Abstract
Molecular mechanisms mediating the progression of ductal carcinoma in situ (DCIS) to invasive breast cancer (IBC) remain largely unknown. We used gene expression profiling of human DCIS (n=53) and IBC (n=51) to discover uniquely expressed genes that may also regulate progression. There were 470 total differentially expressed genes (≥ 2 fold; p<0.05). Elevated expression of genes involved in synthesis and organization of extracellular matrix was particularly prominent in the epithelium of IBC. The degree of overlap of the genes with nine similar studies in the literature was determined to help prioritize their potential importance, resulting in 74 showing overlap in ≥ 2 studies (average 3.6 studies/gene; range 2-8 studies). Using hierarchical clustering, the 74-gene profile was able to correctly categorize 96%, 93%, and 85% of samples in this study and two similar independent studies, respectively. To study the progression of DCIS to IBC in vivo, we introduced human DCIS cell lines engineered to express specific genes into a “mammary intraductal DCIS” (MIND) xenograft model. Progression of xenografts to IBC was dramatically increased by suppressing four genes that were usually elevated in clinical samples of DCIS, including a protease inhibitor (CSTA) and genes involved in cell adhesion and signaling (FAT1, DST, and TMEM45A), strongly suggesting that they normally function to suppress progression. In summary, we have identified unique gene expression profiles of human DCIS and IBC, which include novel genes regulating tumor progression. Targeting some of these genes may improve the detection, diagnosis, and therapy of DCIS.
Keywords: Breast cancer, ductal carcinoma in situ, tumor progression, gene expression profile, tumor suppressor gene
Introduction
The majority of human invasive breast cancer (IBC) is the end result of a usually decades-long evolution of increasingly abnormal premalignant stages (1, 2). Ductal carcinoma in situ (DCIS) is a late stage and the immediate precursor of most IBCs. Approximately 60,000 new cases of DCIS were diagnosed in the US in 2011 (3). Undetected, at least a third would progress to IBCs during an average lifespan (4-6). About 200,000 new cases of IBC were also diagnosed (3), and a large majority evolved from pre-existing DCIS which were not detected. Ten percent of DCIS which are detected still recur and/or progress to IBC despite modern therapy (4, 6). The incidence of IBC could be dramatically reduced by improving our abilities to detect, diagnose, and treat DCIS. Progress will be based on detailed understanding of molecular mechanisms responsible for the development and progression of DCIS to IBC, which is currently quite limited (4, 7).
This study used microarray technology to identify differentially expressed genes in clinical samples of human DCIS and IBC which might be involved in tumor progression. The genes were compared to results from previous studies comparing DCIS to IBC to help prioritize potential importance in tumor progression based on the degree of overlap (8-16). The ability of selected genes to influence tumor progression was evaluated in a novel xenograft model involving injecting human DCIS cells into the primary duct of intact mammary glands of immune-suppressed mice, referred to as the “mammary intraductal DCIS” (MIND) model(17). Cells were genetically modified prior to injection to mimic changes in gene expression observed in clinical samples. The cells grow within ducts in a manner histologically very similar to human DCIS, and may progress to IBC depending on the function of the modulated genes.
Methods
Human Tumor Samples
The human samples used in this study are summarized in Supplementary Table S1. Samples were harvested anonymously with appropriate IRB approvals (Baylor College of Medicine; H-10493 and H-12585).
Evaluation of Gene Expression by Microarrays
RNA from each sample was extracted, purified, amplified, and evaluated for gene expression as previously described (2, 18). Affymetrix U95Av2 microarrays were utilized with samples in groups 1, 2, and 3, and U133-X3P microarrays with samples in groups 4e and 4s.
Validation of Gene Expression by qRT-PCR
Selected genes (n=12) differentially expressed between DCIS and IBCs by microarray analyses were validated by qRT-PCR as previously described (2, 18), using new samples of total RNA from group 4e. Sequences for the probes and primers were designed using Primer Express Software v2.0 (Applied Biosystems), and synthesized by a commercial vendor (Eurogentec, San Diego CA).
Cell Lines
This study used three DCIS-like human breast epithelial cell lines. One, referred to as DCIS.COM, is widely available to the research community and has been used in nearly all previous functional studies of human DCIS (17, 19, 20). It was developed from ras-transformed MCF10AT cells originally cultured from benign breast tissue (fibrocystic disease). The second, referred to as SUM225, was developed from a primary culture of a chest wall recurrence in a patient with a history of DCIS treated by mastectomy (21). The third, referred to as h.DCIS.01, was developed in our laboratory from a primary culture of hyperplastic breast epithelial cells (columnar cell hyperplasia), which is an important early precursor of breast cancer (22). Early passages (<25) developed into small hyperplastic lesions in MIND xenografts. The cells spontaneously transformed in culture and later passages (>30) developed into DCIS in MIND xenografts, similar to DCIS.COM. The related identity of early and late passages was confirmed by array CGH. All cell lines were maintained in humidified incubators at 37°C and 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM)/F12 (50%:50%) media with 5% horse serum and 1% penicillin/streptomycin. 293T cells were grown in DMEM media containing 10% fetal-calf serum and 1% penicillin/streptomycin.
Inhibiting Gene Expression with shRNAi
Transductions of shRNAi against selected genes were performed as described by Stewart (23), utilizing an RNAi vector (pLKO.1puro) containing an RNAi library against the entire human genome. Briefly, 293T cells were infected with a lentiviral vector containing the U6-driven stem-loop directed against candidate genes, and red fluorescent protein (RFP), using TransIT-LT1 (Mirius, Madison, WI). Virus was harvested 48 hours later and DCIS cells were infected with virus in the presence of 10 μg/ml of protamine sulfate overnight, and re-infected 24 hours later. Transduced cells were selected for in 2 μg/ml puromycin. The pLKO.1 bearing short hairpins targeting genes of interest and RFP, were produced by co-transfection with pCMVDR8.2 and pCMV-VSV-G (8:1 ratio). Knockdown efficiency was assessed by qRT-PCR and western blot (when suitable antibodies were available) (Supplementary Figure S2). To control for off-target effects, three or more separate hairpins were transduced independently for each gene, and identical results were obtained with at least two.
MIND Xenografts
The MIND xenograft studies were performed as recently described (17). Briefly, 20,000 cells were injected through the nipple into the primary duct of intact mammary glands (2 per animal) of 6-8 week old female SCID-Beige mice. Over a period of 10 weeks, variable proportions (depending on the cell line) develop intra-ductal tumors which are histologically very similar to human DCIS, and a proportion progress to IBCs. The developing tumors were measured weekly, and all animals were euthanized by 10 weeks. Immediately after being euthanized, mammary glands were excised and processed for further study (including whole-mounts, routine histology, immunohistochemistry, and stored fresh frozen for later use).
In Vitro Analyses of Cell Growth, Migration, and Invasion
Cell Proliferation: Proliferation was measured as previously described by Pazolli (24). Briefly, DCIS cell lines (±shRNAi) cells were seeded into 96-well plates in culture media. At 24, 48, 72 and 96 hours, the cells were washed with PBS, and re-suspended in cell-lysis buffer with luciferase substrate (Promega, Madison, WI). Relative luminescence was measured 5 minutes later.
3-Dimensional (3D) Cell Cultures: 3D growth was evaluated in 24-well culture plates coated with Matrigel (100%) or Matrigel:collagen-1 (200 ul at 1:1 ratio) seeded with 5000 DCIS cells (±shRNAi) in regular culture media, and allowed to grow for 2-4 weeks (re-fed with fresh culture media every 2 days). These assays are not easily quantified, and the characteristics and changes in cell growth were described in a qualitative manner, including formation of multi-cellular spheres and tubules; attachment, growth, and spreading on the gel surface; and invasion into the gels (subjective scale relative to control: “-” = none; “+” = low; “++” = intermediate; “+++” = high). Cultures were photographed at 1, 3, 5, 8, and 15 days, after which gels were fixed and processed for histological evaluation in a routine manner.
