Abstract
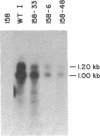
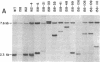
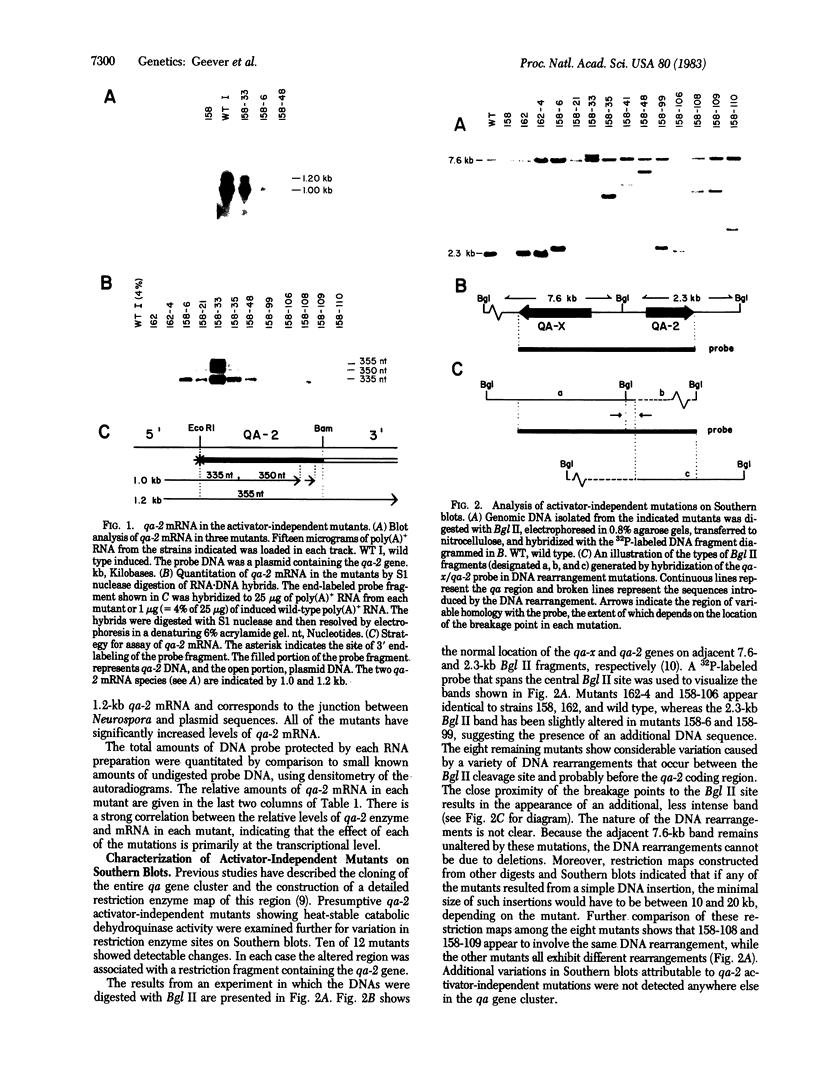
Expression of the qa-2 gene of Neurospora crassa normally requires a functional activator protein encoded by qa-1F. Twelve transcriptional mutants of the qa-2 gene have been isolated in qa-1F- strains, and these allow partial expression of qa-2 (1-45% of induced wild type) in the absence of functional activator protein. All 12 mutants have been characterized by genomic (Southern) blot hybridization and the DNAs of 5 have been cloned and sequenced. Eight mutations consist of large DNA rearrangements within a 500-base-pair region 5' to the qa-2 gene. One large rearrangement mutation, located 378 base pairs before the normal site of transcription initiation, causes exceptional levels of qa-2 transcription (45% of induced wild type) from near the normal initiation site. Two of the other four mutations cloned involve tandem duplications (68 and 84 base pairs) of the same upstream region (centered at nucleotide - 145), and two involve "point" mutations (at nucleotides -200 and -95) that closely flank the duplicated region. With one possible exception, none of the mutations appears to involve changes directly associated with RNA polymerase II binding and hence they differ from analogous mutations in comparable prokaryotic systems. The overall results suggest that at least some of the large DNA rearrangement mutations may be acting as upstream activator elements, possibly by juxtaposing enhancer-like sequences, whereas the duplications and point mutations may define a region of qa-2 regulation, for instance at the level of RNA polymerase II access.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alton N. K., Buxton F., Patel V., Giles N. H., Vapnek D. 5'-Untranslated sequences of two structural genes in the qa gene cluster of Neurospora crassa. Proc Natl Acad Sci U S A. 1982 Mar;79(6):1955–1959. doi: 10.1073/pnas.79.6.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alwine J. C., Kemp D. J., Stark G. R. Method for detection of specific RNAs in agarose gels by transfer to diazobenzyloxymethyl-paper and hybridization with DNA probes. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5350–5354. doi: 10.1073/pnas.74.12.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case M. E., Giles N. H. Genetic evidence on the organization and action of the qa-1 gene product: a protein regulating the induction of three enzymes in quinate catabolism in Neurospora crassa. Proc Natl Acad Sci U S A. 1975 Feb;72(2):553–557. doi: 10.1073/pnas.72.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cass L. G., Horwitz A. H., Wilcox G. The araIc mutation in Escherichia coli B/r. J Bacteriol. 1981 Jun;146(3):1098–1105. doi: 10.1128/jb.146.3.1098-1105.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagert M., Ehrlich S. D. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene. 1979 May;6(1):23–28. doi: 10.1016/0378-1119(79)90082-9. [DOI] [PubMed] [Google Scholar]
- Englesberg E., Sheppard D., Squires C., Meronk F., Jr An analysis of "revertants" of a deletion mutant in the C gene of the L-arabinose gene complex in Escherichia coli B-r: isolation of initiator constitutive mutants (Ic). J Mol Biol. 1969 Jul 28;43(2):281–298. doi: 10.1016/0022-2836(69)90268-x. [DOI] [PubMed] [Google Scholar]
