Abstract
Two biosynthetic pathways for ascorbate (l-ascorbic acid [AsA]; vitamin C) in plants are presently known, the mannose/l-galactose pathway and an l-GalUA pathway. Here, we present molecular and biochemical evidence for a possible biosynthetic route using myo-inositol (MI) as the initial substrate. A MI oxygenase (MIOX) gene was identified in chromosome 4 (miox4) of Arabidopsis ecotype Columbia, and its enzymatic activity was confirmed in bacterially expressed recombinant protein. Miox4 was primarily expressed in flowers and leaves of wild-type Arabidopsis plants, tissues with a high concentration of AsA. Ascorbate levels increased 2- to 3-fold in homozygous Arabidopsis lines overexpressing the miox4 open reading frame, thus suggesting the role of MI in AsA biosynthesis and the potential for using this gene for the agronomic and nutritional enhancement of crops.
l-Ascorbic acid (AsA) is the major antioxidant in plant cells. Since AsA was first isolated, there have been numerous reports on its role regulating redox potential during photosynthesis, environment-induced oxidative stress (ozone, UV, high light, SO2, etc), and wound- and pathogen-induced oxidative processes. In both plants and animals, AsA is important as a cofactor for numerous key enzymes such as hydroxylases and dioxygenases, some of which are involved in the biosynthesis of phytohormones and secondary metabolites or in the hydroxylation of specific peptidyl-prolyl and peptidyl-lisyl residues (Loewus and Loewus, 1987; Smirnoff et al., 2001; Arrigoni and De Tullio, 2002). Current data indicate that AsA is a major substrate for synthesis of l-tartaric, l-threonic, l-glyceric, and l-oxalic acids used for calcium regulation via calcium oxalate formation (Loewus, 1999). There is emerging evidence that AsA is involved in photoprotection, metal and xenobiotic detoxification, the cell cycle, cell wall growth, and cell expansion (Smirnoff, 2000; Smirnoff and Wheeler, 2000; Franceschi and Tarlyn, 2002). Interestingly, a recent study indicates that leaf AsA content can also modulate the expression of genes involved in plant defense and regulate genes that control development through hormone signaling (Pastori et al., 2003). The antioxidant property of AsA also is one of its major functions in humans (Homo sapiens), who are unable to synthesize their own vitamin C. Because AsA can neither be produced nor stored in the body, the vitamin must be acquired regularly from dietary sources, primarily from plants rich in AsA. Because ascorbate levels in plants vary widely, the ability to increase the level of this vitamin in crops would increase their nutritive value, shelf life, and ability to withstand oxidative stress during the growing season.
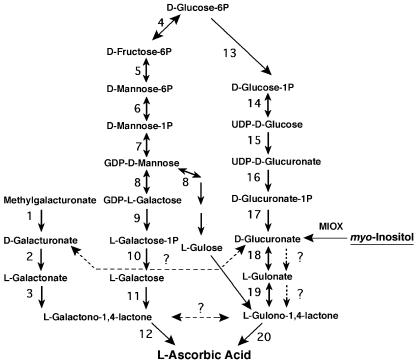
AsA biosynthetic pathways differ between animals and plants (Fig. 1). In animals, d-Glc is converted to AsA via d-glucuronate, l-gulonate, and l-gulono-1,4-lactone (l-GulL), which is then oxidized to AsA. In plants, AsA biosynthesis proceeds via GDP-Man, GDP-l-Gal, l-Gal-1-phosphate, l-Gal, and l-GalL (Wheeler et al., 1998). Although the AsA biosynthetic pathway proposed by Wheeler et al. is consistent with most available data, there is growing evidence indicating the existence of other pathways operating in plants that contribute to the AsA pool. Tracer and feeding studies have shown conversion of methyl-d-galacturonate and d-glucuronolactone to AsA in detached leaves of several plant species (Loewus, 1963) and Arabidopsis cell cultures (Davey et al., 1999). Recently, the cloning of a d-GalUA reductase from strawberry (Fragaria × ananassa) fruit, and its expression in Arabidopsis, provided molecular evidence of the use of d-galacturonate as a precursor for AsA biosynthesis (Agius et al., 2003). A 4- to 7-fold increase in the AsA content was obtained in lettuce (Lactuca sativa) and tobacco (Nicotiana tabacum) plants after constitutive expression of the rat (Rattus norvegicus) gene encoding l-GulL oxidase, the enzyme involved in the final step of the animal pathway (Jain and Nessler, 2000). It is still unclear if this enzyme works on the known plant AsA precursor, l-GalL, or if plants can produce l-GulL. Others have proposed the presence of a C3-epimerase that could catalyze the interconversion of l-GalL and l-GulL (Baig et al., 1970; Davey et al., 1999). Interestingly, recent metabolic profiling analysis has shown the presence of gulonate in Arabidopsis, suggesting the possibility of an AsA pathway in plants that resembles the one in animals (Wagner et al., 2003). Additional evidence indicating the complex network of metabolic pathways leading to AsA comes from the analysis of the vitamin C-deficient Arabidopsis mutants. The vtc2-1, vtc3-1, and vtc4-1 mutants are defective in AsA biosynthesis, but when several of the proposed AsA biosynthetic enzymes were assayed, their activities were not significantly different from those in wild type (Conklin et al., 2000; Smirnoff et al., 2001). None of these mutants appear to turn over AsA more rapidly than normal plants.
