Abstract
IscR is a global transcription regulator responsible for governing various physiological processes during growth and stress responses. The IscR-mediated regulation of the Pseudomonas aeruginosa isc operon, which is involved in iron-sulphur cluster ([Fe-S]) biogenesis, was analysed. The expression of iscR was highly induced through the exposure of the bacteria to various oxidants, such as peroxides, redox-cycling drugs, intracellular iron-chelating agents, and high salts. Two putative type 1 IscR-binding sites were found around RNA polymerase recognition sites, in which IscR-promoter binding could preclude RNA polymerase from binding to the promoter and resulting in repression of the isc operon expression. An analysis of the phenotypes of mutants and cells with altered gene expression revealed the diverse physiological roles of this regulator. High-level IscR strongly inhibited anaerobic, but not aerobic, growth. iscR contributes significantly to the bacteria overall resistance to oxidative stress, as demonstrated through mutants with increased sensitivity to oxidants, such as peroxides and redox-cycling drugs. Moreover, the regulator also plays important roles in modulating intracellular iron homeostasis, potentially through sensing the levels of [Fe-S]. The increased expression of the isc operon in the mutant not only diverts iron away from the available pool but also reduces the total intracellular iron content, affecting many iron metabolism pathways leading to alterations in siderophores and haem levels. The diverse expression patterns and phenotypic changes of the mutant support the role of P. aeruginosa IscR as a global transcriptional regulator that senses [Fe-S] and directly represses or activates the transcription of genes affecting many physiological pathways.
Introduction
Iron-sulphur clusters ([Fe-S]) are important components of many enzymes involved in diverse cellular processes, such as DNA repair, gene regulation, RNA modifications, biosynthetic pathways, aerobic and anaerobic respirations, and nitrogen and carbon metabolism [1]. Approximately 5% of the total proteins in Escherichia coli are [Fe-S]-containing proteins [1]. Three different types of bacterial [Fe-S] biosynthetic systems, Isc (iron-sulphur cluster), Suf (sulphurformation) and Nif (nitrogen fixation), have been characterised [1], [2]. The Isc and Suf systems are involved in the maturation of most [Fe-S]-containing proteins in the cell, while Nif contributes to the maturation of nitrogenase in certain nitrogen-fixing bacteria [3], [4]. In many bacteria, the Isc machinery is encoded by the conserved iscRSUA-hscBA-fdx (iscR, iron-sulphur cluster assembly transcription factor; iscS, cysteine desulphurase; iscU, [Fe-S] assembly scaffold; iscA, [Fe-S] assembly protein; hscB, [Fe-S] protein assembly co-chaperone; hscA, [Fe-S] protein assembly chaperone; and fdx, ferredoxin) gene cluster in the isc operon in a housekeeping pathway for [Fe-S] biogenesis [5].
Iron is important for bacterial growth and survival. The iron concentration and availability are crucial; too much iron leads to severe oxidative stress, and too little iron severely hinders bacterial growth. Bacteria have evolved mechanisms to maintain a precise intracellular iron concentration, including the iron storage proteins (such as ferritins) and a general ferric iron buffering system (such as haem and [Fe-S] cluster) [6], [7]. IscR acts as a sensor of cellular [Fe-S] levels and a global transcription regulator for [Fe-S] biogenesis under both physiological and stressful conditions [8], [9]. In Escherichia coli, IscR exists in two major forms: apo-IscR lacks the [2Fe-2S] cluster and [2Fe-2S]-IscR has the [2Fe-2S] ligated to IscR. In addition, two IscR binding motifs, type 1 and type 2, have been characterised [10]. [2Fe-2S]-IscR binds to both types of binding motifs, whereas apo-IscR only binds to the type 2 motif [11]. In E. coli, IscR is a transcriptional repressor of the isc operon [12]. [2Fe-2S]-IscR binds to the type 1 IscR-binding motif (5′ATASYYGACTRWWWYAGTCRRSTAT3′) and represses the transcription of the isc operon [8], [13]. A proposed model was showing that the level of [2Fe-2S]-IscR is reduced during oxidative stress and iron limiting conditions, leading to derepression of the isc operon [12]. In E. coli, the post-transcriptional regulation of the isc gene expression is mediated through the Fur (ferric uptake regulator)-regulated small non-coding RNA RyhB [14]. Moreover, the expression of the suf operon (sufABCDSE gene cluster) is activated by IscR [9]. suf operon expression has been shown to be induced by stresses implying it plays a role in adaptation to stressful conditions [15].
Pseudomonas aeruginosa is a major opportunistic human pathogen that causes serious infections in hospitalised patients, particularly patients with cancer, cystic fibrosis, and burn injuries. The P. aeruginosa PAO1 genome contains a homologue of the isc operon [16] but lacks the putative suf operon [16], [17]. The iscR gene is required for H2O2 protection in P. aeruginosa PA14 [18]. The inactivation of iscR reduced KatA catalase activity and attenuated virulence in Drosophila melanogaster and mouse peritonitis models [19]. Nonetheless, the regulation of the P. aeruginosa isc operon has not been characterised. P. aeruginosa IscR has been implicated as a global regulator of many cellular responses, including oxidative stress [20]. In the present study, we showed that IscR regulated the isc operon for [Fe-S] biogenesis under both physiological and stress-induced conditions. In addition, iscR is also important in maintaining overall iron homeostasis and its inactivation leads to a phenotype that indicates iron limitation.
Results and Discussion
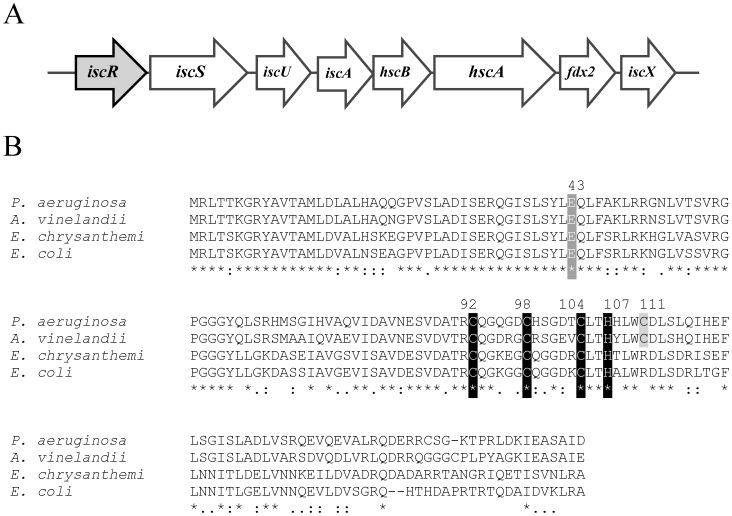
The analysis of the P. aeruginosa PAO1 genome sequence [16] revealed the homologues of the isc gene cluster iscRSUA-hscBA-fdx2-iscX (PA3815-PA3808) that could be involved in [Fe-S] biogenesis. The PAO1 isc operon gene organisation shares many similarities to that of the E. coli isc operon [8], [12]. The minor differences are the lack of rhyB binding site between iscR and iscS and additional iscX (Figure 1A). Northern blot analysis, probing with either the coding sequence of iscR or fdx2, showed a similar positively hybridised band of approximately 5 kb, suggesting that this gene cluster is transcribed as an operon (data not shown). No homologues of the suf operon were identified in the PAO1 genome. IscR regulates the isc operon in E. coli [12], [21]. P. aeruginosa IscR shares 78%, 65% and 67% identity with the IscR from Azotobacter vinelandii, Erwinia chrysanthemi and E. coli, respectively [12], [22], [23]. The residues involved in [Fe-S] ligation (C92, C98, C104, H107) [13] and E43, which prevents the apo-IscR from binding to the type 1 binding site [11], are all conserved (Figure 1B). Nonetheless, no putative ryhB or P. aeruginosa sRNA prrF (a rhyB analogue) binding motifs were identified between the iscR and iscS intergenic sequences.
Figure 1. Gene organisation of isc gene cluster and multiple alignments of P. aeruginosa IscR.
(A) Gene organisation of isc gene cluster in P. aeruginosa PAO1 [16]. (B) Alignments of IscR from P. aeruginosa with IscR sequences from other bacteria. The alignments were performed using the CLUSTALW algorithm. Black and grey boxes indicate the amino acids responsible for the iron-sulphur cluster ligation, and C111, respectively. The asterisk, colon, and period symbols indicate identical residues, conserved substitutions, and semi-conserved substitutions, respectively. The number on top of the alignments indicates the position of the indicated amino acid.
