Abstract
Endo-β-mannanase (EC 3.2.1.78) is involved in cell wall disassembly and the weakening of plant tissues by degrading mannan polymers in the cell walls. Endo-β-mannanase genes are expressed in tomato (Lycopersicon esculentum) seeds (LeMAN1 and LeMAN2) and fruits (LeMAN3 and LeMAN4). A novel endo-β-mannanase gene (termed LeMAN5) was found in the tomato genome by genome-walking PCR and bacterial artificial chromosome library screening. The 5′-upstream region of this endo-β-mannanase gene contained four copies of the pollen-specific cis-acting elements POLLEN1LELAT52 (AGAAA). A GUS-reporter gene driven with the putative LeMAN5 promoter (-543 to +38) was activated in anthers and pollen of transgenic Arabidopsis, with the highest β-glucuronidase activity detected in pollen. β-Glucuronidase expression was detected in mature pollen retained in sporangia, discharged pollen, and elongating pollen tubes in transgenic Arabidopsis. Consistently, expression of LeMAN5 mRNA and endo-β-mannnanase activity was detected in tomato anthers and pollen. In anthers, the highest mRNA expression and endo-β-mannanase activity were detected during late stages of anther development, when pollen maturation occurred. Endo-β-mannanase activity was present in discharged pollen, which was easily eluted in a buffer, indicating that the enzyme proteins are probably secreted from, and deposited on, the surface of pollen. These data suggest that the LeMAN5 endo-β-mannanase is associated with anther and pollen development.
Cell wall disassembly plays significant roles in plant growth and development, including embryogenesis, seed germination, shoot growth, leaf formation, flower development, and fruit ripening. Primary cell walls consist of cellulose microfibrils and xyloglucans, which are rearranged most likely by the action of expansins and xyloglucan endotransglycosylases when cells expand and grow (Cosgrove, 1999). Primary cell walls are often reinforced by deposition of pectins and hemicelluloses such as β-1,3-glucan (callose) and mannans, which constitute secondary cell walls (Carpita and Gibeaut, 1993). Endo-β-mannanase (EC 3.2.1.78) is involved in degradation of Man polymers, such as galacto-, gluco-, and galactoglucomannans, and modifies the properties of cell walls in different tissues (Bewley et al., 1997, 2000). Degradation of cell wall mannans is also important for reserve mobilization in plant tissues (e.g. fruits and seeds; Daud and Jarvis, 1992; Bewley and Black, 1994). Despite the potential involvement of endo-β-mannanase in many aspects of plant growth and development by changing the mechanical properties of cell walls and providing nutrition, characterization of this enzyme is limited to only a few plant tissues.
Endo-β-mannanases in seed endosperm have been relatively well characterized. The first endo-β-mannanase gene isolated from tomato (Lycopersicon esculentum) was from germinated seeds (LeMAN1; Bewley et al., 1997). This gene is expressed in the whole endosperm of tomato seeds after germination and is involved in reserve mobilization of mannans stored in endosperm cell walls (Bewley et al., 1997; Nonogaki et al., 2000). Another mannanase gene, LeMAN2, is expressed in germinating tomato seeds. Expression of this gene occurs exclusively in the micropylar region of the endosperm (endosperm cap), which provides a mechanical resistance to radicle expansion. LeMAN2 endo-β-mannanase is involved in weakening of the endosperm cap to allow radicle emergence from seeds (Nonogaki et al., 2000). Endo-β-mannanase activity is also detected in germinating and germinated seeds of a wide range of plants (Halmer et al., 1976; Sánchez and de Miguel, 1997; Marraccinni et al., 2001; Williams et al., 2001; Homrichhausen et al., 2003). Thus, expression of endo-β-mannanase in seeds is widespread.
At least two different endo-β-mannanase genes, LeMAN3 and LeMAN4, are expressed in tomato fruit during ripening. Endo-β-mannanase activity is detected mainly in the skin of tomato fruit, where the enzyme proteins exist in active or inactive form (Bewley et al., 2000; Banik et al., 2001). The biochemical properties of the fruit-associated endo-β-mannanase proteins are well characterized (Bourgault and Bewley, 2002). The endo-β-mannanase detected in fruit is possibly involved in fruit softening through modification of cell walls, although a cause-and-effect relationship between the enzyme and ripening has not been established (Bewley et al., 2000).
Compared with seeds and fruits, endo-β-mannanase expression in other plant tissues is not well investigated. We isolated and characterized expression of a new endo-β-mannanase gene, LeMAN5, from the tomato genome. The putative promoter of this gene drove expression of a reporter transgene in anthers and pollen of Arabidopsis. The potential roles of this new endo-β-mannanase in tomato anther and pollen will be discussed.