Figure 1.
Proposed biosynthetic network for AsA synthesis in plants (reactions 1–12), and animals (reactions 13–20). Four potential branch pathways operating in plants, the Man pathway, galacturonate pathway, the gulose pathway, and the MI pathway, are shown. Enzymes catalyzing the numbered reactions are: 1, methyl-esterase; 2, d-galacturonate reductase; 3, aldono-lactonase; 4, Glc-6-phosphate isomerase; 5, Man-6-phosphate isomerase; 6, phosphomannomutase; 7, GDP-Man pyrophosphorylase; 8, GDP-Man-3,5-epimerase; 9, phosphodiesterase; 10, sugar phosphatase; 11, l-Gal-1-dehydrogenase; 12, l-galactono-1,4-lactone (l-GalL) dehydrogenase; 13, phosphoglucomutase; 14, UDP-Glc pyrophosphorylase; 15, UDP-Glc dehydrogenase; 16, glucuronate-1-phosphate uridylyltransferase; 17, glucurono kinase; 18, glucuronate reductase; 19, aldono lactonase, and 20, guluno-1,4-lactone dehydrogenase. The reaction catalyzed by MIOX, and the possible pathway from MI to AsA (dashed arrows), are also shown. Question marks indicate enzymatic reactions that have not been demonstrated in plants.
Here, we used molecular tools to investigate the contribution of myo-inositol (MI) as a precursor of AsA biosynthesis in Arabidopsis. An ORF encoding a putative MI oxygenase (MIOX) was identified in chromosome 4 (miox4) of Arabidopsis. A PCR fragment encoding miox4 was expressed in Escherichia coli, and MIOX activity was confirmed in the affinity-purified recombinant protein. To our knowledge, this constitutes the first MIOX enzyme of plant origin that has been cloned and characterized. RNA gel-blot analysis in Arabidopsis shows that miox4 is expressed predominantly in flowers and leaves. Constitutive expression of miox4 enhanced the AsA content of the leaves 2- to 3-fold, demonstrating the feasibility of using this gene to engineer increased levels of this important antioxidant in plants. These results provide the first molecular evidence, to our knowledge, indicating the possible use of MI as a precursor of AsA biosynthesis in any organism.
RESULTS
Cloning and Characterization of an ORF Encoding a MIOX from Arabidopsis
A full-length cDNA that encodes a MIOX (EC 1.13.99.1) from pig (Sus scrofa) was isolated recently (GenBank accession no. AF401311) and characterized (Arner et al., 2001). We searched the Arabidopsis genome (at The Arabidopsis Information Resource; Huala et al., 2001) with the BLAST (Altschul et al., 1997) algorithm (TBLASTN version) for similar sequences. This resulted in five matching ORFs: At1g14520, At4g26260, At2g19800, At5g56640, and At5g08200, none of which have assigned functions. Alignment of these sequences with the pig miox cDNA revealed two segments for which the Arabidopsis sequences have very high similarity: 68 to 112 and 200 to 220. This high level of conservation suggests that these regions might be functional domains. Then, we used these two conserved subsequences of the pig miox to interrogate Inter Pro (Apweiler et al., 2000), the integrated protein documentation resource. Subsequence 68 to 112 matched the ProDom (Corpet et al., 2000) domain PD037591, whereas 200 to 220 had no match. The protein family containing domain PD037591 includes five Arabidopsis sequences and one mRNA from Pinus radiata D. Don embryo (GenBank accession no. AF049069) that also has high similarity to the original pig miox gene. The domain is annotated as “kidney specific,” presumably because of the origin of a number of animal sequences containing the domain. The five Arabidopsis sequences in this protein family correspond to four ORFs: At5g56640, At4g26260, At2g19800, and At1g14520 (in two separate BAC sequences). At5g56640 and At4g26260 have the same domain structure, containing additional domains PD330223 and PD348512. At1g14520 also contains PD330223 and PD354868. At2g19800 contains only the common domain (PD037591), as does the P. radiata sequence.