Stress-induced Expression Profile and Promoter Analysis of the isc Operon
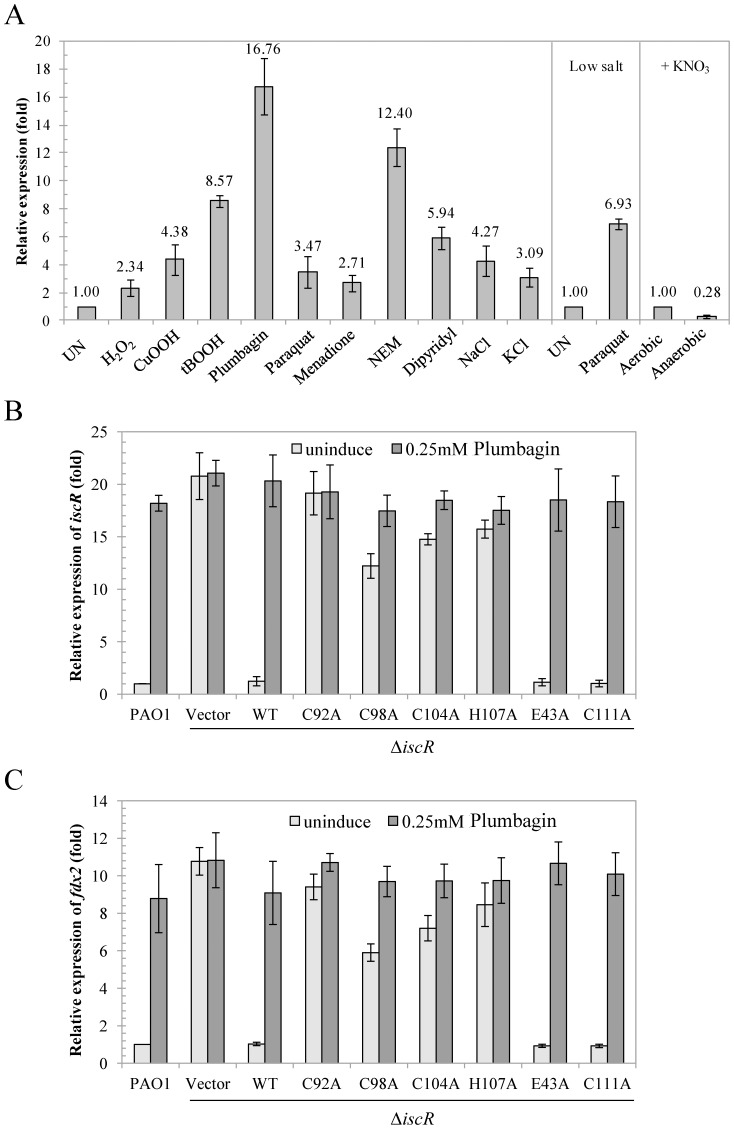
The lack of the suf operon in P. aeruginosa suggests that the isc operon functions in [Fe-S] biosynthesis under both housekeeping and stressful conditions. IscR is a global regulator involved in the regulation of genes from diverse physiological functions, hence the expression of IscR is likely important for the ability of P. aeruginosa to adapt to changing conditions. qRT-PCR was performed to determine the expression levels of the genes in the isc operon (iscR and fdx2) in response to an intracellular iron-chelating agent (1 mM 2,2′-dipyridyl) and various stresses, including hydrogen peroxide (1 mM H2O2), organic hydroperoxides (1 mM cumene hydroperoxide [CuOOH] or t-butyl hydroperoxide [tBOOH]), redox-cycling drugs (0.5 mM plumbagin, paraquat, or menadione), thiol-depleting agent (0.1 mM N-ethylmaleimide [NEM]), and high salt conditions (0.5 M NaCl or KCl). The results of the expression analysis illustrated that iscR expression was highly induced (4.4–16.8-fold) upon treatment with plumbagin, organic hydroperoxides, dipyridyl, and NEM and to a lesser extent (2.3–4.3-fold) through paraquat, menadione, H2O2 and high salt treatments (Figure 2A). The induction of iscR through paraquat is affected by the salt concentration in the media. A high salt concentration inhibits paraquat transportation into the cells; thus, lowering the salt concentration in the medium enhances the paraquat-mediated induction of iscR expression (Figure 2A). The expression of the fdx2 (PA3809) gene was used to monitor gene expression in the isc operon. The fdx2 expression profiles in response to plumbagin-induced stress were comparable to that of iscR (Figure 2B and C). These observations indicate that the expression of the isc operon is induced through iron depletion (dipyridyl treatment), high salts, thiol depletion and oxidative stresses (peroxide and redox-cycling agents). These conditions affect not only the iron availability but also damage [Fe-S], resulting in the overall reduced availability of [Fe-S]. Thus, increased [Fe-S] biogenesis and the up-regulation of the isc operon are required to maintain [Fe-S] homeostasis under stressful conditions.
Figure 2. Expression levels of the isc operon in response to stresses.
(A) Analysis of the iscR expression during normal growth and in response to oxidants after inducing the culture with 1 mM H2O2, 1 mM CuOOH, 1 mM tBOOH, 0.5 mM paraquat, 0.5 mM menadione, 0.5 mM plumbagin, 0.1 mM NEM, 1 mM dipyridyl, 0.5 M NaCl, 0.5 M KCl, LB medium without NaCl (low salt) without (UN) and 0.5 mM paraquat for 15 min under aerobic and anaerobic conditions supplemented with nitrate (+KNO3). (B) and (C) show the expression profiles of iscR and fdx2, respectively, in uninduced (grey bars) and 0.25 mM plumbagin-induced (closed bars) cultures of P. aeruginosa PAO1 and ΔiscR mutant harbouring pBBR1MCS-4 plasmid (vector), plasmid expressing wild-type IscR (WT), and mutated IscR (C92A, C98A, C104A, H107A, E43A, or C111A). qRT-PCR was performed as described in the Methods. The data are presented as the means ± SD from three independent experiments. The relative expression was analysed using the 16S rRNA gene as the normalising gene and expressed as fold expression relative to the level of the uninduced PAO1.
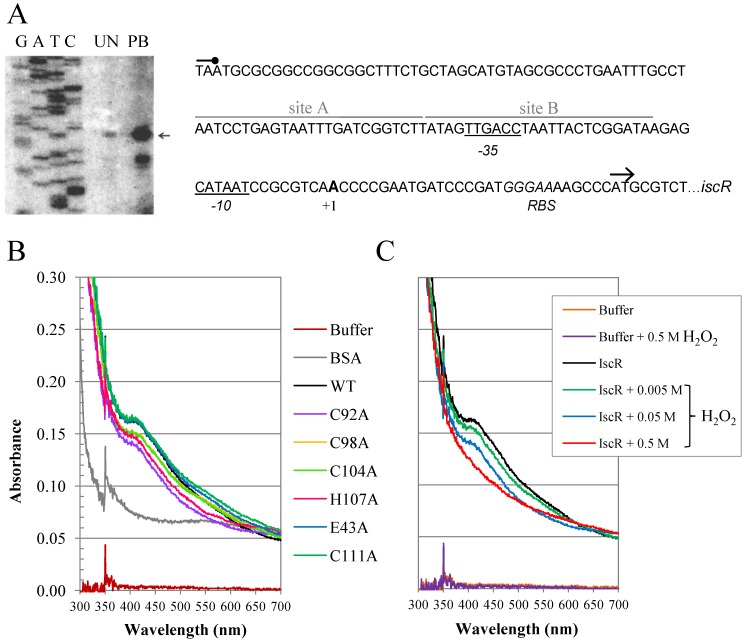
Next, a primer extension experiment was performed to determine the transcriptional start site (+1) of the isc operon. The +1 was mapped to A residue located at 29 bases upstream of the ATG translation start codon of iscR (Figure 3A). The putative −35 and −10 promoter elements, separated by 18 bases, were identified as TTGACC and CATAAT, respectively. The results of the iscR primer extension showed several-fold more products in plumbagin-induced samples. Thus, these results confirmed the qRT-PCR analysis, implicating plumbagin as a potent inducer of iscR expression (Figure 2B). In addition, these results show that the stress induced up-regulation of iscR results from increased transcription initiation from the promoter.
Figure 3. Characterisation of the iscR promoter.
(A) The primer extension assay was performed using 32P-labelled primer BT3577 and RNA extracted from PAO1 grown under uninduced (UN) and 0.25 mM plumbagin-induced conditions. The extension products were separated on an 8% acrylamide-7 M urea sequencing gel. G, A, T, and C represent the DNA ladder sequence prepared using 32P-labelled primer BT3577 and putative iscR promoter fragments as templates. The arrowhead indicates the transcription start site (+1). The −10 and −35 elements are underlined. The consensus sequence of type 1 E. coli IscR binding site is aligned above the sequence line in corresponding letters, and the homologous bases are marked with asterisks. The putative ribosome-binding site (RBS) is indicated in italic type. (B) UV-visible absorption spectra of various IscR variants (C92A, C98A, C104A, H107A, E43A, or C111A). Purified IscR proteins in 50 mM phosphate buffer (10 µM) were used in the experiments. BSA (10 µM) was used as non-[Fe-S] protein control. (C) UV-visible absorption spectra of wild-type IscR treated with the indicated concentrations of H2O2.
Analysis of sequences upstream of the iscR transcription start revealed two putative type 1 IscR-binding motifs denoted site A (5′AATCCTGAGTAATTTGATCGGTCTT3′) and site B (5′ATAGTTGACCTAATTACTCGGATAA3′) located between positions −18 to −67 (Figure 3A). These two motifs had 68% and 76% identity, respectively, to the consensus sequence for the E. coli type 1 IscR-binding motif (5′ATASYYGACTRWWWYAGTCRRSTAT3′) [10]. This suggests autoregulation of iscR-expression in response to varying [Fe-S] demand. The location of these two type 1 sites overlaps with the RNA polymerase recognition motif (Figure 3A). Thus, the binding of IscR to these sites would be expected to prevent RNA polymerase binding to the promoter and inhibit the transcription of isc operon. A recent report indicates that E. coli has three IscR binding sites upstream of the iscR promoter that are involved in regulation of the iscR operon [8]. The possible role of the IscR binding sites within the P. aeruginosa iscR promoter region is being investigated.
