982; Lupas et al., 1991). More recently, the combination of coiled-coil prediction algorithms such as MultiCoil (Wolf et al., 1997) with whole-genome information has permitted the mining of all coiled-coil proteins of an organism. Using this approach on a total yeast genome translation, approximately 300 two-stranded and 250 three-stranded coiled-coils have been identified (Newman et al., 2000). Over one-half of these open reading frames represent proteins of unknown function. An investigation of a number of structural motifs in several whole genomes showed independently that the human (Homo sapiens), fruitfly, Caenorhabditis elegans, and yeast genomes contain roughly 10% coiled-coil proteins (Liu and Rost, 2001).
We report here the identification of all long coiled-coil proteins from Arabidopsis and the establishment of a novel searchable database, ARABI-COIL (http://www.coiled-coil.org/arabidopsis). In the future, as more fully annotated plant genomes such as rice (Oryza sativa) and C. reinhardtii become available, our analysis pipeline will be applied to these species as well, and the data will be added to the database.
RESULTS
Genome-Wide Screen for Coiled-Coil Proteins
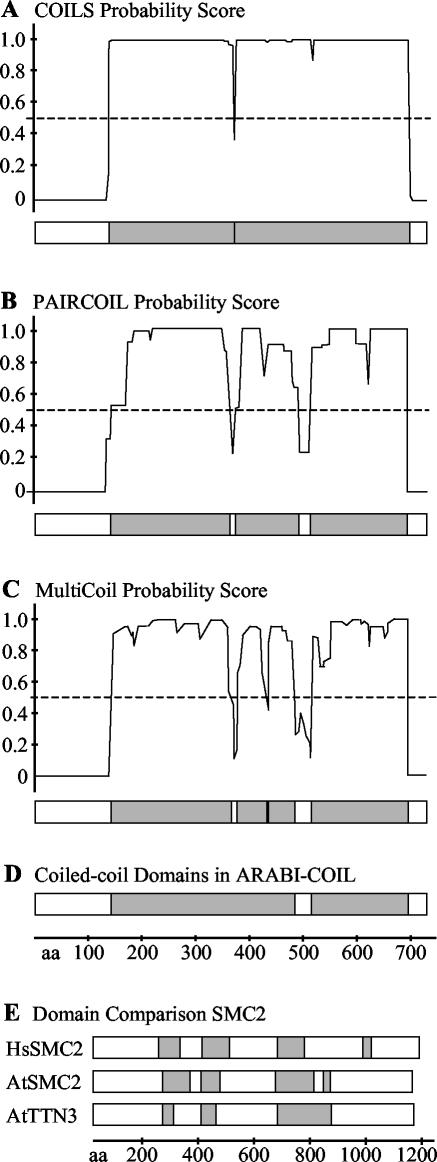
Arabidopsis long coiled-coil proteins were identified using the algorithm MultiCoil (Wolf et al., 1997). MultiCoil is capable of predicting two-stranded and three-stranded coiled-coils with significantly less false positives than earlier prediction methods (Wolf et al., 1997). Figure 1 shows a comparison of MultiCoil performance with older prediction methods, using Arabidopsis MFP1 (Harder et al., 2000; Jeong et al., 2003) as an example. MultiCoil offers the highest stringency of the methods tested. The program is available as a Web resource allowing prediction of individual sequences online. With more than 25,000 sequences requiring analysis, the single sequence submission through the Web was not a tractable approach; therefore, the MultiCoil program was installed on a local multiprocessor system to run the Arabidopsis proteome sequence set. After confirming the consistency of results between the locally installed version of MultiCoil and the available Web resource with a small subset of test sequences, the entire Arabidopsis predicted proteome (http://www.ebi.ac.uk/proteome/ARATH/) was processed. Using a cutoff value of 20 amino acids minimum length for a coiled-coil domain and 0.5 for the probability score, 5.6% of all Arabidopsis sequences (about 1,500 proteins) were identified as coiled-coil proteins. Of these sequences (1.5% of the genome), 386 were predicted to have coiled-coil domains of 50 or more amino acids in length.
Figure 1.
Comparison of different algorithms for coiled-coil domain prediction. At3g16000 (AtMFP1, GenBank accession no. BAB02666; Harder et al., 2000; Jeong et al., 2003) is shown as an example. A to C, Probability scores plotted against the length of the protein in amino acids (aa) and bar diagram generated from plots. The dashed lines mark the cutoff score of 0.5. Coiled-coil domains are shown in gray in the bar diagram. A, COILS; B, PAIRCOIL; C, MultiCoil. D, Bar diagram generated after processing of MultiCoil data through ExtractProp (“Materials and Methods”) to eliminate short gaps in coiled-coil domain predictions. E, Comparison of domain predictions by MultiCoil using the same score and length cutoff parameters for human and Arabidopsis structural maintenance of chromosomes 2 proteins. HsSMC2, NP_006435; AtSMC2, NP_190330; AtTTN3, NP_201047.
Selection of Proteins with Long Coiled-Coil Domains
To focus on proteins potentially involved in structural aspects of the cells and to exclude shorter coiled-coil domains like Leucine zippers, the output from the original MultiCoil run was further processed and filtered. A software package (ExtractProp Suite, see “Materials and Methods”) was developed to automate the processing of data and selection of sequences. In this process, small gaps shorter than 25 amino acids between predicted coiled-coil domains were ignored and the domains treated as a single, larger coiled-coil (Fig. 1D). The relative consistency of the prediction between Arabidopsis and animal sequences was tested by comparing family members of the conserved SMC proteins. SMC proteins typically contain two clusters of coiled-coil domains separated by a central linker domain. Figure 1E shows that this domain distribution was observed for human SMC2 and its two Arabidopsis homologs.
Because a high-stringency algorithm like MultiCoil often predicts long stretches of coiled-coil domains with significant intradomain gaps (as shown in Fig. 1), a filter was introduced to include only proteins with at least one coiled-coil domain of at least 70 amino acids, two domains and a minimal domain length of 50 amino acids, or three domains and a minimal domain length of 30 amino acids in the final data set. This strategy isolated 286 sequences with long or multiple coiled-coil domains while excluding 97% of the known Arabidopsis bZIP proteins (Jakoby et al., 2002). Table I shows the distribution of the maximum length of predicted coiled-coil domains per protein in the ARABI-COIL database. The total percentage of the residues per protein sequence predicted to be in a coiled-coil region is summarized in Table II.
Table I.
Distribution of lengths of long coiled-coil domains in the ARABI-COIL database CC, Coiled-coil.
| Maximum CC Length | No. of Proteins |
|---|---|
| >400 | 7 |
| 250-399 | 12 |
| 150-249 | 57 |
| 100-149 | 67 |
| <100a | 143 |
No. includes only proteins with maximum domain length/domain no. of at least 70/1, 50/2, or 30/3
Table II.
Distribution of percentage coiled-coil coverage per protein in the ARABI-COIL database CC, Coiled-coil.
| CC Coverage | No. of Proteins |
|---|---|
| % | |
| >80 | 2 |
| 79-60 | 13 |
| 59-40 | 48 |
| 39-20 | 112 |
| <20 | 111 |
The coiled-coil property information presented and searchable in ARABI-COIL is summarized for a single protein example in Table III. It includes the predicted number of coiled-coil domains, length of the largest coiled-coil domain, percentages of the total sequence and the N-terminal, middle, and C-terminal one-third of the sequence predicted to be in a coiled-coil, and the highest prediction score over the whole sequence. The ARABI-COIL database search form allows for searches limited to a certain length of protein and/or coiled-coil domain and percentage coverage over the whole length and/or the N-terminal, middle, and C-terminal one-third of the sequence. A second output table summarizes the detailed positions of all predicted coiled-coil domains and the length of the longest intradomain gap for each given domain (Table IV). A graphical representation of the predicted coiled-coil structures was included (Fig. 1D). Links to National Center for Biotechnology Information (NCBI) GenBank sequence entries are provided in ARABI-COIL to retrieve the underlying sequence information for each database entry.