RESULTS
Isolation of a Novel Mannanase Gene LeMAN5 from Tomato
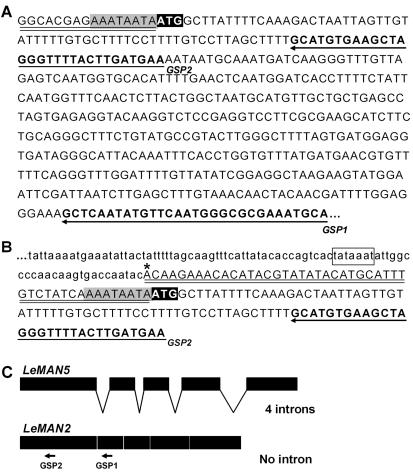
While attempting to isolate and clone the genomic DNA-flanking LeMAN2 (using genome-walking PCR coupled with a combination of adapter- and gene-specific primers [GSP, Fig. 1]), we cloned a portion of a putative new endo-β-mannanase gene in the tomato genome (cv Moneymaker) as follows. The first PCR amplification of genomic DNA, using GSP1 and an adaptor-specific primer, produced an approximately 1.3-kb PCR product. A second PCR amplification of the first-round PCR product with nested GSP2 and adaptor primers yielded a relatively shorter DNA fragment (approximately 0.7 kb) that was cloned and sequenced. The GSP2 primer sequence at the 3′ end of this fragment flanked DNA sequence identical to the 5′ region of LeMAN2 cDNA (Fig. 1B). A putative TATA box (Fig. 1B, square) and a transcription initiation site (Fig. 1B, asterisk) were predicted in the 5′-upstream region of this DNA fragment by the Neural Network Promoter Prediction (http://www.fruitfly.org/seqtools/promoter.html; Reese and Eeckman, 1995). These results initially suggested that the isolated fragment contained the promoter and the coding regions of LeMAN2. However, although the coding region and the 3′ end of the untranslated regions (Fig. 1, A and B, gray shades) of this fragment were identical to their respective regions of LeMAN2 cDNA (Fig. 1, A and B), unique sequences were found in the 5′-untranslated regions of this gene fragment (Fig. 1, A and B, underlines). The 5′ sequence of this gene matched none of the other known endo-β-mannanase genes in tomato (LeMAN1, Bewley et al., 1997; LeMAN3, GenBank accession no. AF290893; LeMAN4, Bourgault and Bewley, 2002). Taken together, this suggested that the amplified gene fragment represented a novel tomato endo-β-mannanase gene.
Figure 1.
A, Sequence of the 5′ region of tomato seed germinationassociated endo-β-mannanase LeMAN2 cDNA. The first ATG in the coding region is highlighted by reverse shading. The 5′-untranslated region is indicated by double underlining. The sequences used to design GSP1 and GSP2 for genome-walking PCR are highlighted in bold, and their positions are indicated by arrows. The sequence that was also conserved in the untranslated region of genome-walking PCR product (see below) is highlighted by gray shading. B, DNA sequence of a portion of the 0.7-kb product obtained by genome-walking PCR. The predicted TATA-box and transcription initiation site are highlighted by a square and an asterisk, respectively. The DNA sequence in the untranscribed region is shown in small letters and the untranslated region in the putative cDNA is shown by double underlining. The sequence highlighted by gray shading (AAATAATA) was also found in the 3′ end of untranslated region LeMAN2 cDNA (Fig. 1A). The 5′-upstream sequence of the promoter region is not shown. C, The structure of LeMAN2 and LeMAN5 genes. The positions of GSP1 and GSP2 primers in LeMAN2 gene are shown with arrows.
To obtain the genomic DNA sequence of this putative endo-β-mannanase, a bacterial artificial chromosome (BAC) library LA483 of the wild tomato (Lycopersicon cheesmanii; obtained from the National Science Foundation Tomato Genome Project, Cornell University, Ithaca, NY) was screened using labeled LeMAN2 cDNA as a probe. Fifteen positive clones were obtained from the BAC library screening. Clone 241 P6 contained a DNA sequence identical with the sequence of the 0.7-kb DNA fragment isolated by genome walking PCR. By sequencing portions of this BAC clone with LeMAN2-specific primers, the DNA sequence of the 3′ end of the endo-β-mannanase gene was obtained. Primers specific for the 5′ and the 3′ end of the endo-β-mannanase gene in wild tomato were used to PCR amplify the whole gene from the genomic DNA of tomato cv Moneymaker, yielding an approximately 3.7-kb PCR product that was sequenced. The sequence of the 5′ region of this gene was identical to the 0.7-kb fragment isolated by genome-walking PCR. This putative endo-β-mannanase gene encoded four putative introns in contrast to the LeMAN2 gene, which does not encode any introns (Fig. 1C). The predicted cDNA showed 98% identity to LeMAN2 cDNA, and the entire coding region of this gene was identical to that of LeMAN2. This gene was termed LeMAN5 and the genomic DNA sequence was deposited in the GenBank (accession no. AY102168).
Characterization of LeMAN5 Promoter in Arabidopsis
A search for cis-acting elements in the 5′-upstream region of LeMAN5 (-543 to +38) using plant cis-acting regulatory DNA elements (http://www.dna.affrc.go.jp/htdocs/PLACE/; Higo et al., 1999) indicated that the putative promoter contained four copies of the pollen-specific motifs (AGAAA) (POLLEN1LELAT52, Bate and Twell, 1998) and some other motifs associated with seed storage protein genes (CAATBOX1, Shirsat et al., 1989; EBOXBN-NAPA, Stalberg et al., 1996; and CANBNNAPA, Ellerstrom et al., 1996). To characterize tissue specificity of LeMAN5 expression, the 5′-upstream region of this gene (-543 to +38) was cloned into a promoterless vector encoding a GUS gene to create a reporter vector. Arabidopsis plants were transformed with Agrobacterium tumefaciens harboring this reporter-gene cassette and more than 20 T1 transformants were obtained after screening for kanamycin resistance. By examining the segregation patterns of kanamycin-resistant to kanamycin-sensitive plants in subsequent generations, lines homozygous for a single transgene locus were isolated and used for further assay. The presence of the transgene in these plants was confirmed by PCR with transgene-specific primers.