The coding region of the miox cDNA in chromosome 4 (miox4, ORF At4g26260) of Arabidopsis was isolated by PCR and sequenced (for details, see “Materials and Methods”). A BLAST (Altschul et al., 1997) search with the 957-bp PCR product revealed three changes at bases 233, 759, and 901 when compared with the published sequence. Two of those changes caused a substitution at the amino acid level (Q78 to R and K300 to E, GenBank accession no. AY232552). The molecular mass based on the translated amino acid sequence for MIOX4 was calculated to be 37 kD with a theoretical pI of 4.83.
MIOX is an enzyme-containing non-heme iron and catalyzes a four-electron oxidation with the transfer of only one atom of oxygen into the product d-glucuronate. This enzyme is of considerable interest physiologically because it catalyzes the first step of MI catabolism in plants (Loewus and Murthy, 2000), some yeast species (Molina et al., 1999), and also in the kidney and lens of several animals (Arner et al., 2001). The identity of MIOX4 was confirmed by expressing the candidate ORF in bacteria. The pET32a:miox4 construct was transformed into E. coli, and fusion protein expression was observed after 1 h of induction with isopropyl β-d-thiogalactoside by SDS-PAGE analysis. Supplementing the Luria-Bertani (LB) medium with a cocktail of folding promoting agents (250 mm Suc, 250 mm NaCl, 2 mm glutathione, and 1 mm Pro) increased the amount of soluble recombinant protein facilitating further purification steps. MIOX4 was purified from the soluble fraction of cell lysates by metal affinity chromatography; then, the fusion partner was removed by enterokinase digestion (for details, see “Materials and Methods”).
The MIOX4 specific activity obtained using the orcinol-based assay (Table I) is slightly higher than the activity of the only other MIOX enzyme expressed in bacteria, the enzyme from pig (Arner et al., 2001). Control reactions where an equivalent amount of protein of the empty vector (pET32a) or boiled MIOX4 enzyme gave no activity (Table I).
Table I.
Specific activity of recombinant MIOX4 from E. coli and MIOX enzymes from other organisms
| Protein Sample | Specific Activitya | Reference |
|---|---|---|
| Empty vector (pET32a) | n.d. | This work |
| MIOX4 | 2,174 ± 220 | This work |
| MIOX4 boiled | n.d. | This work |
| MIOX (pig) | 1,546 | Arner et al. (2001) |
| MIOX (yeast) | 198 | Molina et al. (1999) |
| MIOX (oat [Cryptococcus neoiformans Avena sativa]) | 57 | Koller et al. (1976) |
Specific activity in nanomoles glucuronic acid per minute per milligram of protein, mean value ± sd, n = 3. n.d., Not detectable
miox4 Is Predominantly Expressed in Flowers and Leaves, Tissues with a High AsA Concentration
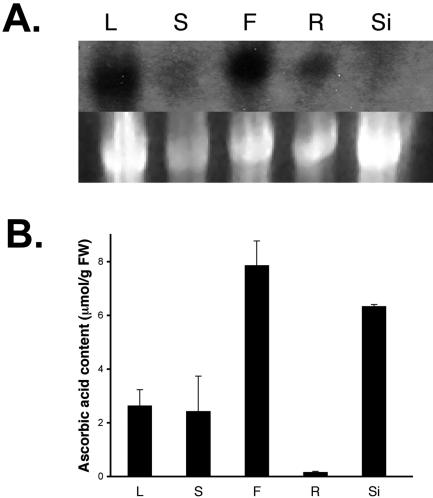
To gain a better understanding of the function of the miox4 gene in the physiology of Arabidopsis plants, hybridization experiments were carried out. Gel-blot analysis performed with total RNA purified from different tissues of Arabidopsis plants showed that miox4 is predominantly expressed in flowers and leaves and to a lesser extent in roots (Fig. 2A). The AsA content of Arabidopsis flowers has been found here (Fig. 2B) and by others (Conklin et al., 2000) to be 2 to 3 times greater than the AsA content of the leaves.
Figure 2.
The miox4 message is present in flowers and leaves, tissues with high concentration of AsA. A, RNA expression of miox4 in leaf, flower, and root tissues of 6-week-old Arabidopsis wild-type plants. rRNA of the samples stained with ethidium bromide is shown as a loading control in the lower panel. B, Mean ascorbic acid content of the corresponding plant tissues (n = 3). Bars = one sd. L, Leaf; S, stem; F, flower; R, root; Si, silique.