Table III.
Coiled-coil features as presented in ARABI-COIL Example prediction data for At3g16000 (GenBank accession no. BAB02666; compare with Figure 1, C and D, and Table IV).
| Computed Coiled-Coil Property | Value |
|---|---|
| No. of coiled-coil domains in sequence | 2 |
| Length of largest coiled-coil domain | 338 |
| % of entire sequence predicted as coiled-coil | 70 |
| % of N-terminal one-third predicted as coiled-coil | 40 |
| % of middle one-third predicted as coiled-coil | 99 |
| % of C-terminal one-third predicted as coiled-coil | 72 |
| Highest coiled-coil probability score | 1.0 |
Table IV.
Coiled-coil domain data as presented in ARABI-COIL Example prediction data for At3g16000 (GenBank accession no. BAB02666; compare with Figure 1, C and D, and Table III).
| Start | End | Property | Value |
|---|---|---|---|
| 145 | 482 | Length of domain | 338 |
| 145 | 482 | Maximum intradomain gap | 14 |
| 518 | 692 | Length of domain | 175 |
| 518 | 692 | Maximum intradomain gap | 0 |
Functional Categories of Arabidopsis Long Coiled-Coil Proteins
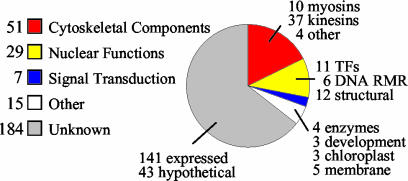
Only 10% of the 286 proteins in ARABI-COIL have been characterized so far by experimental data, with about one-half of these falling into the categories kinesin or myosin motors or SMC proteins. For a preliminary estimate of protein functions, annotations were assigned manually. They are based on available publications (refs. linked to PubMed are available in ARABI-COIL), annotations in NCBI RefSeq (http://www.ncbi.nlm.nih.gov/RefSeq/), The Arabidopsis Information Resource (http://www.arabidopsis.org/), The Institute for Genomic Research (http://www.tigr.org/tdb/e2k1/ath1/), and the Munich Information Center for Protein Sequences (http://mips.gsf.de/proj/thal/db/), and conserved domains outside of the coiled-coil domain. The ARABI-COIL database can be searched for keywords within these annotations. Figure 2 summarizes the functional annotations of the proteins in ARABI-COIL and shows that two main fractions of the annotated proteins are involved in either cytoskeletal or nuclear functions. The putative function of 66% of the sequences in ARABI-COIL remains unknown. The percent of uncharacterized ORFs increases with the percentage coverage from 60% unknown proteins with less than 50% coiled-coil to 86% unknown proteins with 50% or more coiled-coil coverage. Seventy-five percent of the proteins with unknown function matched known expressed sequence tags and were annotated as “expressed proteins.” The remaining proteins without expressed sequence tag data were annotated as “hypothetical proteins.” Table V lists all Arabidopsis proteins of at least 500 amino acids in length with a predicted coiled-coil coverage of more than 25% for which published data are presently available.
Figure 2.
Putative functions of proteins in ARABI-COIL based on annotations. TFs, Transcription factors; DNA RMR, DNA recombination, modification, and repair.
Table V.
Previously investigated long coiled-coil proteins in Arabidopsis Proteins listed are of at least 500 amino acids in length with at least 25% predicted to form coiled-coils.
| AGI Locus | Protein | Protein Length | Maximum CC Length | Total CC Coverage | Putative Function | References |
|---|---|---|---|---|---|---|
| % | ||||||
| At5g41790 | CIP1 | 1,305 | 1,060 | 81 | Cytoskeleton, COP1 signal transduction | Matsui et al. (1995) |
| At3g16000 | MFP1 | 727 | 338 | 70 | Chloroplast DNA-binding protein | Jeong et al. (2003) |
| At1g77580 | FPP1 | 629 | 236 | 55 | Unknown | Gindullis et al. (2002) |
| At1g21810 | FPP2 | 639 | 196 | 50 | Unknown | Gindullis et al. (2002) |
| At3g05270 | FPP3 | 603 | 208 | 40 | Unknown | Gindullis et al. (2002) |
| At1g13220 | NMCP1 like | 1,128 | 200 | 39 | Nuclear matrix | Masuda et al. (1997)a |
| At1g19835 | FPP4 | 1,024 | 201 | 38 | Unknown | Gindullis et al. (2002) |
| At1g67230 | NMCP1 like | 1,166 | 198 | 38 | Nuclear matrix | Masuda et al. (1997)a |
| At2g18540 | preproMP73 | 699 | 253 | 36 | Storage protein | Mitsuhashi et al. (2001)a |
| At3g12020 | T21B14.15 | 956 | 109 | 31 | Kinesin | Reddy and Day (2002b) |
| At1g47900 | FPP6 | 1,054 | 110 | 30 | Unknown | Gindullis et al. (2002) |
| At4g21270 | KatA, ATK1 | 793 | 198 | 30 | Kinesin | Marcus et al. (2002, 2003) |
| At4g27180 | KatB, ATK2 | 744 | 138 | 30 | Kinesin | Mitsui et al. (1994, 1996) |
| At4g38070 | pHLH131 | 1,496 | 122 | 30 | Transcription factor | Heim et al. (2003) |
| At5g48600 | SMC4 | 1,241 | 108 | 30 | Condensin | Liu et al. (2002) |
| At1g68790 | NMCP1 like | 1,085 | 173 | 29 | Nuclear matrix | Masuda et al. (1997)a |
| At3g10180 | F14P13.22 | 1,348 | 276 | 29 | Kinesin | Reddy and Day (2002b) |
| At5g65770 | NMCP1 like | 1,042 | 122 | 29 | Nuclear matrix | Masuda et al. (1997)a |
| At3g47460 | SMC2 | 1,171 | 141 | 28 | Condensin | Liu et al. (2002) |
| At4g36120 | FPP5 | 981 | 125 | 28 | Unknown | Gindullis et al. (2002) |
| At5g54670 | KatC, ATK3 | 746 | 94 | 26 | Kinesin | Mitsui et al. (1994, 1996) |
| At5g61460 | MIM1 | 1,057 | 76 | 25 | DNA repair | Mengiste et al. (1999), Hanin et al. (2000) |
| At5g62410 | TTN3 | 1,175 | 202 | 25 | Condensin | Liu et al. (2002) |
Published data only available for homologs from other plant species (NMCP1, carrot; and preproMP73, pumpkin). bHLH, basic Helix-Loop-Helix; CC, coiled-coil; CIP, COP1 Interactive Protein; MFP, MAR-binding filament-like Protein; MIM, hypersensitive to MMS, irradiation, and MMC; NMCP, nuclear matrix constituent protein; SMC, structural maintenance of chromosomes; TTN, TITAN
The Arabidopsis genome appears to encode only one protein with a continuous coiled-coil domain of more than 1,000 amino acids. This protein, CIP1, has been characterized as a component of the cytoskeleton and functions as a binding site for the photomorphogenesis regulator COP1 (Matsui et al., 1995). Another characterized protein with a high coiled-coil coverage is AtMFP1, a DNA-binding chloroplast thylakoid protein (Jeong et al., 2003). Of the remaining proteins in Table V, six have been described as a family of filament-like proteins (FPPs; Gindullis et al., 2002), five contain a kinesin motor domain, and eight have functions suggesting their localization in the nucleus.