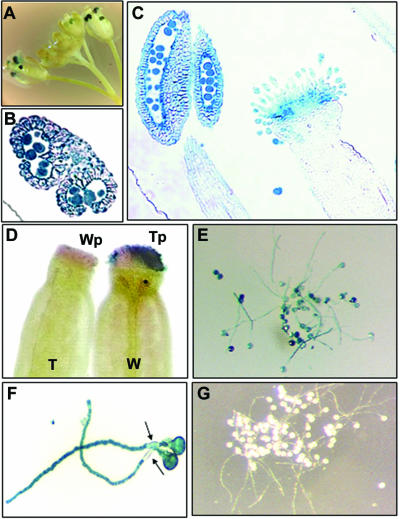
Expression of the GUS-reporter gene was examined in different tissues of the transgenic Arabidopsis. GUS activity was not detected in roots, stems, leaves, and siliques (data not shown). GUS staining was observed in some germinated seeds, but the intensity of staining was variable and inconsistent among seeds (data not shown). Intense GUS staining was observed in anthers of developing and opened flowers of the transformants (Fig. 2A), whereas wild-type Arabidopsis plants did not show GUS staining in flowers (data not shown). The highest GUS activity was detected at late stages of flower development (Fig. 2A). These results suggested that activation of the LeMAN5 promoter in anthers was developmentally regulated. Histochemical analysis was conducted to localize LeMAN5-promoter-GUS expression in Arabidopsis flowers in detail. GUS activity was detected in pollen retained in sporangia and anther walls, the activity in pollen being higher than in anther walls (Fig. 2B). Mature pollen released from anthers also showed strong GUS activity (data not shown). GUS staining in the stigma was also visible in tissue sections (Fig. 2C). To determine whether GUS activity emanated from the stigma itself or if the apparent stigmatic activity derived from transferred pollen, reciprocal pollination experiments were conducted between wild-type and transgenic plants. When anthers were removed from transgenic flowers, GUS staining was no longer observed in stigmas (data not shown). When stigmas of wild-type Arabidopsis were pollinated with transgenic pollen, GUS activity was detected in stigmas (Fig. 2D, W-Tp), indicating that the GUS signal on stigmas was most likely from GUS activity in pollen grains. However, it was possible that the stimulus caused by pollination might induce GUS expression in transgenic stigmatic tissue. To address this, wild-type Arabidopsis pollen was transferred onto stigmas of transgenic plant. No blue staining was detected in this experiment (Fig. 2D, T-Wp), confirming that GUS was not produced in the stigma itself but came from pollen. Not only pollen grains, but also elongated pollen tubes, showed intense staining (Fig. 2, E and F). The positions occupied by callose plugs did not stain blue (Fig. 2F, arrows), but growing pollen tube tips showed strong GUS activity, suggesting that GUS proteins are continuously present after the deposition of callose plugs. Pollen grains from wild-type Arabidopsis did not show GUS staining (Fig. 2G).
Figure 2.
Activation of LeMAN5 promoter in Arabidopsis flowers. Expression of the GUS-reporter gene driven with the 5′-upstream region (-543 to +38) of LeMAN5 was characterized. A, GUS staining of inflorescence of transgenic Arabidopsis. B, GUS staining in a cross section of anther of transgenic Arabidopsis. C, GUS staining in a longitudinal section of anther and pistil of transgenic Arabidopsis. D, GUS staining of the pistils of transgenic (T) and wild-type (W) Arabidopsis cross-pollinated with wild-type (Wp) and transgenic pollen (Tp), respectively. E and F, GUS staining of germinated pollen of transgenic Arabidopsis. Note that callose plugs are not stained (arrows). G, Germinated pollen of wild-type Arabidopsis in the GUS staining solution.
Expression of LeMAN5 mRNA and Endo-β-Mannanase Activity in Tomato Anther and Pollen
Characterization of the 5′-upstream sequence of LeMAN5 in Arabidopsis strongly suggested that this gene is expressed in anthers and pollen. Therefore, LeMAN5 gene expression was investigated in tomato anthers during floral development.
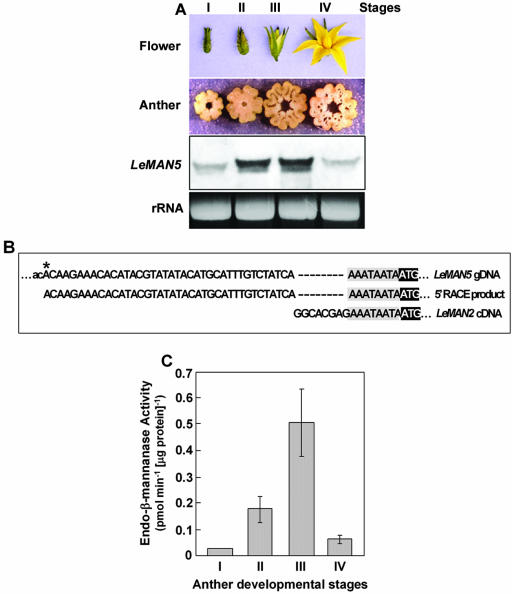
First, four different developmental stages of tomato flowers were characterized (stage I, unopened bud 5–9 mm; stage II, unopened bud 10–15 mm; stage III, opening flower; and stage IV, fully opened flower). The diameters of the cross-sections of anther cores at stages I through IV were approximately 2.0, 2.5, 3.0, and 3.0 mm, respectively (Fig. 3A, top two panels). The detailed morphology of tomato anthers was examined by tissue sectioning and toluidine blue staining and shown in the “Supplemental Data” online.
Figure 3.