Constitutive Expression of miox4 Increases AsA Content in Arabidopsis
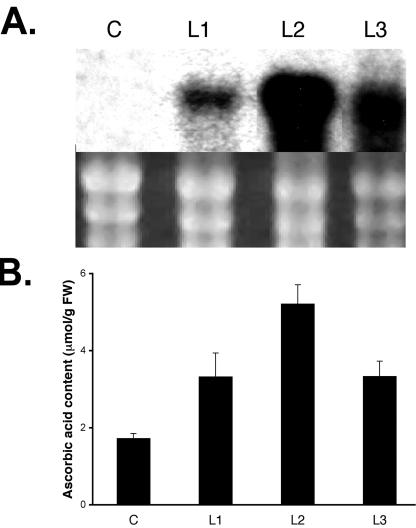
To study the potential contribution of MI as a precursor of AsA biosynthesis, the miox4 ORF was expressed under the control of the strong constitutive 35S promoter in Arabidopsis plants. Analysis of three independent homozygous lines (L1–L3) revealed a 2- to 3-fold increase in the AsA content of the leaves compared with wild type (Fig. 2B) and a line transformed with the empty vector pCAMBIA1380 growing under identical conditions (Fig. 3B). The increase in leaf AsA content in the overexpressers correlated with the amount of miox4 message detected by northern analysis (Fig. 3A).
Figure 3.
Constitutive expression of miox4 in Arabidopsis increases the ascorbic acid content of the leaves. A, RNA expression of miox4 in control (labeled as C, pCAMBIA1380), and three homozygous transgenic lines, numbered L1 to L3. rRNA of the samples stained with ethidium bromide is shown as a loading control in the lower panel. B, Mean ascorbic acid content in the leaf extract of the corresponding control (C) and transgenic lines L1 to L3 (n = 3). Bars = one sd.
DISCUSSION
We have demonstrated that the ORF identified on chromosome 4 of Arabidopsis encodes an enzyme that catalyzes the conversion of MI into the product d-glucuronate. To our knowledge, this is the first MIOX enzyme of plant origin that has been cloned and characterized. Based on sequence similarity and domain organization, miox4 appears to be part of a four-member family in the Arabidopsis genome.
The specific activity of the recombinant MIOX4 (Table I) was approximately 11 times greater than MIOX purified from the yeast Cryptococcus neoformans (Molina et al., 1999) and 38 times more than the activity purified from oat seedlings (Koller et al., 1976). This variation in specific activity could be because of differences in the methods of purification, differences in the properties of the enzymes (the molecular mass reported for the oat MIOX is 62 kD), or the poor stability of the oat enzyme preparation (Koller et al., 1976).
The native MIOX protein from pigs is found in a complex with glucuronate reductase (E.C. 1.1.1.19), the enzyme responsible for the second step of MI catabolism. This reductase prefers the acyclic form of glucuronate. From this, it is presumed that MIOX can transfer the acyclic form of the substrate directly to the reductase in the complex (Reddy et al., 1981). It is an open question if MIOX enzymes form part of a multienzyme complex in Arabidopsis.
The observed expression pattern for miox4 (Fig. 2A) correlated with the high concentration of AsA in the flowers (Fig. 2B). This pattern also correlated with the observation that AsA is transported from source leaf phloem to root tips, shoots, and floral organs (Franceschi and Tarlyn, 2002). Ascorbate transport is likely because of a greater demand for AsA in reproductive and actively growing tissues, which have higher metabolic rates and, therefore, a higher demand for antioxidants. Such tissues have increased rates of cell expansion and division, and AsA is thought to have a role in these processes (Smirnoff et al., 2001; Arrigoni and De Tullio, 2002). The amount of AsA detected in siliques did not correlate well with the expression pattern of miox4. It is not known if other miox gene family members are expressed in this tissue and, thus, contribute to elevated AsA levels (Fig. 2B).
The analysis of three independent homozygous lines (L1–L3) of Arabidopsis constitutively expressing miox4 revealed a 2- to 3-fold increase in the AsA content of the leaves compared with controls (Fig. 3B). This increase in the leaf AsA content correlates with the amount of miox4 message detected by northern analysis (Fig. 3A).
