Abstract
The earliest events leading to autoimmune type 1 diabetes (T1D) are not known in any species. A T-cell receptor (TCR)-variable region, TCR-Vβ13, is required for susceptibility to autoimmune diabetes in rats, and selective depletion of Vβ13+ T cells with an allele-specific monoclonal antibody prevents disease in multiple rat strains. To investigate the role of Vβ13 early in diabetes, we examined islet T-cell transcripts in susceptible (LEW.1WR1) and resistant (LEW.1W and Wistar Furth) strains induced with polyinosinic:polycytidylic acid. Vβ13+ T cells displayed antigenic focusing in LEW.1WR1 islets 5 days postinduction and were characterized by a substantial decrease in complementarity determining region 3 diversity. This occurred prior to significant islet T-cell accumulation (day 7) or frank diabetes (days 10–14). Vβ13+ transcripts increased in LEW.1WR1 islets during diabetes progression, but not in resistant rats. We also analyzed transcript clonality of rat TCR-Vα5, an ortholog of the dominant TCR-Vα chain found on insulin B:9-23–reactive T cells in nonobese diabetic rat islets. We observed clonal expansion of Vα5+ transcripts in prediabetic LEW.1WR1 islets, suggesting that rat Vα5 is also an important component of islet autoantigen recognition. These data provide additional evidence that genome-encoded TCR sequences are important determinants of genetic susceptibility to T1D.
Introduction
Type 1A diabetes (T1D) is an autoimmune disease characterized by T cell–mediated destruction of insulin-producing pancreatic β-cells. T1D is thought to arise through the interaction of genetic and environmental factors, with progressive loss of β-cells occurring over months to years in the presence of circulating islet autoantibodies; clinical diabetes occurs after ∼80% of insulin secretory capacity is lost. Because T1D typically develops over the course of years, and because tissue biopsy is not possible, little is known about early events that underlie T1D. There are no proven therapeutics to prevent, halt, or reverse established diabetes (1), and thus a better understanding of disease onset and progression is necessary.
We have developed rat models of diabetes with no or low incidence of spontaneous disease in which immune perturbation induces diabetes. Inducible animal models (e.g., LEW.1WR1 rats) demonstrate autoimmune pathology that reproduces human disease with considerable fidelity (2,3). We have taken advantage of these models to study early events in diabetes pathogenesis and its genetic control.
T1D is common in inbred rat strains with a high-risk MHC-II haplotype (RT1.B/Du). We previously reported that Iddm14 is a dominant rat diabetes susceptibility locus (4–6) harboring T-cell receptor (TCR) β-chain–variable region (TCR-Vβ) genes. An allele of one TCR-Vβ gene (TcrbV13S1A1) is expressed in six diabetes-susceptible RT1.B/Du rat strains (including LEW.1WR1), whereas three strains that are resistant to, or confer resistance to, diabetes express either TcrbV13S1A2 (e.g., Wistar Furth [WF]) or the nonfunctional TcrbV13S1A3P (F344) gene (7). These polymorphisms are of interest because preferential usage of the TcrbV13S1A1 allelic gene product, termed TCR-Vβ13a, by CD4+ but not CD8+ cells has been reported (8). We confirmed that the gene encoding TCR-Vβ13 is Iddm14 when we prevented both induced (polyinosinic:polycytidylic acid [poly I:C] or poly I:C + Kilham Rat Virus) and spontaneous diabetes by depleting TCR-Vβ13a+ T cells with an allele-specific monoclonal antibody (17D5) (9).
The trimolecular complex is the interface of the TCR, autoantigenic peptide, and major histocompatibility complex (TCR-pMHC), each with validated roles in diabetes pathogenesis. Recent studies highlight the trimolecular complex as a prime therapeutic target for halting diabetes in rodents (9–12). There is a well-established association between MHC-II alleles and T1D susceptibility in humans and rodents. The diabetes-predisposing MHC-II alleles in mice (I-Abg7) (13), rats (RT1.Bbu) (14), and humans (HLA-DQB1*0201/*0302/*0304) (15) share conserved polymorphisms in the antigen-binding groove at position 57 of the β-chain. In nonobese diabetic (NOD) mice, I-Ag7 molecules bind peptides poorly (16–18). This weak binding depends largely on the Serβ57 residue of I-Abg7 (16,18) and leads to increased numbers of autoreactive T cells (17).
In addition, substantial evidence indicates that insulin is a key autoantigen recognized by islet-infiltrating T cells in mice and in people (10,19–21). Consistent with this view, the insulin B:9-23 epitope binds weakly to I-Ag7, limiting thymic negative selection of insulin-reactive T cells (22–24). RT1.Bbu in rats also encodes Serβ57, suggesting that similar binding affinities could apply to the RT1:insulin peptide complex.
