Abstract
Hox genes are classically ascribed to function in patterning the anterior-posterior axis of bilaterian animals; however, their role in directing molecular mechanisms underlying morphogenesis at the cellular level remains largely unstudied. We unveil a non-classical role for the zebrafish hoxb1b gene, which shares ancestral functions with mammalian Hoxa1, in controlling progenitor cell shape and oriented cell division during zebrafish anterior hindbrain neural tube morphogenesis. This is likely distinct from its role in cell fate acquisition and segment boundary formation. We show that, without affecting major components of apico-basal or planar cell polarity, Hoxb1b regulates mitotic spindle rotation during the oriented neural keel symmetric mitoses that are required for normal neural tube lumen formation in the zebrafish. This function correlates with a non-cell-autonomous requirement for Hoxb1b in regulating microtubule plus-end dynamics in progenitor cells in interphase. We propose that Hox genes can influence global tissue morphogenesis by control of microtubule dynamics in individual cells in vivo.
Keywords: Hoxb1b, Hoxa1, Cell polarity, In vivo tissue morphogenesis, Neural tube formation, Oriented cell division, Zebrafish
INTRODUCTION
Hox genes are an evolutionarily conserved cluster of homeodomain-containing transcription factors traditionally implicated in patterning the anterior-posterior body axis of animals (Duboule, 2007; Iimura and Pourquié, 2007) and crucial for a set of developmental programs. Many studies focused on the role of Hox genes in determining cell identity and revealed the concepts of Hox code and Hox co-linearity. Few Hox-controlled ‘realisator’ genes - the proposed downstream targets of Hox genes that control morphogenesis (García-Bellido, 1975) - have been identified, and our understanding of whether and how Hox genes impact the basic cell properties, such as cell shape or polarization, that govern tissue level morphogenesis are so far rudimentary.
Control of cell polarity is essential for the generation of three-dimensional tissues and organs of correct size and shape. A fundamental goal is to understand how genetic control of individual cell polarization determines coherent morphogenetic events at the tissue level in vivo. Here, we focus on the zebrafish hindbrain neural tube lumen formation as a paradigm of how single cell polarization determines global tissue morphogenesis in vertebrates in vivo. In the immature neuroepithelium of the neural keel, the apical surfaces of elongated progenitor cells from the left and right sides of the neuroepithelium interdigitate at the presumptive midline. This period is characterized by stereotyped polarized mitoses, with mitotic cells first assembling a mitotic spindle in planar orientation relative to the tissue, followed by a stereotyped 90° spindle rotation into apico-basal orientation (Geldmacher-Voss et al., 2003). This polarized spindle orientation is required to distribute daughter cells to either side of the midline and for the timely formation of a straight, single neural tube lumen (Ciruna et al., 2006; Tawk et al., 2007; Quesada-Hernández et al., 2010; Žigman et al., 2011).
How spindle orientation is developmentally determined is less clear. In fission yeast, intrinsic control of the rod-shape cell geometry is thought to impose interphase microtubule (MT) alignment as the determinant for alignment of the duplicated spindle pole bodies and consequently the orientation of the mitotic spindle and cell division axis (Vogel et al., 2007; Daga and Nurse, 2008). How cell division orientation is coordinated in a cell embedded within a tissue is more complex, as mitotic spindle positioning can be influenced by extrinsic cues, including cell shape, cell-cell adhesion, morphogen signals and extracellular matrix organization (Minc and Piel, 2012).
Here, we identify a non-classical role for zebrafish Hoxb1b, which shares ancestral functions with mammalian Hoxa1 (McClintock et al., 2001), in controlling the cytoskeletal organization and mitotic spindle orientation of neuroepithelial progenitors as a regional determinant for morphogenesis of the neural tube.
RESULTS
A zebrafish hoxb1b mutant has classical homeotic transformations of the anterior hindbrain segments and derived pharyngeal arch structures
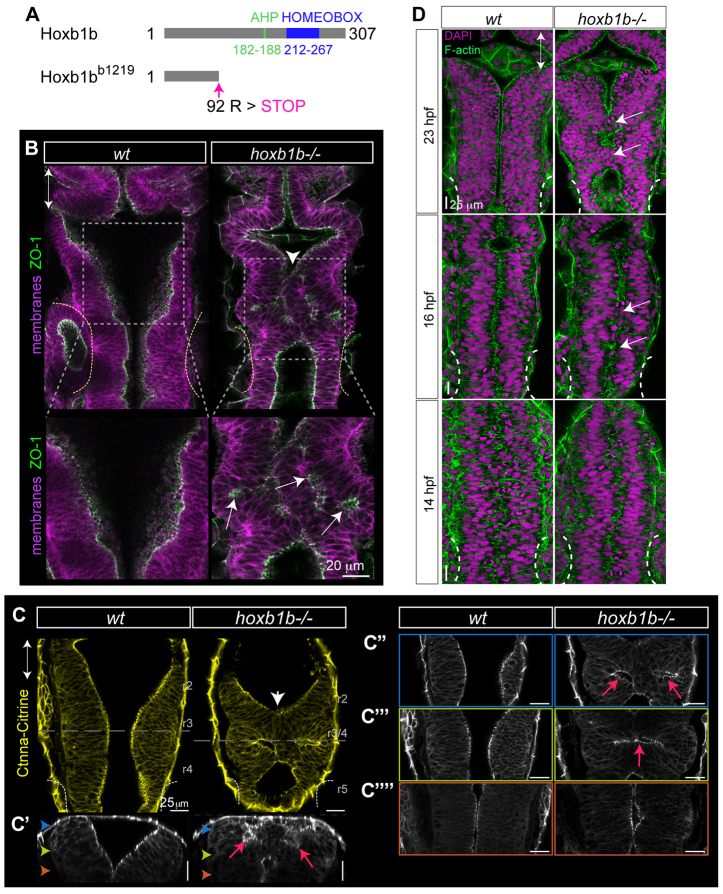
We identified a zebrafish hoxb1b mutant, hoxb1bb1219, by TILLING (Draper et al., 2004) that carries a point mutation in the first exon of hoxb1b. The A-to-T transversion at position 274 leads to a premature stop codon at amino acid 92 (R92X) of this 307 amino acid protein, upstream of the essential DNA-binding homeobox motif (Fig. 1A). This is the only nonsense mutation in a 6.5 Mb interval linked to the phenotype (Miller et al., 2013).
Fig. 1.
Hoxb1b is required for hindbrain lumen morphogenesis. (A) Schematic of wild-type (Hoxb1b) and mutant (Hoxb1bb1219) protein domains of Danio rerio hoxb1b, with Antenapedia-type hexapeptide (AHP), DNA-binding homeobox domain and designated premature stop codon in Hoxb1bb1219 mutant protein. (B-C′′) Abnormal anterior hindbrain lumen morphology at r3/4 in hoxb1b-/- compared with wild type at 22 hpf. (B) ZO-1 (apical tight junction) immunostaining; (C) Gt(Ctnna-Citrine)ct3a (adherens junctions; live embryos). C′-C′′ are progressively deeper z-sections of the dorsal view in C; C′ is a sagittal reconstruction at r3/4. Note duplicated lumina in the dorsal hindbrain (red arrows). Colored arrowheads in C′ indicate planes of subsequent panels. White arrowheads and white arrows in B and C indicate neural tube lumen abnormalities. In B, lower panels are higher magnifications of boxed areas in upper panels. (D) Onset of r3/4 hindbrain morphology defects at 14, 16 and 23 hpf in hoxb1b-/- compared with wild-type hindbrain development. White arrows point to lumen discontinuities. Anterior-posterior axis marked by double arrows in all panels.
As expected, the hoxb1bb1219/b1219 (henceforth referred to as hoxb1b-/-) has phenotypes associated with Hoxa1 mutation in the mouse and hoxb1b morpholino knockdown in the zebrafish (Chisaka and Capecchi, 1991; Lufkin et al., 1991; Carpenter et al., 1993; Gavalas et al., 1998; Rossel and Capecchi, 1999; Barrow et al., 2000; McClintock et al., 2001). These include a small otic vesicle, absence of hoxa2 (hoxa2b - Zebrafish Information Network) expression in the second pharyngeal arch neural crest cells (supplementary material Fig. S1A), fusion of first and second arch neural crest streams (supplementary material Fig. S1B) and consequent fusion of the first and the second branchial arch cartilages (supplementary material Fig. S1C).
Hox genes are crucial for establishing segmental patterning of the vertebrate hindbrain into rhombomeres (r) and for the formation of segment boundaries (Lumsden and Krumlauf, 1996; Iimura and Pourquié, 2007). We found that hoxb1b-/- embryos lack proper segmental organization between r2 and r5 (supplementary material Fig. S1D,E). Consistent with this, the egr2 transgenic reporter Tg(egr2b:KalTA4) (Distel et al., 2009) in hoxb1b-/- embryos is ectopically activated in neuroepithelial progenitors outside of r3 and r5 (supplementary material Fig. S1E). Moreover, the large Mauthner interneurons that lie in r4 of wild-type embryos are absent in hoxb1b-/- embryos (supplementary material Fig. S1F) (McClintock et al., 2002).