Tracer studies previously performed with detached ripening strawberry fruits and parsley (Petroselinum hortense Hoffm.) leaves have clearly established the presence of MIOX activity in those tissues but failed to detect formation of AsA from MI (Loewus, 1963). This may be because of the relatively low amount of label (0.7 μCi) used in those experiments (Loewus, 1963). Feeding studies performed with labeled MI and Arabidopsis cell cultures also failed to detect conversion of MI to AsA (Davey et al., 1999); however, it is not known if miox genes are expressed in undifferentiated cells.
Recent findings, which appeared while this manuscript was in review (Wolucka and Van Montagu, 2003), have identified a new branch in the plant AsA network that results in the formation of l-gulose (Fig. 1, step 8). Exogenous l-gulose and l-GulL were shown to serve as direct precursors of AsA in Arabidopsis cell cultures (Davey et al., 1999). There are just two enzymatic reactions required for the conversion of d-glucuronate, the MIOX4 product, to l-GulL. The initial reaction is most likely catalyzed by an aldehyde reductase (E.C. 1.1.1.19); however, there are no reports in the literature regarding the characterization of such an enzyme in plants. Nevertheless, there is indirect evidence suggesting that plants have the machinery to catalyze the conversion of d-glucuronate into l-gulonate as indicated by a recent metabolic profiling analysis of Arabidopsis (Wagner et al., 2003).
The oxidation of free MI to d-glucuronate, the reaction catalyzed by MIOX, has been shown to be an important determinant of MI levels in vivo. In plants, this reaction plays a key role in both the MI oxidation pathway and the sugar nucleotide oxidation route (for review, see Loewus and Murthy, 2000). These processes are involved in the biogenesis of uronosyl and pentosyl units of pectin, hemicelluloses, and related structures in plant cell walls.
Here, we have reported that constitutive expression of an ORF encoding the MIOX4 enzyme caused a 2- to 3-fold increase in the AsA content of the leaves of Arabidopsis plants compared with controls. Other strategies that have been successful in increasing the leaf AsA content of plants are the overexpression of two biosynthetic enzymes: a rat l-GulL oxidase in lettuce and tobacco (Jain and Nessler, 2000) and a d-GalUA reductase from strawberry in Arabidopsis (Agius et al., 2003) and the overexpression of an enzyme involved in the recycling of AsA: a dehydroascorbate reductase from wheat (Triticum aestivum) in tobacco and maize (Zea mays; Chen et al., 2003). These studies have demonstrated an increase of 2- to 7-fold in the leaf AsA content of various plant species. In the case of Arabidopsis, different strategies seem to coincide in a 2- to 3-fold increase of the leaf AsA content, possibly reflecting the action of a feedback or some other regulatory mechanism that keeps a constant pool of AsA as has been suggested in previous turnover studies performed with pea (Pisum sativum) seedlings (Pallanca and Smirnoff, 2000).
Although it is clear that MIOX enzymes can participate in the biogenesis of pectin, hemicelluloses, and other plant cell wall components (Loewus and Murthy, 2000), our results indicate that MI can also be used as a precursor of AsA. Experiments are in progress to clone and characterize the remaining members of the miox family and to test the potential contribution of each member to AsA biosynthesis using T-DNA-tagged knockout lines.
MATERIALS AND METHODS
Isolation of a MIOX Insert from Arabidopsis
Specific primers for the putative miox gene in chromosome 4 (miox4) were designed with NcoI and BamHI sites added to the forward (MX4-5 CCCATGGCGATCTCTGTTGAG) and reverse (MX4-3 CCGGATCCTCACCAC CTCAAG) primers to facilitate subcloning. A 25-μL PCR reaction containing 3 μL of an Arabidopsis mixed tissue cDNA library (CD4-7) from the Arabidopsis Biological Resource Center (Columbus, OH) as template was performed with proofreading polymerase (Pfu Turbo DNA polymerase, Stratagene, La Jolla, CA). After denaturation at 94°C for 5 min, amplification was performed by 30 cycles of 1 min at 94°C, 1 min at 50°C, and 2 min at 72°C, followed by 5 min at 72°C. The 957-bp PCR fragment was cloned into the pGEM-T Easy vector (Promega, Madison, WI), amplified in Escherichia coli DH5α, and sequenced in both directions with T7 and SP6 primers using the ABI PRISM BigDye Terminator Cycle Sequencing Kit (PE Applied Biosystems, Foster City, CA).