Furthermore, our genetic studies in rats and the work of others on diabetogenic T cells in mice (21,25) led us to conclude that germline-encoded elements of the TCR (complementarity determining region [CDR] 1 and CDR2) are critical for recognition of autoantigen + MHC (21,26). This implies that genomically encoded elements in the TCR-variable chain region (CDR1 and CDR2) are critical determinants of autoimmunity, predisposing certain T cells to recognize islet autoantigens. This in turn suggests that targeting T cells expressing those elements can be used, as we have shown, to prevent autoimmune diabetes (9).
LEW.1WR1 and LEW.1W rats are genetically identical for all components of the trimolecular complex (TCR-Vβ13a, TCR-Vα5, insulin, and RT1.B/Du) but differ at the Iddm37 susceptibility locus (5). Iddm37 significantly modifies diabetes penetrance to poly I:C + Kilham Rat Virus infection in (LEW.1WR1xWF)F2 rats harboring RT1.B/Du and Iddm14 (5). LEW.1WR1 rats are highly susceptible to induction of diabetes by injection of 1 μg/g poly I:C (80–100% become diabetic in 3 weeks) (2), whereas LEW.1W rats are relatively resistant (∼10% become diabetic using the same protocol within 3 weeks; (R.A.E. and L.C., unpublished data). WF rats (RT1.B/Du) are highly resistant to diabetes even at very high doses of poly I:C (27) because the expression of the TcrbV13S1A2 resistance allele (4,7). WF rats, like LEW.1W rats, also carry resistance alleles at Iddm37 (5).
Diabetes induction in LEW.1WR1 rats is well-documented (2), but little is known about the timing of islet T-cell infiltration during progression to diabetes or the temporal incidence of islet autoimmunity. In the current study, we hypothesized that if Vβ13a is required for the development of diabetes in LEW.1WR1 rats, then Vβ13a+ T cells should accumulate early in the prediabetic islet and display antigen-specific clonal expansion.
Additionally, we tested the hypothesis that Iddm14 is required for the initiation of diabetes in LEW.1WR1 rats, whereas Iddm37 modifies disease progression. If only Iddm14 is required for disease initiation, then we would predict that Vβ13a+ T cells will infiltrate LEW.1WR1 and LEW.1W islets. Conversely, if the expression of a permissive allele at Iddm37 were required prior to islet T-cell infiltration, then we should not observe islet infiltration in LEW.1W or WF rats.
We analyzed the timing of islet T-cell infiltration and anti-islet autoimmunity by T cells bearing critical germline-encoded TCR elements (Vβ13a and Vα5). We conclude that Vβ13a+ T cells are a critical component of the diabetogenic trimolecular complex; they display islet-specific clonal expansion and increase in abundance throughout diabetes progression. Furthermore, Iddm37 modifies disease progression in LEW.1W rats by delaying and dampening islet infiltration of diabetogenic Vβ13a+ T cells, minimizing overall islet infiltration and preventing disease.
Research Design and Methods
Animals
LEW.1WR1 and LEW.1W rats were obtained from BRM (Worcester, MA). WF rats were purchased from Harlan Laboratories (Dublin, VA). Animals were housed at Drexel University College of Medicine or the University of Massachusetts in viral antibody–free conditions, confirmed monthly to be serologically free of rat pathogens, and maintained in accordance with institutional and national guidelines. No differences in the incidence or latency of diabetes induced in these rat strains have been observed at the two sites.
Diabetes Induction
Spontaneous diabetes incidence in LEW.1WR1 rats is ∼2.5%. Treatment with poly I:C, 1 μg/g i.p. three times weekly, increases diabetes frequency to ∼100% (2). Poly I:C is a ligand of Toll-like receptor 3 and IFIH1 (interferon induced with helicase C domain 1, also known as MDA5). WF rats are resistant to diabetes induction both at this dose and much higher doses (27). Diabetes was induced in rats of either sex starting at 21–28 days of age with poly I:C (Sigma, St. Louis, MO). All rats were injected intraperitoneally with 1 μg/g body weight three times per week (e.g., Monday, Wednesday, and Friday) until takedown. Typically, ∼85–100% of susceptible animals become diabetic within 10–18 days of the first injection.