IscR is a Transcriptional Regulator of the isc Operon
The iscR mutant was constructed through gene deletion. The expression of iscR and fdx2 was used to monitor the expression of the isc operon in PAO1 and a ΔiscR mutant. qRT-PCR analysis of iscR was performed using forward primers EBI120 (located at position +1 to +18 of iscR transcript [Figure 3A], hence outside a ribosome binding site and ATG of the IscR and the gene deletion site of the ΔiscR mutant) and BT3613 (located in coding region of iscR). These primers facilitate the analysis of ΔiscR mutants carrying Tn7TiscR, pBBRiscR or various site-directed iscR mutations. The results showed that under uninduced conditions, the expression of iscR and fdx2 in the ΔiscR mutant was 20.8- and 10.8-fold higher than the levels in PAO1, respectively. Plumbagin treatments did not further enhance the expression of either iscR or fdx2 in the ΔiscR mutant (Figure 2B and C). The expression of iscR from a Tn7T vector in the mutant led to the repression of iscR and fdx2 expression to levels similar to those observed in PAO1 (Figure 2B and C). Furthermore, the plumbagin-induced expression of both genes in the complemented strain was restored to wild-type levels (Figure 2B and C).
Additional experiments were performed to determine the form of IscR and type of binding site involved in the repression of the isc operon. Two major forms of the transcription regulator, apo-IscR and [2Fe-2S]-IscR have been characterised [13]. Apo-IscR binds to type 2-binding sites with greater affinity than type 1 sites. This is thought to be due to the inhibitory effect of residue E43 during apo-IscR binding to type 1 sites based on the observation that an E43A mutation enhances the binding of apo-IscR to type 1 sites in the absence of [2Fe-2S] ligation [11]. The residue E43 is conserved in P. aeruginosa IscR. Experiments were performed to test the effect of wild-type IscR and IscR-E43A on the regulation of iscR operon expression in the ΔiscR mutant. Our results showed that, similar to the wild-type IscR, IscR-E43A repressed iscR and fdx2 expression in the ΔiscR mutant (Figure 2B and C). IscR-E43A repressed iscR and fdx2 transcription to levels similar to those observed in a wild-type IscR background, this indicated that that E43 is not required for IscR-mediated repression of the iscR promoter.
The IscR residues, C92, C98, C104, and H107, which serve as ligands for [2Fe-2S] cluster ligation [13] are conserved in P. aeruginosa IscR. Experiments to determine whether [2Fe-2S]-IscR is involved in the regulation of the isc operon were performed through mutating C92, C98, C104, and H107 residues to alanine (A) residues in P. aeruginosa IscR. The plasmids containing the mutagenised iscR, i.e., pBBRiscR-C92A, pBBRiscR-C98A, pBBRiscR-C104A, and pBBRiscR-H107A, were transferred into the iscR mutant, and the restoration of plumbagin-induced iscR and fdx2 expression was tested. The concentration of plumbagin was reduced to 0.25 mM due to the plumbagin hypersensitive phenotype of the ΔiscR mutant. The expression of iscR fully complemented the constitutive high and oxidant non-inducible expression of iscR and fdx2 (Figure 2B and C). The ΔiscR mutant harbouring pBBRiscR repressed expression of iscR (16.9-fold) and fdx2 (10.5-fold) compared with the iscR mutant harbouring empty vector. The plumbagin treatment of these cells induced the expression of both iscR and fdx2 to the levels attained in PAO1 harbouring the vector control (Figure 2B and C). The expression of mutated C92A and H107A iscR did not repress the expression of these genes while the IscR-C98A and IscR-C104A mutants showed 1.6- and 1.4-fold repression of iscR expression compared with the 16.9-fold expression observed with wild-type IscR. Similar expression patterns were obtained for the fdx2 analysis. Nonetheless, plumbagin treatment induced the expression of target genes to levels comparable to those obtained with wild-type IscR. The inability of mutant IscR-C92A and IscR-H107A to repress iscR and fdx2 expression suggests that these mutations prevent the ligation of [2Fe-2S] to IscR, which is sufficient to prevent the repression of the isc operon. Surprisingly, IscR-C98A and IscR-C104A produced intermediate repression levels and plumbagin inducible expression patterns, implying that these IscR mutants might weaken the attachment of [2Fe-2S] to IscR. Under aerobic conditions, the weakened [2Fe-2S] attachment to mutated IscR that might render [2Fe-2S]-IscR more susceptible to aerobic oxidation, resulting in a mixture comprised primarily of apo-IscR and low amounts of [2Fe-2S]-IscR within the cell. This could explain the observed partial repression and plumbagin-induced expression of target genes. The mutation of the non-conserved C111 near the ligand ligation site generated a target gene expression pattern similar to that of wild-type IscR.
The analysis of the IscR-binding motifs on the iscR promoter indicates that these motifs could belong to the type 1 motif class (Figure 3A). Taken together, these findings indicate that [2Fe-2S]-IscR binding at the type 1 site is responsible for the repression of isc operon expression. Thus, apo-IscR is likely not involved in the regulation of the isc operon [8].
Characterisation of IscR Residues Involved in Iron-sulphur Cluster Ligation
Apo-IscR atypically ligates a [2Fe-2S] cluster with three cysteines and one histidine residue [13]. To address the importance of these [2Fe-2S] cluster ligands, 6xHis-tagged IscR wild-type and IscR variants carrying amino acid substitutions at residues thought to be involved in ligation (C92A, C98A, C104A, or H107A) were purified and equal amounts of the mutant proteins were then subjected to UV-visible spectroscopy as described in Methods. Both IscR-E43A and IscR-C111A had significant absorption at 415 nm similar to the wild-type protein (Figure 3B). By contrast, the IscR variant proteins (C98A, C104A, H107A, or C92A) had lower absorption at 415 nm relative to the wild-type protein indicating a reduced amount of [2Fe-2S] clusters ligated to these IscR variants (Figure 3B). The IscR-C92A had the lowest absorption at 415 nm, indicating that [2Fe-2S] clusters bind less well to this protein. These results implicate residues C92, C98, C104, and H107 in [2Fe-2S] binding to IscR and support the qRT-PCR data (Figure 2B and C) indicating that defects in [2Fe-2S] cluster ligation affect the ability of IscR to repress transcription of the isc operon.
To determine the effect of ROS on [2Fe-2S] cluster integrity, purified wild-type IscR was incubated with various concentrations of hydrogen peroxide (H2O2) for 10 minutes prior to UV-visible spectroscopy. The results showed decreases in IscR absorbance at 415 nm that were H2O2 concentration-dependent, suggesting that the [2Fe-2S] cluster ligated to IscR were targets for H2O2-mediated oxidation (5–50 mM) that resulted in destabilization of [Fe-S] bound to the protein. Treatment of the protein with a high concentration (0.5 M) of H2O2 led to a total loss of [2Fe-2S] clusters bound to IscR as shown in Figure 3C. These results, together with the qRT-PCR results showing the derepression of isc operon transcription in the presence of plumbagin (Figure 2B and C), suggest that during oxidative stress, IscR-bound [2Fe-2S] clusters are damaged by ROS resulting in a decrease in [2Fe-2S]-IscR levels leading to derepression of isc operon transcription.
High-level Expression of iscR Impairs Anaerobic Growth
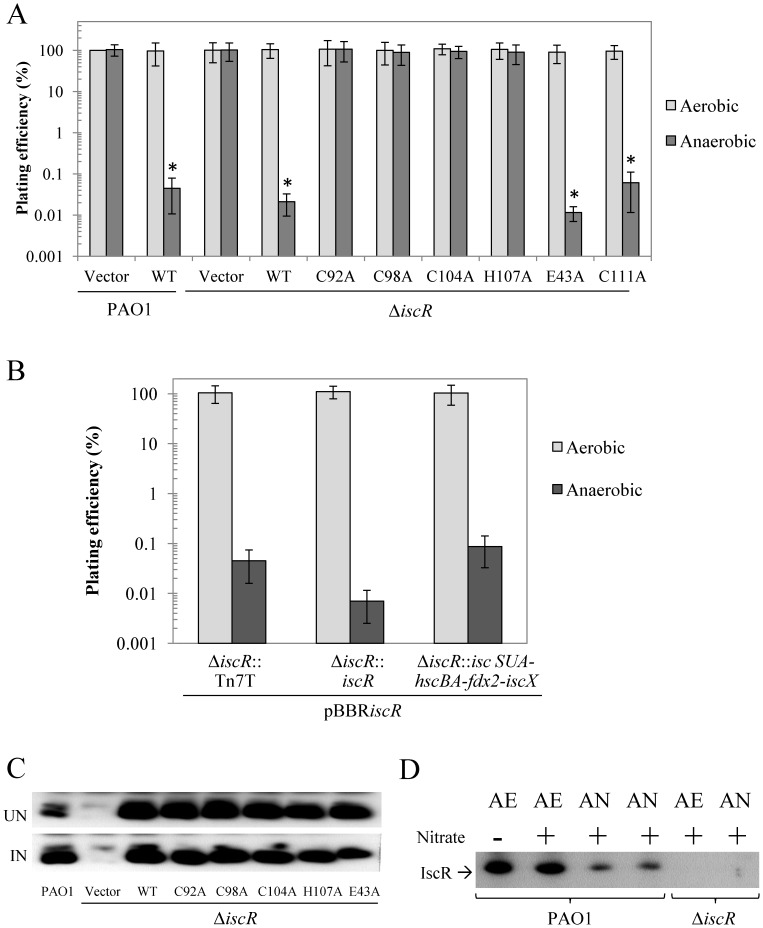
The roles of iscR in aerobic and anaerobic growth were assessed. The deletion of iscR did not affect growth, as shown through comparable aerobic and anaerobic growth between ΔiscR mutant and PAO1 (Figure 4A). However, the expression of iscR from an expression vector strongly inhibited (5 × 103-fold reduction in % plating efficiency) the anaerobic growth of both PAO1 and the ΔiscR mutant, while aerobic growth remained unaffected (Figure 4A). A key question is whether the levels of IscR produced from expression vectors are physiologically relevant. Western blot analysis was performed to determine the levels of wild-type IscR and mutated IscR variants (C92A, C98A, C104A, H107A, or E43A) expressed from plasmids in the ΔiscR mutant. The results show similar intensities of the IscR-specific bands in the P. aeruginosa ΔiscR mutant strains expressing wild-type IscR and the IscR variants (Figure 4C), indicating similar production and stability levels. The level of IscR in each strain was approximately 30-fold higher than the IscR level in PAO1 harbouring an empty vector as determined by Western blot comparison of IscR band intensity in two-fold serial dilutions of protein extracts from an IscR over-producing strain and PAO1 containing vector only (data not shown). The levels of IscR in strains harbouring expression vectors were similar to the level of IscR in wild-type PAO1 exposed to an inducer. Thus, these IscR levels are physiological relevant.