In vitro cell migration and invasion were also measured with the HTS FluoroBlok chamber assay (BD Biosciences, Bedford, MA), following manufacturer’s instructions. Briefly, cells suspended in serum-free media were seeded onto the porous membrane of the upper chamber. The bottom chamber was filled with serum-containing media and the cells were incubated for 48 hours. Cells migrating and/or invading through the membrane into the bottom chamber were stained with Calcein AM dye (Invitrogen, USA) and quantified using a bioluminescence reader. Traversing a bare membrane was defined as migration. Traversing a membrane coated with 50 μl Matrigel or Matrigel + COL1 (1:1) was defined as invasion
Data Analyses
Microarray results were evaluated using dChip software (http://www.dchip.org) for estimating gene expression and class comparisons as previously described (2, 18). Normalization of raw data at PM- and MM-probe level was by the invariant set normalization method and expression was estimated by the perfect match only model. Differentially expressed genes were identified using the t-test and the empirical false discovery rate (FDR) was estimated by permutation. The filtered genes have the “value of standard deviation/mean” of expression intensity between 0.5 and 1000 (default setting in dChip software). On average, about 2,500 genes in each group of samples met these criteria.
Each of the five groups of samples was evaluated by unsupervised hierarchical clustering utilizing dChip software and large numbers of genes (average = 2866) expressed in the samples selected by identical normalization and filtering criteria (20% present calls; 0.5<standard deviation/mean<1000 variation across samples).
Gene ontologies (GO) associated with the progression of DCIS to IBC were inferred from differentially expressed genes utilizing Ontologizer 2.0 software (25). The hierarchical tree of GO terms were ranked Term-for-Term, and by the Bonferonni method to correct to multiple comparisons. Details are available on the Ontologizer website URL : (http://compbiol.charite.de/index.pht/ontologizer2.html).
Differences in the size of MIND xenografts were determined by averaging tumor size along the seven time points (week 0 3 5 6 8 9 10), after square root transformation, was then fitted by a mixed model for repeated measure, with Group (gene knockout), time (centered at the mid time point, week 6) and their interaction in the model. In the mixed model, the tumor size under each experiment condition (gene knockout) over time was each modeled as a linear line with its own intercept and slope. To test if a mean growth line under a gene knock out differs from that of the reference (RFP), we tested if the slope of each line was different from that of the reference group (RFP) and similarly on the intercept. As time is centered at week 6, the comparison of a group’s intercept will indicate any difference between the group and the reference at week 6 while the difference in slopes indicates different tumor growth rate.
Results
Gene Expression Profiles of DCIS and IBC
There have been several hundred studies profiling gene expression in fully-developed IBCs, providing many insights into important molecular and clinical features of the disease. Our goal was to determine whether expression profiling could provide similar insights into the progression of DCIS to IBC, which represents a critical step in the evolution of a non-lethal to potentially lethal disease. Therefore, we performed comprehensive expression profiling on clinical samples of DCIS and IBC, including separate samples of tumor epithelium and adjacent stroma prepared by laser-capture micro-dissection (LCM).
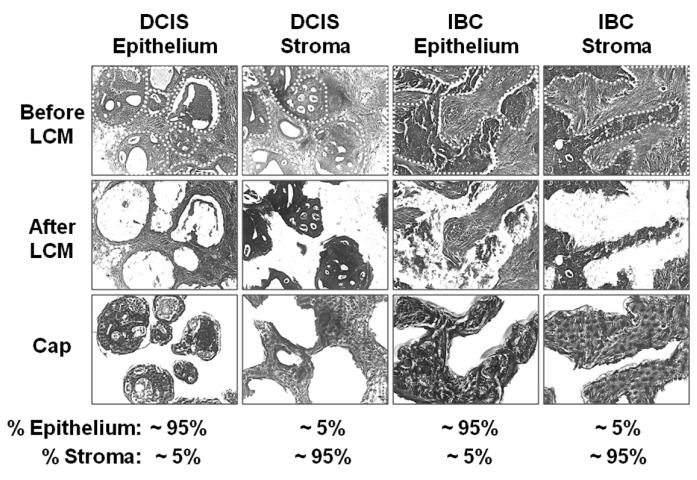
Briefly, samples were obtained from clinical cases of DCIS and IBC, distributed in five groups, which were profiled independently (Supplementary Table S1). Group 1 consisted of 26 samples of DCIS and 24 samples of IBC enriched for tumor epithelial cells (>75% total cells) by manual macro-dissection. Groups 2, 3, and 4e consisted of 7, 9, and 11 samples each of DCIS and IBCs enriched for tumor epithelial cells (>95%) by LCM (Figure 1). Group 4s was composed of samples of near-pure (>95%) stroma immediately adjacent to tumor epithelium in 4e, also prepared by LCM (Figure 1). The samples of DCIS in groups 1, 2, and 4 were harvested from breasts without IBC. The samples of DCIS in group 3 were harvested from breasts with adjacent IBC.
Figure 1. LCM of samples in Group 4.
Tumor epithelial cells and immediately adjacent stroma were independently isolated by laser-capture micro dissection (LCM) to a purity of about 95%.
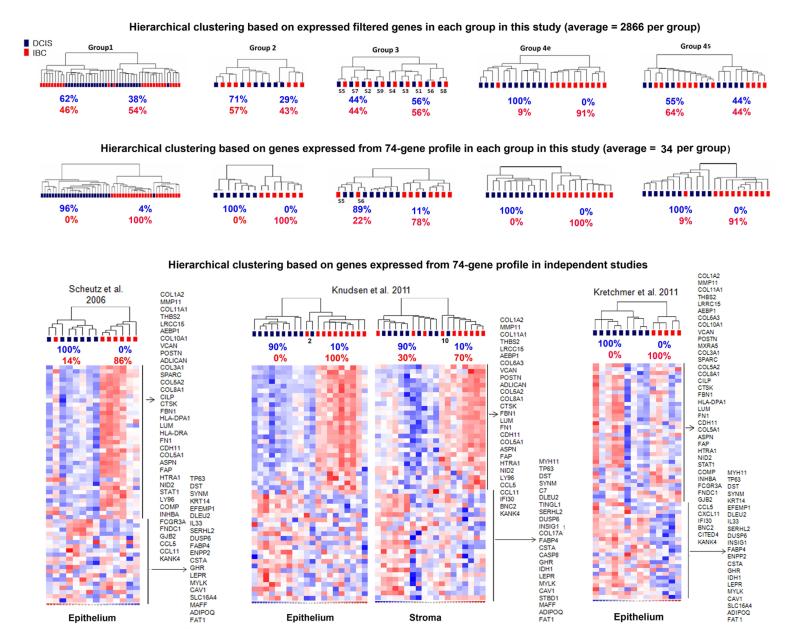
The results from each group were evaluated by unsupervised hierarchical clustering using variably expressed genes selected by identical normalization and filtering criteria, averaging 2866 genes/group. As shown in Figure 2 (top row), there was a wide range in the degree that similar types of tumors clustered together between groups. For example, in group 3, composed of DCIS and IBC isolated from the same breasts, analysis categorized patients, but not tumor type, suggesting that synchronous DCIS and IBC are more similar than different, which might be expected since the former evolved directly into the latter. In contrast, analysis of group 4e revealed that pure DCIS and IBC harvested from separate patients are very different (100% and 91% clustering accuracy, respectively), indicating that there are also unique patterns of gene expression in these two stages of breast cancer.
Figure 2. Hierarchical clustering.
The top row shows dendrograms from unsupervised hierarchical clustering analyses based on genes expressed within each group of samples in this study (average=2866). The center row shows results from hierarchical clustering of the same samples using the refined profile of 74 differentially expressed genes determined in this study. The bottom row show results (dendrograms, heat maps, and genes) using the 74-gene profile in two independent cohorts from previous gene expression profiling studies comparing human DCIS and IBC. Note: analysis of the Scheutz study was restricted to 7 of the 9 total samples each of DCIS and IBC that were all evaluated using the same platform (U133A Plus 2.0 microarray chips).