Islet Isolation
Islet isolation by collagenase perfusion of the common bile duct and islet culture was adapted from Jarchum et al. (28). The common bile duct was cannulated with a 27-gauge needle. Pancreata were perfused with 7–9 mL of 0.85 mg/mL cold collagenase P (Roche, Indianapolis, IN) dissolved in Hanks’ balanced salt solution. Inflated pancreata were removed, placed in RPMI 1640, incubated at 37°C for 20 min then homogenized with a 14-gauge needle in ice-cold RPMI containing 1% horse serum. Islets were separated by a Histopaque 1077 gradient, removed from the interface, washed in ice-cold 5% horse serum RPMI and placed in TRIzol (Invitrogen, Carlsbad, CA).
Culture of Islet T Cells
Isolated islets pooled from eight rats were cultured with interleukin (IL)-2 to promote the expansion of activated T cells. Culture medium contained RPMI 1640 with 10% FBS, 1 mmol/L Na pyruvate, nonessential amino acids, 28 μmol/L β-mercaptoethanol, and 50 units/mL recombinant human IL-2 (PeproTech, Rocky Hill, NJ). Fifty islets per well were cultured in 24-well plates (Greiner Bio-One) in 5% CO2 at 37°C. On day 7, islets and infiltrating cells were collected and passed through a 40-μm strainer to retain the islets. T cells were pelleted and resuspended in TRIzol (28).
Quantitative RT-PCR
Islets were isolated from individual rats and analyzed separately. Total RNA was isolated from islets in TRIzol according to the manufacturers’ protocol, treated with Turbo DNA-free (Applied Biosystems, Carlsbad, CA) to prevent genomic DNA contamination and reverse transcribed into cDNA (High Capacity cDNA Reverse Transcription Kit; Applied Biosystems). Quantitative PCR was performed with an ABI 7900HT Sequence Detector using SYBR Green PCR Mix (Applied Biosystems), 0.5 μmol/L primers and 40 amplification cycles (95.0°C for 0:15; 60.0°C for 1:00). Amplification data were collected and analyzed using ABI SDS2.2 software. Fold change in transcript abundance was normalized to either glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or TCR β-chain constant region (TCR-Cβ) by the ΔΔCt method and analyzed by Student t test (two comparators) or one-way ANOVA (three comparators) to determine significant differences in transcript abundance between strains or time points. Primers are listed in Supplementary Table 1.
Analysis of TCR-Vβ13 and TCR-Vα5 CDR3 Clonality
We determined the clonality of TCRs by examining the clone-specific CDR3 region in each of them. To analyze CDR3 sequence variants in Vβ13+ and Vα5+ transcripts, islet or spleen RNA was isolated and reverse transcribed into cDNA as described above. TCR-Vβ13+ and TCR-Vα5+ transcripts were PCR-amplified with variable region-specific forward primers and constant region-specific reverse primers (Supplementary Table 1) using platinum high-fidelity Taq DNA polymerase (Invitrogen). Amplicons were cloned into the pCR4-TOPO vector (TOPO TA cloning kit; Invitrogen). Transformants were plated on Luria broth (LB)-agar-kanamycin plates. Single clones were 1) grown in LB-kanamycin overnight and plasmids were isolated (QIAprep spin mini-prep kit; Qiagen, Valencia, CA) or 2) arrayed onto LB-kanamycin plates. Vβ13 and Vα5 plasmids or bacterial clones were sequenced by GeneWiz (Plainfield, NJ) with the T3 primer. Sequences were aligned with a multiple sequence alignment program (http://www.genome.jp/tools/clustalw/).
Vα5 Confirmation Analysis
Transcript abundance of Vα5 or Vα68 joined to one of three Jα regions was analyzed by quantitative RT-PCR (qRT-PCR). Vα-Jα abundance was normalized to total Jα abundance, as further described in the legend to Supplementary Fig. 4.