Figure 4. Determination of growth and IscR production in P. aeruginosa strains.
(A) and (B) show the plating efficiencies of P. aeruginosa strains grown under aerobic and anaerobic conditions. (A) The PAO1 and ΔiscR mutants harbouring pBBR1MCS-4 (vector) along with plasmids expressing either wild-type IscR (WT), or a mutated IscR (C92A, C98A, C104A, H107A, E43A, or C111A), (B) ΔiscR mutants harbouring plasmids expressing wild-type IscR, as well as a chromosomal TN7 expression cassette carrying either iscR or iscSUA-hscBA-fdx2-iscX, were spread onto solid medium containing oxidants and incubated under aerobic and anaerobic conditions. Similar to the plate sensitivity assay, observations were also obtained in the absence of oxidant. The plating efficiency is defined as the number of CFU under anaerobic conditions divided by the number of CFU obtained with PAO1 harbouring an empty vector under aerobic conditions. The asterisk indicates statistically significant differences (paired t-test, P>0.05) compared with aerobic conditions. (C) and (D) Western blot analyses to determine IscR levels in P. aeruginosa strains. (C) Crude extracts were prepared from PAO1 and the ΔiscR mutant expressing wild-type and various mutated iscR (C92A, C98A, C104A, H107A, E43A, or C111A) grown under either uninduced (UN) conditions or induced (IN) conditions (i.e. 0.5 mM plumbagin for 30 min). (D) Crude extracts were prepared from PAO1 and ΔiscR mutant cultures cultivated under aerobic (AE) and anaerobic (AN) atmospheres with (+) or without (−) KNO3 (1%) supplementation. Electrophoresis was carried out using 15 µg protein and 12.5% SDS-PAGE.
The mechanism of IscR-mediated regulation can vary with respect to both the specific target gene and the particular organism. It should be possible to use the various IscR variants to identify the physiologically relevant forms of IscR and their target genes. Phenotypic analyses of ΔiscR mutant strains expressing the various mutated IscR variants could provide information on the roles for the regulator in both its apo- and [Fe-S] cluster bound forms. The anaerobic growth defect phenotype was not detected in strains producing [2Fe-2S] ligation mutants (C92A, C98A, C104A, or H107A), as shown by the similar plating efficiency in anaerobic growth (Figure 4A). Moreover, the iscR-E43A expression in the ΔiscR mutant showed an anaerobic growth defect phenotype, indicating that E43 is not required for this phenotype. The ΔiscR mutant producing IscR-C111A, a non-conserved cysteine, also showed growth defects under anaerobic conditions similar to those producing wild-type IscR. Taken together, these results suggest that the iscR multi-copy inhibition of anaerobic growth likely reflects the binding of [2Fe-2S]-IscR to the type 1 binding sites of target genes. The results indicate that high levels of [2Fe-2S]-IscR adversely affect the anaerobic growth of P. aeruginosa. They also indicate that the ligation of [2Fe-2S] to IscR is required for inhibition of the anaerobic growth phenotype. Thus, it is unlikely that apo-IscR is responsible for this phenotype.
The inhibition of anaerobic growth by high levels of IscR could arise from repression of the isc operon. One of the targets for [2Fe-2S]-IscR type 1 binding is the isc operon, where high levels of [2Fe-2S]-IscR lead to strong repression of the iscR promoter that inhibit the expression of the isc operon, leading to the overall reduction in [Fe-S] availability in the cell. Alternatively, mis-regulation of other genes in the IscR regulon may be responsible for the phenotype. We attempted to differentiate between these possibilities by reasoning that if overly strong repression of the isc operon was responsible for the phenotype then unregulated expression of the genes in the isc operon, but not iscR, from a single-copy expression cassette, should alleviate the anaerobic growth inhibition phenotype. Results showed that expression of (iscSUA-hscBA-fdx2-iscX) from a chromosomal Tn7 expression cassette did not complement the impaired anaerobic growth of cells highly expressing iscR from a vector (Figure 4B). Hence, it is likely that high levels of IscR caused mis-regulation of genes in the IscR regulon other than those in the isc operon.
The expression levels of iscR under aerobic and anaerobic conditions in P. aeruginosa were determined using qRT-PCR. The results showed that the iscR expression level during anaerobic growth conditions was approximately 2.8-fold lower than the level attained during aerobic growth (Figure 2A). Furthermore, Western blots determined that the level of IscR showed a 4-fold decrease in anaerobically grown cultures compared to those grown aerobically (Figure 4D). This decrease matched the observed decrease in transcription of iscR under anaerobic conditions (Figure 2A). This anaerobic reduction was not simply due to the addition of nitrate since nitrate addition to aerobic cultures had no effect on the IscR level (Figure 4D). These results are consistent with the idea that the presence of oxygen reduces the proportion of [2Fe-2S]-IscR through oxidative damage to the [2Fe-2S] cluster, thereby derepressing isc operon transcription. This effect is amplified by increased competition for iron-sulphur cluster insertion between IscR and other damaged [Fe-S] cluster-containing proteins [21]. Under anaerobic conditions, the increased repression of the operon reflects the reduced competition for, and turnover of, [Fe-S] clusters resulting in higher [2Fe-2S]-IscR levels and increased isc operon repression.
The iscR Mutant is Hypersensitive to Oxidants
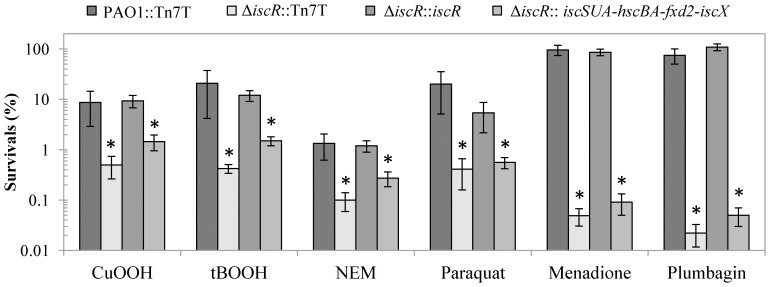
Previous studies have shown that the P. aeruginosa iscR mutant is susceptible to H2O2 and paraquat. The former phenotype results from decreased catalase activity [19]. Here, we extended the investigation into phenotypic changes of the ΔiscR mutant to oxidative stresses. The plate sensitivity assay was used to determine the sensitivity level toward redox-cycling agents and organic hydroperoxides. The results showed that the ΔiscR mutant is 40- to 50-fold less resistant to organic hydroperoxides (tBOOH and CuOOH). In addition, the mutant exhibited greater than 103-fold reduced resistance to redox-cycling drugs, such as plumbagin and menadione, and 50-fold reduced resistance to paraquat (Figure 5). The sensitive phenotype against both the organic hydroperoxides and redox-cycling drugs could be complemented by the expression of a single copy of iscR in Tn7-borne expression (Figure 5), indicating that IscR plays an important role in the oxidative stress response. In PAO1, IscR directly regulated tpx, a thiol peroxidase gene encoding a broad peroxide substrate thiol peroxidase [20]. It is likely that other as yet unidentified genes, directly or indirectly regulated through IscR, could be responsible for the increased sensitivity to organic hydroperoxides. In addition, a ΔiscR mutant containing Tn7TiscSUA-hscBA-fdx2-iscX did not show significant changes in oxidant resistance levels compared to the ΔiscR mutant suggesting mis-regulation of other genes in the IscR regulon is likely to be responsible for the increased oxidant sensitivity of the ΔiscR mutant. Furthermore, altered oxidant resistance in the ΔiscR mutant is likely a direct result of the loss of IscR, which reduces the capacity of the mutant to respond to oxidative stress.
Figure 5. Determination of the resistance levels against stresses in P. aeruginosa strains.