Putative Membrane-Associated Long Coiled-Coil Proteins in Arabidopsis
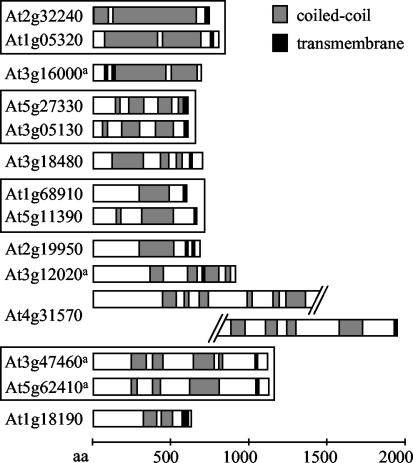
In addition to coiled-coil domain prediction, transmembrane domain prediction data from several programs (see Table VI) were incorporated in the database, including the number of predicted transmembrane domains in the ARAMEMNON database (http://aramemnon.botanik.uni-koeln.de; Schwacke et al., 2003). The ARABI-COIL search page allows for limiting searches to coiled-coil proteins with a certain number of predicted transmembrane domains in combination with specific coiled-coil properties. Cross-references to the more comprehensive details pages in ARAMEMNON, which include graphic comparisons of a larger number of transmembrane prediction programs, are provided with the output details for proteins with an entry in that database. Fourteen proteins were identified that are at least 500 amino acids long, have at least 25% coiled-coil coverage, and contain at least one transmembrane domain according to ARAMEMNON. Figure 3 shows a schematic representation of these proteins. Four proteins in this category have been characterized previously. AtMFP1 is a thylakoid membrane protein (Jeong et al., 2003). At3g12020 contains a kinesin motor domain, suggesting that it might function as a membrane-bound microtubule motor (Reddy and Day, 2001b). The Arabidopsis SMC2 homologs (Liu et al., 2002) are predicted to contain a transmembrane domain in their C-terminal domain. All novel proteins in this category contain a C-terminal predicted transmembrane domain.
Table VI.
Programs used for sequence analysis and targeting prediction cTP, Chloroplast targeting peptide; mTP, mitochondrial targeting peptide; NLS, nuclear localization signal; TMH, transmembrane helix; SP, signal peptide (ER, secretory pathway).
| Program | Predicted Feature | URL | Reference |
|---|---|---|---|
| MultiCoil 1.0 | Coiled-coil | http://theory.lcs.mit.edu/multicoil | Wolf et al. (1997) |
| ChloroP 1.1 | cTP | http://www.cbs.dtu.dk/services/ChloroP/ | Emanuelsson et al. (1999) |
| Predotar 0.5 | cTP, mTP | http://www.inra.fr/predotar/ | |
| TargetP 1.01 | cTP, mTP, SP, other | http://www.cbs.dtu.dk/services/TargetP/ | Nielsen et al. (1997a), Emanuelsson et al. (2000) |
| MitoProt II | mTP | http://www.mips.biochem.mpg.de/cgi-bin/proj/mcdgcn/mitofilter | Claros et al. (1996) |
| PredictNLS | NLS | http://maple.bioc.columbia.edu/predictNLS/ | Cokol et al. (2000) |
| PSORT | NLS, TMH | http://psort.nibb.ac.jp/ | Nakai and Horton (1999) |
| SignalP 2.0 HMM | SP | http://www.cbs.dtu.dk/services/SignalP-2.0/ | Nielsen and Krogh (1998) |
| SignalP 2.0 NN | SP | http://www.cbs.dtu.dk/services/SignalP-2.0/ | Nielsen et al. (1997b) |
| TMHMM 2.0 | TMH | http://www.cbs.dtu.dk/services/TMHMM/ | Sonnhammer et al. (1998), Krogh et al. (2001) |
| HMMTOP1.1 | TMH | http://www.enzim.hu/hmmtop/ | Tusnády and Simon (1998) |
| Several | TMH | http://aramemnon.botanik.uni-koeln.dc | Schwacke et al. (2003) |
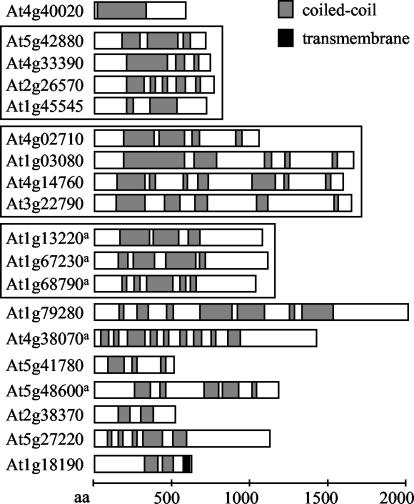
Figure 3.
Putative membrane proteins with high coiled-coil coverage. Proteins are of at least 500 amino acids in length and at least 25% coiled-coil coverage, sorted from top to bottom by decreasing percentage of coiled-coil coverage. Bar diagrams show the coiled-coil domain structure as represented in the ARABI-COIL database; gray boxes, coiled-coil domains; black boxes, transmembrane domains according to ARAMEMNON. Proteins belonging to gene families are boxed together. a, Proteins are characterized by published data (see Table V for comparison).
Long Coiled-Coil Proteins Are Predicted in All Cellular Compartments Investigated
The ARABI-COIL sequence set was further analyzed using a battery of programs to predict putative subcellular targeting of the proteins (Table VI). Two (NLSs) or three (N-terminal targeting signals) prediction scores were included in the database for each targeting signal. The ARABI-COIL search options allow limiting searches to coiled-coil proteins with a certain predicted localization in addition to transmembrane prediction and selected coiled-coil features. The results returned include all proteins with at least one program resulting in a prediction for that location above a probability cutoff of 0.5. The reliability of the prediction scores is color-coded for easier reference on the online result details page by using yellow for lower probability (0.50-0.74) and red for higher probability (0.75-1.00). Table VII shows an example for the detailed prediction output, which also illustrates how predicting the localization of individual proteins can be ambiguous.
Table VII.
Targeting signal predictions summarized in ARABI-COIL for At3g16000 The scores shown in the table were acquired using GenBank accession no. BAB02666. cTP, Chloroplast targeting peptide; mTP, mitochondrial targeting peptide; SP, signal peptide for secretory pathway.
| Predicted Signal | Program | Score |
|---|---|---|
| cTP | TargetP | 0.78 |
| cTP | ChloroP | 0.96a |
| cTP | Predotar | 0.88 |
| mTP | Predotar | 0.15 |
| mTP | Mitoprot | 0.72 |
| mTP | TargetP | 0.16 |
| SP | TargetP | 0.00 |
| SP | SignalP HMM | 0.70 |
| SP | SignalP NN | 0.46 |
| NLS | PredictNLS | 1.00b |
| NLS | PSORT | 0.07 |
| No signal | TargetP | 0.06 |
ChloroP prediction scores were normalized to a 0 to 1 scale
PredictNLS generates a `yes' = 1/`no' = 0 prediction
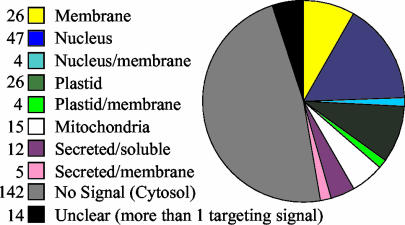
To summarize the predicted targeting for all proteins, the cross-program average of the scores for each type of targeting signal were computed and probability values of 0.5 and higher counted as positive. Figure 4 shows the computationally predicted distribution of the ARABI-COIL proteins in the cell using this method. Only proteins with an entry in ARAMEMNON were counted as transmembrane proteins for this analysis. The result shows that proteins with high coiled-coil coverage are predicted to be present in all compartments of the plant cell for which targeting signals were predicted computationally.
Figure 4.
Summary of subcellular targeting predictions of proteins in ARABI-COIL. The mean values of all prediction programs used (see Table VI) were computed and localization predictions with a mean value above a probability cutoff score of 0.5 were counted as positive for that location. Proteins with mean values above cutoff for two or more compartments of the cell were labeled “unclear.”