Expression of LeMAN5 mRNA and endo-β-mannanase activity in developing tomato anther. A, Northern blot of total RNA extracted from anther at stages I through IV (top two panels show developmental stages of tomato flowers and anthers). The same amounts (5 μg) of total RNA were applied and the blot was probed with LeMAN2 riboprobe. Ethidium bromide-stained ribosomal RNA bands are shown under the RNA gel blots to indicate RNA loading in each lane. B, Comparison of the DNA sequence of 5′-RACE product obtained from stage III anther with LeMAN5 genomic DNA and LeMAN2 cDNA sequences. Only 5′ regions of the DNA sequences are shown. The first ATG in the coding region is highlighted by reverse shading. The sequence conserved in the three sequences is highlighted by gray shading. An asterisk in LeMAN5 gene indicates the predicted transcription initiation site. The DNA sequence in the untranscribed region is shown in small letters. C, Specific activities of endo-β-mannanase in developing anther during stages I through IV. The diameters of clearing zones in gel diffusion assay were measured and quantified using a commercial fungal (Aspergillus niger) mannanase (Megazyme, Wicklow, Ireland) as a standard. The data represent the averages of the two experiments. Vertical bars represent variations between two data points.
Total RNA was extracted from stage I through IV anthers, and RNA gel blotting was conducted using a LeMAN2 riboprobe. In these samples, the probe hybridized to a approximately 1.5-kb band, consistent with the predicted mRNA size of LeMAN2 and LeMAN5. Although the mRNA was barely detectable at stage I, mRNA expression increased during stages II and III, followed by a decrease at stage IV (Fig. 3A). A minor band under the major band was detected in the blot. It was not clear if this represented a partial degradation product, another endo-β-mannanase mRNA, or a nonspecific binding of the probe.
As predicted, LeMAN5 cDNA shared 98% sequence identity with LeMAN2 cDNA, the source of the riboprobe for RNA gel blotting. Therefore, it was impossible to distinguish whether the detected signal was indicative of LeMAN2 or LeMAN5 mRNA. To address this question, reverse transcriptase (RT)-PCR and 5′-RACE were performed using total RNA from stage III anthers with primers designed for the coding region of LeMAN5 (identical to that of LeMAN2). The sequence of the RT-PCR product matched the LeMAN2/LeMAN5 coding region (data not shown). To obtain the 5′-untranslated region of the cDNA, 5′-RACE was performed starting with the same stage III anther-derived total RNA. The sequence of the 5′-RACE product perfectly matched the LeMAN5 genomic DNA sequence (Fig. 3B), starting from the transcription initiation site predicted in the LeMAN5 genomic DNA (Fig. 3B, asterisk), but differed from the LeMAN2 sequence. The same result was obtained for total RNA extracted from stage II anthers and mature pollen as well (data not shown). These results indicated that LeMAN5 is expressed in tomato anthers and pollen. LeMAN2 transcripts were not found in these experiments.
Endo-β-mannanase activity in developing tomato anthers was measured by a gel diffusion assay (Downie et al., 1994). Enzyme activity was detected at all stages (I–IV) with a large increase between stages II and III. The activity decreased at stage IV (Fig. 3C), when pollen grains were already discharged and lost. The overall pattern of changes in endo-β-mannanase activity in developing anthers was consistent with that of endo-β-mannanase mRNA expression in this tissue (Fig. 3, A and C).
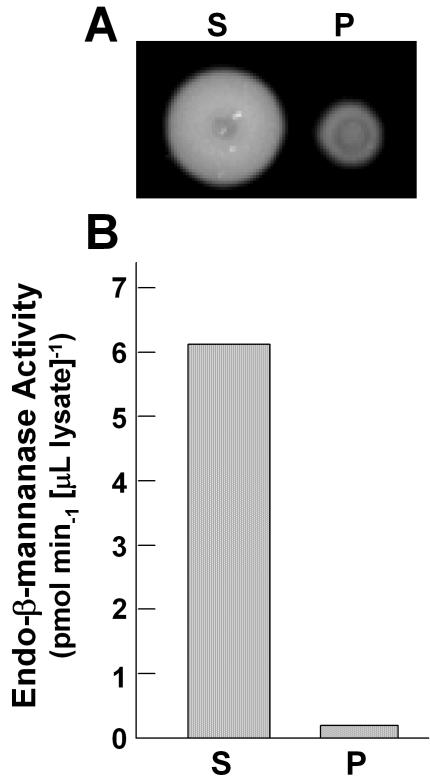
To examine whether endo-β-mannanase activity is present in tomato pollen, mature pollen grains were extracted from stage IV anthers using forceps. When the extracted pollen was suspended in buffer and applied to a gel diffusion assay plate, endo-β-mannanase activity was detected (data not shown), consistent with GUS expression in the transgenic Arabidopsis and detection of the LeMAN5 mRNA in tomato pollen. To determine if the enzyme proteins were retained in the pollen or present on its surface, a pollen suspension was centrifuged and the activities in the supernatant and the resuspended pollen were analyzed. Most endo-β-mannanase activity was present in the supernatant (Fig. 4). It was unlikely this activity was due to disruption of pollen grains by centrifugation because the precipitated pollen grains looked intact and germinated normally in pollen growth medium (data not shown). The enzyme protein is likely secreted from, and deposited on, the surface of pollen.
Figure 4.
Endo-β-mannanase activity in pollen. A, Gel diffusion assay. Pollen grains were extracted from stage IV tomato anthers by forceps and were then suspended in 50 mm K-phosphate buffer, pH 6.8. The sample was centrifuged at 10,000g for 2 min. The supernatant (S) and the resuspended pollen (P) were frozen, thawed, and applied to a gel diffusion assay plate. B, Endo-β-mannanase activities in the supernatant and pollen fraction were calculated as described above.
DISCUSSION
The predicted amino acid sequence of LeMAN5 is exactly the same as that of LeMAN2, which is expressed in tomato seeds. The recombinant LeMAN2 proteins expressed in bacteria show capacity to degrade galactomannans (Nonogaki et al., 2000). Therefore, LeMAN5 also encodes a functional enzyme. In fact, LeMAN5 mRNA and endo-β-mannanase activity were detected in tomato anther and pollen. To our knowledge, this is the first report on the expression of endo-β-mannanase in anthers and pollen.