Plate sensitivity assays were performed in PAO1 and ΔiscR mutant transposed with Tn7T vector (PAO1::Tn7T and ΔiscR::Tn7T), the iscR complemented strain (ΔiscR::iscR), and the isc operon-overexpressed ΔiscR strain (ΔiscR::iscSUA-hscBA-fdx2-iscX) using plates containing 1.6 mM CuOOH, 1 mM tBOOH, 280 µM NEM, 250 µM paraquat, 4 mM menadione, and 1 mM plumbagin. Survival (%) is defined as the percentage of colony-forming units (CFU) on plates containing oxidant over the number of CFU on plates without oxidant. The data are presented as the means ± SD from three independent experiments. The asterisk indicates statistically significant (paired t-test, P<0.05) compared with the PAO1::Tn7T treated with the same condition.
IscR Modulates Intracellular Iron
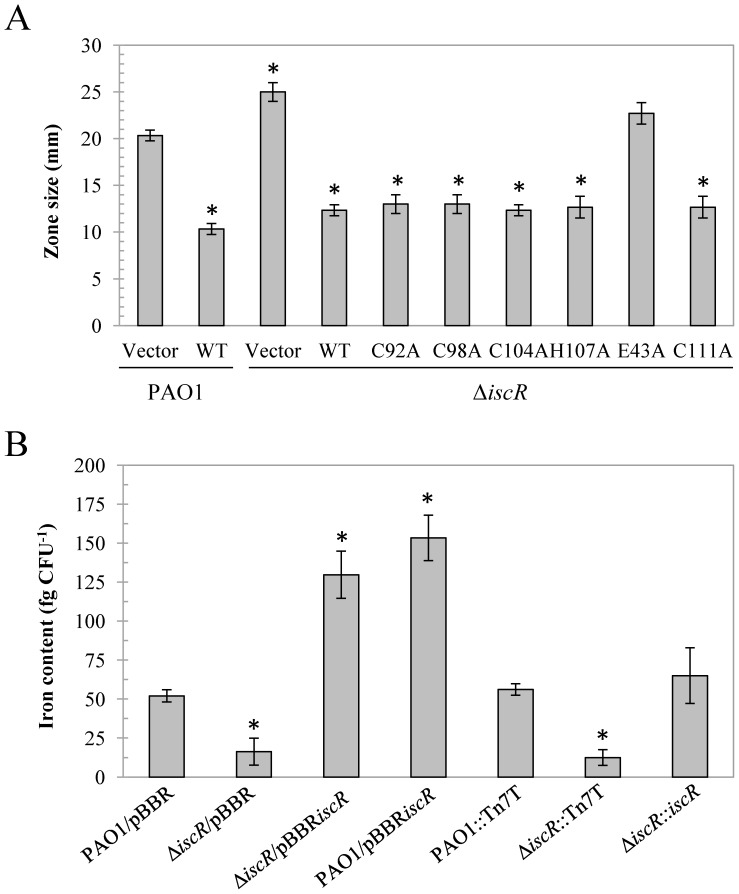
The availability of intracellular iron is finely balanced and controlled. The roles of iscR in iron homeostasis and the iron status of the mutant were investigated. Here, we have shown that the deletion of iscR resulted in the high-level expression of [Fe-S] biogenesis genes in the isc operon. Decreased intracellular labile iron in the cell could be a consequence of increased levels of [Fe-S] biosynthesis. The ΔiscR mutant could sense this condition as iron starvation and respond accordingly. The increased production of siderophores is one of the initial responses to low intracellular iron [24], [25]. Hence, the production of siderophores was measured in PAO1 and the ΔiscR mutant using a CAS assay (Figure 6A). The results showed that the ΔiscR mutant produced comparatively higher levels of siderophore than PAO1, based on a wider zone diameter in the CAS assay. In accordance with this observation, the over-expression of iscR led to the hyper-repression of siderophore expression (25 mm zone diameter for ΔiscR mutant compared with 10 mm zone for the mutant harbouring pBBRiscR) (Figure 6A). Here, we have shown that [2Fe-2S]-IscR binding to the type 1 binding site is responsible for the repression of the isc operon. The investigation was extended to determine whether [2Fe-2S] ligation to IscR and the E43 residue are required to complement the increased siderophore phenotype of the ΔiscR mutant harbouring various site-directed mutagenesis of amino acid residues involved in [2Fe-2S] ligation and the E43A mutation of iscR. The results illustrated that the ΔiscR mutants harbouring mutated iscR that produced IscR variants (C92A, C98A, C104A, or H107A) exhibited similar lower levels of siderophore production compared with the strain producing wild-type IscR (Figure 6A). However, the mutant expressing IscR-E43A expressed siderophores at a similar level as the ΔiscR mutant (Figure 6A), suggesting that the E43 residue of IscR is required for the transcription modification of genes leading to complement of the mutant increased siderophore production phenotype. Altogether, these results suggest that apo-IscR binds to the type 2 binding motif upstream of unidentified IscR target genes and activates/represses gene expression, resulting in the modulation of siderophore production. Furthermore, the results suggest that the phenotype is not a simple effect of the repression of the isc operon through [2Fe-2S]-IscR. Additional direct or indirect alterations in the expression of IscR-regulated genes subsequently alter siderophore production. Further experiments are currently underway to determine whether IscR directly or indirectly regulates the genes that produce this phenotype.
Figure 6. Siderophore production and intracellular iron content in P. aeruginosa strains.
(A) Siderophore production in P. aeruginosa PAO1, the ΔiscR mutant, harbouring pBBR1MCS-4 (vector), and plasmids expressing wild-type IscR (WT) and mutated IscR (C92A, C98A, C104A, H107A, E43A, or C111A) was determined using chrome azural S (CAS) agar plates. Siderophore production is directly associated with the yellow halo zone size and the data shown are the means ± SD from three independent experiments. (B) ICP-MS was conducted as described in the Methods to measure the intracellular iron content in the indicated P. aeruginosa strains. The data are presented as the means ± SD from three independent experiments. The asterisk indicates statistically significant difference (P<0.05) compared with the PAO1 harbouring empty vector.
The total intracellular iron content in P. aeruginosa strains was measured using ICP-MS. Figure 6B shows that the total intracellular iron content of the ΔiscR mutant (12.5 fg CFU−1) was approximately 4-fold lower than the level observed in wild-type PAO1 (56.07 fg CFU−1). This reduced iron content phenotype could be restored through the expression of single copy iscR (65.01 fg CFU−1). The increased expression of iscR from a multi-copy vector (pBBRiscR) in PAO1 and the ΔiscR mutant increased the intracellular iron content to the levels of 129.64 fg CFU−1, respectively compared with PAO1 carrying the vector control (52.05 fg CFU−1) (Figure 6B). The ΔiscR mutant exhibits reduced total iron and higher siderophore levels than PAO1 (Figure 6A and B). The high-level expression of iscR increased the total iron content and significantly reduced siderophore production. The inactivation of iscR reduced the total intracellular iron content even though the production of siderophores, and presumably other iron uptake system components, was significantly increased. This suggests that changes other iron metabolism pathways, such as uptake, iron storage, or efflux, are affected through iscR inactivation leading to a reduction in the total intracellular iron content. Interestingly, increased siderophore production was also in a PAO1 fur mutant, which displayed high siderophore production but was defective in siderophore mediated iron uptake [25].
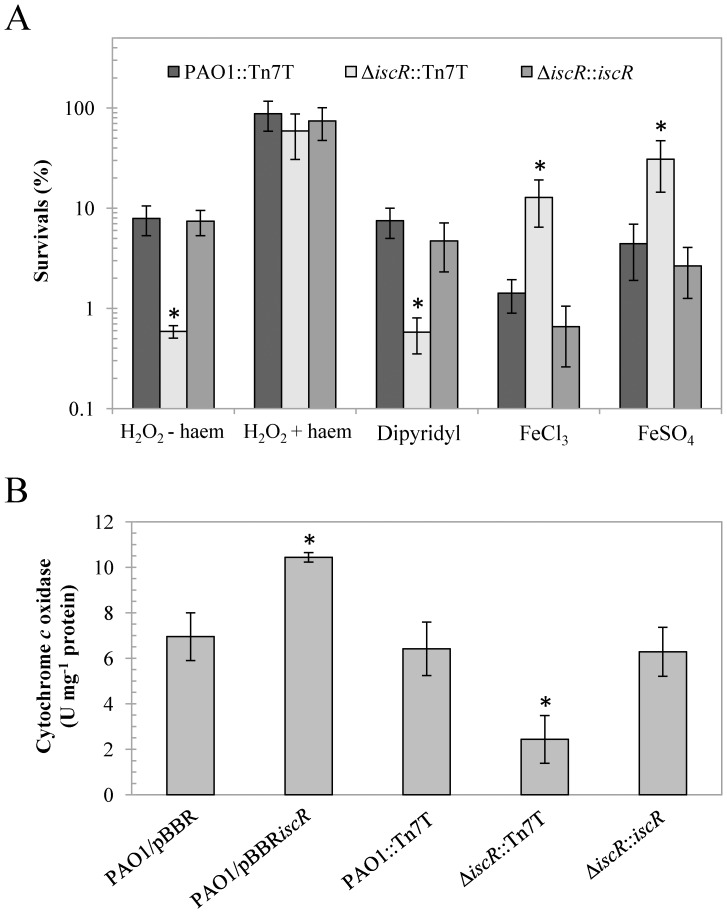
The ability to cope with either iron deprivation or iron excess could reflect the intracellular iron status of the cells. The resistance levels against iron repletion and depletion conditions in the ΔiscR mutant were determined. The ΔiscR mutant showed decreased resistance (approximately 40-fold) to an intracellular iron chelator dipyridyl (1.2 mM), but increased resistance to the addition of both FeCl3 (5.5 mM) and FeSO4 (4 mM) (12-fold and 7-fold, respectively) relative to PAO1 (Figure 7A). Both phenotypes in ΔiscR mutant were fully complemented through the expression of iscR from a Tn7T vector. These results indicate that the ΔiscR mutant copes less well with iron deprivation stress, such as dipyridyl treatment than PAO1 and vice versa with excess iron conditions. These observations support the low intracellular iron conditions in ΔiscR mutant. The inactivation of P. aeruginosa iscR lowers KatA catalase activity at a posttranslational level [19]. Thus, the ΔiscR mutant is sensitive to H2O2 treatment [18]. KatA is a haem-containing enzyme that detoxifies H2O2 to water and oxygen and is crucial for peroxide resistance [26]. Since ΔiscR mutant exhibits reduced intracellular labile iron, the depletion of haem could be one of the factors responsible for the reduced KatA activity in the ΔiscR mutant. We observed that haem supplementation in the culture medium restored the H2O2-sensitive phenotype of the ΔiscR mutant (Figure 7A) and increased catalase activity (data not shown). However, it was possible that haem supplementation could complex iron thereby affecting the activity of other iron-containing enzymes involve in oxidative stress defense. In order to test this, the level of SodB, a non-haem iron-containing enzyme was measured using SOD activity gel assays, in the P. aeruginosa strains cultured in LB-medium supplemented with the divalent metal chelator, EDTA, or the iron-specific chelator, dipyridyl. In both cases, haem supplementation had no significant effect on SodB activity, indicating that haem was not acting as a source of free iron (data not shown). No significant difference in SodB activity was observed between the strains. This observation, combined with the fact that haem supplementation increases H2O2 resistance (Figure 7A), suggest that haem could be directly incorporated into haem-containing enzymes.