Putative Nuclear Long Coiled-Coil Proteins in Arabidopsis
About 10% of the annotations in ARABI-COIL suggest a nuclear function, and Figure 4 illustrates that 16% of the proteins in ARABI-COIL are predicted to be nuclear. The ARABI-COIL search functions were used to single out putative nuclear proteins of more than 500 amino acids in length with coiled-coil coverages above 25%. The resulting group of 37 proteins was manually checked for consistency of the predictions as described for Figure 4 to exclude proteins with only weak nuclear prediction or with ambiguous predictions (“unclear” in Fig. 4). The domain structures of the remaining 19 putative nuclear long coiled-coil proteins are summarized in Figure 5. The proteins with the highest predicted coiled-coil coverage are functionally uncharacterized so far. Three of the four Arabidopsis homologs of the carrot nuclear matrix protein NMCP1 (Masuda et al., 1997) are predicted as nuclear proteins. Other published proteins in the nuclear-predicted fraction include the putative transcription factor bHLH131 (Heim et al., 2003) and the condensin SMC4 protein (Liu et al., 2002).
Figure 5.
Putative nuclear proteins with high coiled-coil coverage. Proteins are of at least 500 amino acids in length and at least 25% coiled-coil coverage, sorted from top to bottom by decreasing percentage of coiled-coil coverage. Bar diagrams, Coiled-coil domain structure as represented in the ARABI-COIL database; gray boxes, coiled-coil domains; black boxes, transmembrane domains according to ARAMEMNON. Proteins belonging to gene families are boxed together. a, Proteins are characterized by published data (see Table V for comparison).
Putative Organellar Long Coiled-Coil Proteins in Arabidopsis
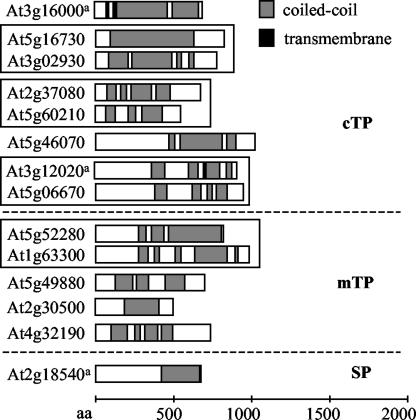
Searching ARABI-COIL for proteins with N-terminal targeting signals such as mitochondrial or plastid targeting or secretory signal peptides, 52 proteins matching the criteria used for Table V and Figure 3 were identified. Twenty-seven were predicted by at least one method to target to the chloroplasts, 23 to the mitochondria, and two to the secretory pathway. Disregarding proteins with cross-program average scores below the cutoff or strong ambiguous predictions (“unclear” in Fig. 4), the remaining proteins with clear targeting predictions are summarized in Figure 6. Of the eight proteins predicted to target to plastids, only the localization of AtMFP1 has been characterized experimentally (Jeong et al., 2003). None of the five proteins predicted as mitochondrial has been characterized. The only protein with a clear prediction to follow the secretory pathway shows significant similarity to the pumpkin (Cucurbita maxima) protein preproMP73, a protein targeted to storage vacuoles (Mitsuhashi et al., 2001).
Figure 6.
Proteins with high coiled-coil coverage and putative N-terminal targeting signals. Proteins are of at least 500 amino acids in length and at least 25% coiled-coil coverage, sorted from top to bottom by decreasing percentage of coiled-coil coverage. Bar diagrams, Coiled-coil domain structure as represented in the ARABI-COIL database; gray boxes, coiled-coil domains; black boxes, transmembrane domains according to ARAMEMNON. Proteins belonging to gene families are boxed together. a, Proteins are characterized by published data (see Table V for comparison). cTP, Chloroplast targeting peptide; mTP, mitochondrial targeting peptide; SP, signal peptide for secretory pathway.
Putative Cytoplasmic Long Coiled-Coil Proteins in Arabidopsis
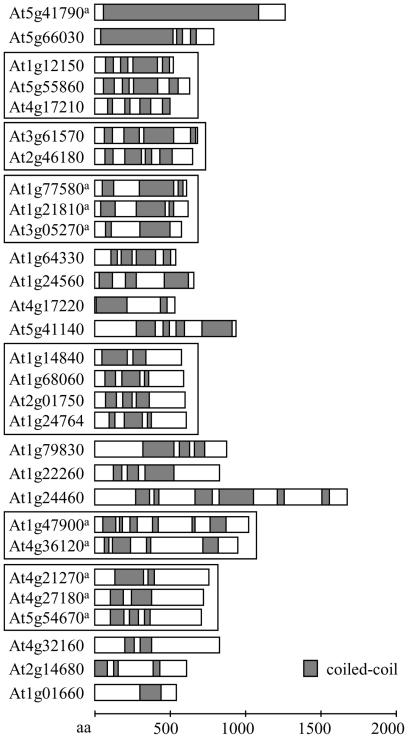
Of the proteins longer than 500 amino acids with at least 25% coiled-coil coverage, 29 fall into the group defined as cytoplasmic in Figure 4. These proteins are summarized in Figure 7. The cytoskeletal protein CIP1 (Matsui et al., 1995), having the longest continuous coiled-coil domain in Arabidopsis predicted by MultiCoil in our screen, falls into this group. Other proteins include members of the family of FPPs (Gindullis et al., 2002) and the kinesin family of KatA, KatB, and KatC (Mitsui et al., 1994, 1996; Marcus et al., 2002, 2003).
Figure 7.
Putative cytosolic proteins with high coiled-coil coverage. Proteins are of at least 500 amino acids in length and at least 25% coiled-coil coverage, sorted from top to bottom by decreasing percentage of coiled-coil coverage. Bar diagrams, Coiled-coil domain structure as represented in the ARABI-COIL database; gray boxes, coiled-coil domains. Proteins belonging to gene families are boxed together. a, Proteins are characterized by published data (see Table V for comparison).
DISCUSSION
Increasing experimental evidence demonstrates the importance of long coiled-coil proteins for the spatial organization of cellular processes. Although several protein classes with long coiled-coil domains have been studied in animals and yeast, our knowledge about plant long coiled-coil proteins is very limited. The repeat nature of the coiled-coil sequence motif makes it almost impossible to identify homologs of animal coiled-coil proteins without highly conserved non-coiled-coil domains. As a consequence, counterparts of many animal proteins with long coiled-coil domains, like lamins, golgins, or microtubule organization center components, have not been identified yet in plants. The ARABI-COIL database was created to provide the research community with a tool to sort and browse Arabidopsis long coiled-coil proteins to facilitate the identification and selection of candidate proteins of potential interest for specific research areas.
Coiled-Coil Prediction and Selection Criteria
To predict coiled-coil structures based on amino acid sequence, several programs with differing performance rates are available. COILS and NEWCOILS (Lupas et al., 1991), based on Parry's algorithm (Parry, 1982), have become the standard for coiled-coil prediction and are used widely in published literature. However, COILS generates a high number of false positives by predicting non-coiled-coil alpha-helical regions as coiled-coil structures (Berger et al., 1995; Lupas, 1997). In tests on the PDB database of solved protein structures, two-thirds of the sequences predicted by COILS did not contain coiled-coils (Berger and Singh, 1997). Thus, this program would generate a high number of false hits if used for a genome-wide screen. The PAIRCOIL program takes pair-wise residue correlations within the heptad repeat into account and performs significantly better than COILS in avoiding false positives. However, PAIRCOIL often fails to predict antiparallel or multistranded coiled-coils (Lupas, 1997). MultiCoil, based on data of two-stranded and three-stranded coiled-coils, is capable of predicting both types of structures while achieving a similar low rate of false predictions as PAIRCOIL (Wolf et al., 1997). Therefore, MultiCoil was applied as the program of choice to define coiled-coil proteins from the Arabidopsis genome for this analysis. A probability cutoff of 0.5 was used, which is the default suggested by the program developers. Because MultiCoil is already more stringent than the older programs, using this moderate cutoff leads to a prediction of coiled-coil structures that are more comparable with those often found in the literature (see Fig. 1).
In a genome-wide screen using the MultiCoil program, 5.6% of all Arabidopsis sequences (about 1,500) were identified as coiled-coil proteins. This number is lower than those found for other eukaryotic genomes (about 10%; Liu and Rost, 2001). However, the older studies did not describe using a cutoff in length. Because MultiCoil has no internal length cutoff and the formation of coiled-coil structures requires a minimum number of residues, we believe the setting of a minimal domain size more significant than a high per-residue probability cutoff. Studies using synthetic peptides showed that a minimum length of three to four heptads or six to eight helical turns is required for peptides to form stable coiled-coils (Lumb et al., 1994; Su et al., 1994; Litowski and Hodges, 2001). The cutoff of 20 amino acids minimal length for a coiled-coil domain used in our primary screen allows for the formation of about six helical turns in the secondary structure of the protein.