To analyze whether hoxb1b is required for segment identity, we transplanted labeled hoxb1b-/- mutant cells unilaterally into the presumptive hindbrain of wild-type Tg(egr2b:KalTA4) hosts (supplementary material Fig. S1G). We find that whereas wild-type progenitor cells contribute evenly throughout the hindbrain, hoxb1b-/- cells sort-out from r3, r4 and r5, but contribute normally to more anterior and posterior segments. Thus, hoxb1b-/- cells cannot properly assume r3-r5 rhombomere identities. This cell-autonomous requirement for hoxb1b in r3 is unexpected, given that hoxb1b was not previously thought to be expressed anterior to the r3/r4 boundary (McClintock et al., 2001). However, a recent lineage study in the mouse has shown that Hoxa1 expression extends into r3 (Makki and Capecchi, 2011). Our observation of a cell-autonomous requirement for hoxb1b in the specification of r3 identity thus supports these findings in mouse. Altogether, we reveal the same phenotypes in hoxb1b-/- as previously described in hoxb1b morphants and Hoxa1 mouse mutants, indicative of classical Hox gene loss-of-function phenotypes.
hoxb1b mutants have a regionally restricted defect in neuroepithelial morphogenesis
The mammalian and teleost neural tube forms via folding of the neural plate. Whereas in zebrafish, neural plate cells invaginate to form the neural keel and the neural tube lumen opens secondarily, in mammals the lateral folds of the neural plate rise up and fuse dorsally to form a neural tube de novo (Lowery and Sive, 2004). Hoxa1 mouse mutants show defects in neural tube closure (Lufkin et al., 1991); however, no effect of Hox gene abrogation on neural tube morphogenesis in zebrafish has been described. In addition to the expected hoxb1b mutant phenotypes described above, we unexpectedly found that hoxb1b-/- embryos exhibit abnormal hindbrain architecture with a discontinuous lumen at the level of presumptive r3 and r4 (therefore termed r3/4) (Fig. 1A,B). In dorsal and transverse optical sectioning of live hoxb1b-/-;Gt(Ctnna-Citrine)ct3a embryos (Fig. 1C), we mapped the tissue structure defects specifically to the dorsal part of r3/4, often resulting in duplicated small lumina at lateral positions.
To uncover the critical time window for Hoxb1b function in normal hindbrain morphogenesis, we analyzed F-actin and nuclei in whole-mount samples (Fig. 1D). The onset of abnormalities is between 14 and 16 hours post fertilization (hpf), when defects in cellular orientation become apparent. This is at the time when neuroepithelial progenitors undergo stereotypic, symmetrical oriented cell divisions. hoxb1b-/- embryos at 11 hpf or earlier are indistinguishable from their wild-type siblings. To verify whether these effects on hindbrain morphogenesis are due to mutation in the hoxb1b gene, we ectopically expressed full-length hoxb1b in wild type and hoxb1b-/-. While this approach is complicated by the fact that constitutive hoxb1b expression causes brain morphogenesis defects, we found that injection of hoxb1b mRNA could rescue otherwise abnormal r3/4 neuroepithelial cell organization in about one quarter of hoxb1b-/- embryos, confirming that the morphogenesis defect in hoxb1b-/- is due to loss of functional Hoxb1b (supplementary material Fig. S1H,H′). Thus, the neuroepithelial morphogenesis defect we observe is due to hoxb1b loss of function.
Hoxb1b does not disrupt classical apico-basal or planar neuroepithelial cell polarization
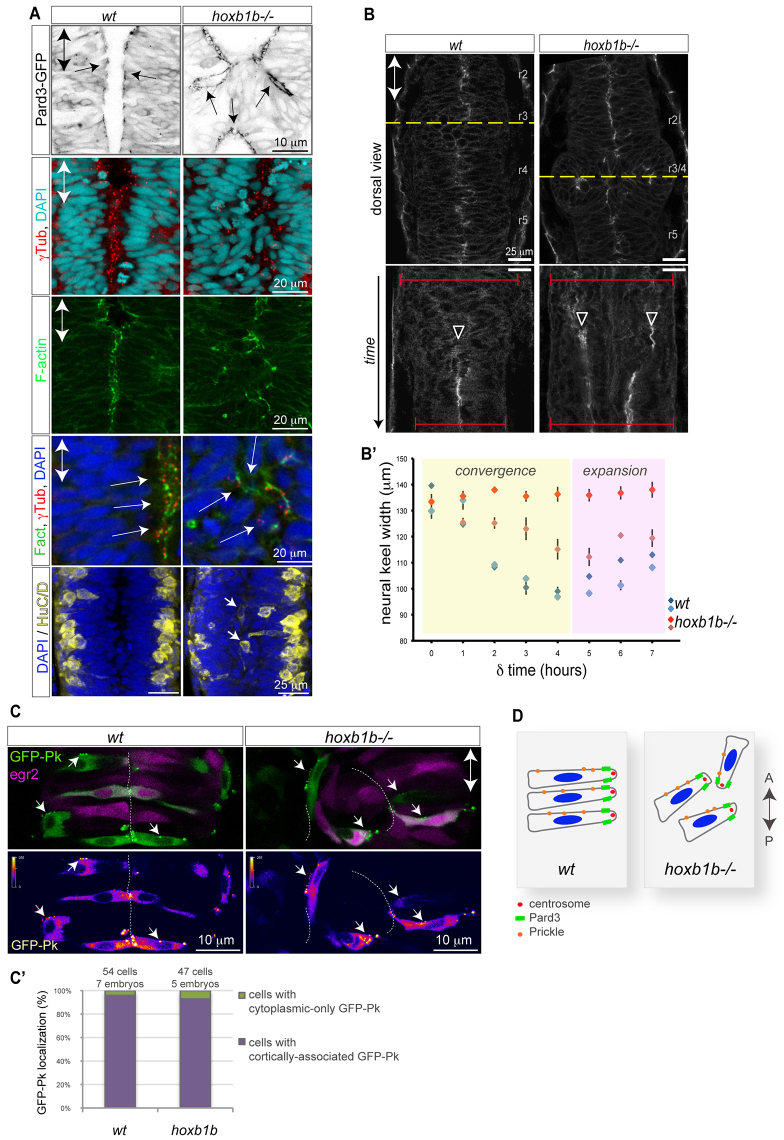
As the apical polarity Par components are required for proper lumen formation (Tawk et al., 2007; Munson et al., 2008; Buckley et al., 2013), Hoxb1b could have an instructive role in the localization of the Par complex in r3-r5. Pard3-GFP (Tawk et al., 2007) localizes to the subapical cortex in wild type and is not altered in hoxb1b-/- mutants at the single-cell level (Fig. 2A). The subapical localization of the tight junctional component ZO-1, the apical position of centrosomes and the apical enrichment of F-actin fibers are all normal at the cellular level in hoxb1b-/- embryos (Fig. 1B; Fig. 2A). However, the cellular orientation of neuroepithelial progenitors was strikingly abnormal in r3/4 of hoxb1b-/- embryos (Fig. 2A). These results suggest that although the hoxb1b mutation does affect individual cell orientation and thereby tissue architecture, it does not affect apico-basal or tight junction-based lumen control.
Fig. 2.
Normal cell polarity but disrupted convergence in the hoxb1b-/- neural keel. (A) Normal neuroepithelial polarity at the single-cell level in hoxb1b-/- based on sub-apical Pard3-GFP (black) at 21 hpf, apical centrosomes (γ-tubulin, red) and apically enriched F-actin (green), all analyzed at 19 hpf. Neuronal differentiation occurs at wild-type levels but generates HuC/D-positive neurons (yellow) in abnormal medial postitions in hoxb1b-/- at 20 hpf. (B) Dynamics of neural keel convergence in time-lapses of wild-type and hoxb1b-/- transgenic Gt(Ctnna-Citrine)ct3a embryos with multiphoton imaging. Upper panels show a single z-section in dorsal view one-third of the way through the time lapse, corresponding to about the 10-somite stage. Lower panels: kymograph of the position marked by a yellow dotted line, beginning at the 3-somite stage (11 hpf) and continuing until the 18 somite stage (18 hpf). A single lumen appears at the midline in wild type (arrowhead) whereas a duplicated midline appears at about the same stage in hoxb1b-/-. The width of the neural keel is depicted as red brackets. (B′) Quantification of neural keel width measured at three positions in r3/4 over the period in the kymograph. Convergence is significantly slowed in hoxb1b-/- compared with wild-type siblings (n=2 embryos for each genotype). Data are mean ± s.e.m. (C) Assessment of planar cell polarity by GFP-Prickle (green) localization at 18 hpf. Tg(egr2b:KalTA4) transgene (purple) is included to identify rhombomere 3. Mosaically expressed GFP-Pk localizes in puncta, which in wild type lie on anterior progenitor cell membranes (white arrows). In hoxb1b-/-, GFP-Pk localizes to puncta that lie on one side of mis-oriented progenitor cells. Lower panels show GFP-Pk signal presented in pseudocolors. (C′) Quantitative analysis of GFP-Prickle subcellular localization. (D) Schematic of cell polarity in hoxb1b-/-. The anterior-posterior animal axes are marked by double arrows in all panels.
hoxb1b could have a more general effect on progenitor cell proliferation or neurogenesis. Indeed, knockdown of HOXA1 in human fibroblasts impairs progression through G1 phase (Trapnell et al., 2013). We tested whether Hoxb1b is required for proper generation of Huc/D+ neurons within the hindbrain. In wild-type controls as well as in hoxb1b-/-, Huc/D+ neurons are generated in comparable amounts and positioned laterally; however, interestingly, hoxb1b-/- embryos reveal ectopic Huc/D+ neurons at medial positions consistent with the locally duplicated neuroepithelial structure (Fig. 2A). Furthermore, we detect no evidence of decreased cell proliferation in the r3/4 region of hoxb1b-/- (Fig. 3G). Thus, Hoxb1b is not generally required in neuropeithelial progenitors to proliferate or to generate differentiated neurons.