Figure 7. Survival under iron depletion and repletion conditions and cytochrome c oxidase activity in P. aeruginosa strains.
(A) Determination of the resistance against agents affecting iron homeostasis in P. aeruginosa PAO1 and the ΔiscR mutant. Plate sensitivity assays against 0.5 mM H2O2 in the absence (−) and presence (+) of 100-µM haem, iron depletion (dipyridyl, 1.2 mM), and iron repletion (5 mM FeCl3 and 4 mM FeSO4) were performed. The asterisk indicates statistically significant (P<0.05) compared with the PAO1::Tn7T treated with the same condition. (B) Cytochrome c oxidase activity was measured in the indicated P. aeruginosa strains as described in the Methods. The data are presented as the means ± SD from three independent experiments. The asterisk indicates statistically significant (P<0.05) compared with the PAO1 harbouring empty vector.
The observed increased H2O2 sensitivity suggests haem deficiency in the ΔiscR mutant. Therefore, we measured cytochrome c oxidase activity, another haem-containing enzyme in the electron transport chain. The results illustrated that the cytochrome c oxidase activity in the ΔiscR mutant (2.4 U mg−1 protein) was approximately 3-fold less than that in the wild-type PAO1 (6.4 U mg−1 protein), and this reduced enzyme activity could be restored through iscR in a complemented strain (6.3 U mg−1 protein) (Figure 7B). Taken together, the results indicate defects in the activity of haem-containing enzymes that could reflect overall reduced haem levels of the ΔiscR mutant.
Our data showed that iscR plays an important role in iron homeostasis. Inactivation of iscR resulted in the increased expression of the isc operon containing genes encoding enzymes for [Fe-S] biogenesis. This, in turn diverts the pool of labile iron towards [Fe-S] biosynthesis. Moreover, the amount of total intracellular iron was also reduced in the ΔiscR mutant. Other parameters of iron deprivation conditions, such as increased siderophores output, increased sensitivity to iron depletion (dipyridyl treatment), increased resistance to iron excess conditions (addition of either FeCl3 or FeSO4) and reduced haem availability as shown by reduction in haem containing enzymes levels, were observed in the ΔiscR mutant. The observed low total iron content in the ΔiscR mutant could not be correct by increased siderophore production. These results support important roles of IscR in iron homeostasis. In P. aeruginosa, IscR regulated target genes involved in iron homeostasis have not been characterised. Although, Fur is a regulator of iron homeostasis that regulates more than a hundred genes that participate in iron uptake, metabolism, and efflux [27]. fur is a possible target for IscR regulation in PAO1. Preliminary data suggest that there could be interplay between these two global regulators and their effects on target genes and overall iron homeostasis is currently being investigated.
Methods
Bacterial Strains and Growth Conditions
All bacterial strains and plasmids used in this study are listed in Table 1. P. aeruginosa PAO1 and E. coli strains were aerobically cultivated in Luria-Bertani (LB) broth at 37°C with shaking at 180 rpm unless otherwise stated. The medium for E. coli growth were supplemented with 100 µg ml−1 ampicillin (Ap) or 15 µg ml−1 gentamicin (Gm) as required, whereas the medium for P. aeruginosa were supplemented with 200 µg ml−1 carbenicillin (Cb), 25 µg ml−1 chloramphenicol (Cm) or 30 µg ml−1 Gm as required. To produce synchronous growth, an overnight culture was inoculated into fresh LB medium to give an optical density at 600 nm (OD600 nm) of 0.1. Exponential phase cells (OD600 nm of 0.6, after 3 h of growth) were used in all experiments.
Table 1. Bacterial strains and plasmids used in this study.
| Strain or Plasmid | Relevant characteristic(s) | Source |
| P. aeruginosa | ||
| PAO1 | Wild-type strain | ATCC15692 |
| PAO1/pBBR | PAO1 harbouring pBBR1MCS-4 | This study |
| PAO1::Tn7T | PAO1 transposed with pUC18-mini-Tn7T-Gm-LAC | This study |
| ΔiscR | PAO1 ΔiscR mutant | This study |
| ΔiscR/pBBR | ΔiscR mutant harbouring pBBR1MCS-4 | This study |
| ΔiscR::Tn7T | ΔiscR mutant transposed with pUC18-mini-Tn7T-Gm-LAC | This study |
| ΔiscR::iscR | ΔiscR mutant transposed with pTn7TiscR | This study |
| ΔiscR::iscSUA-hscBA-fdx2-iscX | ΔiscR mutant transposed with pTn7TiscSUA-hscBA-fdx2-iscX | This study |
| E. coli | ||
| DH5α | φ80d lacZΔM15, recA1, endA1, gyrA96, thi-1, hsdR17(rk −, mk +), supE44, relA1, deoR, Δ(lacYMA-argF)U169 | Stratagene Inc.(USA) |
| Plasmid | ||
| pUC18-mini-Tn7T-Gm-LAC | pUC18 containing mini-Tn7T::Plac site, Apr, Gmr | [29] |
| pTn7TiscR | pUC18-mini-Tn7T-Gm-LAC containing iscR | This study |
| pBBR1MCS-4 | Medium-copy-number expression vector, Apr | [32] |
| pBBRiscR | pBBR1MCS-4 containing iscR | This study |
| pBBRiscR-C92A | pBBRiscR with C92A mutagenesis | This study |
| pBBRiscR-C98A | pBBRiscR with C98A mutagenesis | This study |
| pBBRiscR-C104A | pBBRiscR with C104A mutagenesis | This study |
| pBBRiscR-H107A | pBBRiscR with H107A mutagenesis | This study |
| pBBRiscR-E43A | pBBRiscR with E43A mutagenesis | This study |
| pBBRiscR-C111A | pBBRiscR with C111A mutagenesis | This study |
| pCM351 | vector containing the loxP-Gmr-loxP region, Gmr | [30] |
Molecular Techniques
General molecular techniques including DNA and RNA preparation, DNA cloning, PCR amplification, Southern and Northern analyses and E. coli transformation were performed using standard protocols [28]. Transformation of plasmids into P. aeruginosa strains was carried out using electroporation as previously described [29]. The oligonucleotide primers used are listed in Table 2.