- Englesberg E., Squires C., Meronk F., Jr The L-arabinose operon in Escherichia coli B-r: a genetic demonstration of two functional states of the product of a regulator gene. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1100–1107. doi: 10.1073/pnas.62.4.1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faye G., Leung D. W., Tatchell K., Hall B. D., Smith M. Deletion mapping of sequences essential for in vivo transcription of the iso-1-cytochrome c gene. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2258–2262. doi: 10.1073/pnas.78.4.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geever R. F., Wilson L. B., Nallaseth F. S., Milner P. F., Bittner M., Wilson J. T. Direct identification of sickle cell anemia by blot hybridization. Proc Natl Acad Sci U S A. 1981 Aug;78(8):5081–5085. doi: 10.1073/pnas.78.8.5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L., Yocum R. R., Gifford P. A GAL10-CYC1 hybrid yeast promoter identifies the GAL4 regulatory region as an upstream site. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7410–7414. doi: 10.1073/pnas.79.23.7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight S. L., Kingsbury R. Transcriptional control signals of a eukaryotic protein-coding gene. Science. 1982 Jul 23;217(4557):316–324. doi: 10.1126/science.6283634. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Montgomery D. L., Boss J. M., McAndrew S. J., Marr L., Walthall D. A., Zitomer R. S. The molecular characterization of three transcriptional mutations in the yeast iso-2-cytochrome c gene. J Biol Chem. 1982 Jul 10;257(13):7756–7761. [PubMed] [Google Scholar]
- Patel V. B., Schweizer M., Dykstra C. C., Kushner S. R., Giles N. H. Genetic organization and transcriptional regulation in the qa gene cluster of Neurospora crassa. Proc Natl Acad Sci U S A. 1981 Sep;78(9):5783–5787. doi: 10.1073/pnas.78.9.5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer M., Case M. E., Dykstra C. C., Giles N. H., Kushner S. R. Identification and characterization of recombinant plasmids carrying the complete qa gene cluster from Neurospora crassa including the qa-1+ regulatory gene. Proc Natl Acad Sci U S A. 1981 Aug;78(8):5086–5090. doi: 10.1073/pnas.78.8.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone A. E., Arditti R. R., Magasanik B. Catabolite-insensitive revertants of lac promoter mutants. Proc Natl Acad Sci U S A. 1970 Jul;66(3):773–779. doi: 10.1073/pnas.66.3.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Struhl K. The yeast his3 promoter contains at least two distinct elements. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7385–7389. doi: 10.1073/pnas.79.23.7385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler B. M., Cowman A. F., Adams J. M., Harris A. W. Generation of long mRNA for membrane immunoglobulin gamma 2a chains by differential splicing. Nature. 1981 Oct 1;293(5831):406–408. doi: 10.1038/293406a0. [DOI] [PubMed] [Google Scholar]
- Vapnek D., Hautala J. A., Jacobson J. W., Giles N. H., Kushner S. R. Expression in Escherichia coli K-12 of the structural gene for catabolic dehydroquinase of Neurospora crassa. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3508–3512. doi: 10.1073/pnas.74.8.3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiher H., König M., Gruss P. Multiple point mutations affecting the simian virus 40 enhancer. Science. 1983 Feb 11;219(4585):626–631. doi: 10.1126/science.6297005. [DOI] [PubMed] [Google Scholar]