In fruit and seeds, endo-β-mannanase is most likely associated with disassembly of cell walls to weaken or digest specific tissues (Bewley et al., 1997, 2000; Nonogaki et al., 2000; Banik et al., 2001). In developing tomato anthers, degradation of cell walls of the septum and weakening of the stomium occurs before anther dehiscence. The prominent morphological changes in tomato anthers take place at late stages of flower development (stages III and IV), which correspond with the timing of expression of LeMAN5 mRNA and endo-β-mannanase activity in this tissue. The activation of the putative promoter of the LeMAN5 gene in transgenic Arabidopsis flowers is also developmentally regulated with the highest activation at late stages of flower development. It is possible that LeMAN5 endo-β-mannanase is responsible for the weakening of the anther wall at the septum and dehiscence at the stomium.
It appears that the LeMAN5 endo-β-mannanase is also associated with pollen development and maturation because in Arabidopsis transformed with LeMAN5-promoter-driven GUS, high GUS activity is detected in pollen retained in sporangia as well as the tapetal layers, because strong GUS activity is continuously detected in discharged pollen in transgenic Arabidopsis, and because in tomato pollen, endogenous endo-β-mannanase activity is present after discharge. The decrease in LeMAN5 mRNA and endo-β-mannanase activity observed in stage IV tomato anthers may reflect the loss of pollen grains discharged from the upper part of the anther cores, which were already ruptured at this stage.
Endo-β-mannanase activity in mature pollen might be involved in germination and pollen tube elongation. In tomato seeds, endo-β-mannanase is localized exclusively at the endosperm cap (where the radicle emerges), and its activity degrades cell wall mannans, weakening the surrounding tissue and eventually permitting radicle protrusion (Groot et al., 1988). The pollen tube wall (intine) is bipartite: The inner sheath of callose is covered by an outer fibrillar layer composed of pectin with hemicellulose and cellulose (Taylor and Hepler, 1997). It is plausible that weakening of cell walls is also required in pollen, especially at an aperture, the site of pollen tube emergence. Interestingly, the same sets of cell wall-associated genes are expressed in seed and pollen germination (polygalacturonases, expansins, extensins, and endo-β-mannanases; Brown and Crouch, 1990; Rubinstein et al., 1995; Cosgrove et al., 1997; Sitrit et al., 1999; Chen and Bradford, 2000; Dubreucq et al., 2000; Nonogaki et al., 2000; this study). However, pollen and pollen tubes typically contain Glc-based polymers and it is not known whether their cell walls contain mannans. Furthermore, endo-β-mannanase activity was not retained in pollen cell walls, but was detected in the supernatant of washed pollen grains. This suggests that rather than playing a role in pollen germination or pollen tube elongation, the physiological role(s) of pollen endo-β-mannanase resides outside pollen itself. The endo-β-mannanase deposited on the surface of pollen might be involved in degradation of the cell walls of female tissues, such as the stigma or transmitting tissue in the pistil, being involved in the delivery of male gametes to female gametophytes in the ovules.
Although the causal relationship between endo-β-mannanase expression and anther and pollen development is yet to be established by using RNA interference or knockout plants, this opens a new insight into the functions of endo-β-mannanase in plant reproduction. Furthermore, the promoter of LeMAN5, which drives strong expression of a reporter gene in a stage-specific manner, is a potential tool to manipulate expression of foreign genes in anther and pollen, in addition to the previously described pollen-specific promoters (LAT52, Muschietti et al., 1994; DC3, Touraev et al., 1995). The LeMAN5 promoter might be used for pollen-fertility control, important for enhancing F1 hybrid seed production and preventing gene flow from transgenic plants.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Tomato (Lycopersicon esculentum cv Moneymaker) plants were grown in a greenhouse at 27°C with a 14-h light period. Arabidopsis (Colombia-0 ecotype) plants were grown at 22°C under cool-white fluorescent lights with a 12-h light period.
Genome Walking
Genomic DNA was extracted from tomato seedlings using phenol extraction according to the QUICK-PREP method described at http://www.biotech.wisc.edu/NewServicesandResearch/Arabidopsis/ and was used for genome-walking experiments. Genome-walking PCR was performed using a Genome Walker kit (Clontech Laboratories, Palo Alto, CA) according to the manufacturer's manual. Briefly, tomato (cv Moneymaker) genomic DNA was digested by DraI. Adaptor DNA provided in the kit was ligated to the DraI-digested genomic DNA fragments. The first PCR was conducted using an adaptor primer (5′-GTAATACGACTCACTATAGGGC-3′) provided by the kit and a GSP1 (5′-TGCATTTCGCGCCCATTGAACATATTGAGC-3′) that was complementary to the 5′ region of LeMAN2 cDNA. Second-round PCR was done using the nested adaptor primer (5′-ACTATAGGGCACGCGTGGT-3′) and GSP2 (5′-TTCATCAAGTAAAACCCTAGCTTCACATGC-3′) complementary to the upstream sequence of LeMAN2. The conditions used for the first and second PCR were one cycle at 94°C (for 4 min), one cycle at 80°C (for 2 min), seven cycles at 94°C (for 25 s) and at 72°C (for 3 min), 32 cycles at 94°C (for 25 s) and at 67°C (for 3 min) followed by one cycle at 67°C (for 7 min), one cycle at 94°C (for 4 min), one cycle at 80°C (for 2 min), five cycles at 94°C (for 25 s) and at 72°C (for 3 min), and 20 cycles at 94°C (for 25 s) and at 67°C (for 3 min) followed by one cycle at 67°C (for 7 min), respectively. The DNA amplified by PCR was cloned into TOPO pCR4.0 vector (Invitrogen, Carlsbad, CA) and was sequenced.