Fig. 3.
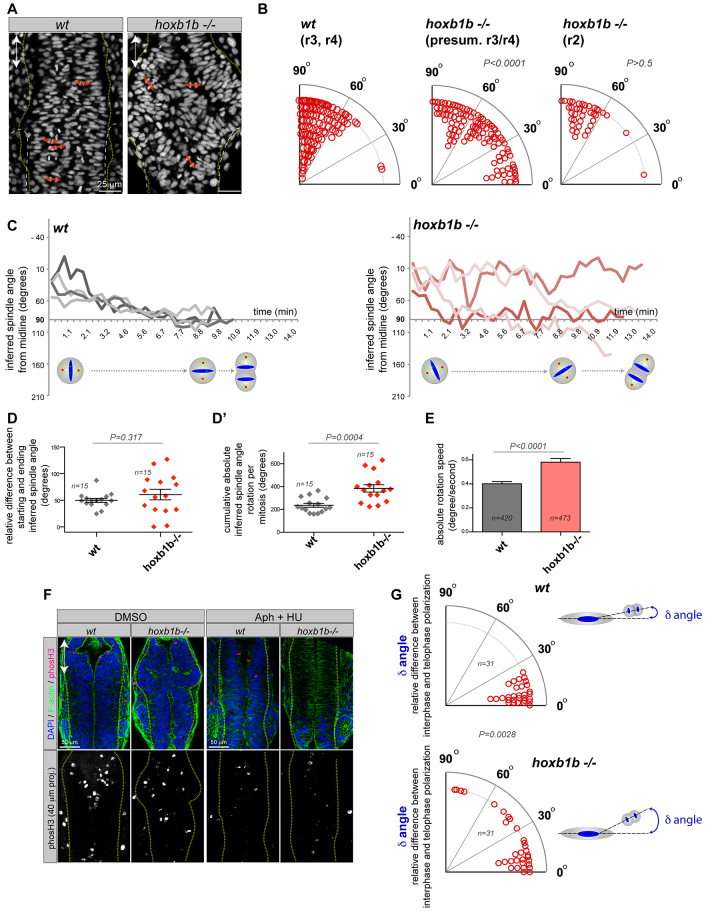
Hoxb1b regulates oriented cell division in the neural keel. (A) Tg(h2a.f/z-GFP) localizes to chromosomes that at anaphase reflect the orientation of cell division in the hindbrain (red arrows). (B) Quantification of mitosis orientation relative to the animal A/P axis (0°) in wild-type (n=153 cells, two embryos) and hoxb1b-/- (n=142 cells in r3/4 of hoxb1b-/- mutants with significantly abnormal mitosis orientation and n=41 cells in unaffected hoxb1b-/- r2 hindbrain region, three embryos for each). Imaging acquired at neural keel (12-15 hpf). (C) Representation of inferred spindle rotation relative to the A/P axis (0°) from time-lapse analysis of four wild-type and four hoxb1b-/- randomly chosen progenitors; additional tracks are shown in supplementary material Fig. S3A; the quantitation shown in D, D′ and E is based on n=15 single cell time lapses from each of three wild-type and three mutant embryos. Time interval is 21 seconds. (D) Relative difference between starting and ending inferred spindle orientation in wild-type and hoxb1b-/- mutant progenitors. (D′) Cumulative absolute spindle rotation (inferred from metaphase plate orientation) in wild type versus hoxb1b-/-. (E) Increased mitotic spindle rotation velocity (change in angle over time) in hoxb1b-/- compared with wild-type mitoses. (D-E) Graphs show the mean value with s.e.m. Hoxb1b is required for single-cell chromosome rotation independently of the prior interphase positioning. Quantitation of single cell behavior tracked over time when analyzed for the angle between the longest interphase nuclear axis prior to cell division and the axis of chromosome separation. (F) Rescue of abnormal lumen morphology by aphidicolin (Aph) and hydroxyurea (HU) incubation to block cell division in hoxb1b-/- (eight out of 11 hoxb1b-/- embryos) compared with control DMSO-treated hoxb1b-/- (seven out of eight hoxb1b-/- embryos). The lower panel shows the inhibition of proliferation caused by the drugs as verified by phosho-H3 staining. Double arrows mark the anterior-posterior axis. (G) Relative difference in the orientation (angle, degrees) between interphase and anaphase measured in individual wild-type and hoxb1b-/- cells, as schematized on the right.
Cadherin-based neuroepithelial cell-cell adhesion is required for proper neural tube formation (Hong and Brewster, 2006) and we previously described that disorganization of the neural tube lumen correlates with reduced cell-cell adhesion in the keel using a transgenic α-Catenin gene-trap reporter, Ctnna-Citrine (Žigman et al., 2011). To examine whether hoxb1b-/- cells have abnormal levels of cell-cell adhesion, we analyzed Ctnna-Citrine in wild-type and hoxb1b-/-; Gt(Ctnna-Citrine)ct3a transgenic embryos (supplementary material Fig. S2A). We could not detect significant differences in Ctnna-Citrine levels or localization in hoxb1b-/- embryos compared with sibling controls at the cell or tissue level in the neural keel (supplementary material Fig. S2A). Moreover, if Hoxb1b has an effect on cell-cell adhesion properties, a putative adhesion impairment would be detectable in genetic mosaics (Lele et al., 2002). Although hoxb1b-/- cells sorted out of r3-r5 (supplementary material Fig. S1G), cell clumping and cell group segregation away from wild-type host cells, typical of loss of cell-cell adhesion molecules (Žigman et al., 2011), was not detectable. Thus, Hoxb1b is unlikely to be required for neuroepithelial morphogenesis via regulation of cell-cell adhesion properties.
Planar cell polarization (PCP) is required for proper neural tube lumen formation (Ciruna et al., 2006; Tawk et al., 2007; Quesada-Hernández et al., 2010). A duplication of the neural tube lumen has been associated with loss of PCP signaling in zebrafish, which results in delayed convergence of the neural keel (Tawk et al., 2007). We detected locally duplicated lumen structures at r3/4 in hoxb1b-/- (Fig. 1B,C; supplementary material Movie 1). Therefore, we set out to analyze quantitatively the velocity of neural keel convergence using four-dimensional time-lapse imaging (Fig. 2B,B′). Measuring the width of the neural keel at r3/4 positions over time (Fig. 2B, lower panels), we find that the r3/4 hoxb1b-/- neuroepithelium converges more slowly than more anterior or more posterior unaffected regions of hoxb1b-/- mutants or the r3/4 region of wild type (Fig. 2B′). After the neural tube lumen is formed, inflation of the hindbrain ventricle causes the hindbrain to widen rapidly (Fig. 2B′, pink area). This expansion is significantly faster in wild type compared with hoxb1b-/- (Fig. 2B′). Because delayed neural keel convergence has been thought to be a major contributor to neural tube defects in PCP mutants (Tawk et al., 2007), our results suggest that local tissue convergence abnormalities in hoxb1b-/- hindbrain may contribute to the later neural tube abnormalities. However, the delay in neuroepithelial convergence that we observe in hoxb1b mutants is milder than in PCP mutants and occurs later than the convergence defect of PCP mutants (Ciruna et al., 2006; Tawk et al., 2007).
To address directly the role of Hoxb1b in regulating PCP, we assessed the subcellular localization of an established PCP marker, GFP-Prickle (GFP-Pk) (Tawk et al., 2007; Ciruna et al., 2006; Žigman et al., 2011). In wild-type embryos, GFP-Pk localizes to puncta on the anterior membranes of neuroepithelial cells (Ciruna et al., 2006) (Fig. 2C). GFP-Pk localization is cytoplasmic in hindbrain cells of PCP mutants (Ciruna et al., 2006; Žigman et al., 2011). In hoxb1b-/- embryos, GFP-Pk localizes in clearly defined foci in individual misoriented hoxb1b-/- cells, just as in wild-type controls (Fig. 2C,C′), indicating that PCP signaling is intact in the neural keel progenitors when cellular abnormalities in hoxb1b-/- become detectable (Fig. 2C,C′). Furthermore, as we show below, hoxb1b has effects on MT dynamics that are not observed in PCP mutants (Žigman et al., 2011) (supplementary material Fig. S4). Therefore, Hoxb1b is unlikely to be involved in regulation of either apico-basal or classical PCP signaling (Fig. 2D).
Morphogenesis defects are not the result of loss of segment boundaries or of the r4 organizer
Hox-1 genes in zebrafish are known to be required for the establishment of an r4 FGF signaling center in zebrafish (Waskiewicz et al., 2002; Maves and Kimmel, 2005) (supplementary material Fig. S2B). We asked whether the loss of the r4-FGF-signaling hindbrain organizer is responsible for the morphogenesis defects in hoxb1b mutant embryos and found that this is not the case. Embryos treated with the FGF inhibitor SU5402 or embryos expressing an inducible dnFGFR1 (Lee et al., 2005; Lee et al., 2005; Lee et al., 2005) have no hindbrain morphogenesis defect in spite of having an array of other well-established FGF signaling associated phenotypes (supplementary material Fig. S2C).