The goal of the ARABI-COIL database creation was to provide a searchable selection of proteins with high coiled-coil coverage and long coiled-coil domains putatively involved in structural functions. Many long coiled-coil domains, for example that of AtMFP1 (Fig. 1), contain small gaps and disruptions in the overall coiled-coil structure predicted by MultiCoil. To identify the complete length of the long but discontinuous coiled-coil domains of such proteins, a feature was included to ignore small gaps of less than 25 amino acids between predicted coiled-coil structures, thus fusing the predictions for these domains to a single larger coiled-coil as exemplified in Figure 1D. Subsequently, a subset of proteins containing long coiled-coil regions was selected while trying to exclude shorter coiled-coil motifs such as Leucine zippers. The criteria chosen succeeded in excluding 97% of the known Arabidopsis bZIPs (Jakoby et al., 2002), thus providing a stringent selection against the inclusion of Leu-zipper-containing proteins. The bZIP factors included in ARABI-COIL, such as ATB2, contain unusually long coiled-coil domains for this protein family (Rook et al., 1998). However, MultiCoil prediction data for shorter domains are available and integrated into the ARABI-COIL database environment. Future enhancements of the database could include making data for the currently excluded short coiled-coil proteins available to users by offering a choice of additional selection parameter combinations that incorporate proteins with shorter domains.
ARABI-COIL Search Functions and Prediction Data Interpretation
The search features provided to browse the database allow users to select for proteins of a certain coiled-coil length and coverage. By providing coiled-coil percentages predicted for the N-terminal, middle, and C-terminal domains of the protein, the database allows for a crude search for coiled-coil domain configurations. This facilitates the identification of proteins with similar coiled-coil domain structures without detectable sequence homology.
The incorporation of transmembrane and targeting signal prediction data allows the user to specify searches for putative chloroplast, mitochondria, secretory pathway, nuclear, and transmembrane proteins. This helps to identify subsets of coiled-coil proteins predicted to localize to a certain cell compartment that are of enhanced interest for further functional studies.
However, the comparison of localization prediction results from different programs and with available experimental data shows that computationally retrieved targeting predictions are ambiguous (Table VII; also see Emanuelsson and von Heijne, 2001; Schwacke et al., 2003). ARABI-COIL searches return results if at least one of the incorporated predictions scores above the cutoff of 0.5, with the goal to provide the user with the largest group possible from which to select candidates for further analysis. These prediction results need to be evaluated critically on a case-by-case basis, which is aided by color-coding of low-probability predictions (0.5-0.74, yellow) and high-probability predictions (0.75-1) on the results display. In general, the reliability of computational targeting predictions is lower for plant sequences than for non-plant sequences and varies from about 85% overall correct predictions by TargetP to about 70% for PSORT (Emanuelsson et al., 2000). Predict-NLS works on the basis of a database of known NLS motifs and their variations and was found to correctly predict 43% of known nuclear proteins (Cokol et al., 2000), whereas PSORT searches for consensus patterns, thus potentially creating higher numbers including false-positive NLS predictions. Predotar frequently generates false positives by predicting proteins with signal peptides as putative mitochondrial or chloroplast proteins. In some cases, this might reflect a true dual targeting to the ER and organelle as has been observed for cytochrome b5 (Zhao et al., 2003). MitoProt and ChloroP are less efficient than Predotar in distinguishing between mitochondrial and plastid targeting sequences and occasionally predict high scores for both types of organellar targeting sequences, as can be seen in the high MitoProt score for the example of the chloroplast protein MFP1 in Table VII. Such predictions could also reflect true dual targeting to both organelles. Yeast mitochondrial targeting sequences have been shown to target proteins to both organelles in plants (Huang et al., 1990), and isolated plant mitochondria are capable of importing a range of chloroplast protein precursors (Cleary et al., 2002). Dual targeting is being observed for an increasing number of plant proteins (Peeters and Small, 2001; Rudhe et al., 2002; Goggin et al., 2003), thus making computational predictions difficult. Each ARABI-COIL details page provides a normalized list of prediction scores that allows the user to compare and evaluate the results from a number of prediction programs without having to submit the sequence to the individual prediction servers. However, experimental data will have to prove whether the predicted targeting actually occurs in the cell.
Future Directions of the ARABI-COIL Database
Future enhancements of the ARABI-COIL database and Web site will include the incorporation of additional prediction data and adding the capability of BLAST searches against the sequences populating the database. As more fully annotated plant genomes become available, the ARABI-COIL database will serve as a template for the addition of other genomes, enabling comparative analyses between different plant species. Flexibility and expandability were fundamental criteria for the underlying MySQL database and schema. The ability to add results from additional programs and sources is key to the successful viability of the database over the long term. Essentially, ARABI-COIL is a warehouse of annotated and computed information, with relatively few update transactions relative to the number of queries. For increased availability to the scientific community, the ARABI-COIL data will be made accessible through existing data mining and distribution tools, such as for, example, The Arabidopsis Information Resource (Rhee et al., 2003) and MOBY Central (Wilkinson and Links, 2002).
Arabidopsis Coiled-Coil Proteins Identified Using ARABI-COIL
The ARABI-COIL database was used to select groups of candidate proteins of at least 500 amino acids in length and more than 25% coiled-coil coverage in combination with other features that could be of potential interest for future research. The length cutoff for this analysis was chosen based on the lengths of animal and yeast coiled-coil proteins with known structural functions in the cell that range from about 600 amino acids (for example, lamin A/C, golgin-67) to more than 3,000 (for example, giantin).
Several long coiled-coil proteins of unknown function with transmembrane domains at the C terminus were identified (Fig. 3). This domain structure is characteristic of a subgroup of animal golgins including golgin-84, golgin-67, giantin, and CASP (Bascom et al., 1999; Jakymiw et al., 2000; Misumi et al., 2001; Gillingham et al., 2002). Three of the identified Arabidopsis proteins contain similarity to golgins: At3g18480 to CASP and At1g18190 and At2g19950 to golgin-84 (Gillingham et al., 2002). Thus, the identified Arabidopsis proteins are promising candidates for plant integral membrane golgins or proteins with endosomal functions. No plant golgins have been characterized in the literature so far.
Another group of potentially interesting proteins is comprised of nuclear long coiled-coil proteins of unknown function (Fig. 5). In animal cells, intermediate filament proteins such as the lamins and NuMA play an important role in the structural organization of the nuclear matrix and the lamina underlying the inner surface of the nuclear envelope. Early immunocyto-logical evidence pointed at the possible existence of similar proteins in plant cells (McNulty and Saunders, 1992; Mínguez and Moreno Díaz de la Espina, 1993; Yu and Moreno Díaz de la Espina, 1999). However, with the exception of NMCP1 from carrot (Masuda et al., 1997), no lamin- or NuMA-like protein sequences have been identified from plants so far. Several candidates for nuclear intermediate filament proteins with high coiled-coil coverage could be identified using ARABI-COIL. These include homologs of the carrot protein NMCP1 (Masuda et al., 1997) and three proteins of similar length to lamin A/C (about 650 amino acids). Future experiments will have to reveal whether these proteins localize to the nuclear envelope in plant cells and whether they are involved in forming the elusive plant nuclear lamina.
MATERIALS AND METHODS
Sequence Sources
The Arabidopsis proteome sequence set (all nonredundant SWISS-PROT and TrEMBL entries) was downloaded from the European Bioinformatics Institute proteome analysis database (http://www.ebi.ac.uk/proteome/ARATH/). The initial set of 26,945 sequences at the time of download (June 2002) was updated to reflect the NCBI RefSeq database (http://www.ncbi.nlm.nih.gov/RefSeq/) sequences.