Results
TCR Transcript Accumulation in the Prediabetic Islet
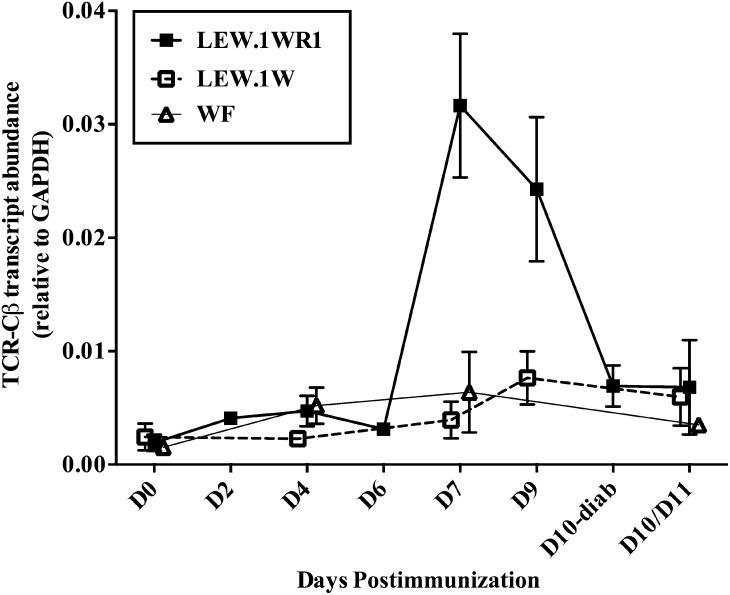
To understand early events in T1D at the islet level, we first measured islet T-cell accumulation after induction. TCR-Cβ transcript abundance, indicating T-cell infiltration of the islet, was measured by qRT-PCR. LEW.1WR1 islets had a nearly 10-fold increase in TCR transcripts on day 7 relative to LEW.1W or WF rats on the same day and compared with untreated islets (Fig. 1). Elevated TCR-Cβ transcript abundance persisted at 9 days after poly I:C infusion but was diminished by day 10, even in the diabetic islets, likely because β-cell loss limits available antigen for T cells, as is suggested by the loss of insulin expression at this time (Fig. 5). There was no significant increase in T-cell transcripts in the islets of either strain of resistant rats at any time point (Fig. 1). We also analyzed TCR-α constant region transcripts to confirm our results. As expected, Cα and Cβ transcript abundance was strongly correlated in all samples analyzed (R2 = 0.90, P < 0.0001; data not shown).
Figure 1.
T-cell transcripts accumulate in the islets of prediabetic LEW.1WR1 rats but not in resistant rats. Rats of each strain were injected with 1 μg/g poly I:C three times weekly (e.g., days 0, 2, 4, 7, 9, and 11) or until the day of takedown, as indicated. Day 0 rats are untreated. To determine the extent of T-cell infiltration into the islets of poly I:C–treated rats, we measured the transcript abundance of the TCR-Cβ by qRT-PCR. Transcript abundance was normalized to GAPDH, and the mean ± SE is plotted above. TCR-Cβ abundance in LEW.1WR1 islets is significantly elevated on day 7, as determined by one-way ANOVA. Overall F(2,7) = 8.554, P = 0.0132. The LEW.1WR1 group was significantly different from both other groups, P < 0.05. At no time point were Cβ transcripts increased in the islets of resistant rat (by Student t test, all days compared with untreated). N = 3–5 LEW.1WR1/LEW.1W or N = 2–4 WF per time point for a total of 29 LEW.1WR1, 18 LEW.1W, and 13 WF rats. D, day; diab, diabetes.
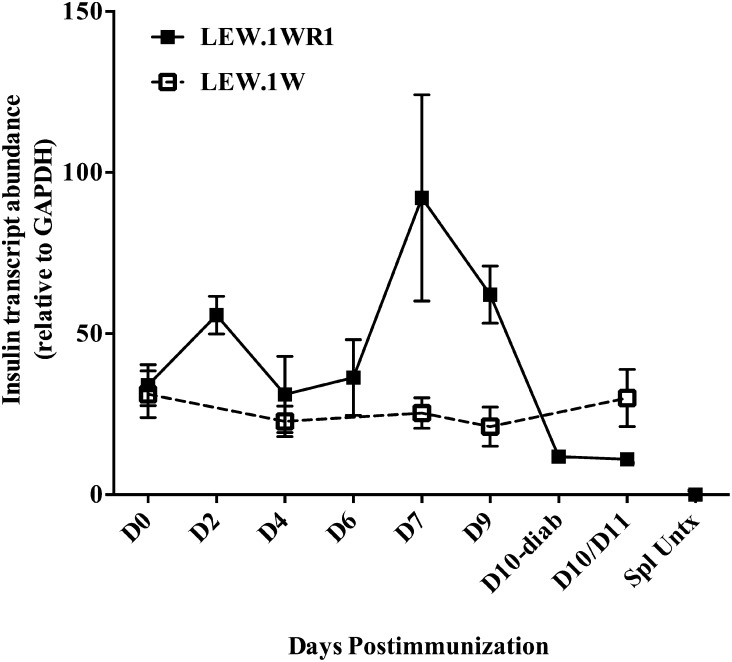
Figure 5.