Segment boundaries can organize epithelial morphogenesis (Lee et al., 2005; Zallen and Wieschaus, 2004; Lee et al., 2005; Zallen and Wieschaus, 2004; Zallen and Wieschaus, 2004). Because the loss of hindbrain segment identity in hoxb1b-/- results in disrupted segment boundaries in the anterior hindbrain, we asked whether neuroepithelial disorganization is a general outcome in mutants that lack segment boundaries. However, this is not the case: both egr2b/krox20fh227 mutants and morphants for the essential Hox co-factors pbx2 and pbx4 lack hindbrain segment boundaries, but neither have neuroepithelial morphogenesis defects like those in hoxb1b-/- embryos (supplementary material Fig. S2D,E). Furthermore, because pbx2;pbx4 morphants lack all Hox activity during hindbrain development (Waskiewicz et al., 2002), the lack of a broad neuroepithelial morphogenesis defect in pbx2;pbx4 morphants suggests that orchestrating neuroepithelial morphogenesis on a segment-by-segment basis is not a general role of Hox genes.
Loss of Hoxb1b affects mitotic orientation
Finally, normal neural tube lumen formation depends on orientation of cell divisions in the neural keel (Quesada-Hernández et al., 2010; Žigman et al., 2011). We investigated whether Hoxb1b could be acting as a regional determinant of neural tube morphogenesis by controlling the orientation of mitoses in the neural keel by analyzing cell divisions in Tg(h2a.z/f-GFP) transgenic embryos. Cells in the wild-type neural keel divide at an angle that is within 30° of perpendicular to the anterior-posterior (A/P) axis (Geldmacher-Voss et al., 2003) (apico-basal cell division; Fig. 3A,B; supplementary material Movie 2), as do cells within the unaffected r2 region of hoxb1b-/- (Fig. 3B). However, within the r3/4 region of hoxb1b-/- mutants, cells divide at random angles relative to local tissue axis (Fig. 3A,B; supplementary material Movie 2). This suggests a very specific local effect on mitosis orientation within the region that develops lumen architecture defects.
To better understand the effect of Hoxb1b on the orientation of cell division, we used analysis of dividing cells in live Tg(h2a.z/f-GFP) transgenics to quantify the kinetics of metaphase plate rotation, from which we infer the mitotic spindle orientation (Geldmacher-Voss et al., 2003; Žigman et al., 2011). In wild-type embryos at the neural keel stage, the orientation of cell division is highly stereotyped. The mitotic spindle is set up at prometaphase approximately parallel to the anterior-posterior axis and rotates into a medio-lateral (apico-basal) orientation by anaphase (Fig. 3C shows select traces; supplementary material Fig. S3C shows 15 traces). By contrast, the r3/4 region of hoxb1b-/- embryos metaphase plate rotation is abnormal (Fig. 3C-E; supplementary material Fig. S3A). In hoxb1b-/-, the difference in inferred spindle angle between prometaphase and anaphase is widely variable (Fig. 3D), the absolute amount of spindle rotation is significantly higher than for wild-type mitoses (Fig. 3D′), and the absolute spindle rotation speed is significantly faster, with more extensive rocking movements (Fig. 3C,E). Importantly, these results suggest either the absence of cell contact-mediated cues for spindle orientation or the cell-intrinsic ability to detect them (see below).
The final spindle orientation of wild-type neuroepithelial progenitors in the neural keel is parallel to the long axis of the cell in interphase (Fig. 3F). Because single cells within hoxb1b-/- neural keel tissue are misoriented in interphase, we considered whether single hoxb1b-/- cells were undergoing proper mitotic spindle rotation relative to their long axis, but aberrant spindle rotation relative to the embryo A/P axis. Therefore, we traced single cells over time and compared the interphase long axis of cells immediately before chromosome condensation with the orientation of cell division. We defined the long axis of the cell in interphase based on the shape of the Tg(h2a.z/f-GFP)-positive nucleus, which is elongated along the same axis (Fig. 3A,G). If mitotic spindle orientation defects in hoxb1b-/- are due only to the abnormal orientation of single interphase cells within the tissue plane, then we would expect that individual progenitors divide ‘intrinsically correctly’ with a final orientation close to the interphase cell axis. In fact, we observed that the mean difference between interphase orientation and anaphase orientation (δ angle) is significantly larger in hoxb1b-/- compared with wild type (Fig. 3G). However, we find that the initial orientation of the metaphase plate in both wild type and hoxb1b-/- mutants is parallel to the orientation of the cell at interphase. This suggests that neuroepithelial cells in r3/4 in hoxb1b mutants not only divide with abnormal orientation due to abnormal interphase positioning within the tissue plane, but additionally have a decreased ability to properly position their mitotic spindle by rotation relative to the initial interphase cell axis.
If the defects in mitotic orientation are the major cause of the abnormalities in lumen morphogenesis, then inhibiting cells from going through mitosis should rescue the morphogenesis phenotype. Cell cycle blockage using established small molecule inhibitors (aphidicolin and hydroxyurea) (Ciruna et al., 2006; Tawk et al., 2007; Žigman et al., 2011) revealed that significant inhibition of mitoses, verified by absence of phos-H3 staining, rescues the hoxb1b-/- r3/4 morphogenesis defect (Fig. 3F). Thus, Hoxb1b is regulating both neural keel convergence as well as cell polarization in mitosis, and it is likely that both contribute to proper hindbrain lumen morphogenesis.
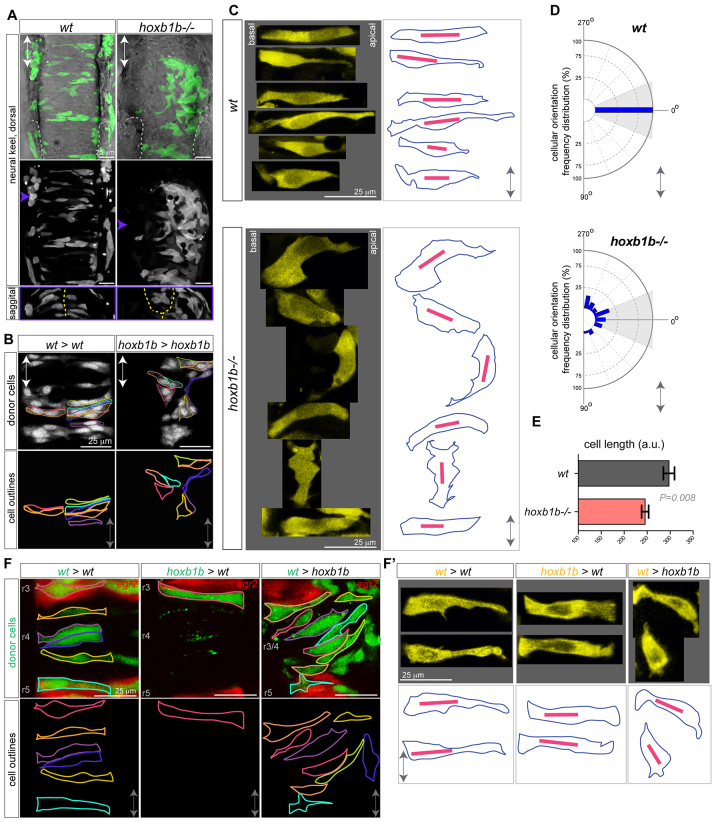
In the neural keel, apico-basal mitotic orientation results in the bilateral distribution of cellular progeny (Ciruna et al., 2006; Tawk et al., 2007; Quesada-Hernández et al., 2010; Žigman et al., 2011). Given the abnormally oriented mitoses we observe in hoxb1b-/- r3/4, we predicted that the bilateral distribution of cells in the neural keel would be affected. We marked populations of cells unilaterally during gastrulation and visualized their distribution in the neural tube, after apico-basal divisions are complete. Indeed, whereas in wild-type embryos neural progenitors are distributed bilaterally, labeled cells in hoxb1b-/- mutants lie unilaterally in the r3/4 territory (Fig. 4A,B). Thus, Hoxb1b is essential for bilateral daughter distribution in the r3/4 territory.
Fig. 4.
Hoxb1b-dependent cell shape changes. (A) Mosaic unilateral labeling (green) of wild-type and hoxb1b-/- embryos results in a bilateral cell distribution in wild-type but unilateral, abnormal cell distribution in hoxb1b-/- embryos. Top and middle panels, dorsal view; bottom panels, sagittal optical section at the level of r3/4 at ∼16 hpf. White dashed lines outline otic vesicles; yellow dashed lines indicate midline. (B) Unilaterally transplanted wild-type and hoxb1b-/- donor cells in wild-type and hoxb1b-/- hosts, respectively. Single cells are outlined and are bilaterally distributed in wild type but unilateral and disorganized in hoxb1b-/- r3/4 at ∼17 hpf. (C) Cell shape as visualized in maximal intensity projections of single cells from r3, r4 of wild-type and hoxb1b-/- embryos at 15-17 hpf (n=3 embryos per genotype). Projections from single cells were isolated and arranged as originally embedded within the animal axis, with basal to left and more apical side to the right. Right panels: outlines of corresponding single cells (blue) from the left, with the longest diagonal of each cell presented (red line). Note elongated wild-type cells and broader, misoriented hoxb1b-/- cells. (D) Cellular orientation (based on the longest diagonal of each cell) relative to the A/P embryo axis of wild-type and hoxb1b-/- cells. The A/P axis is vertical with anterior at 0°. Note strong bias towards orientation perpendicular to the A/P in wild-type cells compared with significantly abnormal orientation of hoxb1b-/- cells. (E) Length of individual cell longest diagonal in wild-type and hoxb1b-/- r3/4 territory presented as mean ± s.e.m. Note that wild-type cells are significantly longer compared with hoxb1b-/-. (F) Cell shape of individual neuroepithelial progenitors (green) in genetic mosaics expressing egr2 transgenic reporter (red) visualizing r3 and r5 at 17 hpf. (F′) Zoomed in maximal intensity projections of single cells (yellow) from r3, r4 of transplanted cells at 15-17 hpf (n=3 embryos for all genotypes). Projections from single cells were isolated and arranged as noted for C Lower panels: outlines of corresponding single cells (blue) with the longest diagonal of each cell presented (red line). Anterior-posterior axis marked by double arrow in all panels.