Coiled-Coil Domain Prediction and Data Generation
The MultiCoil program version suitable to run on Silicon Graphics systems was downloaded from http://theory.lcs.mit.edu/multicoil and installed on a 32-processor SGI Origin 2000 system. Sequence files in FASTA format were transferred to the SGI system and processed through the locally installed MultiCoil program using the default settings of the program (cutoff score of 0.5, window size 28). A Java-based program suite, ExtractProp, was developed to post-process and extract relevant computed properties from the aggregate computed MultiCoil program output. (The ExtractProp Suite continues to be enhanced and is available upon request.) Gaps of less than 25 amino acids between predicted coiled-coil domains were ignored and the domains fused. The minimum domain length was defined as 20 amino acids, and predicted coiled-coils shorter than 20 residues were disregarded. Proteins having domain numbers and maximum domain length values of at least one of 70, two of 50, or three of 30 were selected to populate the ARABI-COIL database. The sequences for these selected proteins were extracted and summarized in FASTA format for further analyses. XML was selected as the medium for representing the extracted data. Coiled-coil information for inclusion in the database was extracted from this output, such as lengths and positions of coiled-coil regions, and percentages of amino acids were predicted to form a coiled-coil for the complete sequences and the N-terminal, middle, and C-terminal thirds of the sequence.
Computational Sequence Analysis
Sequences were analyzed using a battery of structural and subcellular targeting signal prediction programs (see Table VI). Predotar, MitoProt, and HMMTOP were installed and integrated into the existing basic bioinformatics research environment. A Sun Grid Engine Portal was used to provide Web-based submission of the analysis tasks for these programs with the ExtractProp suite employed to recover the desired properties from the computed output. The remaining programs were applied through their respective Web sites, and the data were compiled into delimited text tables and subsequently processed by the ExtractProp suite for conversion to XML and incorporation in the underlying MySQL database. Hits in the Predict-NLS database were given a score of 1, and no hits were counted as 0. ChloroP scores (0.4-0.6 range in raw output) were normalized to a 0 to 1 scale to match the range for the remaining prediction scores.
Database and Web Site Development
MySQL was selected as a database engine to support the Web site. For maximum flexibility and expandability, a denormalized table definition was adopted. The computed output previously translated to XML was converted subsequently to SQL and used to populate the MySQL database. The population of the database is staged, enabling updates, additions, deletions, and minor edits to be done with a high level of automation. The database and its Web interface are hosted on servers maintained by the Ohio Supercomputer Center.
Acknowledgments
We thank the Ohio Supercomputer Center for providing computer usage time for this analysis and Heather Wang and Tszyeung Ching for collection of PSORT data for input in the database.
This work was supported by the National Science Foundation 2010 Project (grant no. NSF 0209339 to I.M.).
References
- Barr FA, Short B (2003) Golgins in the structure and dynamics of the golgi apparatus. Curr Opin Cell Biol 15: 405-413 [DOI] [PubMed] [Google Scholar]
- Bascom RA, Srinivasan S, Nussbaum RL (1999) Identification and characterization of golgin-84, a novel golgi integral membrane protein with a cytoplasmic coiled-coil domain. J Biol Chem 274: 2953-2962 [DOI] [PubMed] [Google Scholar]
- Berger B, Singh M (1997) An iterative method for improved protein structural motif recognition. J Comput Biol 4: 261-273 [DOI] [PubMed] [Google Scholar]
- Berger B, Wilson DB, Wolf E, Tonchev T, Milla M, Kim PS (1995) Predicting coiled coils by use of pairwise residue correlations. Proc Natl Acad Sci USA 92: 8259-8263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhard P, Stetefeld J, Strelkov SV (2001) Coiled coils: a highly versatile protein folding motif. Trends Cell Biol 11: 82-88 [DOI] [PubMed] [Google Scholar]
- Chen XP, Yin H, Huffaker TC (1998) The yeast spindle pole body component Spc72p interacts with Stu1p and is required for proper microtubule assembly. J Cell Biol 141: 1169-1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claros MG, Vincens P (1996) Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem 241: 779-786 [DOI] [PubMed] [Google Scholar]
- Cleary SP, Tan FC, Nakrieko KA, Thompson SJ, Mullineaux PM, Creissen GP, von Stedingk E, Glaser E, Smith AG, Robinson C (2002) Isolated plant mitochondria import chloroplast precursor proteins in vitro with the same efficiency as chloroplasts. J Biol Chem 277: 5562-5569 [DOI] [PubMed] [Google Scholar]
- Cokol M, Nair R, Rost B (2000) Finding nuclear localization signals. EMBO Rep 1: 411-415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton DA, Szilak I, Cleveland DW (1992) Primary structure of NuMA, an intranuclear protein that defines a novel pathway for segregation of proteins at mitosis. J Cell Biol 116: 1395-1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick FH (1952) Is alpha-keratin a coiled coil? Nature 170: 882-883 [DOI] [PubMed] [Google Scholar]
- Diviani D, Scott JD (2001) AKAP signaling complexes at the cytoskeleton. J Cell Sci 114: 1431-1437 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005-1016 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8: 978-984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, von Heijne G (2001) Prediction of organellar targeting signals. Biochim Biophys Acta 1541: 114-119 [DOI] [PubMed] [Google Scholar]
- Esteban MR, Giovinazzo G, de la Hera A, Goday C (1998) PUMA1: a novel protein that associates with the centrosomes, spindle and centromeres in the nematode Parascaris. J Cell Sci 111: 723-735 [DOI] [PubMed] [Google Scholar]
- Fava F, Raynaud-Messina B, Leung-Tack J, Mazzolini L, Li M, Guillemot JC, Cachot D, Tollon Y, Ferrara P, Wright M (1999) Human 76p: a new member of the γ-tubulin-associated protein family. J Cell Biol 147: 857-868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flory MR, Moser MJ, Monnat RJ Jr, Davis TN (2000) Identification of a human centrosomal calmodulin-binding protein that shares homology with pericentrin. Proc Natl Acad Sci USA 97: 5919-5923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T, Mikami Y, Nishihashi A, Regnier V, Haraguchi T, Hiraoka Y, Sugata N, Todokoro K, Brown W, Ikemura T (2001) CENP-H, a constitutive centromere component, is required for centromere targeting of CENP-C in vertebrate cells. EMBO J 20: 4603-4617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingham AK, Pfeifer AC, Munro S (2002) CASP, the alternatively spliced product of the gene encoding the CCAAT-displacement protein transcription factor, is a golgi membrane protein related to giantin. Mol Biol Cell 13: 3761-3774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gindullis F, Rose A, Patel S, Meier I (2002) Four signature motifs define the first class of structurally related large coiled-coil proteins in plants. BMC Genomics 3: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goggin DE, Lipscombe R, Fedorova E, Millar AH, Mann A, Atkins CA, Smith PMC (2003) Dual intracelluar localization and targeting of aminoimidazole ribonucleotide synthetase in cowpea. Plant Physiol 131: 1033-1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman RD, Gruenbaum Y, Moir RD, Shumaker DK, Spann TP (2002) Nuclear lamins: building blocks of nuclear architecture. Genes Dev 16: 533-547 [DOI] [PubMed] [Google Scholar]