Differentially Expressed Genes Between DCIS and IBC
To identify differentially expressed genes that may be associated with and/or regulate the progression of DCIS to IBC, the microarray results were evaluated by supervised hierarchical clustering at a threshold of ≥ 2 fold/p ≤ 0.05. The numbers of genes meeting these criteria were 117, 76, 30,165, and 184 in groups 1, 2, 3, 4e, and 4s, respectively (Supplementary Table S2). These results were distilled to 470 genes differentially expressed in at least one group, which was further refined to 74 genes showing similar profiles in 2 or more groups (Table 1). The 74 genes were then compared with results of nine similar studies in the literature to help prioritize potential importance based on degree of overlap (8-16, 26). Forty-nine of the 74 genes (66%) overlapped with one or more previous study (range 2 to 8 studies/gene; average = 3.7 studies/gene). The relative expression levels of selected genes (n=12) were evaluated by qRT-PCR in the same study samples, validating the original microarray results for all of the genes tested (Supplementary Figure S1). The majority (78%) of the 74 genes have not been studied in the progression of human DCIS to IBC beyond the original microarray studies mentioned above. The exceptions include genes previously shown to be expressed at different levels in DCIS and IBC by other techniques (COL1A2, MMP11, THBS2, VCAN, CTSK, FN1, FAP, GJB2, CCL5, MYH11, TP63, KRT14, CASP8, NGFR, CAV1, and FAT1), and/or functionally involved in the progression of DCIS to IBC (MMP11, CTSK, TP63, and CAV1) – the latter providing some reassurance that this vetting strategy is helpful in identifying potentially important genes.
Table 1.
Genes (n=74) showing increased (n=42) and decreased (n=32) expression in invasive breast cancers (IBCs) versus ductal carcinoma in situ (DCIS) in at least 2 groups of samples in this study, and overlap with previous studies comparing human IBCs and DCIS.
| Differentially Expressed Genes |
Groups of Samples in this Study (Fold-Change) |
Previous Studies (Direction Significant Differences) |
Studies in Common | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Symbol | Locus Li nk | Group 1 | Group 2 | Group 3 | Group 4 Epithelium | Group 4 Stroma | Average p-value | Porter 2001 | Ma 2003 | Porter 2003 | Abba 2004 | Allinen 2004 | Hanneman 2006 | Scheutz 2006 | Ma 2009 | Knudsen 2011 | |
| Increased in IBC vs. DCIS | |||||||||||||||||
| COL1A21 | 1278 | 2.27 | 5.23 | 3.2 | 4.66 | 0.0024 | ↑ | ↑ | ↑ | ↑ | 8 | ||||||
| MMP111,2 | 4320 | 3.33 | 3.19 | 4.17 | 0.0090 | ↑ | ↑ | ↑ | ↑ | ↑ | 8 | ||||||
| COL11A13 | 1301 | 5.43 | 11.54 | 8.95 | 5.49 | 0.0009 | ↑ | ↑ | 6 | ||||||||
| THBS21 | 7058 | 2.05 | 7.25 | 4.17 | 5.67 | 0.0137 | ↑ | ↑ | 6 | ||||||||
| LRRC153 | 131578 | 2.68 | 5.09 | 2.06 | 2.43 | 0.0055 | ↑ | ↑ | 6 | ||||||||
| AEBP13 | 165 | 2.02 | 4.19 | 2.35 | 0.0137 | ↑ | ↑ | ↑ | 6 | ||||||||
| COL6A33 | 1293 | 3.56 | 4.1 | 5.55 | 0.0086 | ↑ | ↑ | ↑ | 6 | ||||||||
| COL10A13 | 1300 | 2.77 | 5.13 | 3.34 | 0.0095 | ↑ | ↑ | ↑ | 6 | ||||||||
| VCAN1 | 1462 | 3.59 | 2.75 | 3.12 | 0.0152 | ↑ | ↑ | ↑ | 6 | ||||||||
| POSTN4 | 10631 | 3.86 | 3.35 | 2.99 | 0.0186 | ↑ | ↑ | ↑ | 6 | ||||||||
| MXRA53 | 25878 | 3.57 | 2.27 | 2.64 | 0.0101 | ↑ | ↑ | ↑ | 6 | ||||||||
| COL3A13 | 1281 | 3.58 | 3.29 | 4.87 | 0.0190 | ↑ | ↑ | ↑ | 6 | ||||||||
| SPARC4 | 6678 | 3.04 | 2.22 | 0.0076 | ↑ | ↑ | ↑ | 5 | |||||||||
| COL5A23 | 1290 | 2.41 | 3.92 | 3.65 | 0.0086 | ↑ | ↑ | 6 | |||||||||
| COL8A13 | 1295 | 2.28 | 2.47 | 2.16 | 0.0154 | ↑ | ↑ | 6 | |||||||||
| CILP3 | 8483 | 2.05 | 16.09 | 4.75 | 0.0091 | ↑ | ↑ | 6 | |||||||||
| CTSK1,2 | 1513 | 3.8 | 3.08 | 0.0264 | ↑ | ↑ | ↑ | 6 | |||||||||
| FBN13 | 2200 | 4.82 | 2.89 | 0.0157 | ↑ | ↑ | ↑ | 6 | |||||||||
| HLA-DPA14 | 3113 | 2.02 | 2.02 | 0.0300 | ↑ | ↑ | ↑ | 5 | |||||||||
| LUM4 | 4060 | 4.57 | 3 | 0.0160 | ↑ | ↑ | ↑ | 5 | |||||||||
| HLA-DRA3 | 3122 | 2.17 | 2.07 | 0.0294 | ↑ | ↑ | 4 | ||||||||||
| FN11 | 2335 | 2.89 | 2.75 | 0.0101 | ↑ | ↑ | 4 | ||||||||||
| CDH114 | 1009 | 3.8 | 2.01 | 0.0290 | ↑ | ↑ | 4 | ||||||||||
| COL5A13 | 1289 | 3.18 | 2.26 | 0.0298 | ↑ | ↑ | 4 | ||||||||||
| ASPN4 | 54829 | 7.18 | 2.7 | 0.0026 | ↑ | ↑ | 4 | ||||||||||
| FAP1 | 2191 | 6.61 | 4.58 | 0.0082 | ↑ | ↑ | 4 | ||||||||||
| HTRA14 | 5654 | 2.39 | 2.14 | 0.0233 | ↑ | ↑ | 4 | ||||||||||
| NID23 | 22795 | 2.05 | 2.89 | 0.0307 | ↑ | ↑ | 4 | ||||||||||
| STAT14 | 6772 | 2.11 | 2.66 | 0.0076 | ↑ | 3 | |||||||||||
| LY963 | 23643 | 3.07 | 2.14 | 0.0112 | ↑ | 3 | |||||||||||
| COMP4 | 1311 | 2.38 | 3.11 | 0.0254 | ↑ | 3 | |||||||||||
| INHBA4 | 3624 | 2.13 | 2.59 | 0.0092 | ↑ | 3 | |||||||||||
| FCGR3A3 | 2214 | 2.1 | 3 | 0.0052 | ↑ | 3 | |||||||||||
| FNDC13 | 84624 | 4.54 | 3.27 | 0.0108 | 2 | ||||||||||||
| GJB21 | 2706 | 4.16 | 4.25 | 0.0091 | 2 | ||||||||||||
| HLA-G4 | 3135 | 2.28 | 2.23 | 0.0078 | 2 | ||||||||||||
| CCL51 | 6352 | 2.04 | 2.25 | 0.0273 | 2 | ||||||||||||
| CXCL114 | 6373 | 3.52 | 4.52 | 0.0157 | 2 | ||||||||||||
| IFI303 | 10437 | 2.21 | 3.08 | 0.0134 | 2 | ||||||||||||
| BNC23 | 54796 | 2.22 | 2.7 | 0.0026 | 2 | ||||||||||||
| CITED44 | 163732 | 4.05 | 3.3 | 0.0207 | 2 | ||||||||||||
| KANK43 | 163782 | 3.27 | 2.46 | 0.0175 | 2 | ||||||||||||
| Decreased in IBC vs. DCIS | |||||||||||||||||
| MYH111 | 4629 | −4.2 | −3.1 | −2 | −2.7 | 0.0097 | ↓ | ↓ | 6 | ||||||||
| TP631,2 | 8626 | −2.5 | −2.2 | 0.0263 | ↓ | ↓ | ↓ | 5 | |||||||||
| DST3 | 667 | −5.8 | −4.6 | −5.3 | 0.0233 | ↓ | 4 | ||||||||||
| SYNM4 | 23336 | −2.7 | −2 | −2.5 | 0.0206 | ↓ | 4 | ||||||||||