Expression, Purification, and Assay of Bacterially Expressed MIOX4
The NcoI/BamHI fragment corresponding to the coding region of the miox4 cDNA was subcloned into the pET32a(+) expression vector (Novagen, Madison, WI), placing it in frame with the 3′ end of the E. coli thioredoxin gene and a linker that includes a His-Tag sequence and the recognition sequence for protease enterokinase. For expression, the pET32a: miox4 construct was transformed into E. coli BL21-Codon Plus(DE3) (Stratagene), and a positive colony was grown overnight in LB/ampicillin (100 mg L-1) medium at 37°C and 250 rpm. This starter culture (10 mL) was used to inoculate 500 mL of LB/ampicillin medium supplemented with 250 mm Suc, 250 mm NaCl, 2 mm glutathione, and 1 mm Pro and grown at 37°C until OD600 reached 0.5. The culture was induced with 0.5 mm isopropyl β-d-thiogalactoside for 1 h, and the cells were recovered by centrifugation at 10,000g. The pellet obtained was suspended and sonicated in 50 mm Tris-HCl (pH 7.0), 500 mm NaCl, 10% (v/v) glycerol, 10 mm β-mercaptoethanol, 5 mm imidazole, and 0.1 mg mL-1 lysozyme, and the resulting lysate was separated into soluble and insoluble fractions by centrifugation. The fusion protein was purified using BD-Talon cobalt-based affinity chromatography resin (BD Biosciences CLONTECH, Palo Alto, CA) according to the protocol supplied by the manufacturer. Eluted fractions were collected, dialyzed against 50 mm Tris-HCl (pH 7.2), 50 mm KCl, and 1 mm glutathione, and MIOX4 was then cleaved away from the fusion protein by digestion with enterokinase (1 units per 50 μg of recombinant protein, Sigma, St. Louis). The purity and Mr of the proteins in the column fractions were examined by 10% (w/v) SDS-PAGE. Protein concentration was determined using the Bradford assay and bovine serum albumin as a standard (Bradford, 1976). MIOX activity was measured using an orcinol-based assay as described previously (Reddy et al., 1981). The standard assay mixture (1 mL) contained 100 mm K-phosphate (pH 7.2), 2 mm l-Cys, 1.0 mm ferrous ammonium sulfate, 60 mm MI, and appropriate quantities of enzyme. Before initiating the reaction by adding an aliquot of the substrate, the other reagents were incubated at room temperature for 15 min. After initiation, the reaction was allowed to proceed at room temperature for 30 min and terminated by boiling. After removal of the precipitated protein by centrifugation at 15,000g for 15 min, the amount of d-glucuronate formed was determined after adding 1 mL of fresh orcinol reagent (28 mm orcinol and 3 mm anhydrous ferric chloride in concentrated HCl), boiling for 30 min, and measuring the A660 (Charalampous and Lyras, 1957).
Plant Material and Growth Conditions
Seeds of Arabidopsis (ecotype Columbia), both wild-type and transgenic homozygous lines, were grown in Sunshine Mix Number 1 (Wetzel, Harrisonburg, VA) in a greenhouse during the months of June and July of 2003. The greenhouse was equipped with supplemental light (mercury vapor) and a heat pump to keep temperature and relative humidity conditions as follows: a 16-:8-h photoperiod, photon flux density of 950 μmol m-2 s-1, temperature of 26°C:18°C ± 2.5°C (day:night), and relative humidity of 50%:70% ± 10% (day:night).
RNA Gel-Blot Analysis
Total RNA was extracted from above ground organs and roots by either TRI Reagent (Sigma) or RNeasy kit (Qiagen USA, Valencia, CA) following the instructions provided by the manufacturers and from green siliques by a SDS/phenol method (Takahashi et al., 1992). RNA was suspended in water and precipitated twice with 7 m ammonium acetate and 100% (v/v) ethanol. RNA yield was quantified spectrophotometrically. For northern analysis, 24 μg of total RNA (tissues) or 8 μg of total RNA (overexpressers) was separated on 1.2% (w/v) denaturing (formaldehyde) agarose gels and transferred to nylon membranes (Hybond-N+, Amersham, Piscataway, NJ). Membranes were prehybridized for 2 h at 65°C and hybridized overnight at 65°C in 500mm sodium phosphate buffer (pH 7.2) containing 7% (w/v) SDS, 1% (w/v) bovine serum albumin, and 1 mm EDTA. The miox4 insert was excised from pGEM-T using NcoI/BamHI, purified after gel electrophoresis, and labeled with 32P using Primer-It RmT Random Primer Labeling Kit (Stratagene). After hybridization, filters were washed five times for 30 min at 65°C in 20 mm sodium phosphate buffer (pH 7.2), 0.1% (w/v) SDS, 33 mm NaCl, and 1 mm EDTA and subjected to autoradiography (Kodak X-Omat AR film, Kodak, Rochester, NY) between intensifying screens for 20 h at -80°C.