Altered β-cell function in LEW.1WR1 rats follows Vβ13-specific autoimmunity and is concurrent with significant islet T-cell infiltration. To analyze β-cell dysfunction in the prediabetic islet, we analyzed insulin transcript abundance from poly I:C–treated LEW.1WR1 (susceptible) and LEW.1W (resistant) islets. Insulin transcripts were normalized to GAPDH and the mean ± SE are displayed. LEW.1WR1 islets show significantly elevated insulin transcripts on days 7 and 9 (P = 0.02 and P = 0.01, respectively, vs. untreated), which is severely diminished in the diabetic islet, compared with naïve, day 2, day 7, or day 9 after poly I:C induction (P = 0.001, P < 0.001, P = 0.046, and P = 0.002, respectively). LEW.1W insulin levels do not vary during the course of poly I:C treatment. One of five LEW.1W day 11 islet samples was an outlier (∼2× SD) and was excluded from the above data set, but may be biologically relevant given that 1) a small proportion of LEW.1W rats become diabetic with poly I:C treatment and 2) Vβ13+ T cells begin to accumulate in the islets of some LEW.1W rats on days 9–11. Spleen samples from untreated LEW.1WR1 rats were analyzed as a negative control for insulin transcript production. N = 3–5 rats per time point, 29 LEW.1WR1, 18 LEW.1W total. D, day; diab, diabetes; Spl Untrx, untreated spleen.
Vβ13+ T Cells Accumulate in the Islets of Prediabetic LEW.1WR1 Rats
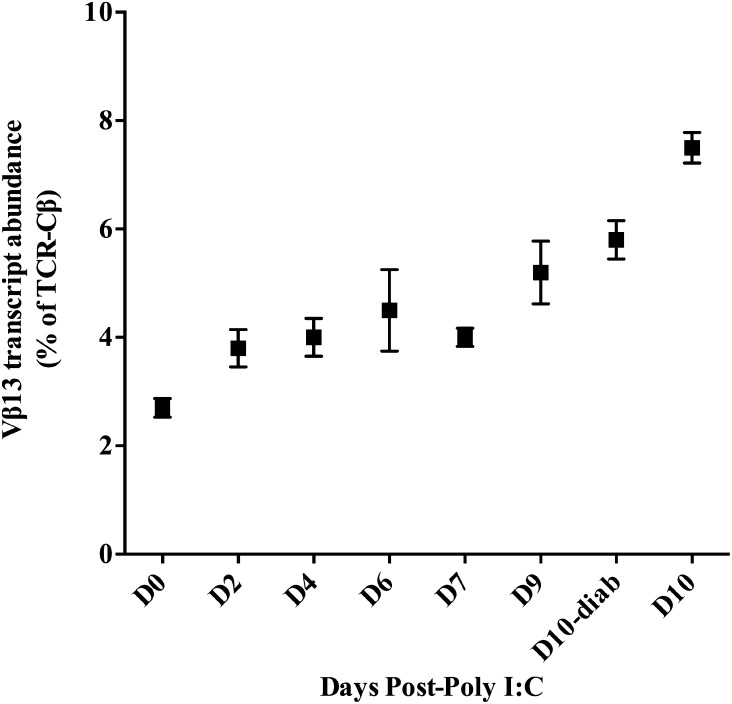
Having established that susceptible, but not resistant, rats accumulate T cells in their islets early in diabetes progression, we hypothesized that Vβ13+ T cells would be abundant in LEW.1WR1 islets. TCR-Vβ13+ transcripts from poly I:C–treated LEW.1WR1 islets were analyzed by qRT-PCR and expressed as a percentage of total TCR transcripts. We find that while Vβ13+ transcripts compose a small proportion of islet T-cell transcripts in untreated rats, their abundance increases approximately threefold during diabetes progression, to ∼7.5% of TCR transcripts in the diabetic islet (Fig. 2). Given the requisite role for TCR-Vβ13+ T cells in diabetes pathogenesis, the relatively low proportion of this population of T cells in the islet was unexpected, although when the absolute increase of islet-infiltrating T cells on days 7–9 is considered, the actual number of islet Vβ13+ T cells is substantial.
Figure 2.
TCR-Vβ13+ T cells increase in abundance during diabetes progression in LEW.1WR1 islets. To determine the relative abundance of Vβ13+ T cells in the islet during progression to diabetes, we measured TCR-Vβ13 transcript abundance and normalized this to the abundance of TCR-Cβ transcripts. This measure gives us the percentage of TCR transcripts that are Vβ13+. There is a strong positive trend in Vβ13 transcript abundance with a significant increase on day 10 in both the diabetic (P = 0.02) and nondiabetic (P = 0.001) islet. N = 3–5 rats per time point, 29 rats total. D, day; diab, diabetes.