| C73 | 730 | −2.8 | −3.9 | 0.0144 | ↓ | ↓ | 4 | ||||||||||
| KRT141 | 3861 | −2.7 | −4.3 | 0.0187 | ↓ | ↓ | 4 | ||||||||||
| EFEMP14 | 2202 | −2 | −2.4 | 0.0032 | ↓ | 3 | |||||||||||
| DLEU24 | 8847 | −2.6 | −2.2 | 0.0371 | ↓ | 3 | |||||||||||
| TINAGL14 | 64129 | −2.2 | −2.4 | 0.0133 | ↓ | 3 | |||||||||||
| IL334 | 90865 | −2.4 | −4.8 | 0.0092 | ↓ | 3 | |||||||||||
| SERHL23 | 253190 | −4.2 | −3 | −2.5 | 0.0017 | ↓ | 3 | ||||||||||
| DUSP64 | 1848 | −2.6 | −2 | 0.0099 | ↓ | 3 | |||||||||||
| INSIG14 | 3638 | −2.1 | −2.5 | 0.0280 | ↓ | 3 | |||||||||||
| COL17A13 | 1308 | −2.1 | −2.3 | 0.0131 | ↓ | 3 | |||||||||||
| FABP44 | 2167 | −2.2 | −4.5 | 0.0243 | ↓ | 3 | |||||||||||
| ENPP24 | 5168 | −2.1 | −2.4 | 0.0110 | ↓ | 3 | |||||||||||
| ATP2A23 | 488 | −2.1 | −2.2 | 0.0146 | 2 | ||||||||||||
| CSTA4 | 1475 | −9.4 | −3.8 | 0.0324 | 2 | ||||||||||||
| CYP51A14 | 1595 | −2.2 | −2.2 | 0.0257 | 2 | ||||||||||||
| CASP81 | 841 | −2.1 | −2.1 | 0.0033 | 2 | ||||||||||||
| GHR4 | 2690 | −2.6 | −2.8 | 0.0275 | 2 | ||||||||||||
| IDH14 | 3417 | −2.1 | −2.3 | 0.0261 | 2 | ||||||||||||
| LEPR4 | 3953 | −2.1 | −2.4 | 0.0416 | 2 | ||||||||||||
| MYLK4 | 4638 | −2.2 | −2.2 | 0.0127 | 2 | ||||||||||||
| NGFR1 | 4804 | −2.2 | −3.7 | 0.0127 | 2 | ||||||||||||
| CAV11,2 | 857 | −2.2 | −2.5 | 0.0090 | 2 | ||||||||||||
| STBD13 | 8987 | −2.6 | −2.1 | 0.0328 | 2 | ||||||||||||
| SLC16A44 | 9122 | −2.1 | −2.9 | 0.0228 | 2 | ||||||||||||
| ATP6V1G13 | 9550 | −4.1 | −2.5 | 0.0288 | 2 | ||||||||||||
| MAFF4 | 23764 | −2.2 | −3.2 | 0.0197 | 2 | ||||||||||||
| ADIPOQ4 | 9370 | −3.2 | −7.0 | 0.0175 | 2 | ||||||||||||
| FAT11 | 2195 | −4.9 | −2.8 | 0.0080 | 2 | ||||||||||||
| Number in 74 | 32 | 35 | 21 | 46 | 37 | ||||||||||||
| Percent in 74 | 43.2% | 47.3% | 28.4% | 62.2% | 50% | ||||||||||||
Prevously studied in DCIS, primarily comparing expression levels with IBCs.
Prevously studied in DCIS, including functional involvement in progression to IBC.
Not previously studied in breast cancer.
Prevously studied in IBC.
Classification of DCIS and IBC Based on Differentially Expressed Genes
Each group of tumor samples was re-analyzed by hierarchical clustering using the 74-gene profile to determine its ability to classify DCIS and IBC (Figure 2, middle row). There was considerable improvement over initial analyses based on all filtered genes expressed within each group (Figure 2, top row). The 74-gene profile correctly categorized 97% of all DCIS and 95% of all IBCs. A high degree of accuracy was expected because the 74 genes were derived from analysis of the same samples considered collectively. However, each group contributed only a subset of the genes, ranging from 28% (21/74) in group 3 to 62% (46/74) in group 4e (average = 47%; 34/74).
The ability of the 74-gene profile to classify DCIS versus IBC was further evaluated in cohorts from three similar independent studies (14, 16, 26) (Figure 2, bottom row). In the study by Scheutz (14), which involved 7 paired samples of DCIS and IBC epithelium (purified by LCM) from the same breasts, 100% and 86% of the samples were classified accurately, respectively - which is promising considering how similar gene expression profiles are in samples of this nature (8-10). In the study by Knudsen (16), involving 10 samples of DCIS and IBC from different breasts, accuracy was 100% and 90% in the epithelial samples, and 90% and 70% in the stromal samples (both independently purified by LCM). Classification of DCIS and IBC in the study by Kretchmer (26), was 100% accurate for both types of samples. However, the sample (9 DCIS and 5 IBC) in the latter study were not micro-dissected and, therefore, each contained varying unknown amounts of tumor epithelium and stroma. The expression profile (heat map) of up-regulated genes from the 74-gene profile in this analysis showed some inconsistencies with the other cohorts (e.g., elevation and suppression of several collagens in three DCIS and two IBC samples, respectively), although the majority were consistent (especially involving down-regulated genes). Overall, the combined results strongly suggest that some of these genes may be useful clinically for molecular classification and diagnostic targeted imaging of DCIS, which will require additional studies.
Ontologies and Pathways Associated with Differentially Expressed Genes
To gain a general sense of biological processes, molecular functions, and cellular structures involved in the progression of DCIS to IBC, ontologies and pathways associated with differentially expressed genes were evaluated using Ontologizer 2.0 software (Table 2 and Supplementary Table S3) (25). Analysis of the 470 unique differentially expressed genes revealed many highly significant associations (adjusted p-value <0.01), including migration, proliferation, cell-cell adhesion, cell-substrate adhesion, angiogenesis, response to wounding, synthesis of extracellular matrix (ECM), organization of ECM, skin development, and response to glucocorticoids. Analysis of the 74-gene signature re-enforced many of the same functions. Based on analyses of groups 4e and 4s, ECM-related ontologies were primarily restricted to tumor epithelial cells, and a majority of genes involved were significantly elevated in IBCs versus DCIS. In contrast, genes involved in skin development were suppressed in the epithelium of IBCs. Changes in genes involved in angiogenesis and response to gluccocorticoids were primarily restricted to tumor stroma, and the majority of genes were suppressed in IBCs.
Table 2.