BAC Library Screening
The BAC filters of the library LA483 of wild tomato (Lycopersicon cheesmanii) were obtained from the National Science Foundation Tomato Genome Project (Cornell University) and were screened by hybridization with LeMAN2 cDNA probe labeled with enhanced chemiluminescence-labeling reagents (Amersham Bioscience, Piscataway, NJ). Overnight hybridization at 42°C in enhanced chemiluminescence gold buffer, including 5% (w/v) blocking reagent (Amersham Bioscience) and 0.5 m NaCl, followed a 1-h prehybridization at the same temperature. After hybridization, the membranes were washed twice for 20 min each at 42°C with 6 m urea, 0.5% (w/v) SDS, and 0.5× SSC, and twice for 5 min each at room temperature with 2× SSC. Detection was done using chemiluminescent substrate provided in the kit and with x-ray film. Bacterial stocks of the 15 positive clones were obtained from the Tomato Genome Project. LeMAN5 sequence was identified in a clone 241 P6, which was further sequenced to obtain the 5′-upstream and 3′-downstream sequence of this gene. The primers designed for the 5′ region (5′-TCGATCCCATAGACTAGAGA-3′) and 3′ region (5′-GTCTATAGCGAATCTAGTGAC-3′) of this sequence were used to amplify LeMAN5 gene from the genomic DNA of cv Moneymaker tomato.
Computer Analysis of DNA Sequences
The structure of LeMAN5 gene was analyzed using GENSCAN (http://genes.mit.edu/GENSCAN.html; Burge and Karlin, 1997). Predictions of eukaryotic promoter and transcription initiation sites were performed using the Neural Network Promoter Prediction (http://www.fruitfly.org/seqtools/promoter.html; Reese and Eeckman, 1995). Plant-specific promoter elements were identified using the software plant cis-acting regulatory DNA elements (Higo et al., 1999). Sequence alignment was done using Lasergene, v. 5.1 (DNASTAR, Madison, WI).
Construction of the Reporter Gene Cassette
The putative promoter in the 5′-upstream region of LeMAN5 gene (-543 to +38) was amplified using forward (5′-CTGAGCTCAAATGTCTACAAAGCTTA-3′) and reverse (5′-CAGTCGACTGATAGACAAATGCATGT-3′) primers, which contained SacI and SalI restriction enzyme sites at their 5′ ends, respectively. The PCR products were digested with SacI and SalI and were cloned to the SacI and SalI sites in a shuttle vector pRJG23 (Grebenok et al., 1997) that contained the uidA (GUS) gene. The promoter-GUS construct (3.9 kb) in pRJG23 was cleaved out with SacI and SpeI and was subcloned into SacI and XbaI sites in pGPTV-KAN binary vector (Becker et al., 1992).
Arabidopsis Transformation and Screening
For transformation, 200 mL of 1% (w/v) yeast extract, 1% (w/v) peptone, and 0.5% (w/v) NaCl medium (Weigel and Glazebrook, 2002) containing 50 μg mL-1 kanamycin, 50 μg mL-1 streptomycin, and 50 mm MOPS buffer, pH 6.0 was inoculated with 5 mL overnight culture of Agrobacterium tumefaciens EHA 105 strain (Hood et al., 1993) harboring the binary vector, which contained the LeMAN5 promoter-reporter construct, and was grown for an additional 16 h at 28°C with vigorous shaking. Cells were harvested by centrifugation at 6,000g with a centrifuge (Sorval RC-5B; DuPont Instruments, Wilmington, DE) at ambient temperature, resuspended in 400 mL of 5% (w/v) Suc solution containing 0.02% (v/v) of Silwet L-77 detergent (Lehle Seeds, Round Rock, TX), and used for transformation, which was performed using a floral dip method as described previously (Clough and Bent, 1998). Seeds were harvested and stored at room temperature. For screening, seeds were sterilized in 95% (v/v) ethanol for 1 min and 50% (v/v) bleach solution containing 0.1% (v/v) Tween 20 for 10 min, followed by several washes with sterile water. Kanamycin-resistant plants were selected by incubating plants for 14 d on 1% (w/v) phytoagar (Invitrogen, Rockville, MD) plates containing 4.3 mg mL-1 Murashige Skoog salts, 3% (w/v) Suc, and 25 μg mL-1 kanamycin.
GUS Staining of Arabidopsis Tissues
GUS staining of Arabidopsis tissues was performed as previously described (Weigel and Glazebrook, 2002). Plant tissues were incubated at 37°C overnight in 100 mm sodium phosphate buffer, pH 7.0, containing 0.1% (v/v) Triton X-100 and 2 mm 5-bromo-4-chloro-3-indolyl-β-GlcU, cyclohexyl ammonium salt (RPI Co., Mount Prospect, IL). Staining solution was subsequently replaced with 25%, 30%, 50%, and 70% (v/v) ethanol. For sectioning, GUS-stained tissues were dehydrated in ethanol and acetone series and were embedded into resin (Technovit 7100; Technovit, Kuzler Heraus, Germany) according to the manufacturer's instruction.