Islet Vβ13+ T Cells Clonally Expand During Diabetes Progression
Vβ13+ T cells comprise a small proportion of islet T cells during diabetes progression (Fig. 2), but they do represent a significant portion of CD4+ T cells expanded ex vivo with IL-2 (from day 5 islets, ∼20% of the CD4+ T cells) (9). Therefore, we hypothesized that islet Vβ13+ T cells clonally expand in response to autoantigen. To analyze Vβ13 clonality, we performed CDR3 sequence analysis of islet Vβ13+ transcripts. If Vβ13+ T cells become focused in islet antigens, they will clonally expand and we will observe fewer CDR3 sequences, each at high frequency. If there is not antigen-specific clonal expansion, many different Vβ13+ transcripts will be observed, each at low frequency. We analyzed Vβ13-CDR3 sequences from LEW.1WR1 islets 4, 5, and 7 days after poly I:C induction and from T cells cultured ex vivo from islets 5 days after poly I:C induction. Vβ13+ transcripts from untreated LEW.1WR1 spleens serve as controls for T cells outside the target organ. Sequence analysis of islet-homing TCRs was not performed for resistant rats (LEW.1W and WF rats) because there were few Vβ13+ transcripts in their islets (Supplementary Fig. 1).
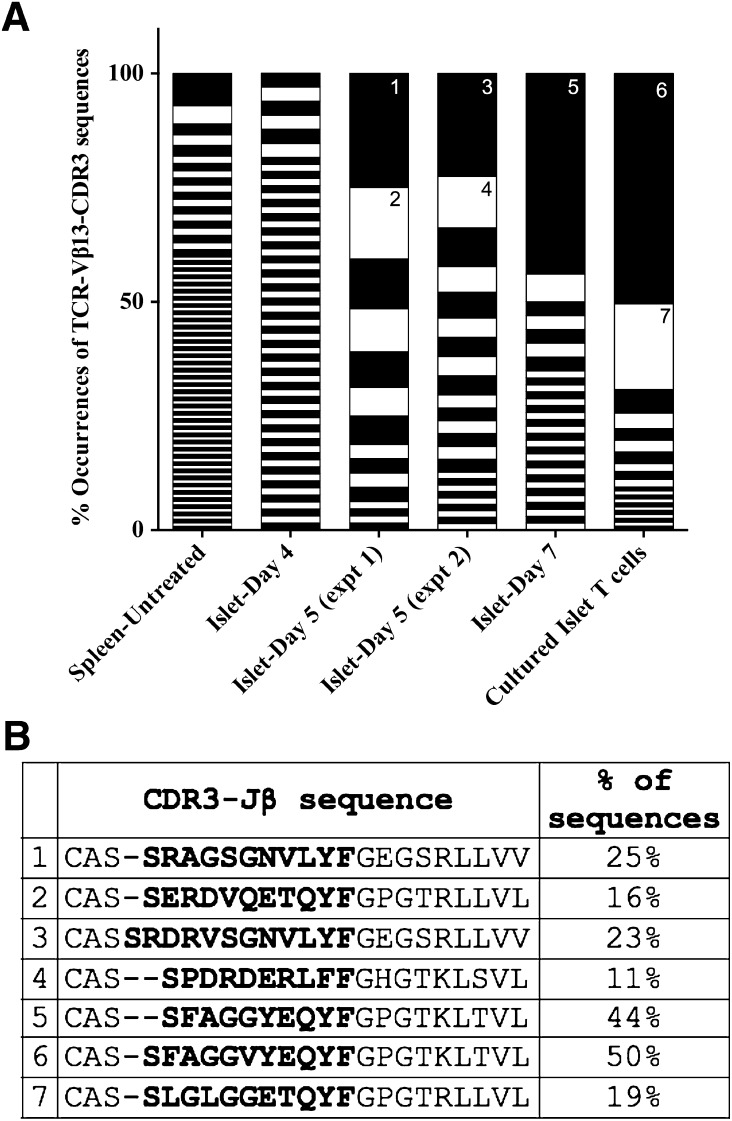
We aligned Vβ13-CDR3 sequences to cluster identical sequences then calculated the frequency of each unique sequence (Fig. 3). As expected, T cells in the naïve periphery are not clonally expanded, displaying substantial CDR3 sequence diversity with no sequence observed at high frequency (Fig. 3). Islet Vβ13+ transcripts were not clonally expanded 4 days postinduction, but by day 5 we observe oligoclonal expansion of Vβ13+ transcripts: there are fewer unique sequences each at a higher frequency. By day 7, there is substantial clonal expansion of one CDR3 sequence in 44% of all Vβ13+ sequences. This sequence was equally represented in each of the two strains rats analyzed. Vβ13+ transcripts analyzed from T cells cultured ex vivo with IL-2 displayed large expansions of a few Vβ13+ clonotypes. Two dominant clonotypes by themselves represent ∼70% of all sequences.