Selected ontologies associated with the progression of DCIS to IBC based on differentially expressed genes in this study.
| GO ID | GO Term | 470 Gene Profile |
74 Gene Profile |
Group 4 Epithelium |
Group 4 Stroma |
|---|---|---|---|---|---|
| Adjusted p-Value | |||||
| GO:0005578 | proteinaceous extracellular matrix | 1.8E-15 | 7.6E-18 | 7.3E-13 | ns |
| GO:0030198 | extracellular matrix organization | ns | 1.1E-06 | 7.7E-03 | ns |
| GO:0007155 | cell adhesion | 2.1E-09 | 1.3E-06 | ns | ns |
| GO:0016477 | cell migration | 1.0E-04 | ns | ns | ns |
| GO:0042127 | regulation of cell proliferation | 2.9E-03 | ns | ns | ns |
| GO:0019838 | growth factor binding | 3.1E-05 | 1.7E-02 | ns | ns |
| GO:0001568 | blood vessel development | 1.4E-05 | ns | ns | 1.3E-02 |
| GO:0009611 | response to wounding | 1.4E-05 | 1.7E-02 | ns | ns |
| GO:0043588 | skin development | 7.1E-03 | 9.5E-06 | 8.4E-04 | ns |
| GO:0051384 | response to glucocorticoid stimulus | 2.1E-03 | ns | ns | 7.1E-02 |
| Group 4 Epithelium | Differentially Expressed Genes in IBC vs. DCIS (Up and Down) | ||||
| proteinaceous extracellular matrix |
ASPN, CILP, COCH, COL10A1, COL11A1, COL1A2, COL3A1, COL5A1, COL6A3, COL8A1, CTHRC1, DCN, FBLN1, FBN1, LUM, MMP11, MMP12, OMD, POSTN, SPARC, SPOCK1, SPON1, VCAN, VEGFA |
||||
| extracellular matrix organization |
ASPN, CILP, COCH, COL10A1, COL11A1, COL1A2, COL3A1, COL5A1, COL6A3, COL8A1, CTHRC1, DCN, FBLN1, FBN1, LUM, MMP11, MMP12, OMD, POSTN, SPARC, SPOCK1, SPON1, VCAN, VEGFA |
||||
| skin development | COL1A2, COL3A1, COL5A1, NGFR, SFN, TP63 | ||||
| Group 4 Stroma | |||||
| blood vessel development |
C1GALT1, CASP8, CAV1, CDH5, COL5A1, COL8A1, FLT1, LMO2, MAPK1, MEOX2, PDGFA, ROBO4, SEMA5A, TGFBR2 |
||||
| response to glucocorticoid stimulus |
ADIPOQ, ADM, CAV1, CCL2, FABP4, GHR, SDC1 | ||||
In Vivo Functional Studies of Differentially Expressed Genes
The high degree of overlap between studies, impressive classification accuracy, and types of ontologies associated with the 74-gene profile suggest that some of the genes may be functionally important in the progression of DCIS to IBC. We investigated the function of selected genes using human DCIS-like cell lines and the MIND xenograft model (17). In this model, the cells grow as authentic DCIS (i.e. growing within ducts of intact mammary glands), whereas most previous studies utilized subcutaneous xenografts to draw conclusions about DCIS (12, 19).
Our initial studies focused on genes expressed at relatively high levels in samples of DCIS versus IBCs, hypothesizing that they normally suppress tumor progression. Ten total candidate suppressor genes from chosen for these studies, taken from the 74-gene profile (CHD1, CSTA, FAT1, DST, SERHL2, SYNM) or 470 gene list (KRT10, PHLDA2, SFN, TMEM45A). All have known functions which could reasonably be involved in the progression of DCIS (proteases, adhesion molecules, etc) and none except SFN and CDH1 have been studied in DCIS. The latter have both been shown to be suppressed in clinical IBC versus DCIS (27, 28). The DCIS-like cell lines we utilized included DCIS.COM and SUM225, which are the only widely available to the research community (20, 29), and a third, referred to as h.DCIS.01, which we recently developed in a manner similar to that used to develop DCIS.COM. DCIS.COM has been used almost exclusively in previous studies addressing gene function in human DCIS (20).
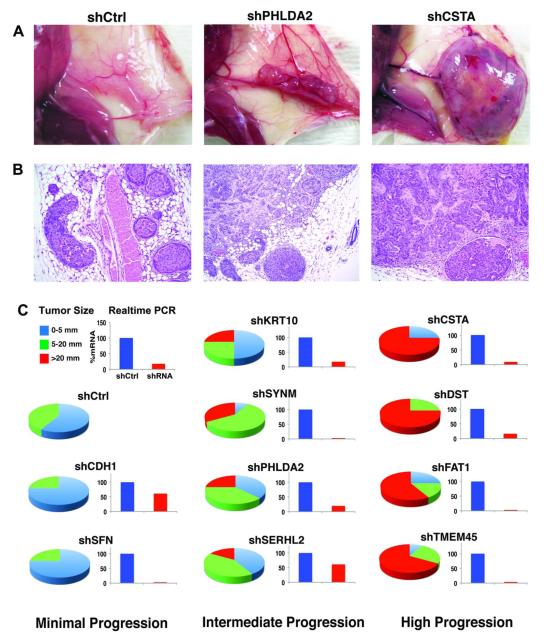
DCIS.COM cells expressed relatively high levels of all 10 candidate suppressor genes, and we predicted that knockdown would promote the progression of DCIS.COM to IBC MIND xenografts. Therefore, the cells were independently transduced with shRNAi corresponding to each gene (3 to 5 hairpins/gene), which decreased expression averaging about 80% for all genes (demonstrated with ≥ 2 hairpins/gene) compared to control cells (Supplementary Figure S2). Controls were transduced with shRNAi to red fluorescent protein (RFP), which was not expressed in the cells, to partially control for off-target effects, and to facilitate in vitro bioluminescence growth assays. As shown in Figure 3 and Supplementary Table S4, all transduced cell lines were able to grow and progress from DCIS to IBC in MIND xenografts, although at varying rates compared to control cells (parental and shRFP): two (shSFN and shCDH1) showed similar rates as controls; four (shKRT10, shSYNM, shPHLDA2, and shSERHL2) showed moderately increased rates; and four (shCSTA, shDST, shFAT1, and shTMEM45A) showed highly increased (p < 0.001) rates at 10 weeks. Xenografts ≤ 5 mm were about equally divided between DCIS alone and DCIS+IBC. All xenografts > 5 mm were composed of DCIS+IBC.
Figure 3. Effect of Modulating Gene Expression in DCIS.COM Cells on MIND Xenografts.
DCIS.COM cells were transduced with shRNAi corresponding to ten candidate invasion-suppressor genes (the cells originally expressed high levels of all genes). Four of the most efficiently suppressed genes (CSTA, DST, FAT1, TMEM45A) resulted in a highly significant increase in the progression of DCIS to IBC, consistent with the hypothesis that these genes function normally to suppress tumor invasion.
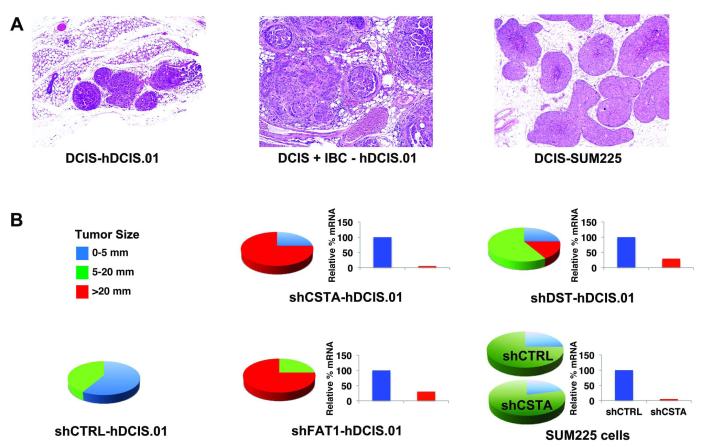
Similar experiments were conducted using h.DCIS.01 and SUM225 DCIS cells lines (Figure 4). h.DCIS.01 expressed high levels of CSTA, FAT1, and DST. It was transduced with the single most suppressive hairpin observed for each gene in DCIS.COM cells, which significantly reduced expression (average about 80%), and significantly (p<0.001) increased the growth and progression of DCIS to IBC xenografts, nearly identical to the results obtained with DCIS.COM. SUM225 cells expressed very low levels of CSTA, FAT1, and DST, and we predicted that further suppression would not influence tumor progression. Transducing shFAT1 and shDST was lethal in SUM225, so they could not be further evaluated. However, shCSTA resulted in about 50% suppression and, as predicted, growth and progression were not increased, although we cannot exclude the possibility that the level of suppression was inadequate.
Figure 4. Effect of Modulating Gene Expression in h.DCIS.01 and SUM225 Cells on MIND Xenografts.
h.DCIS.01 cells expressed high levels of CSTA, DST, and FAT1, and were transduced with the corresponding shRNAi which were most effective in suppressing expression in DCIS.COM cells. All three genes were highly suppressed, which resulted in significantly increased progression of DCIS to IBC xenografts, confirming results obtained with DCIS.COM. SUM225 cells expressed very low levels of all three genes. Further suppression using the same hairpins for CSTA and FAT1 were lethal, and further analysis was not possible. Further suppression of DST was not lethal and did not increase in the progression of DCIS to IBC xenografts.