RNA Extraction and Gel Blotting
Total RNA was extracted from tomato anther cores using standard phenol-SDS extraction (Sambrook et al., 1989). Equal amounts (5 μg) of total RNA were separated on a 1.3% (w/v) agarose gel containing 7% (v/v) formaldehyde, transferred to a positively charged nylon membrane (Hybond-N+; Amersham Biosciences), and UV-cross linked. To make the RNA probe, LeMAN2 cDNA in pBSIIKS vector (Stratagene, La Jolla, CA) was transcribed using a digoxygenin-labeled NTP mixture (Roche Applied Science, Indianapolis) and T7 RNA polymerase (Ambion, Austin, TX). Overnight hybridization was done at 60°C in hybridization buffer containing 50% (v/v) deionized formamide, 4% (w/v) blocking reagent (Roche Applied Science), 0.2% (w/v) SDS, 5× SSC, and approximately 100 ng mL-1 RNA probe following 15 min of prehybridization at the same temperature. The membranes were washed once for 25 min with 2× SSC and 0.1% (w/v) SDS at 60°C and twice for 25 min with 0.2× SSC and 0.1% (w/v) SDS at 60°C. They were then blocked for 30 min with 5% (w/v) nonfat milk in 0.1 m maleic acid buffer, pH 7.5, containing 0.15 m NaCl and 0.3% (v/v) Tween 20 (buffer A) and were incubated with alkaline phosphate-conjugated antidigoxygenin antibody (1:15,000 dilution) for 1 h at 25°C. After washing with buffer A, the membranes were subjected to chemiluminescence detection. The signal was detected on x-ray film after about 30 min of exposure.
RT-PCR and 5′-RACE
Two micrograms of total RNA extracted from stage II and III anthers was used for RT with a RETROscript Kit (Ambion). The RT product was amplified by PCR using Ex TaqDNA polymerase (Takara, Madison, WI), the primers (5′-GCTAGGGTTTTACTTGATGAA-3′ and 5′-CTTATTTTCTTCCATGTCCTC-3′) and the following conditions: the initial denaturation at 94°C (for 4 min), 25 cycles at 94°C (for 1 min), at 50°C (for 1 min), and at 72°C (for 2 min) followed by extension at 72°C (for 7 min). The cDNA amplified by PCR was cloned into TOPO pCR2.1 vector (Invitrogen) and sequenced. A 5′-RACE was conducted using a RLM-RACE kit (Ambion) according to the manufacturer's manual.
Endo-β-Mannanase Activity Assay
Proteins were extracted from tomato anthers with 50 mm potassium phosphate buffer, pH 6.8. The extract was centrifuged at 10,000g for 2 min and the supernatant was used for gel diffusion assays of endo-β-mannanase activity (Downie et al., 1994). Enzyme activity was quantified using a commercial fungal (Aspergillus niger) mannanase (Megazyme) as a standard. Total soluble protein in crude extracts was measured according to Bradford (1976), using a protein assay kit (Bio-Rad, Hercules, CA).
Pollen Germination
Arabidopsis pollen was collected from stage IV flowers and was grown in a medium containing 0.29 m Suc,0.4 mm boric acid, and 1 mm calcium nitrate at 25°C for 6 h. Germinated pollen was collected by centrifugation at 900g and was used for GUS staining.
Acknowledgments
We thank the National Science Foundation Tomato Genome Project (DBI–9872617, Steve Tanksley, Gregory Martin, James Giovannoni, and Catharine Ronning, Cornell University, Ithaca, NY) for providing the BAC library and clones for our research. We also thank Kathy Cook, (Department of Botany and Plant Pathology, Oregon State University) for her technical assistance for tissue sectioning, and Jessica Hewitt (Department of Biochemistry and Biophysics, Oregon State University) and Tanja Homrichhausen (Department of Chemical Engineering, Oregon State University) for their assistance in preparing the manuscript.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.035998.
This work was supported by the National Science Foundation (grant no. IBN–0237562).
References
- Banik M, Bourgault R, Bewley JD (2001) Endo-β-mannanase is present in an inactive form in ripening tomato fruits of the cultivar Walter. J Exp Bot 52: 105-111 [PubMed] [Google Scholar]
- Bate N, Twell D (1998) Functional architecture of a late pollen promoter: Pollen-specific transcription is developmentally regulated by multiple stage-specific and co-dependent activator elements. Plant Mol Biol 37: 859-869 [DOI] [PubMed] [Google Scholar]
- Becker D, Kemper E, Schell J, Masterson R (1992) New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol Biol 20: 1195-1197 [DOI] [PubMed] [Google Scholar]
- Bewley JD, Banik M, Bourgault R, Feurtado JA, Toorop P, Hilhorst HWM (2000) Endo-β-mannanase activity increases in the skin and outer pericarp of tomato fruits during ripening. J Exp Bot 51: 529-538 [DOI] [PubMed] [Google Scholar]
- Bewley JD, Black M (1994) Seeds: Physiology of Development and Germination, Ed 2. Plenum Press, New York
- Bewley JD, Burton RA, Morohashi Y, Fincher GB (1997) Molecular cloning of a cDNA encoding a (1→4)-β-mannan endohydrolase from the seeds of germinated tomato (Lycopersicon esculentum). Planta 203: 454-459 [DOI] [PubMed] [Google Scholar]
- Bourgault R, Bewley JD (2002) Variation in its C-terminal amino acids determines whether endo-β-mannanase is active or inactive in ripening tomato fruits of different cultivars. Plant Physiol 130: 1254-1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of proteins utilizing the principle of protein-dye binding. Anal Biochem 2: 248-254 [DOI] [PubMed] [Google Scholar]
- Brown SM, Crouch ML (1990) Characterization of a gene family abundantly expressed in Oenothera organensis pollen that shows sequence similarity to polygalacturonase. Plant Cell 2: 263-274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge C, Karlin S (1997) Prediction of complete gene structures in human genomic DNA. J Mol Biol 268: 78-94 [DOI] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM (1993) Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J 3: 1-30 [DOI] [PubMed] [Google Scholar]