Hoxb1b is limiting for correct cell shape acquisition
Cell shape and size, geometry and retraction fibers have all been shown to bias mitosis polarization in cultured cells (Minc and Piel, 2012), but whether and how interphase cell shape affects mitotic orientation is not understood in three-dimensional tissues in vivo (Marshall et al., 2012) due to a lack of accessible paradigms. We asked whether Hoxb1b-dependent control of cell shape in the developing hindbrain underlies the establishment of cell polarization. Quantitative individual cell shape analysis (Fig. 4C,D) revealed that whereas wild-type interphase neuroepithelial cells display elongated morphology with their long (apico-basal) axis perpendicular to the embryonic A/P axis, hoxb1b-/- cells in r3/4 lie in abnormal orientations with their longest axis oblique or even parallel to the A/P axis. hoxb1b-/- cells also display a variety of aberrant cell shapes (Fig. 4C,D). Specifically, the long axes of hoxb1b-/- cells were significantly shorter and cell shape more spread compared with those of the wild-type cells in r3/4. This additionally distinguishes hoxb1b-/- from PCP mutants, in which neuroepithelial cell length is not affected (Ciruna et al., 2006; Tawk et al., 2007). Interestingly, abnormal cell shape of hoxb1b-/- cells was rescued when mutant cells were transplanted into wild-type hosts at the gastrula stage and imaged in the early neural tube (Fig. 4F,F′). Moreover, when wild-type cells were transplanted into hoxb1b-/- hosts, they obtained abnormal cell shape and cellular orientation in r3/4 (Fig. 4F,F′). Thus, we uncover a new role for Hoxb1b in cell shape regulation possibly acting via a cell-extrinsic mechanism during in vivo tissue morphogenesis.
Requirement of Hoxb1b for proper microtubule dynamics in anterior hindbrain segments
As Hoxb1b is required for proper cell shape acquisition in interphase, we set out to analyze the intrinsic cytoskeletal polarity of these cells by assessing MT dynamics. We examined MT plus-end directed growth in interphase cells by quantifying EB3-mCherry (Du and Macara, 2004; Stepanova et al., 2003). Live imaging of EB3-mCherry comets in wild type indicated MT-end growth in directed, predominantly apical-to-basal fashion, emanating from the MT organizing center (localized apically) towards the cell periphery (Fig. 5A). EB3-mCherry comets moving from the cell periphery towards the microtubule organizing center (MTOC) were detectable only occasionally. MT plus-end growth directionality in the wild-type cells is along polarized arrays but is unpolarized in hoxb1b-/- cells (Fig. 5A,B; supplementary material Movie 3). This demonstrates that not only individual cell shape and cell axis orientation but also the directionality of MT plus-end polymerization is affected in the absence of Hoxb1b. Furthermore, individual EB3 comet track velocity was reduced in hoxb1b-/- cells compared with wild-type cells, demonstrating kinetic impairment of hoxb1b-/- cells (Fig. 5C). When hoxb1b-/- cells were transplanted into wild-type hosts they regained normal EB3 comet velocity. Moreover, when wild-type cells were transplanted into hoxb1b-/- hosts, their MT dynamics were also significantly affected, though not to the degree observed in hoxb1b-/- mutants (Fig. 5C).
Fig. 5.
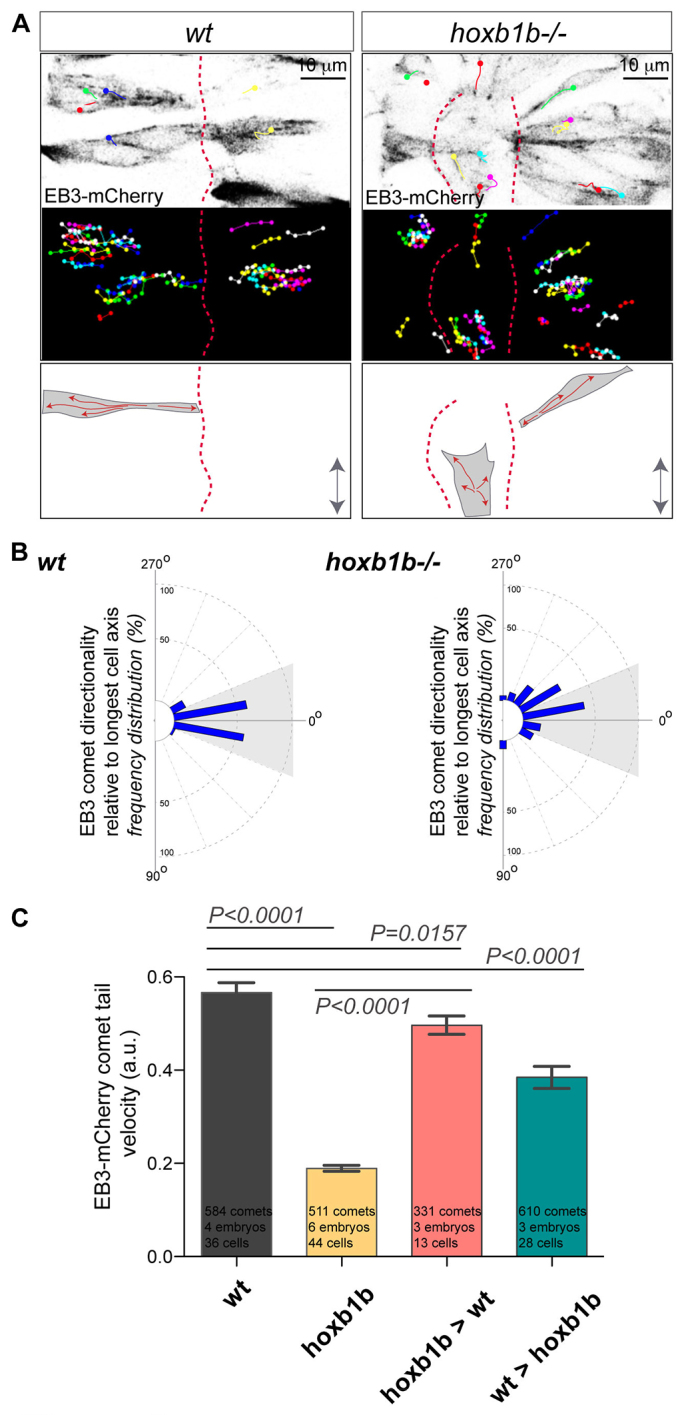
Hoxb1b is essential for plus end microtubule dynamics. (A) Live imaging of neuroepithelial cells of EB3-mCherry mRNA-injected wild type and hoxb1b-/- at r3/r4 with single EB3-mCherry comets marked as colored dots (presenting last position) connected with lines (movement over time) between 15 and 17 hpf. Top panels show a single time point, middle panels show tracks of individual EB3-mCherry comets projected over time revealing the effect on Hoxb1b loss on the comet directionality. MT plus-end dynamics are schematized at the bottom. Double arrows indicate the embryo anterior-posterior axis. Red dashed lines indicate midline. (B) Quantitation of single EB3-mCherry comets when tracked over time in respect to the A/P axis (0°) demonstrating quantitative defects in MT growth in hoxb1b-/- (n=6 embryos) compared with wild-type controls (n=4 embryos). (C) Decreased velocity of individual EB3-mCherry comets in hoxb1b-/- compared with wild type. Note rescue of EB3-mCherry comet dynamics in hoxb1b-/- cells when transplanted into wild-type hosts and partial (but significant) decrease in comet velocity in wild-type cells when transplanted into hoxb1b-/- embryos. Results presented as mean ± s.e.m. The data used for quantification of wild type and hoxb1b-/- in B and C originate from the same data set.
The rescue of both cell shape and MT dynamics in hoxb1b-/- cells that develop in a wild-type neuroepithelial environment suggests that Hoxb1b has an indirect role in organizing the cell cortex in the r3/4 region, possibly via a cell-contact-mediated mechanism that is transduced into regulation of MT dynamics. This function may carry through to dividing cells, affecting the interactions between MT plus-ends and the cell cortex, required for spindle orientation. Collectively, our data unveil a new role for Hoxb1b in the oriented cell division-dependent morphogenesis of the hindbrain neuroepithelium. Based on abnormalities in MT dynamics in interphase cells in hoxb1b mutants, we hypothesize that Hoxb1b-dependent MT dynamics during oriented cell division may regulate neuroepithelial morphogenesis in the anterior hindbrain.