Collectively, our microarray results in clinical samples, and in vivo results with human DCIS cell lines, provide compelling evidence that CSTA, FAT1, and DST normally suppress the progression of DCIS to IBC.
In Vitro Functional Studies of Suppressor Genes
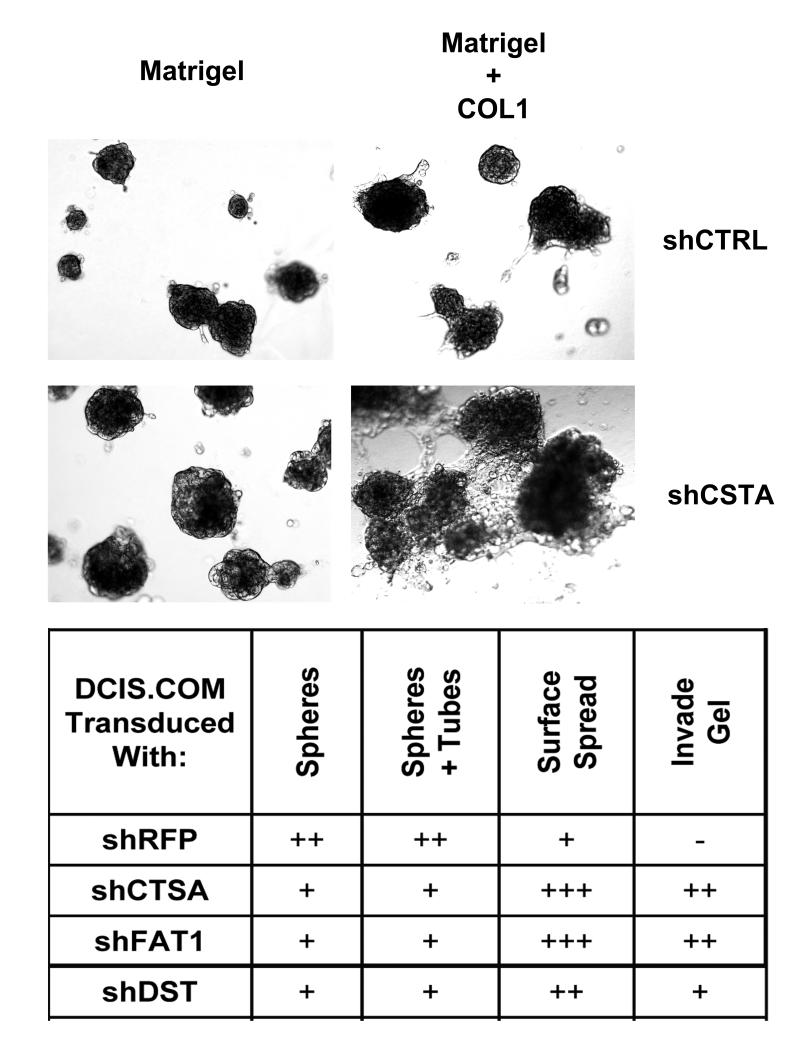
The progression of DCIS to IBC is a complex process involving many types of cell behaviors, including growth, migration, and invasion. To further characterize the functions of CSTA, FAT1, and DST observed in MIND xenografts, DCIS.COM.sh cells were evaluated in 3-dimensional cultures composed of Matrigel ± COL1 (Figure 5). All cells on Matrigel alone grew as small spheres. Control cells on Matrigel + COL1 formed small spheres which were occasionally connected by branching tubules. In contrast, shCSTA, shFAT1, and shDST cells spread and piled up on the gel surface, and invaded into the gels. Interestingly, COL1, which was necessary to reveal these altered behaviors, was expressed at highly elevated levels in clinical samples of human IBC versus DCIS, suggesting that it may be particularly important in enabling or promoting tumor invasion.
Figure 5. Effects of Modulating Gene Expression in DCIS.COM Cells in 3-Dimensional (3D) Culture.
To further characterize the functions of CSTA, FAT1, and DST observed in MIND xenografts, DCIS.COM.sh cells were evaluated in 3-dimensional cultures composed of Matrigel ± COL. All cells on Matrigel alone grew as small spheres. Control cells on Matrigel + COL1 formed small spheres which were occasionally connected by branching tubules. In contrast, shCSTA, shFAT1, and shDST cells spread and piled up on the gel surface, and invaded into the gels.
Other more quantifiable methods were also utilized to assess the growth (2-dimension culture), migration and invasion (both in Boydon chambers assays) of DCIS.COM and h.DCIS.01 cells transduced with shCSTA, shFAT1, and shDST (Supplementary Figure S3). Migration and invasion were significantly increased in both cell lines transduced with shFAT1. All other results were similar to controls, or highly variable and difficult to interpret. However, despite the negative or ambiguous results of some of these in vitro studies, especially involving shCSTA and shDST cells, tumor cell growth, migration, and invasion were unequivocally elevated in all corresponding MIND xenografts.
Discussion
Gene expression profiling has taught us a great deal about the progression of fully-developed IBC, and our study used this approach as a starting point to learn more about the progression of DCIS to IBC. We are aware of at least nine previous studies comparing expression between DCIS and IBC, publishing results which could be compared to ours (8-16, 30). Collectively, these studies included 130 cases of DCIS and 126 of IBC, which pales in comparison to previous expression profiling studies IBC alone involving thousands of cases. Our study increases the number of samples comparing DCIS to IBC by about 50%, which is a meaningful contribution given the relatively small numbers of cases overall addressing this important question.
A proportion of samples from this (17%) and the previous (37%) studies were paired DCIS and IBC from the same breasts, which may not be the ideal setting (at least exclusively) to identify invasion-regulating genes because some portion of these DCIS have already progressed to IBC, and may already possess some alterations associated with progression. Consistent with this, unsupervised hierarchical clustering based on expressed filtered genes of Group 3 in our study, composed of paired samples of DCIS and IBC from the same breasts, was able to distinguish the pairs, but not tumor type. A majority of samples from this (83%) and previous (63%) studies were IBC and DCIS from different breasts, and the DCIS were not associated with IBC in the same breast. Similar analyses of the samples in our study from different breasts (groups 1, 2, and 4) were more accurately categorized, presumably due to biological differences which may be related to tumor progression. The 74-gene profile was able to correctly classify 96% of all samples in this study, and about 90% of samples from two independent studies. Thus, some of these genes are promising targets for the development of improved methods of detection and diagnosis of DCIS (e.g. targeted imaging).
Identifying genes important in tumor progression from static measurements of expression by microarrays is challenging. The main criterion defining “important” in this study was the degree of overlap of specific genes between multiple studies comparing DCIS and IBC. Similarly, inferring ontologies and pathways important in tumor progression from differentially expressed genes is also challenging. Analyses of the 470 unique differentially expressed genes across all groups in this study, and the refined subset of 74 overlapping genes, converged on essentially the same ontologies and pathways. Nearly all have been implicated in the progression of many types of cancers over the years, including growth, adhesion, and motility. Our results support the obvious importance of these behaviors in the progression of DCIS to IBC, and point to many new genes which may be involved in these behaviors
Group 4 in this study enabled independent analyses of tumor epithelium and adjacent stroma, and both tissue compartments were implicated in many of the same ontologies - with a few interesting exceptions. For example, changes in genes associated with synthesis and organization of extracellular matrix (especially fibrillar collagens) were particularly prominent in IBC epithelial cells, consistent with epithelial-to-mesenchymal transformation (EMT), and adding to the molecular definition of EMT. Previous similar studies by Emory (31) and Knudsen (16) also observed elevated expression of ECM-related genes in IBC epithelium (31). In particular, the Knudsen study, identified and prioritized important ontologies in a manner almost identical to our study, which is scientifically reassuring, although the specific genes utilized for identification overlapped with our group 4 only about 20% and 10% in the epithelium and stroma, respectively.