- Chen F, Bradford KJ (2000) Expression of an expansin is associated with endosperm weakening during tomato seed germination. Plant Physiol 124: 1265-1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735-743 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ (1999) Enzymes and other agents that enhance cell wall extensibility. Annu Rev Plant Physiol Plant Mol Biol 50: 391-417 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ, Bedinger P, Durachko DM (1997) Group I allergens of grass pollen as cell wall-loosening agents. Proc Natl Acad Sci USA 94: 6559-6564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daud MJ, Jarvis MC (1992) Mannan of oil palm kernel. Phytochemistry 31: 463-464 [Google Scholar]
- Downie B, Hilhorst HWM, Bewley JD (1994) A new assay for quantifying endo-β-mannanase activity using Congo Red dye. Phytochemistry 36: 829-835 [Google Scholar]
- Dubreucq B, Berger N, Vincent E, Boisson M, Pelletier G, Caboche M, Lepiniec L (2000) The Arabidopsis AtEPR1 extensin-like gene is specifically expressed in endosperm during seed germination. Plant J 23: 643-652 [DOI] [PubMed] [Google Scholar]
- Ellerstrom M, Stalberg K, Ezcurra I, Rask L (1996) Functional dissection of a napin gene promoter: identification of promoter elements required for embryo and endosperm-specific transcription. Plant Mol Biol 32: 1019-1027 [DOI] [PubMed] [Google Scholar]
- Grebenok RJ, Pierson E, Lambert GM, Gong FC, Afonso CL, Haldeman- Cahill R, Carrington JC, Galbraith DW (1997) Green-fluorescent protein fusions for efficient characterization of nuclear targeting. Plant J 11: 573-586 [DOI] [PubMed] [Google Scholar]
- Groot SPC, Kieliszewska-Rokicka B, Vermeer E, Karssen CM (1988) Gibberellin-induced hydrolysis of endosperm cell walls in gibberellin-deficient tomato seed prior to radicle protrusion. Planta 174: 500-504 [DOI] [PubMed] [Google Scholar]
- Halmer P, Bewley JD, Thorpe TA (1976) An enzyme to degrade lettuce endosperm cell walls: appearance of a mannanase following phytochrome- and gibberellin-induced germination. Planta 130: 189-196 [DOI] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res 27: 297-300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homrichhausen TM, Hewitt JR, Nonogaki H (2003) Endo-β-mannanase activity is associated with the completion of embryogenesis in imbibed carrot (Daucus carota L.) seeds. Seed Sci Res 13: 219-227 [Google Scholar]
- Hood EE, Gelvin SB, Melchers S, Hoekema A (1993) New Agrobacterium helper plasmids for gene transfer to plants (EHA105). Trans Res 2: 208-218 [Google Scholar]
- Marraccinni P, Rogers WJ, Allard C, André M-L, Caillet V, Lacoste N, Lausanne F, Michaux S (2001) Molecular and biochemical characterization of endo-β-mannanases from germinating coffee (Coffea arabica) grains. Planta 213: 296-308 [DOI] [PubMed] [Google Scholar]
- Muschietti J, Dircks L, Vancanneyt G, McCormick S (1994) LAT52 protein is essential for tomato pollen development: pollen expressing antisense LAT52 RNA hydrates and germinates abnormally and cannot achieve fertilization. Plant J 6: 321-328 [DOI] [PubMed] [Google Scholar]
- Nonogaki H, Gee OH, Bradford KJ (2000) A germination-specific endo-β-mannanase gene is expected in the micropylar endosperm cap of tomato seeds. Plant Physiol 123: 1235-1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese MG, Eeckman FH (1995) Novel Neural Network Algorithms for Improved Eukaryotic Promoter Site Recognition. Presented at the Seventh International Genome Sequencing and Analysis Conference, September 16–20, Hilton Head Island, South Carolina
- Rubinstein AL, Broadwater AH, Lowrey KH, Bedinger PA (1995) Pex1, a pollen-specific gene with an extensin-like domain. Proc Natl Acad Sci USA 92: 3086-3090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Sánchez RA, de Miguel L (1997) Phytochrome promotion of mannan-degrading enzyme activities in the micropylar endosperm of Datura ferox seeds requires the presence of embryo and gibberellin synthesis. Seed Sci Res 7: 27-33 [Google Scholar]
- Shirsat A, Wilford N, Croy R, Boulter D (1989) Sequences responsible for the tissue specific promoter activity of a pea legumin gene in tobacco. Mol Gen Genet 215: 326-331 [DOI] [PubMed] [Google Scholar]
- Sitrit Y, Hadfield KA, Bennett AB, Bradford KJ, Downie B (1999) Expression of a polygalacturonase associated with tomato seed germination. Plant Physiol 121: 419-428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalberg K, Ellerstom M, Ezcurra I, Ablov S, Rask L (1996) Disruption of an overlapping E-box/ABRE motif abolished high transcription of the napA storage-protein promoter in transgenic Brassica napus seeds. Planta 199: 515-519 [DOI] [PubMed] [Google Scholar]
- Taylor LP, Hepler PK (1997) Pollen germination and tube growth. Annu Rev Plant Physiol Plant Mol Biol 48: 461-491 [DOI] [PubMed] [Google Scholar]
- Touraev A, Fink CS, Stoger E, Heberle-Bors E (1995) Pollen selection: a transgenic reconstruction approach. Proc Natl Acad Sci USA 92: 12165-12169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J (2002) Arabidopsis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Williams HA, Bewley JD, Greenwood JS, Bourgault R, Mo B (2001) The storage cell walls in the endosperm of Asparagus officinalis L. seeds during development and following germination. Seed Sci Res 11: 293-303 [Google Scholar]