DISCUSSION
Here, we uncover a novel role for a classical homeotic gene, hoxb1b. Due to different routes of Hox gene subfunctionalization after the divergence of fish and mammals, zebrafish hoxb1b has been identified as the Hox gene that carries out the ancestral functions of mouse Hoxa1 (McClintock et al., 2001). Hoxa1 has been previously described to regulate segmental patterning of hindbrain compartments and their neural crest derivatives in the head periphery via specification of segment-specific cell fates (Lufkin et al., 1991; Mark et al., 1993; Carpenter et al., 1993). We find that a zebrafish hoxb1b mutant exhibits these classical Hoxa1 phenotypes, but unexpectedly that hoxb1b is also required for proper neural tube lumen morphogenesis in the anterior-most part of its expression domain. The hoxb1b-/- progenitor cells in the r3/4 region become abnormally shaped and lose their correct cellular orientation. This is accompanied by overall slower MT plus-end dynamics in hoxb1b-/- neuroepithelial progenitor cells. Our data indicate a role for Hoxb1b in cell shape control and cytoskeletal organization as determinants for in vivo tissue morphogenesis.
Genetic control of tissue morphogenesis by Hoxb1b
Hox genes have been identified as crucial factors primarily specifying positional identity along the anterior-posterior axis of bilaterian animals (McGinnis and Krumlauf, 1992). Additionally, Hox genes have been shown to be essential for morphogenesis of certain organs in Drosophila (Hombría and Lovegrove, 2003; Roch and Akam, 2000) and in Caenorhabditis elegans vulva (Pellegrino et al., 2011), as well as for mammalian organ morphogenesis such as that of the pulmonary epithelium (Aubin et al., 1997), the gastrointestinal tract (Zacchetti et al., 2007), genitalia (Dollé et al., 1991) or digits (Zákány and Duboule, 1999). These studies predominantly focused on changes in cellular identity (fate) and final global morphology at the level of the organ.
In Drosophila, where Hox genes were initially identified, hypothetical downstream targets of Hox transcription factors have been termed ‘realisator’ genes, and these were postulated to control organ morphogenesis on a cellular level as the elusive effectors of Hox gene function (McGinnis and Krumlauf, 1992; García-Bellido, 1975). Recent work has identified Hox transcriptional targets that might direct morphogenesis in Drosophila and C. elegans (Lovegrove et al., 2006; Castelli Gair Hombría et al., 2009). In recent examples, Drosophila AbdB, an ortholog of vertebrate posterior Hox genes, directly regulates the unconventional myosin gene Myo1D (Myo31DF - FlyBase) to control left-right asymmetric hindgut morphogenesis in the fly (Coutelis et al., 2013), and C. elegans lin-39, a vertebrate Hox4 ortholog, indirectly regulates the transmembrane Semaphorin SMP-1 to control cell adhesion during vulval morphogenesis in the worm (Pellegrino et al., 2011). Despite these recent results, the mechanism by which Hox genes regulate tissue and organ morphogenesis at the cellular level is poorly understood in any system.
Our finding that hoxb1b is required for hindbrain lumen morphogenesis and mitotic spindle orientation specifically in r3/4 is unexpected given that cells all along the anterior-posterior hindbrain axis undergo stereotyped oriented mitoses, and genes required for neural tube lumen formation and progenitor epithelialization, such as Par complex components (Munson et al., 2008) are expressed ubiquitously along the neural axis of the hindbrain. Early mutant analyses of mouse knockouts of Hoxa1 described a delayed hindbrain neural tube closure in the anterior hindbrain, although no analysis of these defects at the cellular level has been reported (Lufkin et al., 1991; Mark et al., 1993). Importantly, because of the different way the neural tube forms in mammals and fish, genes that cause neural tube closure defects in the mouse cause duplicated neural tube phenotypes in zebrafish [e.g. vangl2 (Tawk et al., 2007)], so the local neural tube duplication we observe in hoxb1b mutant zebrafish may be the equivalent of the neural tube closure defect in Hoxa1 mutant mice.
Hoxb1b control of morphogenetic cell behaviors
We uncovered an essential role for hoxb1b in both neuroepithelial convergence and oriented cell division in the r3/4 territory. Neuroepithelial PCP has been shown to drive neuroepithelial convergence and PCP has also been shown to orient cell division in the developing neuroepithelium (Quesada-Hernández et al., 2010). In spite of this, we see no evidence of defects in planar polarity in hoxb1b mutants. Furthermore, loss of core PCP components does not affect neuroepithelial cell length (Ciruna et al., 2006; Tawk et al., 2007), spindle orientation (Žigman et al., 2011) or MT plus-end dynamics (this work) whereas Hoxb1b does. Our own previous work implicated normal cell-cell adhesion downstream of Scribble as a determinant of spindle orientation in the neural keel (Žigman et al., 2011). We could not, however, detect any cell-cell adhesion abnormalities in hoxb1b mutants. Together, these findings show that Hoxb1b may regulate cell behavior in the neural keel independently of an effect on either classical forms of apico-basal or planar cell polarity or cell-cell adhesion. Rather, Hoxb1b may influence a different form of cell polarity by impacting on cell shape and MT dynamics for proper cell division orientation.
Hoxb1b regulation of microtubule-dependent processes
Our data indicate that during cell division, Hoxb1b may function in the spindle-cortex force generation mechanism that is required for the coherent rotation of the mitotic spindle apparatus. Specifically, metaphase plate chromosomes rotate faster and ultimately fail to align perpendicular to the apico-basal axis of the neuroepithelium at anaphase in hoxb1b-/- cells in r3/4. This increased rate of chromosome rotation in hoxb1b-/- neuroepithelial progenitors correlates with slower dynamics of MT plus-ends in interphase. We hypothesize that decreased MT plus-end dynamics could be the underlying cause of uncoordinated force generation and spindle rotation defects in mitosis. This is unlikely to be due to loss of intrinsic apico-basal cell polarity, which is responsible for spindle orientation defects in other contexts, because interphase cell polarity and the initial alignment of the metaphase plate with interphase cell shape is normal in hoxb1b-/- neuroepithelial progenitors. Rotation of the mitotic apparatus is thought to be based on an interaction between astral MTs and the cell cortex, mediated by NuMA and Pins/LGN/AGS3, and force generation involving the Lis1-dynactin complex that pulls the chromosomes into alignment with localized cortical determinants (Siller and Doe, 2008; McNally, 2013). Whatever the intracellular spindle-orienting mechanism, the localization of the cortical landmarks that regulate mitotic spindle orientation in some contexts depends on cell-extrinsic cues such as extracellular matrix components (Lechler and Fuchs, 2005), planar tissue polarity (Gong et al., 2004) or anisotropic tension (Fink et al., 2011). For example, MTs could specify correct spindle orientation by plus-end anchoring to integrin-ILK complexes (Akhtar and Streuli, 2013) and, indeed, integrins are required for correct spindle orientation in mammalian skin progenitors (Lechler and Fuchs, 2005). Furthermore, in the zebrafish neural keel itself, multiple extrinsic cues, including the basement membrane, serve to localize the MTOC in neural progenitor cells (Buckley et al., 2013). As the cellular morphogenesis defects in hoxb1b-/- cells can be rescued by wild-type neighbors, we infer that effects on MT dynamics in both dividing and non-dividing cells are secondary to effects on one or more of these cell-extrinsic influences.
What is the connection between the defects in interphase cell shape we observe in hoxb1b mutants and the defects in cell division orientation? One possibility is that anisotropies in neuroepithelial cell shape before the rounding-up of mitotic cells may provide a memory of previous cell shape through retraction fibers that help properly orient polarized mitoses. Indeed, in cultured cells the mitotic spindle rotates to align with these tension-bearing retraction fibers, which extend from otherwise rounded-up cells in an orientation parallel to the former long axis of the cell (Fink et al., 2011). Likewise, anisotropic extracellular physical forces present in the multicellular milieu could possibly represent polarizing signals in neuroepithelial cells and signal via retraction fibers as shown in culture (Fink et al., 2011). It is possible that such structures would impact on cortical stiffness and anchoring of cortical MTs required for proper dynamics of mitotic spindle rotation in vivo. In this scenario, the primary role of hoxb1b on hindbrain morphogenesis would be on interphase cell shape, where it functions through cell-extrinsic effectors to promote tissue-wide cell alignment. Such safeguarding phenomena may be considered as a response mechanism to noise during tissue morphogenesis, and such mechanisms have likely evolved to ensure tissue-level robustness in multicellular organisms.
Hoxb1b regulation of cell behavior: unanswered questions
How might Hoxb1b regulate the extracellular properties that impinge on MT dynamics in neuroepithelial progenitors? Hox proteins function as transcriptional regulators, so their function in regulating cytoskeletal events is likely to be indirect, via as yet unidentified transcriptional targets. Genome-wide screens for differentially expressed mRNAs in wild-type versus hoxb1b overexpressing zebrafish (Choe et al., 2011) or in r3-r5 of wild-type versus Hoxa1 knockout mice (Makki and Capecchi, 2011), did not reveal changes in the expression of known regulators of cell polarization or MT dynamics. Lack of such transcriptional targets may indirectly indicate a possible non-transcriptional role of Hoxb1b. Although the Hoxb1b nonsense mutation we identified is indeed expected to prevent DNA binding, an additional direct role for Hoxb1b protein in cell shape and MT dynamics cannot be excluded: a recent yeast two-hybrid screen identified non-nuclear Hoxa1 protein interactors in vitro (Lambert et al., 2012), opening a possibility of non-canonical non-transcriptional functions of Hoxb1b in cell-cell interactions regulating cell shape and orientation.