Skin development was another prominent ontology restricted to epithelium, and genes regulating terminal differentiation and polarity of epidermal cells (e.g. SFN and TP63) were suppressed in IBCs. Breasts arise from budding skin appendages during embryonic development (32), and invasion of many types of solid cancers involves re-activation of embryonic pathways. Changes in genes responsive to glucocorticoids were prominent in stroma, and nearly all were suppressed in IBC versus DCIS, although the consequence of these changes on tumor progression is unclear. Angiogenesis as a GO term was restricted to stroma, but many genes regulating growth of endothelial cells were suppressed in IBCs (e.g. VEGFA, FLT1, LMO2, PDGFA, ROBO4), which runs counter to previous studies demonstrating increased angiogenesis in IBCs (33). However, most previous studies observed increased vascular density predominately at the leading edge of IBCs (34). The stroma in this study was adjacent to clusters of invasive epithelium primarily from the interior of tumors (each case was a single piece of tissue harvested prior to the study, so only a single edge, at best, could represent the leading edge) - and perhaps the regulation of angiogenesis is different in these locations. Regardless, this interesting issue requires further study.
Group 4s in this study, and the studies by MA (15) and Knudsen (16), are the only directly comparing transcriptomes of stroma immediately adjacent to tumor epithelial cells of DCIS and IBC. The samples in 4s were from different breasts, while those of Ma were pairs from the same breasts, and there was relatively little overlap of differentially expressed genes (12%), emphasizing the difference in study design. A study by Allinen (12) evaluated stromal gene expression by SAGE in samples from different breasts, and there was also little overlap with 4s (5%). However, this study is difficult to compare because expression was evaluated in purified subtypes of stromal cells rather than intact stroma. Like our study, Knudsen compared purified samples of stroma from IBC and pure DCIS in different breasts. However, based on published results, overlap of specific differentially expressed stromal genes was only about 10%, including COL11A1, TP63, MYH11, FABP4, and ENPP2, which are the only overlapping with our 74-gene profile. The reasons for the low level of overlap overall are unknown at the moment.
Overall, there are relatively few in vivo functional studies addressing the progression of human DCIS to IBCs, and all utilized DCIS.COM cells (20), primarily as subcutaneous xenografts in mice (12, 19). Our study also utilized DCIS.COM to study tumor progression, but in MIND xenografts, which more closely resemble human DCIS (17). In particular, the microenvironment in the mouse mammary ducts (myoepithelial cells, intact basement membrane, vasculature, etc.) is structurally similar to ducts in human breasts. The basement membrane may be particularly important. Interestingly, all 3 cell human DCIS cell lines used in this study somehow displaced the mouse myoepithelial cells, and made their own (Supplementary Figure S4). It is important to keep in mind, however, that all currently utilized human xenografts models have limitations in terms of relevance to human disease. Within these limitations, our studies based on three human DCIS-like cell lines strongly suggest that CSTA, FAT1, and DST normally function to suppress the progression of DCIS to IBC, which underscores the complexity of progression of human DCIS, given the diverse functions of these genes.
CSTA is a protease inhibitor which is thought to primarily inhibit cathepsin B activity (35). A previous study by Kleer (36) showed that in vitro invasion of DCIS.COM cells was significantly increased by co-culturing with fibroblasts expressing high levels of cathepsin K, which is closely related to cathepsin B. Low levels of CSTA, and elevated levels of cathepsin B, have been associated with poor prognosis in prostate (37) and head/neck cancers (35). Zajc (38) observed direct and indirect associations with cathepsin B and CSTA levels, respectively, with the degree of invasiveness of breast cancer cells lines in vitro. Our findings suggest that unchecked cathepsin activity (through loss of CSTA) is important in the progression of clinical DCIS.
DST is a cytoplasmic protein involved in cell adhesion and cytoskeletal organization (39). It regulates the polarity of squamous epithelium and auto antibodies against DST result in bullous pemphigoid, a blistering skin disease (40). DST expression is directly regulated by p63 in normal keratinocytes (41). Myoepithelial cells, which express high levels of p63, play an important role in maintaining differentiation and polarity of luminal epithelial cells in the breast. Hu (42) demonstrated that inhibiting the differentiation of myoepithelium in subcutaneous xenografts of DCIS.COM promoted progression to invasive cancer. Together with our results, this suggests that there may be an important link between p63 and DST in the progression of DCIS.
FAT1 encodes a surface membrane protein involved in cell adhesion, polarity, and signaling (43). It regulates cell proliferation and has tumor suppressor activity in Drosophila (43). Homozygous deletions of the FAT1 gene, and loss of protein expression, are common in human squamous cell carcinomas (44), in line with our results suggesting that suppression of FAT1 promotes the progression of DCIS.
In conclusion, new more effective methods of detecting, diagnosing, and treating DCIS could be developed based on targeting some of the genes identified in this study.
Supplementary Material
Acknowledgments
None
Funding This study was supported by the Breast Cancer Research Foundation through donation by Harry and Lillian Glassman, and NIH P50CA58183.
FINANCIAL SUPPORT: This study was supported by the Breast Cancer Research Foundation through donation by Harry and Lillian Glassman, and NIH P50CA58183.
Footnotes
CONFLICTS OF INTEREST: None to report.
References
- 1.Allred DC. Biological features of human premalignant breast disease and the progression to cancer. In: Harris JR, Lippman ME, Mlorrow M, Hellman S, Osborne CK, editors. Diseases of the Breast. 4th ed Lippincott Williams and Wilkins; Philadelphia: 2009. pp. 323–34. [Google Scholar]
- 2.Allred DC, Wu Y, Mao S, et al. Ductal carcinoma in situ and the emergence of diversity during breast cancer evolution. Clin Cancer Res. 2008;14:370–8. doi: 10.1158/1078-0432.CCR-07-1127. [DOI] [PubMed] [Google Scholar]
- 3.Society AC. Breast Cancer Facts and Figures 2011-2012. 2010.