Figure 3.
TCR-Vβ13+ transcripts are antigen-focused in the LEW.1WR1 islet. A: CDR3 sequence variants in islet Vβ13+ transcripts were analyzed by direct sequence analysis. Vβ13+ transcripts were PCR-amplified with a Vβ13-specific forward primer and a Cβ1/2 reverse primer. Amplicons were cloned and sequenced, and sequences were aligned with a multiple sequence alignment program (ClustalW). The percentage occurrence of each unique CDR3 sequence cluster was calculated and is plotted above. Each black or white segment on the graph represents a unique CDR3 sequence. B: Frequent sequences are annotated on the graph (1–7) and listed. The naïve periphery (spleen untreated) does not display a Vβ13-specific antigen-focused response evident by the large number of sequences all observed at low frequency. Vβ13+ T-cell transcripts do not display an antigen-specific clonal expansion on day 4. Two separate experiments, of pooled islets from eight LEW.1WR1 rats each, indicate antigen-specific expansion of a limited number of CDR3 clonotypes in the islet 5 days after poly I:C. For sequences analyzed, N = 117 cultured islet T cells, 66 islet day 7, 74 islet day 5-experiment (expt) 1, 71 islet day 5-experiment 2, 64 islet day 4, and 127 spleen untreated. Sequences represent results from pooled LEW.1WR1 islets; N = 8 cultured T cells, 2 day 7, 8 day 5-experiment 1, 8 day 5-experiment 2, 2 day 4, 2 untreated spleen. B: The most frequent junctional sequences identified among Vβ13 transcripts are listed here. The entire junction sequence is shown starting with the end of the V region; CDR3 sequences in bold are up to the conserved phenylalanine (F), and in the remainder the Jβ segment is displayed. All sequences identified in the analysis are listed in Supplementary Table 2.
Together, these results strongly suggest an islet antigen-specific response of Vβ13+ T cells in LEW.1WR1 islets beginning 5 days after the start of diabetes induction. Clonal focusing of Vβ13+ transcripts is greatest in cultured islet T cells, which, as we have reported, show a preponderance of Vβ13+CD4+ T cells.
TCR-Vα5 Transcripts Are Antigen-Focused in LEW.1WR1 Islets
We next examined TCR-Vα5 clonality in LEW.1WR1 islets. In NOD mice, islet-homing, insulin-reactive T cells use a dominant TCR-Vα chain (TCR-Vα5D4*04) (29), which recognizes insulin B:9-23 (21). Insulin B:9-23 peptide sequences are identical among mice, rats, and humans, and alignment of published sequences shows that NOD I-Abg7 shares similarities with RT1.Bbu (present in WF, LEW.1WR1, and LEW.1W rats) in the antigen-binding groove, raising the possibility that this peptide is an autoantigen in rats. We analyzed rat TCR-Vα5 orthologs and identified four TCR-Vα5 paralogs harboring identical CDR1 and CDR2 peptide sequences that are >92% identical to NOD TRAV5D4*04 (Supplementary Fig. 2). We hypothesized that if LEW.1WR1 rat T cells target the insulin B:9-23 epitope during diabetes development, then rat TCR-Vα5+ transcripts in the prediabetic LEW.1WR1 islet will be abundant and antigen-focused (i.e., they will have limited CDR3 diversity). We analyzed TCR-Vα5+ transcript abundance in the islets of poly I:C–treated LEW.1WR1 rats by qRT-PCR. Consistent with our hypothesis, we observe transient islet accumulation of Vα5 transcripts that wanes at diabetes onset (Supplementary Fig. 3).