Construction of Transgenic Plants
The miox4 insert was cloned into the NcoI/BamH1 sites of pRTL2 placing it under the control of CaMV 35S promoter with duplicated enhancer between the 5′ tobacco etch virus leader and the 3′ 35S polyadenylation signal (Rastrepo et al., 1990). A PstI fragment including the promoter::miox4::terminator insert was subcloned into the binary vector pCAMBIA1300 and transformed into Agrobacterium tumefaciens strain GV3101. Arabidopsis var Columbia plants were transformed with pCAM-BIA1300:miox4 construct via the floral dip method (Clough and Bent, 1998). Seedlings were selected on Murashige and Skoog (Murashige and Skoog, 1962) plates containing 500 mg L-1 carbenecillin and 25 mg L-1 hygromycin. Both primary transformants and their progeny were used for RNA gel-blot analysis and ascorbic acid assays.
Ascorbic Acid Measurements
Ascorbic acid content was measured by the ascorbate oxidase assay (Rao and Ormrod, 1995). Plant extracts were made from tissue frozen in liquid nitrogen, ground in 6% (w/v) meta-phosphoric acid, and centrifuged at 15,000g for 15 min. Reduced AsA was determined by measuring the decrease in A265 (extinction coefficient of 14.3 mm) after addition of 1 unit of ascorbate oxidase (Sigma) to 1 mL of the reaction medium containing the plant extract and 100 mm potassium phosphate (pH 6.9). Oxidized AsA was measured in a 1-mL reaction mixture plus 1 μL of 2 mm dithiothreitol after incubating at room temperature for 15 min.
Acknowledgments
We thank Christopher Dana and Brenda Winkel for advice in protein purification and Jessica Radzio, Deanna Conquest, and Amy Vance for technical assistance.
Article, publication date, and citation information can be found at http://www.plantphysiol.org/cgi/doi/10.1104/pp.103.033936.
This work was supported by the Interagency Metabolic Engineering Program (National Science Foundation Metabolic Biochemistry and Integrative Plant Biology) (IBN0118612) and USDA/CREES (2002-3S321-11600).
References
- Agius F, González-Lamonthe R, Caballero JL, Muñoz-Blanco J, Botella MA, Valpuesta V (2003) Engineering increased vitamin C levels in plants by over-expression of a d-galacturonic acid reductase. Nat Biotechnol 21: 177-181 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389-3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler R, Attwood TK, Bairoch A, Bateman A, Birney E, Biswas M, Bucher P, Cerutti L, Corpet F, Croning MD et al. (2000) InterPro: an integrated documentation resource for protein families, domains and functional sites. Bioinformatics 16: 1145-1150 [DOI] [PubMed] [Google Scholar]
- Arner RJ, Prabhu S, Thompson JT, Hildenbrandt GR, Liken AD, Reddy CC (2001) myo-Inositol oxygenase: molecular cloning and expression of a unique enzyme that oxidizes myo-inositol and d-chiro-inositol. Biochem J 360: 313-320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrigoni O, De Tullio MC (2002) Ascorbic acid: much more than just an antioxidant. Biochim Biophys Acta 1569: 1-9 [DOI] [PubMed] [Google Scholar]
- Baig MM, Kelly S, Loewus FA (1970) l-Ascorbic acid biosynthesis in higher plants from l-gulono-1,4-lactone and l-galactono-1,4-lactone. Plant Physiol 46: 277-280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248-254 [DOI] [PubMed] [Google Scholar]
- Charalampous FC, Lyras C (1957) Biochemical studies on inositol: IV. Conversion of inositol to glucuronic acid by rat kidney extracts. J Biol Chem 228: 1-13 [PubMed] [Google Scholar]
- Chen Z, Young TE, Ling J, Chang SC, Gallie DR (2003) Increasing vitamin C content of plants through enhanced ascorbate recycling. Proc Natl Acad Sci USA 100: 3525-3530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735-743 [DOI] [PubMed] [Google Scholar]