How Hoxb1b could be regulating extracellular cues that are transduced into the regulation of MT dynamics in neighboring cells, we currently do not understand. Therefore, it will be crucial to integrate knowledge of control mechanisms executed by transcriptional and non-transcriptional Hoxb1b targets along with control principles of mechanical tissue strain to understand how growing tissues dynamically feed back on individual cell shape and MT dynamics to ensure proper organ shape and size during development.
MATERIALS AND METHODS
Zebrafish lines
The hoxb1b b1219 allele was identified by TILLING (Draper et al., 2004) and was genotyped by DdeI restriction digest of PCR products amplified from genomic DNA using primers 5′-GTCGGAGGAAACACTTTCACATCG-3′ and 5′-GTTGATGTCCATAGTCCGAATGAGGAGCTC-3′. The following transgenic lines were crossed into the hoxb1bb1219 background: Tg(egr2b:KalTA4) (an enhancer-trap line) (Distel et al., 2009), Gt(Ctnna-Citrine)ct3a (a gene-trap line) (Žigman et al., 2011) and Tg(h2a.f/z-GFP) (Geldmacher-Voss et al., 2003). A previously described mutant, egr2fh227 (Monk et al., 2009), and pbx2 and pbx4 morpholinos (Waskiewicz et al., 2002) were used.
RNA expression constructs
Capped mRNA was synthesized and 1-2.5 nl microinjected into 1- or 16-cell-stage embryos. GFP-Pk mRNA was used as described (Ciruna et al., 2006). For MT plus-end dynamics, the EB3-mCherry mRNA was used (Stepanova et al., 2003).
Transplants
Transplantation was used to mark cells unilaterally in embryonic hindbrain and to assess cell shape in genetic mosaics. Cells of Alexa488-dextran injected donors were transplanted unilaterally into presumptive hindbrain region of gastrula stage host embryos as described (Kemp et al., 2009).
Small molecule inhibitors
Embryos were incubated in E3 embryo medium with 150 μM aphidicolin (Sigma) and 20 mM hydroxyurea (Sigma) in 4% DMSO as previously described (Ciruna et al., 2006; Žigman et al., 2011). Aphidicolin and hydroxyurea both inhibit DNA synthesis, without direct binding to microtubules. The FGF signaling inhibitor SU5402 was diluted in DMSO and used at 100 μM.
Vital and immunofluorescence labeling
The following primary antibodies were used: mouse anti-ZO-1 (Zymed, 1:200), mouse anti-γ-tubulin (catalog number 6557, Sigma) and rabbit anti γ-tubulin (catalog number 5192, Sigma), rabbit phospho-histone H3 (Ser10) (H3S10ph) (Milipore, 1:1000). Atto488-, Atto568-phalloidin (Sigma) were used to visualize F-actin.
Light microscopy
To image live embryos in 4D, embryos were mounted in 1% low-melting point agarose on glass bottom dishes (Ibidi) submerged in E3 medium. Microscopic measurements were performed on inverted Nikon A1R MP confocal and multiphoton system using Nano-Crystal coated LWD 40× WI Lambda S and 60× WI objectives. For multiphoton measurements, a tunable wavelength Chameleon laser (Coherent) was applied.
Image analysis and quantitation
NIS-Elements Nikon software, Fiji and ImageJ were used to analyze images. The distribution of angles of mitoses and the metaphase plate rotation were quantified as previously described (Žigman et al., 2011). To quantify cell orientation, the orientation of the longest diagonal within each cell surface area was determined by best fitting of an ellipsoid. Cellular orientation was analyzed relative to the anterior-posterior axis of the embryo.
Graphical presentation of data was made using Prism (Graphpad). Radial plots were made by Oriana software. Statistical significance was calculated by non-parametric two-tailed Mann-Whitney testing using Prism.
Supplementary Material
Acknowledgments
We thank B. Ciruna, A. Reugels, U. Engel, Martin Distel, Reinhard Koester and Lisa Maves for providing reagents. Tiffany Su performed the fgf3 in situ hybridization experiments shown in supplementary material Fig. S2B.
Footnotes
Competing interests
The authors declare no competing financial interests.
Author contributions
M.Z. and C.B.M. conceived the project and wrote the manuscript. M.Z. planned and performed the experiments and analyzed the main data. T.T. and J.P. isolated the hoxb1b b1219 allele. Experiments for Fig. 1D and supplementary material Fig. S4 were performed by N.L.-L. and experiments for supplementary material Fig. S1A-D,F and Fig. S2D,E were performed by C.B.M.
Funding
This work was supported by a Heidelberg Academy of Sciences WIN grant (to M.Z.); grants from the National Institutes of Health [NIH RO1 HD37909 to C.B.M. and NIH RO1 HD076585 to C.B.M. and J.P.]. Deposited in PMC for immediate release.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.098731/-/DC1
References
- Akhtar N., Streuli C. H. (2013). An integrin-ILK-microtubule network orients cell polarity and lumen formation in glandular epithelium. Nat. Cell Biol. 15, 17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubin J., Lemieux M., Tremblay M., Bérard J., Jeannotte L. (1997). Early postnatal lethality in Hoxa-5 mutant mice is attributable to respiratory tract defects. Dev. Biol. 192, 432–445 [DOI] [PubMed] [Google Scholar]
- Barrow J. R., Stadler H. S., Capecchi M. R. (2000). Roles of Hoxa1 and Hoxa2 in patterning the early hindbrain of the mouse. Development 127, 933–944 [DOI] [PubMed] [Google Scholar]
- Buckley C. E., Ren X., Ward L. C., Girdler G. C., Araya C., Green M. J., Clark B. S., Link B. A., Clarke J. D. W. (2013). Mirror-symmetric microtubule assembly and cell interactions drive lumen formation in the zebrafish neural rod. EMBO J. 32, 30–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter E. M., Goddard J. M., Chisaka O., Manley N. R., Capecchi M. R. (1993). Loss of Hox-A1 (Hox-1.6) function results in the reorganization of the murine hindbrain. Development 118, 1063–1075 [DOI] [PubMed] [Google Scholar]
- Castelli Gair Hombría J., Rivas M. L., Sotillos S. (2009). Genetic control of morphogenesis - Hox induced organogenesis of the posterior spiracles. Int. J. Dev. Biol. 53, 1349–1358 [DOI] [PubMed] [Google Scholar]
- Chisaka O., Capecchi M. R. (1991). Regionally restricted developmental defects resulting from targeted disruption of the mouse homeobox gene hox-1.5. Nature 350, 473–479 [DOI] [PubMed] [Google Scholar]
- Choe S.-K., Zhang X., Hirsch N., Straubhaar J., Sagerström C. G. (2011). A screen for hoxb1-regulated genes identifies ppp1r14al as a regulator of the rhombomere 4 Fgf-signaling center. Dev. Biol. 358, 356–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciruna B., Jenny A., Lee D., Mlodzik M., Schier A. F. (2006). Planar cell polarity signalling couples cell division and morphogenesis during neurulation. Nature 439, 220–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutelis J.-B., Géminard C., Spéder P., Suzanne M., Petzoldt A. G., Noselli S. (2013). Drosophila left/right asymmetry establishment is controlled by the Hox gene abdominal-B. Dev. Cell 24, 89–97 [DOI] [PubMed] [Google Scholar]
- Daga R. R., Nurse P. (2008). Interphase microtubule bundles use global cell shape to guide spindle alignment in fission yeast. J. Cell Sci. 121, 1973–1980 [DOI] [PubMed] [Google Scholar]
- Distel M., Wullimann M. F., Köster R. W. (2009). Optimized Gal4 genetics for permanent gene expression mapping in zebrafish. Proc. Natl. Acad. Sci. USA 106, 13365–13370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dollé P., Izpisúa-Belmonte J. C., Brown J. M., Tickle C., Duboule D. (1991). HOX-4 genes and the morphogenesis of mammalian genitalia. Genes Dev. 5, 1767–1776 [DOI] [PubMed] [Google Scholar]
- Draper B. W., McCallum C. M., Stout J. L., Slade A. J., Moens C. B. (2004). A high-throughput method for identifying N-ethyl-N-nitrosourea (ENU)-induced point mutations in zebrafish. Methods Cell Biol. 77, 91–112 [DOI] [PubMed] [Google Scholar]
- Du Q., Macara I. G. (2004). Mammalian Pins is a conformational switch that links NuMA to heterotrimeric G proteins. Cell 119, 503–516 [DOI] [PubMed] [Google Scholar]
- Duboule D. (2007). The rise and fall of Hox gene clusters. Development 134, 2549–2560 [DOI] [PubMed] [Google Scholar]
- Fink J., Carpi N., Betz T., Bétard A., Chebah M., Azioune A., Bornens M., Sykes C., Fetler L., Cuvelier D., et al. (2011). External forces control mitotic spindle positioning. Nat. Cell Biol. 13, 771–778 [DOI] [PubMed] [Google Scholar]
- García-Bellido A. (1975). Genetic control of wing disc development in Drosophila. Ciba Found. Symp. 0, 161–182 [DOI] [PubMed] [Google Scholar]