Table 2. List of primers used in this study.
| Primer | Sequence (5′→3′) | Purpose |
| BT2781 | GCCCGCACAAGCGGTGGAG | Forward primer for 16S ribosomal gene |
| BT2782 | ACGTCATCCCCACCTTCCT | Reverse primer for 16S ribosomal gene |
| BT3210 | CGAGGTAGATCGGCAATT | Reverse primer for full-length iscR |
| BT3577 | CCTGCTGTCGGGTAACGC | Reverse primer for primer extension |
| BT3578 | ATCATGCGCGAGGACTCC | Forward primer for iscR deletion |
| BT3579 | GGATCGGCGTTGACCAGC | Reverse primer for iscR deletion |
| BT3555 | CGCAATGGCATCGAGATCGA | Forward primer for fdx2 expression |
| BT3556 | GATAGCCGCGAATCGGGCTC | Reverse primer for fdx2 expression |
| BT3584 | CACCTGTGGGCCGACCTCAGT | Site-directed mutagenesis of IscR-C111A |
| BT3585 | CTGAGGTCGGCCCACAGGTGG | Site-directed mutagenesis of IscR-C111A |
| BT3612 | GAAGATTTCGCCGGAGTCAA | Forward primer for iscR promoter fragment |
| BT3613 | GCGTTCGGAGATATCGGCCAG | Reverse primer for iscR expressionand iscR promoter fragment |
| EBI102 | GCGACCCGCGCCCAGGGGCAG | Site-directed mutagenesis of IscR-C92A |
| EBI103 | CTGCCCCTGGGCGCGGGTCGC | Site-directed mutagenesis of IscR-C92A |
| EBI120 | ACCCCGAATGATCCCGATG | Forward primer for iscR expression |
| EBI121 | GGAAAAGCCCATGCGTCTGA | Forward primer for full-length iscR |
| EBI142 | CAGGGCGATGCCCACTCCGGC | Site-directed mutagenesis of IscR-C98A |
| EBI143 | GCCGGAGTGGGCATCGCCCTG | Site-directed mutagenesis of IscR-C98A |
| EBI144 | GGCGATACCGCTCTGACCCAC | Site-directed mutagenesis of IscR-C104A |
| EBI145 | GTGGGTCAGAGCGGTATCGCC | Site-directed mutagenesis of IscR-C104A |
| EBI148 | TCCTATCTCGCACAGCTGTTC | Site-directed mutagenesis of IscR-E43A |
| EBI149 | GAACAGCTGTGCGAGATAGGA | Site-directed mutagenesis of IscR-E43A |
| EBI192 | TGTCTGACCGCCCACCTGTGG | Site-directed mutagenesis of IscR-H107A |
| EBI193 | CCACAGGTGGGCGGTCAGACA | Site-directed mutagenesis of IscR-H107A |
| M13F | GTAAAACGACGGCCAGT | Universal forward primer for expression vector |
| M13R | AAACAGCTATGACCATG | Universal reverse primer for expression vector |
Strain and Plasmid Constructions
The iscR deletion mutant was constructed using the homologous recombination with an unmarked Cre-loxP antibiotic marker system as previously described [30], [31]. An 1138-bp DNA fragment containing the iscR gene and the sequences flanking in both iscR termini was PCR amplified from PAO1 genomic DNA with the primers BT3578 and BT3579 and cloned into a pUC18 plasmid cut with SmaI, yielding pUCiscR. The KpnI (blunted with T4 DNA polymerase) fragment containing a gentamicin resistance (Gmr) cassette flanked with loxP sequences from pUC18Gm (pUC18 containing loxP-flanked Gmr, which was constructed by inserting SacI-EcoRI fragments containing loxP-flanked Gmr from pCM351 [30] into pUC18 cut with the same enzymes) was cloned into pUCiscR digested with SmaI and EcoRV, generating pUCΔiscR::Gmr. Digestion with SmaI and EcoRV deleted 114 bp of the iscR coding sequence. pUCΔiscR::Gmr was transferred into PAO1, and the putative ΔiscR mutants that arose from a double crossover event were selected for the Gmr and Cbs phenotypes. An unmarked iscR mutant was created using the Cre-loxP system to excise the Gmr gene as previously described [30], and deletion of iscR was confirmed by Southern blot analysis. A pBBRiscR for ectopic expression of iscR was constructed by amplifying the full-length iscR from the PAO1 genomic DNA with primers EBI121 and BT3210. The 553-bp PCR products were cloned into medium-copy-number plasmid pBBR1MCS-4 [32] cut with SmaI, yielding pBBRiscR. A single-copy complementation was done using mini-Tn7 system [29]. The full-length iscR was cloned into pUC18-mini-Tn7T-Gm-LAC prior to transposing into the ΔiscR mutant, generating the complemented strain ΔiscR::iscR. Confirmation of transposition was carried out as previously described [29].
Site-directed Mutagenesis of IscR
Site-directed mutagenesis was performed to convert cysteine (C92, C98, C104, C111), histidine (H107), or glutamic acid (E43) residues to alanine residues through PCR-based mutagenesis as previously described [20], [33]. To construct pBBRiscRC92A for the expression of IscR-C92A, two pairs of primers EBI103 - M13R and EBI102 - M13F, were used in two-step PCR using pBBRiscR as a template. The PCR product was digested with EcoRI and SacI prior to cloning into pBBR1MCS-4, generating pBBRiscR-C92A. pBBRiscR-C98A, pBBRiscR-C104A, pBBRiscR-H107A, pBBRiscR-C111A and pBBRiscR-E43A were constructed using the same protocol with different sets of mutagenic primers: EBI142 and BT143 for C98A, EBI144 and EBI145 for C104A, EBI192 and EBI193 for H107A, BT3584 and BT3585 for C111A, and EBI148 and EBI149 for E43A. The presence of each mutation was verified by DNA sequencing.
Primer Extension
Primer extension experiments were performed using 10 µg of total RNA, 32P-labelled primer BT3577, 200 U Superscript II RNaseH minus M-MLV reverse transcriptase (Life Technologies, USA) and incubated at 42°C for 60 min. The primer extension products were analysed on a sequencing gel (8% polyacrylamide-7 M urea) next to DNA ladders generated using PCR sequencing kit using BT3577 and a 294-bp iscR promoter fragment amplified with primers BT3612 and BT3613 as template.
qRT- PCR
Real-time reverse transcription PCR (qRT-PCR) was performed as previously described [31]. qPCR was performed using 10 ng of cDNA and primer pairs specific for iscR (EBI120 and BT3613), fdx2 (BT3555 and BT3556) and the 16S rRNA gene (BT2781 and BT2782) for 40 cycles of denaturation at 95°C for 10 s, annealing at 60°C for 30 s, and extension at 60°C for 30 s. The iscR forward primer EBI120 corresponds to the sequence from +1 to +19 (transcribed region outside the iscR coding region) of the iscR transcript. Relative expression was shown as fold change relative to the level of wild-type PAO1 grown under uninduced conditions. The experiments were independently repeated three times and the means ± standard deviations (SD) are shown.
Purfication of IscR and UV-visible Spectroscopy
Purification of 6xHis-tagged wild-type and mutant IscR proteins from P. aeruginosa was carried out using the pET-Blue2 expression system as previously described [20]. The purity of IscR protein was more than 90% as judged by a major band corresponding to the 18 kDa protein observed in SDS-PAGE. The UV-visible spectroscopy analysis of various mutated IscR variants were carried out using Shimadzu UV-1800 spectrophotometer. Equal amount of proteins (10 µM) was prepared in 50 mM phosphate buffer containing 300 mM NaCl, pH 7.0. BSA (10 µM) was used as a control. Spectroscopy analysis was immediately done after protein purification step to avoid the oxidation of the iron-sulphur cluster of the IscR proteins.
Preparation of Polyclonal Anti-IscR Antibody
Polyclonal anti-IscR antibody was prepared by immunizing a rabbit with purified 6xHis-tagged P. aeruginosa IscR (conducted by the Biomedical Technology Research Unit, Faculty of Associated Medical Sciences, Chiang Mai University, Thailand).
Western Blot Analysis
Bacterial cell lysates prepared from exponential phase cultures were separated using 12.5% SDS-PAGE and transferred to a polyvinyl difluoride (PVDF) membrane (Bio-Rad). The blotted membrane was blocked with skim milk in Tris-Buffered Saline Tween-20 (TBST) before being probed with a 1∶25,000 dilution of rabbit anti-IscR antibody as the primary antibody. Goat anti-rabbit immunoglobulin G, pAB (horseradish peroxidase [HRP] conjugated) (Enzo Life Sciences) was used as the secondary antibody. Antigen-antibody complexes were detected using chemiluminescent HRP substrate (GE Healthcare, Germany) and Hyperfilm (GE healthcare Life Sciences). Band intensity was measured using an Imagescanner (GE healthcare Life Sciences) and ImageQuant v. 1.2 software (Molecular dynamics).
Inductively Coupled Plasma Mass Spectrometry (ICP-MS)
One milliliter of the exponential phase culture (adjusted to OD600 nm of 1) was washed and resuspended in 50 mM potassium phosphate buffer solution (PBS). The cells were treated 3 times with 100 µM ethylenediamine-N, N′-diacetic acid (EDDA) in PBS prior to treatment with 300 µl of 60% ultrapure nitric acid and heated at 50°C for 18 h. The mixture was adjusted to a total volume of 2.500 ml with autoclaved Milli-Q water (ICP grade). The iron content was measured using ICP-MS. The colony forming units for each strain were determined using the viable count method and expressed as fg iron CFU−1.
Plate Sensitivity Assay
A plate sensitivity assay was performed to determine the oxidant resistance level as previously described [20], [31]. Briefly, exponential phase cells were adjusted to OD600 nm of 0.1 before making 10-fold serial dilutions. Then, 10 µl of each dilution was spotted onto LB agar plates containing appropriate concentrations of testing reagents. The plates were incubated at 37°C for overnight before the surviving colonies were scored. The resistance level against an oxidant was expressed as the % survival, defined as the percentage of the CFU on plates containing oxidant divided by the CFU on plates without oxidant. For anaerobic conditions, the cultured plates were incubated in an anaerobic jar containing an anaerobic gas pack (AnaeroGEN™, Oxoid, UK) for 48 h.
CAS Assay for Siderophore Production
Siderophore production was determined using a Chrome azural S (CAS) agar diffusion assay as previously described [34]. Siderophore production is detected by the presence of a yellow halo zone around bacterial spot.