- 4.Kuerer HM, Albarracin CT, Yang WT, et al. Ductal carcinoma in situ: state of the science and roadmap to advance the field. J Clin Oncol. 2009;27:279–88. doi: 10.1200/JCO.2008.18.3103. [DOI] [PubMed] [Google Scholar]
- 5.Page DL, Dupont WD, Rogers LW, Jensen RA, Schuyler PA. Continued local recurrence of carcinoma 15-25 years after a diagnosis of low grade ductal carcinoma in situ of the breast treated only by biopsy. Cancer. 1995;76:1197–200. doi: 10.1002/1097-0142(19951001)76:7<1197::aid-cncr2820760715>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 6.Virnig BA, Shamliyan T, Tuttle TM, Kane RL, Wilt TJ, editors. Diagnosis and Management of Ductal Carcinoma in Situ (DCIS) Agency for Healthcare Research and Quality, U.S. Department of Health and Human Services; Rockville MD: 2009. [PMC free article] [PubMed] [Google Scholar]
- 7.Allegra CJ, Aberle DR, Ganschow P, et al. State-of-the-Science Conference: Diagnosis and Management of Ductal Carcinoma in Situ (DCIS) J Natl Cancer Inst. 2010;102:161–9. doi: 10.1093/jnci/djp485. [DOI] [PubMed] [Google Scholar]
- 8.Porter D, Lahti-Domenici J, Keshaviah A, et al. Molecular markers in ductal carcinoma in situ of the breast. Mol Cancer Res. 2003;1:362–75. [PubMed] [Google Scholar]
- 9.Porter DA, Krop IE, Nasser S, et al. A SAGE (serial analysis of gene expression) view of breast tumor progression. Cancer Res. 2001;61:5697–702. [PubMed] [Google Scholar]
- 10.Ma XJ, Salunga R, Tuggle JT, et al. Gene expression profiles of human breast cancer progression. Proc Natl Acad Sci U S A. 2003;100:5974–9. doi: 10.1073/pnas.0931261100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abba MC, Drake JA, Hawkins KA, et al. Transcriptomic changes in human breast cancer progression as determined by serial analysis of gene expression. Breast Cancer Res. 2004;6:R499–513. doi: 10.1186/bcr899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allinen M, Beroukhim R, Cai L, et al. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell. 2004;6:17–32. doi: 10.1016/j.ccr.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 13.Hannemann J, Velds A, Halfwerk JB, Kreike B, Peterse JL, van de Vijver MJ. Classification of ductal carcinoma in situ by gene expression profiling. Breast Cancer Res. 2006;8:R61. doi: 10.1186/bcr1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schuetz CS, Bonin M, Clare SE, et al. Progression-specific genes identified by expression profiling of matched ductal carcinomas in situ and invasive breast tumors, combining laser capture microdissection and oligonucleotide microarray analysis. Cancer Res. 2006;66:5278–86. doi: 10.1158/0008-5472.CAN-05-4610. [DOI] [PubMed] [Google Scholar]
- 15.Ma XJ, Dahiya S, Richardson E, Erlander M, Sgroi DC. Gene expression profiling of the tumor microenvironment during breast cancer progression. Breast Cancer Res. 2009;11:R7. doi: 10.1186/bcr2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knudsen ES, Ertel A, Davicioni E, Kline J, Schwartz GF, Witkiewicz AK. Progression of ductal carcinoma in situ to invasive breast cancer is associated with gene expression programs of EMT and myoepithelia. Breast Cancer Res Treat. 2011 doi: 10.1007/s10549-011-1894-3. [DOI] [PubMed] [Google Scholar]
- 17.Behbod F, Kittrell FS, Lamarca H, et al. An intraductal human-in-mouse transplantation model mimics the subtypes of ductal carcinoma in situ. Breast Cancer Res. 2009;11:R66. doi: 10.1186/bcr2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee S, Medina D, Tsimelzon A, et al. Alterations of gene expression in the development of early hyperplastic precursors of breast cancer. Am J Pathol. 2007;171:252–62. doi: 10.2353/ajpath.2007.061010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu M, Yao J, Carroll DK, et al. Regulation of in situ to invasive breast carcinoma transition. Cancer Cell. 2008;13:394–406. doi: 10.1016/j.ccr.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allred DC, Medina D. The relevance of mouse models to understanding the development and progression of human breast cancer. J Mammary Gland Biol Neoplasia. 2008;13:279–88. doi: 10.1007/s10911-008-9093-5. [DOI] [PubMed] [Google Scholar]
- 21.Ignatoski Km Fau - Lapointe AJ, Lapointe Aj Fau - Radany EH, Radany Eh Fau - Ethier SP, Ethier SP. erbB-2 overexpression in human mammary epithelial cells confers growth factor independence. Endocrinology. 1999;140:3615–22. doi: 10.1210/endo.140.8.6939. [DOI] [PubMed] [Google Scholar]
- 22.Feeley L, Quinn CM. Columnar cell lesions of the breast. Histopathology. 2008;52:11–9. doi: 10.1111/j.1365-2559.2007.02890.x. [DOI] [PubMed] [Google Scholar]
- 23.Stewart SA, Dykxhoorn DM, Palliser D, et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. Rna. 2003;9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pazolli E, Luo X, Brehm S, et al. Senescent stromal-derived osteopontin promotes preneoplastic cell growth. Cancer Res. 2009;69:1230–9. doi: 10.1158/0008-5472.CAN-08-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bauer S, Grossmann S, Vingron M, Robinson PN. Ontologizer 2.0--a multifunctional tool for GO term enrichment analysis and data exploration. Bioinformatics. 2008;24:1650–1. doi: 10.1093/bioinformatics/btn250. [DOI] [PubMed] [Google Scholar]
- 26.Kretschmer C, Sterner-Kock A, Siedentopf F, Schoenegg W, Schlag PM, Kemmner W. Identification of early molecular markers for breast cancer. Mol Cancer. 2011;10:15. doi: 10.1186/1476-4598-10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoque MO, Prencipe M, Poeta ML, et al. Changes in CpG islands promoter methylation patterns during ductal breast carcinoma progression. Cancer Epidemiol Biomarkers Prev. 2009;18:2694–700. doi: 10.1158/1055-9965.EPI-08-0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoon NK, Seligson DB, Chia D, et al. Higher expression levels of 14-3-3sigma in ductal carcinoma in situ of the breast predict poorer outcome. Cancer Biomark. 2009;5:215–24. doi: 10.3233/CBM-2009-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gupta PB, Kuperwasser C. Disease models of breast cancer. Drug Discovery Today: Disease Models. 2004;1:9–16. [Google Scholar]
- 30.Castro NP, Osorio CA, Torres C, et al. Evidence that molecular changes in cells occur before morphological alterations during the progression of breast ductal carcinoma. Breast Cancer Res. 2008;10:R87. doi: 10.1186/bcr2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emery LA, Tripathi A, King C, et al. Early dysregulation ofecll adhesion and extracellular matrix pathways in breast cancer progression. Am J Pathol. 2009;175:1292–302. doi: 10.2353/ajpath.2009.090115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cowin P, Wysolmerski J. Molecular mechanisms guiding embryonic mammary gland development. Cold Spring Harb Perspect Biol. 2010;2:a003251. doi: 10.1101/cshperspect.a003251. (online) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kerbel RS. Tumor angiogenesis. N engl J Med. 2008;358:2039–49. doi: 10.1056/NEJMra0706596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uzzan B, Nicolas P, Cucherat M, Perret GY. Microvessel density as a prognostic factor in women with breast cancer: a systematic review of the literature and meta-analysis. Cancer Res. 2004;64:2941–55. doi: 10.1158/0008-5472.can-03-1957. [DOI] [PubMed] [Google Scholar]
- 35.Strojan P, Budihna M, Smid L, et al. Prognostic significance of cysteine proteinases cathepsins B and L and their endogenous inhibitors stefins A and B in patients with squamous cell carcinoma of the head and neck. Clin Cancer Res. 2000;6:1052–62. [PubMed] [Google Scholar]
- 36.Kleer CG, Bloushtain-Qimron N, Chen YH, et al. Epithelial and stromal cathepsin K and CXCL14 expression in breast tumor progression. Clin Cancer Res. 2008;14:5357–67. doi: 10.1158/1078-0432.CCR-08-0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sinha AA, Quast BJ, Wilson MJ, et al. Ratio of cathepsin B to stefin A identifies heterogeneity within Gleason histologic scores for human prostate cancer. Prostate. 2001;48:274–84. doi: 10.1002/pros.1107. [DOI] [PubMed] [Google Scholar]
- 38.Zajc I, Sever N, Bervar A, Lah TT. Expression of cysteine peptidase cathepsin L and its inhibitors stefins A and B in relation to tumorigenicity of breast cancer cell lines. Cancer Lett. 2002;187:185–90. doi: 10.1016/s0304-3835(02)00452-4. [DOI] [PubMed] [Google Scholar]
- 39.Sonnenberg A, Liem RK. Plakins in development and disease. Exp Cell Res. 2007;313:2189–203. doi: 10.1016/j.yexcr.2007.03.039. [DOI] [PubMed] [Google Scholar]
- 40.Hamill KJ, Hopkinson SB, DeBiase P, Jones JC. BPAG1e maintains keratinocyte polarity through beta4 integrin-mediated modulation of Rac1 and cofilin activities. Mol Biol Cell. 2009;20:2954–62. doi: 10.1091/mbc.E09-01-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Osada M, Nagakawa Y, Park HL, et al. p63-specific activation of the BPAG-1e promoter. J Invest Dermatol. 2005;125:52–60. doi: 10.1111/j.0022-202X.2005.23801.x. [DOI] [PubMed] [Google Scholar]
- 42.Hu M, Polyak K. Molecular characterisation of the tumour microenvironment in breast cancer. Eur J Cancer. 2008;44:2760–5. doi: 10.1016/j.ejca.2008.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Katoh Y, Katoh M. Comparative integromics on FAT1, FAT2, FAT3 and FAT4. Int J Mol Med. 2006;18:523–8. [PubMed] [Google Scholar]
- 44.Nakaya K, Yamagata HD, Arita N, et al. Identification of homozygous deletions of tumor suppressor gene FAT in oral cancer using CGH-array. Oncogene. 2007;26:5300–8. doi: 10.1038/sj.onc.1210330. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.