- Hanin M, Mengiste T, Bogucki A, Paszkowski J (2000) Elevated levels of intrachromosomal homologous recombination in Arabidopsis overexpressing the MIM gene. Plant J 24: 183-189 [DOI] [PubMed] [Google Scholar]
- Harder P, Silverstein R, Meier I (2000) Conservation of matrix attachment region-binding filament-like protein 1 among higher plants. Plant Physiol 122: 225-234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim MA, Jacoby M, Werber M, Martin C, Weisshaar B, Bailey PC (2003) The basic helix-loop-helix transcription factor family in plants: a genome wide study of protein structure and functional diversity. Mol Biol Evol 20: 735-747 [DOI] [PubMed] [Google Scholar]
- Hirano T (2002) The ABCs of SMC proteins: two-armed ATPases for chromosome condensation, cohesion, and repair. Genes Dev 16: 399-414 [DOI] [PubMed] [Google Scholar]
- Holaska JM, Wilson KL, Mansharamani M (2002) The nuclear envelope, lamins and nuclear assembly. Curr Opin Cell Biol 14: 257-364 [DOI] [PubMed] [Google Scholar]
- Huang J, Hack E, Thornburg RW, Meyers AM (1990) A yeast mitochondrial leader peptide functions in vivo as a dual targeting signal for both chloroplast and mitochondria. Plant Cell 2: 1249-1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Droge-Laser W, Vincente-Carbajosa J, Tiedemann J, Kroj T, Parcy F (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7: 106-111 [DOI] [PubMed] [Google Scholar]
- Jakymiw A, Raharjo E, Rattner JB, Eystathioy T, Chan EKL, Fujita DJ (2000) Identification and characterization of a novel golgi protein, golgin-67. J Biol Chem 275: 4137-4144 [DOI] [PubMed] [Google Scholar]
- Jeong SY, Rose A, Meier I (2003) MFP1 is a thylakoid-targeted, nucleoid-binding protein with a coiled-coil structure. Nucleic Acids Res 31: 5175-5185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessberger R (2002) The many functions of SMC proteins in chromosome dynamics. Nat Rev Mol Cell Biol 3: 767-778 [DOI] [PubMed] [Google Scholar]
- Kilmartin JV, Dyos SL, Kershaw D, Finch JT (1993) A spacer protein in the Saccharomyces cerevisiae spindle pole body whose transcript is cell cycle-regulated. J Cell Biol 123: 1175-1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer ELL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305: 567-580 [DOI] [PubMed] [Google Scholar]
- Le Masson I, Saveanu C, Chevalier A, Namane A, Gobin R, Fromont-Racine M, Jacquier A, Mann C (2002) Spc24 interacts with Mps2 and is required for chromosome segregation, but is not implicated in spindle pole body duplication. Mol Microbiol 43: 1431-1443 [DOI] [PubMed] [Google Scholar]
- Li Q, Hansen D, Killilea A, Joshi HC, Palazzo RE, Balczon R (2000) Kendrin/pericentrin-B, a centrosome protein with homology to pericentrin that complexes with PCM-1. J Cell Sci 114: 797-809 [DOI] [PubMed] [Google Scholar]
- Liao H, Winkfein RJ, Mack G, Rattner JB, Yen TJ (1995) CENP-F is a protein of the nuclear matrix that assembles onto kinetochores at late G2 and is rapidly degraded after mitosis. J Cell Biol 130: 507-518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litowski JR, Hodges RS (2001) Designing heterodimeric two-stranded alpha-helical coiled-coils: the effect of chain length on protein folding, stability and specificity. J Pept Res 58: 477-492 [DOI] [PubMed] [Google Scholar]
- Liu CM, McElver J, Tzafrir R, Joosen R, Wittich P, Patton D, van Lammeren AA, Meinke D (2002) Condensin and cohesin knockouts in Arabidopsis exhibit a titan seed phenotype. Plant J 29: 405-415 [DOI] [PubMed] [Google Scholar]
- Liu J, Rost B (2001) Comparing function and structure between entire genomes. Protein Sci 10: 1970-1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorson MA, Horvitz HR, von den Heuvel S (2000) LIN-5 is a novel component of the spindle apparatus required for chromosome segregation and cleavage plane specification in Caenorhabditis elegans. J Cell Biol 148: 73-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumb KJ, Carr CM, Kim PS (1994) Subdomain folding of the coiled coil leucine zipper from the bZIP transcriptional activator GCN4. Biochemistry 33: 7361-7367 [DOI] [PubMed] [Google Scholar]
- Lupas A (1997) Predicting coiled-coil regions in proteins. Curr Opin Struct Biol 7: 388-393 [DOI] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J (1991) Predicting coiled coils from protein sequences. Science 252: 1162-1164 [DOI] [PubMed] [Google Scholar]
- Marcus AI, Ambrose JC, Blickley L, Hancock WO, Cyr RJ (2002) Arabidopsis thaliana protein, ATK1, is a minus-end directed kinesin that exhibits non-processive movement. Cell Motil Cytoskelet 52: 144-150 [DOI] [PubMed] [Google Scholar]
- Marcus AI, Li W, Ma H, Cyr RJ (2003) A kinesin mutant with an atypical bipolar spindle undergoes normal mitosis. Mol Biol Cell 14: 1717-1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda K, Xu ZJ, Takahashi S, Ito A, Ono M, Nomura K, Inoue M (1997) Peripheral framework of carrot cell nucleus contains a novel protein predicted to exhibit a long alpha-helical domain. Exp Cell Res 232: 173-181 [DOI] [PubMed] [Google Scholar]
- Matsui M, Stoop CD, von Arnim AG, Wei N, Deng XW (1995) Arabidopsis COP1 protein specifically interacts in vitro with a cytoskeleton-associated protein, CIP1. Proc Natl Acad Sci USA 92: 4239-4243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNulty AK, Saunders MJ (1992) Purification and immunological detection of pea nuclear intermediate filaments: evidence for plant nuclear lamins. J Cell Sci 103: 407-414 [DOI] [PubMed] [Google Scholar]
- Mengiste T, Revenkova E, Bechtold N, Paszkowski J (1999) An SMC-like protein is required for efficient homologous recombination in Arabidopsis. EMBO J 18: 4505-4512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mínguez A, Moreno Díaz de la Espina (1993) Immunological characterization of lamins in the nuclear matrix of onion cells. J Cell Sci 106: 431-439 [DOI] [PubMed] [Google Scholar]
- Misumi Y, Sohda M, Tashiro A, Sato H, Ikehara Y (2001) An essential cytoplasmic domain for the golgi localization of coiled-coil proteins with a COOH-terminal membrane anchor. J Biol Chem 276: 6867-6873 [DOI] [PubMed] [Google Scholar]
- Mitsuhashi N, Hayashi Y, Koumoto Y, Shimada T, Fukasawa-Akada T, Nishimura M, Hara-Nishimura I (2001) A novel membrane protein that is transported to protein storage vacuoles via precursor-accumulating vesicles. Plant Cell 13: 2361-2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui H, Hasezawa S, Nagata T, Takahashi H (1996) Cell cycle-dependent accumulation of a kinesin-like protein, KatB/C in synchronized tobacco BY-2 cells. Plant Mol Biol 30: 177-181 [DOI] [PubMed] [Google Scholar]
- Mitsui H, Nakatani K, Yamaguchi-Shinozaki K, Shinozaki K, Nishikawa K, Takahashi H (1994) Sequencing and characterization of the kinesin-related genes katB and katC of Arabidopsis thaliana. Plant Mol Biol 25: 865-876 [DOI] [PubMed] [Google Scholar]
- Moisoi N, Erent M, Whyte S, Martin S, Bayley PM (2002) Calmodulin-containing substructures of the centrosomal matrix released by microtubule perturbation. J Cell Sci 115: 2367-2379 [DOI] [PubMed] [Google Scholar]
- Nakai K, Horton P (1999) PSORT: a program for detecting the sorting signals of proteins and predicting their subcellular localization. Trends Biochem Sci 24: 34-35 [DOI] [PubMed] [Google Scholar]
- Newman JRS, Wolf E, Kim PS (2000) A computationally directed screen identifying interacting coiled coils from Saccharomyces cerevisiae. Proc Natl Acad Sci USA 97: 13203-13208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G (1997a) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng 10: 1-6 [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G (1997b) A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int J Neural Syst 8: 581-599 [DOI] [PubMed] [Google Scholar]