- Gavalas A., Studer M., Lumsden A., Rijli F. M., Krumlauf R., Chambon P. (1998). Hoxa1 and Hoxb1 synergize in patterning the hindbrain, cranial nerves and second pharyngeal arch. Development 125, 1123–1136 [DOI] [PubMed] [Google Scholar]
- Geldmacher-Voss B., Reugels A. M., Pauls S., Campos-Ortega J. A. (2003). A 90-degree rotation of the mitotic spindle changes the orientation of mitoses of zebrafish neuroepithelial cells. Development 130, 3767–3780 [DOI] [PubMed] [Google Scholar]
- Gong Y., Mo C., Fraser S. E. (2004). Planar cell polarity signalling controls cell division orientation during zebrafish gastrulation. Nature 430, 689–693 [DOI] [PubMed] [Google Scholar]
- Hombría J. C.-G., Lovegrove B. (2003). Beyond homeosis - HOX function in morphogenesis and organogenesis. Differentiation 71, 461–476 [DOI] [PubMed] [Google Scholar]
- Hong E., Brewster R. (2006). N-cadherin is required for the polarized cell behaviors that drive neurulation in the zebrafish. Development 133, 3895–3905 [DOI] [PubMed] [Google Scholar]
- Iimura T., Pourquié O. (2007). Hox genes in time and space during vertebrate body formation. Dev. Growth Differ. 49, 265–275 [DOI] [PubMed] [Google Scholar]
- Kemp H. A., Carmany-Rampey A., Moens C. (2009). Generating chimeric zebrafish embryos by transplantation. J. Vis. Exp. 2009, 1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert B., Vandeputte J., Remacle S., Bergiers I., Simonis N., Twizere J.-C., Vidal M., Rezsohazy R. (2012). Protein interactions of the transcription factor Hoxa1. BMC Dev. Biol. 12, 29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechler T., Fuchs E. (2005). Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature 437, 275–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Grill S., Sanchez A., Murphy-Ryan M., Poss K. D. (2005). Fgf signaling instructs position-dependent growth rate during zebrafish fin regeneration. Development 132, 5173–5183 [DOI] [PubMed] [Google Scholar]
- Lele Z., Folchert A., Concha M., Rauch G.-J., Geisler R., Rosa F., Wilson S. W., Hammerschmidt M., Bally-Cuif L. (2002). parachute/n-cadherin is required for morphogenesis and maintained integrity of the zebrafish neural tube. Development 129, 3281–3294 [DOI] [PubMed] [Google Scholar]
- Lovegrove B., Simões S., Rivas M. L., Sotillos S., Johnson K., Knust E., Jacinto A., Hombría J. C.-G. (2006). Coordinated control of cell adhesion, polarity, and cytoskeleton underlies Hox-induced organogenesis in Drosophila. Curr. Biol. 16, 2206–2216 [DOI] [PubMed] [Google Scholar]
- Lowery L. A., Sive H. (2004). Strategies of vertebrate neurulation and a re-evaluation of teleost neural tube formation. Mech. Dev. 121, 1189–1197 [DOI] [PubMed] [Google Scholar]
- Lufkin T., Dierich A., LeMeur M., Mark M., Chambon P. (1991). Disruption of the Hox-1.6 homeobox gene results in defects in a region corresponding to its rostral domain of expression. Cell 66, 1105–1119 [DOI] [PubMed] [Google Scholar]
- Lumsden A., Krumlauf R. (1996). Patterning the vertebrate neuraxis. Science 274, 1109–1115 [DOI] [PubMed] [Google Scholar]
- Makki N., Capecchi M. R. (2011). Identification of novel Hoxa1 downstream targets regulating hindbrain, neural crest and inner ear development. Dev. Biol. 357, 295–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark M., Lufkin T., Vonesch J. L., Ruberte E., Olivo J. C., Dollé P., Gorry P., Lumsden A., Chambon P. (1993). Two rhombomeres are altered in Hoxa-1 mutant mice. Development 119, 319–338 [DOI] [PubMed] [Google Scholar]
- Marshall W. F., Young K. D., Swaffer M., Wood E., Nurse P., Kimura A., Frankel J., Wallingford J., Walbot V., Qu X., et al. (2012). What determines cell size? BMC Biol. 10, 101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maves L., Kimmel C. B. (2005). Dynamic and sequential patterning of the zebrafish posterior hindbrain by retinoic acid. Dev. Biol. 285, 593–605 [DOI] [PubMed] [Google Scholar]
- McClintock J. M., Carlson R., Mann D. M., Prince V. E. (2001). Consequences of Hox gene duplication in the vertebrates: an investigation of the zebrafish Hox paralogue group 1 genes. Development 128, 2471–2484 [DOI] [PubMed] [Google Scholar]
- McClintock J. M., Kheirbek M. A., Prince V. E. (2002). Knockdown of duplicated zebrafish hoxb1 genes reveals distinct roles in hindbrain patterning and a novel mechanism of duplicate gene retention. Development 129, 2339–2354 [DOI] [PubMed] [Google Scholar]
- McGinnis W., Krumlauf R. (1992). Homeobox genes and axial patterning. Cell 68, 283–302 [DOI] [PubMed] [Google Scholar]
- McNally F. J. (2013). Mechanisms of spindle positioning. J. Cell Biol. 200, 131–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller A. C., Obholzer N. D., Shah A. N., Megason S. G., Moens C. B. (2013). RNA-seq-based mapping and candidate identification of mutations from forward genetic screens. Genome Res. 23, 679–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minc N., Piel M. (2012). Predicting division plane position and orientation. Trends Cell Biol. 22, 193–200 [DOI] [PubMed] [Google Scholar]
- Monk K. R., Naylor S. G., Glenn T. D., Mercurio S., Perlin J. R., Dominguez C., Moens C. B., Talbot W. S. (2009). A G protein-coupled receptor is essential for Schwann cells to initiate myelination. Science 325, 1402–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munson C., Huisken J., Bit-Avragim N., Kuo T., Dong P. D., Ober E. A., Verkade H., Abdelilah-Seyfried S., Stainier D. Y. R. (2008). Regulation of neurocoel morphogenesis by Pard6 gamma b. Dev. Biol. 324, 41–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrino M. W., Farooqui S., Fröhli E., Rehrauer H., Kaeser-Pebernard S., Müller F., Gasser R. B., Hajnal A. (2011). LIN-39 and the EGFR/RAS/MAPK pathway regulate C. elegans vulval morphogenesis via the VAB-23 zinc finger protein. Development 138, 4649–4660 [DOI] [PubMed] [Google Scholar]
- Quesada-Hernández E., Caneparo L., Schneider S., Winkler S., Liebling M., Fraser S. E., Heisenberg C.-P. (2010). Stereotypical cell division orientation controls neural rod midline formation in zebrafish. Curr. Biol. 20, 1966–1972 [DOI] [PubMed] [Google Scholar]
- Roch F., Akam M. (2000). Ultrabithorax and the control of cell morphology in Drosophila halteres. Development 127, 97–107 [DOI] [PubMed] [Google Scholar]
- Rossel M., Capecchi M. R. (1999). Mice mutant for both Hoxa1 and Hoxb1 show extensive remodeling of the hindbrain and defects in craniofacial development. Development 126, 5027–5040 [DOI] [PubMed] [Google Scholar]
- Siller K. H., Doe C. Q. (2008). Lis1/dynactin regulates metaphase spindle orientation in Drosophila neuroblasts. Dev. Biol. 319, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova T., Slemmer J., Hoogenraad C. C., Lansbergen G., Dortland B., De Zeeuw C. I., Grosveld F., van Cappellen G., Akhmanova A., Galjart N. (2003). Visualization of microtubule growth in cultured neurons via the use of EB3-GFP (end-binding protein 3-green fluorescent protein). J. Neurosci. 23, 2655–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawk M., Araya C., Lyons D. A., Reugels A. M., Girdler G. C., Bayley P. R., Hyde D. R., Tada M., Clarke J. D. W. (2007). A mirror-symmetric cell division that orchestrates neuroepithelial morphogenesis. Nature 446, 797–800 [DOI] [PubMed] [Google Scholar]
- Trapnell C., Hendrickson D. G., Sauvageau M., Goff L., Rinn J. L., Pachter L. (2013). Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31, 46–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel S. K., Raabe I., Dereli A., Maghelli N., Tolić-Nørrelykke I. (2007). Interphase microtubules determine the initial alignment of the mitotic spindle. Curr. Biol. 17, 438–444 [DOI] [PubMed] [Google Scholar]
- Waskiewicz A. J., Rikhof H. A., Moens C. B. (2002). Eliminating zebrafish pbx proteins reveals a hindbrain ground state. Dev. Cell 3, 723–733 [DOI] [PubMed] [Google Scholar]
- Zacchetti G., Duboule D., Zakany J. (2007). Hox gene function in vertebrate gut morphogenesis: the case of the caecum. Development 134, 3967–3973 [DOI] [PubMed] [Google Scholar]
- Zákány J., Duboule D. (1999). Hox genes in digit development and evolution. Cell Tissue Res. 296, 19–25 [DOI] [PubMed] [Google Scholar]
- Zallen J. A., Wieschaus E. (2004). Patterned gene expression directs bipolar planar polarity in Drosophila. Dev. Cell 6, 343–355 [DOI] [PubMed] [Google Scholar]
- žigman M., Trinh A., Fraser S. E., Moens C. B. (2011). Zebrafish neural tube morphogenesis requires Scribble-dependent oriented cell divisions. Curr. Biol. 21, 79–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.