Cytochrome c Oxidase Assay
Cytochrome c oxidase activity was spectrophotometrically measured as previously described [35]. One unit of cytochrome c oxidase activity is defined as the amount of enzyme required to oxidize 1 µmol ferrocytochrome c min−1 at 25°C, pH 7.0.
Acknowledgments
We would like to thank Sumontha Nookabkaew for technical assistance, John D. Helmann for valuable suggestions, and James M. Dubbs for his critical reading of the revised manuscript.
Funding Statement
This work was supported by grants Center of Excellence on Environmental Health and Toxicology, Ministry of Education, Thailand (http://www.etm.sc.mahidol.ac.th/index.asp) and by the Royal Golden Jubilee Ph.D. Program (PHD/0294/2550) from the joint funding of Thailand Research Fund and Mahidol University (http://rgj.trf.or.th/eng/rgje11.asp). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Parts of this work are from A.R. dissertation submitted for a Ph.D. degree from Mahidol University.
References
- 1. Py B, Barras F (2010) Building Fe-S proteins: bacterial strategies. Nat Rev Microbiol 8: 436–446. [DOI] [PubMed] [Google Scholar]
- 2. Ayala-Castro C, Saini A, Outten FW (2008) Fe-S cluster assembly pathways in bacteria. Microbiol Mol Biol Rev 72: 110–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jacobson MR, Cash VL, Weiss MC, Laird NF, Newton WE, et al. (1989) Biochemical and genetic analysis of the nifUSVWZM cluster from Azotobacter vinelandii . Mol Gen Genet 219: 49–57. [DOI] [PubMed] [Google Scholar]
- 4. Roche B, Aussel L, Ezraty B, Mandin P, Py B, et al. (2013) Iron/sulfur proteins biogenesis in prokaryotes: formation, regulation and diversity. Biochim Biophys Acta 1827: 455–469. [DOI] [PubMed] [Google Scholar]
- 5. Fontecave M, Ollagnier-de-Choudens S (2008) Iron-sulfur cluster biosynthesis in bacteria: Mechanisms of cluster assembly and transfer. Arch Biochem Biophys 474: 226–237. [DOI] [PubMed] [Google Scholar]
- 6. Yao H, Jepkorir G, Lovell S, Nama PV, Weeratunga S, et al. (2011) Two distinct ferritin-like molecules in Pseudomonas aeruginosa: the product of the bfrA gene is a bacterial ferritin (FtnA) and not a bacterioferritin (Bfr). Biochemistry 50: 5236–5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Heo YJ, Chung IY, Cho WJ, Lee BY, Kim JH, et al. (2010) The major catalase gene (katA) of Pseudomonas aeruginosa PA14 is under both positive and negative control of the global transactivator OxyR in response to hydrogen peroxide. J Bacteriol 192: 381–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Giel JL, Nesbit AD, Mettert EL, Fleischhacker AS, Wanta BT, et al. (2013) Regulation of iron-sulphur cluster homeostasis through transcriptional control of the Isc pathway by [2Fe-2S]-IscR in Escherichia coli . Mol Microbiol 87: 478–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee KC, Yeo WS, Roe JH (2008) Oxidant-responsive induction of the suf operon, encoding a Fe-S assembly system, through Fur and IscR in Escherichia coli . J Bacteriol 190: 8244–8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nesbit AD, Giel JL, Rose JC, Kiley PJ (2009) Sequence-specific binding to a subset of IscR-regulated promoters does not require IscR Fe-S cluster ligation. J Mol Biol 387: 28–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rajagopalan S, Teter SJ, Zwart PH, Brennan RG, Phillips KJ, et al. (2013) Studies of IscR reveal a unique mechanism for metal-dependent regulation of DNA binding specificity. Nat Struct Mol Biol 20: 740–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schwartz CJ, Giel JL, Patschkowski T, Luther C, Ruzicka FJ, et al. (2001) IscR, an Fe-S cluster-containing transcription factor, represses expression of Escherichia coli genes encoding Fe-S cluster assembly proteins. Proc Natl Acad Sci U S A 98: 14895–14900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fleischhacker AS, Stubna A, Hsueh KL, Guo Y, Teter SJ, et al. (2012) Characterization of the [2Fe-2S] cluster of Escherichia coli transcription factor IscR. Biochemistry 51: 4453–4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Desnoyers G, Morissette A, Prevost K, Masse E (2009) Small RNA-induced differential degradation of the polycistronic mRNA iscRSUA . EMBO J 28: 1551–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yeo WS, Lee JH, Lee KC, Roe JH (2006) IscR acts as an activator in response to oxidative stress for the suf operon encoding Fe-S assembly proteins. Mol Microbiol 61: 206–218. [DOI] [PubMed] [Google Scholar]
- 16. Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, et al. (2000) Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406: 959–964. [DOI] [PubMed] [Google Scholar]
- 17. Winsor GL, Lo R, Ho Sui SJ, Ung KS, Huang S, et al. (2005) Pseudomonas aeruginosa genome database and PseudoCAP: facilitating community-based, continually updated, genome annotation. Nucleic Acids Res 33: D338–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Choi YS, Shin DH, Chung IY, Kim SH, Heo YJ, et al. (2007) Identification of Pseudomonas aeruginosa genes crucial for hydrogen peroxide resistance. J Microbiol Biotechnol 17: 1344–1352. [PubMed] [Google Scholar]
- 19. Kim SH, Lee BY, Lau GW, Cho YH (2009) IscR modulates catalase A (KatA) activity, peroxide resistance and full virulence of Pseudomonas aeruginosa PA14. J Microbiol Biotechnol 19: 1520–1526. [DOI] [PubMed] [Google Scholar]
- 20. Somprasong N, Jittawuttipoka T, Duang-Nkern J, Romsang A, Chaiyen P, et al. (2012) Pseudomonas aeruginosa thiol peroxidase protects against hydrogen peroxide toxicity and displays atypical patterns of gene regulation. J Bacteriol 194: 3904–3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Giel JL, Rodionov D, Liu M, Blattner FR, Kiley PJ (2006) IscR-dependent gene expression links iron-sulphur cluster assembly to the control of O2-regulated genes in Escherichia coli . Mol Microbiol 60: 1058–1075. [DOI] [PubMed] [Google Scholar]
- 22. Dos Santos PC, Johnson DC, Ragle BE, Unciuleac MC, Dean DR (2007) Controlled expression of nif and isc iron-sulfur protein maturation components reveals target specificity and limited functional replacement between the two systems. J Bacteriol 189: 2854–2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rincon-Enriquez G, Crete P, Barras F, Py B (2008) Biogenesis of Fe/S proteins and pathogenicity: IscR plays a key role in allowing Erwinia chrysanthemi to adapt to hostile conditions. Mol Microbiol 67: 1257–1273. [DOI] [PubMed] [Google Scholar]
- 24. Cooper SR, McArdle JV, Raymond KN (1978) Siderophore electrochemistry: relation to intracellular iron release mechanism. Proc Natl Acad Sci U S A 75: 3551–3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hassett DJ, Sokol PA, Howell ML, Ma JF, Schweizer HT, et al. (1996) Ferric uptake regulator (Fur) mutants of Pseudomonas aeruginosa demonstrate defective siderophore-mediated iron uptake, altered aerobic growth, and decreased superoxide dismutase and catalase activities. J Bacteriol 178: 3996–4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shin DH, Choi YS, Cho YH (2008) Unusual properties of catalase A (KatA) of Pseudomonas aeruginosa PA14 are associated with its biofilm peroxide resistance. J Bacteriol 190: 2663–2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cornelis P, Matthijs S, Van Oeffelen L (2009) Iron uptake regulation in Pseudomonas aeruginosa . Biometals 22: 15–22. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Russell DW (2001) Molecular Cloning: A Laboratory Manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 29. Choi KH, Schweizer HP (2006) Mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa . Nat Protoc 1: 153–161. [DOI] [PubMed] [Google Scholar]
- 30. Marx CJ, Lidstrom ME (2002) Broad-host-range cre-lox system for antibiotic marker recycling in gram-negative bacteria. Biotechniques 33: 1062–1067. [DOI] [PubMed] [Google Scholar]
- 31. Romsang A, Atichartpongkul S, Trinachartvanit W, Vattanaviboon P, Mongkolsuk S (2013) Gene expression and physiological role of Pseudomonas aeruginosa methionine sulfoxide reductases during oxidative stress. J Bacteriol 195: 3299–3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, et al. (1995) Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166: 175–176. [DOI] [PubMed] [Google Scholar]
- 33. Panmanee W, Vattanaviboon P, Poole LB, Mongkolsuk S (2006) Novel organic hydroperoxide-sensing and responding mechanisms for OhrR, a major bacterial sensor and regulator of organic hydroperoxide stress. J Bacteriol 188: 1389–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jittawuttipoka T, Sallabhan R, Vattanaviboon P, Fuangthong M, Mongkolsuk S (2010) Mutations of ferric uptake regulator (fur) impair iron homeostasis, growth, oxidative stress survival, and virulence of Xanthomonas campestris pv. campestris. Arch Microbiol 192: 331–339. [DOI] [PubMed] [Google Scholar]
- 35. Saenkham P, Vattanaviboon P, Mongkolsuk S (2009) Mutation in sco affects cytochrome c assembly and alters oxidative stress resistance in Agrobacterium tumefaciens . FEMS Microbiol Lett 293: 122–129. [DOI] [PubMed] [Google Scholar]