- Nielsen H, Krogh A (1998) Prediction of signal peptides and signal anchors by a hidden Markov model. Proc Int Conf Intell Syst Mol Biol 6: 122-130 [PubMed] [Google Scholar]
- Parry DA (1982) Coiled-coils in α-helix-containing proteins: analysis of residue types within the heptad repeat and the use of these data in the prediction of coiled-coils in other proteins. Biosci Rep 2: 1017-1024 [DOI] [PubMed] [Google Scholar]
- Peeters N, Small I (2001) Dual targeting to mitochondria and plastids. Biochim Biophys Acta 1541: 54-63 [DOI] [PubMed] [Google Scholar]
- Reddy A, Day I (2001a) Analysis of the myosins encoded in the recently completed Arabidopsis thaliana genome sequence. Genome Biol 2: 0024.1-0024.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy A, Day I (2001b) Kinesins in the Arabidopsis genome: a comparative analysis among eukaryotes. BMC Genomics 2: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee SY, Beavis W, Berardini TZ, Chen G, Dixon D, Doyle A, Garcia-Hernandez M, Huala E, Lander G, Montoya M et al. (2003) The Arabidopsis information resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res 31: 224-228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook F, Weisbeek P, Smeekens S (1998) The light-regulated Arabidopsis bZIP transcription factor gene ATB2 encodes a protein with an unusually long leucine zipper domain. Plant Mol Biol 37: 171-178 [DOI] [PubMed] [Google Scholar]
- Rudhe C, Chew O, Whelan J, Glaser E (2002) A novel in vitro system for simultaneous import of precursor proteins into mitochondria and chloroplasts. Plant J 30: 213-220 [DOI] [PubMed] [Google Scholar]
- Rupp G, Porter ME (2003) A subunit of the dynein regulatory complex in Chlamydomonas is a homologue of a growth arrest-specific gene product. J Cell Biol 162: 47-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaerer F, Morgan G, Winey M, Philippsen P (2001) Cnm67p is a spacer protein of the Saccaromyces cerevisiae spindle pole body outer plaque. Mol Biol Cell 12: 2519-2533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliwa M, Woehlke G (2003) Molecular motors. Nature 422: 759-765 [DOI] [PubMed] [Google Scholar]
- Schramm C, Elliot S, Shevchenko A, Shevchenko A, Schiebel E (2000) The Bbp1p-Mps2p complex connects the SPB to the nuclear envelope and is essential for SPB duplication. EMBO J 19: 421-433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacke R, Schneider A, van der Graff E, Fischer K, Catoni E, Desimone M, Frommer WB, Flügge UI, Kunze R (2003) ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol 131: 16-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann J, Jokitalo E, Pypaert M, Warren G (2000) Matrix proteins can generate the higher order architecture of the Golgi apparatus. Nature 407: 1022-1026 [DOI] [PubMed] [Google Scholar]
- Seemann J, Pyaert M, Taguchi T, Malsam J, Warren G (2002) Partitioning of the matrix fraction of the golgi apparatus during mitosis in animal cells. Science 295: 848-851 [DOI] [PubMed] [Google Scholar]
- Sillibourne JE, Milne DM, Takahashi M, Ono Y, Meek DW (2002) Centrosomal anchoring of the protein kinase CK1δ mediated by attachment to the large, coiled-coil scaffolding protein CG-NAP/AKAP450. J Mol Biol 322: 785-797 [DOI] [PubMed] [Google Scholar]
- Sisson JC, Field C, Ventura R, Royou A, Sullivan W (2000) Lava Lamp, a novel peripheral golgi protein, is required for Drosophila melanogaster cellularization. J Cell Biol 151: 905-917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LG (2002) Plant cytokinesis: motoring to the finish. Curr Biol 12: R206-R209 [DOI] [PubMed] [Google Scholar]
- Sonnhammer EL, von Heijne G, Krogh A (1998) A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol 6: 175-182 [PubMed] [Google Scholar]
- Souès S, Adams IR (1998) SPC72: a spindle pole component required for spindle orientation in the yeast Saccharomyces cerevisiae. J Cell Sci 111: 2809-2818 [DOI] [PubMed] [Google Scholar]
- Starr DA, Saffery R, Li Z, Simpson AE, Choo KHA, Yen TJ, Goldberg ML (2000) Hzwint-1, a novel human kinetochore component that interacts with HZW10. J Cell Sci 113: 1939-1950 [DOI] [PubMed] [Google Scholar]
- Strelkov SV, Herrmann H, Aebi U (2003) Molecular architecture of intermediate filaments. Bioessays 25: 243-251 [DOI] [PubMed] [Google Scholar]
- Su JY, Hodges RS, Kay CM (1994) Effect of chain length on the formation and stability of synthetic alpha-helical coiled coils. Biochemistry 33: 15501-15510 [DOI] [PubMed] [Google Scholar]
- Sugata N, Li S, Earnshaw WC, Yen TJ, Yoda K, Masumoto H, Munekata E, Warburton PE, Todokoro K (2000) Human CENP-H multimers colocalize with CENP-A and CENP-C at active centromere-kinetochore complexes. Human Mol Genet 9: 2919-2926 [DOI] [PubMed] [Google Scholar]
- Sugata N, Munekata E, Todokoro K (1999) Characterization of a novel kinetochore protein, CENP-H. J Biol Chem 274: 27343-27346 [DOI] [PubMed] [Google Scholar]
- Takahashi M, Mukai H, Oishi K, Isagawa T, Ono Y (2000) Association of immature hypophosphorylated protein kinase Cε with an anchoring protein CG-NAP. J Biol Chem 275: 34592-34596 [DOI] [PubMed] [Google Scholar]
- Takahashi M, Shibata H, Shimakawa M, Miyamoto M, Mukai H, Ono Y (1999) Characterization of a novel giant scaffolding protein, CG-NAP, that anchors multiple signaling enzymes to centrosome and the golgi apparatus. J Biol Chem 274: 17267-17274 [DOI] [PubMed] [Google Scholar]
- Takahashi M, Yamagiwa A, Tamako N, Mukai H, Ono Y (2002) Centrosomal proteins CG-NAP and kendrin provide microtubule nucleation sites by anchoring γ-tubulin ring complex. Mol Biol Cell 13: 3235-3245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusnády GE, Simon I (1998) Principles governing amino acid composition of integral membrane proteins: applications to topology prediction. J Mol Biol 283: 489-506 [DOI] [PubMed] [Google Scholar]
- Verde I, Pahkle G, Salanova M, Zhang G, Wang S, Coletti D, Onuffer J, Jin SLC, Conti M (2001) Myomegalin is a novel protein of the golgi/centrosome that interacts with a cyclic nucleotide phosphodiesterase. J Biol Chem 276: 11189-11198 [DOI] [PubMed] [Google Scholar]
- Vinson C, Myakishev M, Acharya A, Mir AA, Moll JR, Bonovich M (2002) Classification of human B-ZIP proteins based on dimerization properties. Mol Cell Biol 22: 6321-6335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson MD, Links M (2002) BioMOBY: an open source biological web service proposal. Brief Bioinform 3: 331-341 [DOI] [PubMed] [Google Scholar]
- Witczak O, Skålhegg BS, Keryer G, Bornens M, Tasken K, Jahnsen T, Ørstavik S (1999) Cloning and characterization of a cDNA encoding an A-kinase anchoring protein located in the centrosome, AKAP450. EMBO J 18: 1858-1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf E, Kim PS, Berger B (1997) MultiCoil: A program for predicting two- and three-stranded coiled coils. Protein Sci 6: 1179-1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CH, Lambie EJ, Snyder M (1992) NuMA: an unusually long coiledcoil related protein in the mammalian nucleus. J Cell Biol 116: 1303-1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Moreno Díaz de la Espina S (1999) The plant nucleoskeleton: ultrastructural organization and identification of NuMA homologues in the nuclear matrix and mitotic spindle of plant cells. Exp Cell Res 246: 516-526 [DOI] [PubMed] [Google Scholar]
- Zhao J, Onduka T, Kinoshita J, Honsho M, Kinoshita T, Shimazaki K, Ito A (2003) Dual subcellular distribution of cytochrome b5 in plant, cauliflower, cells. J Biochem 133: 115-121 [DOI] [PubMed] [Google Scholar]