- Conklin PL, Saracco SA, Norris SR, Last RL (2000) Identification of ascorbic acid-deficient Arabidopsis thaliana mutants. Genetics 154: 847-856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F, Servant F, Gouzy J, Kahn D (2000) ProDom and ProDom-CG: tools for protein domain analysis and whole genome comparisons. Nucleic Acids Res 28: 267-269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey MW, Gilot C, Persiau G, Ostergaard J, Han Y, Bauw GC, Van Montagu MC (1999) Ascorbate biosynthesis in Arabidopsis cell suspension culture. Plant Physiol 121: 535-543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi VR, Tarlyn NM (2002) l-Ascorbic acid is accumulated in source leaf phloem and transported to sink tissues in plants. Plant Physiol 130: 649-656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huala E, Dickerman AW, Garcia-Hernandez M, Weems D, Reiser L, LaFond F, Hanley D, Kiphart D, Zhuang M, Huang W et al. (2001) The Arabidopsis Information Resource (TAIR): a comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res 29: 102-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain AK, Nessler CL (2000) Metabolic engineering of an alternative pathway for ascorbic acid biosynthesis in plants. Mol Breed 6: 73-78 [Google Scholar]
- Koller E, Koller F, Hoffmann-Ostenhof O (1976) myo-Inositol oxygenase from oat seedlings. Mol Cell Biochem 10: 33-39 [DOI] [PubMed] [Google Scholar]
- Loewus FA (1963) Tracer studies on ascorbic acid formation in plants. Phytochemistry 2: 109-128 [Google Scholar]
- Loewus FA (1999) Biosynthesis and metabolism of ascorbic acid in plants and of analogs of ascorbic acid in fungi. Phytochemistry 52: 193-210 [Google Scholar]
- Loewus FA, Loewus MW (1987) Biosynthesis and metabolism of ascorbic acid in plants. CRC Crit Rev Plant Sci 5: 101-119 [Google Scholar]
- Loewus FA, Murthy PPN (2000) myo-Inositol metabolism in plants. Plant Sci 150: 1-19 [Google Scholar]
- Molina Y, Ramos SE, Douglass T, Klig LS (1999) Inositol synthesis and catabolism in Cryptococcus neoformans. Yeast 15: 1657-1667 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog FA (1962) A revised medium for rapid growth and bioassays with tobacco culture. Physiol Plant 15: 473-497 [Google Scholar]
- Pallanca JE, Smirnoff N (2000) The control of ascorbic acid synthesis and turnover in pea seedlings. J Exp Bot 51: 669-674 [PubMed] [Google Scholar]
- Pastori GM, Kiddle G, Antoniw J, Bernard S, Veljovic-Jovanovic S, Verrier PJ, Noctor G, Foyer CH (2003) Leaf vitamin C contents modulate plant defense transcripts and regulate genes that control development through hormone signaling. Plant Cell 15: 939-951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao M, Ormrod DP (1995) Ozone pressure decreases UVB sensitivity in a UVB-sensitive flavonoid mutant of Arabidopsis. Photochem Photobiol 61: 71-78 [DOI] [PubMed] [Google Scholar]
- Rastrepo MA, Freed DD, Carrington JC (1990) Nuclear transport of plant potyviral proteins. Plant Cell 2: 987-998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy CC, Swan JS, Hamilton GA (1981) myo-Inositol oxygenase from hog kidney. J Biol Chem 256: 8510-8518 [PubMed] [Google Scholar]
- Smirnoff N (2000) Ascorbic acid: metabolism and functions of a multifacetted molecule. Curr Opin Plant Biol 3: 229-235 [PubMed] [Google Scholar]
- Smirnoff N, Wheeler GL (2000) Ascorbic acid in plants: biosynthesis and function. Crit Rev Biochem Mol Biol 35: 291-314 [DOI] [PubMed] [Google Scholar]
- Smirnoff N, Conklin PL, Loewus FA (2001) Biosynthesis of ascorbic acid in plants: a renaissance. Annu Rev Plant Physiol Plant Mol Biol 52: 437-467 [DOI] [PubMed] [Google Scholar]
- Takahashi T, Naito S, Komeda Y (1992) Isolation and analysis of the expression of two genes for the 81-kilodalton heath-shock proteins from Arabidopsis. Plant Physiol 99: 383-390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner C, Sefkow M, Kopka J (2003) Construction and application of a mass spectral and retention time index database generated from plant GC/EI-TOF-MS metabolite profiles. Phytochemistry 62: 887-900 [DOI] [PubMed] [Google Scholar]
- Wheeler GL, Jones MA, Smirnoff N (1998) The biosynthetic pathway of vitamin C in higher plants. Nature 393: 365-369 [DOI] [PubMed] [Google Scholar]
- Wolucka BA, Van Montagu M (2003) GDP-mannose-3′,5′-epimerase forms GDP-l-gulose, a putative intermediate for the de novo biosynthesis of vitamin C in plants. J Biol Chem 278: 47483-47490 [DOI] [PubMed] [Google Scholar]