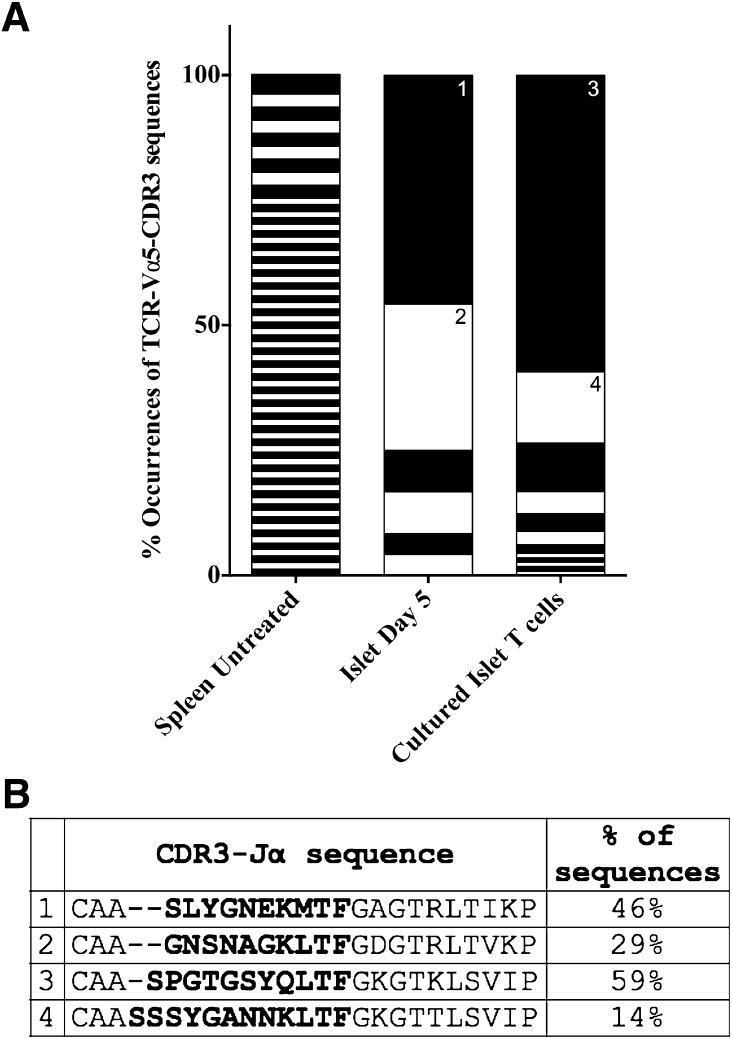
We next performed Vα5-CDR3 sequence analysis from LEW.1WR1 spleen, islets, and ex vivo–cultured islet T cells to determine whether Vα5+ transcripts are clonally expanded. T cells in the periphery are not clonally expanded, displaying substantial CDR3 sequence diversity (Fig. 4). Vα5+ transcripts from LEW.1WR1 islets and cultured islet T cells display clonal expansion of few unique Vα5+ transcripts (Fig. 4), indicating an antigen-specific response of Vα5+ T cells in prediabetic LEW.1WR1 islets. We confirmed this islet-specific skewing of Vα5+ transcripts by qRT-PCR analysis (Supplementary Fig. 4). Novel rat Vα5 paralogs identified in our analysis are listed in Supplementary Fig. 2.
Figure 4.
TCR-Va5+ transcripts are clonally focused in prediabetic LEW.1WR1 islets. CDR3 sequence variants in islet-homing Vα5+ transcripts were analyzed by direct sequence analysis. Vα5+ transcripts were PCR-amplified with a Vα5-specific forward primer and a Cα reverse primer. Amplicons were cloned and sequenced, and sequences were aligned with a multiple sequence alignment program. A: The percentage occurrence of each unique CDR3 sequence was calculated and is plotted above. Each black or white segment on the graph represents a unique CDR3 sequence. Frequent sequences are annotated on the graph (1–4) and listed in B. Islets from eight LEW.1WR1 rats were pooled for islet T-cell culture; three independent samples were analyzed for day 5 islets or untreated spleen. The naïve periphery (splenocytes from untreated rats) does not display a Vα5-specific antigen-focused response, as evidenced by the large number of sequences, all of which were observed at low frequency. There was substantial clonal expansion of Vα5+ transcripts on day 5 and cultured islets indicating an islet antigen-specific expansion. For sequences analyzed, N = 113 cultured islet T cells, 24 islet T cells on day 5, 62 untreated rat splenic T cells. B: The most frequent junctional sequences identified among Vα5 transcripts are listed here. The entire junction sequence is shown starting with the end of the V region. CDR3 sequences are shown in bold up to the conserved phenylalanine (F), and the remainder of the Jβ segment is displayed. All sequences identified in the analysis are listed in Supplementary Table 3.
Insulin Upregulation Is Concurrent With T-Cell Infiltration and Vβ13+ Autoantigen Focusing in Prediabetic LEW.1WR1 Islets
The data presented above suggest that infiltration and clonal expansion of Vβ13+ T cells occur in well-defined stages during T1D progression in poly I:C–treated LEW.1WR1 rats. We reasoned that this stereotypic sequence of events should also allow us to detect evidence of altered β-cell function due to Vβ13+ T-cell infiltration. To interrogate β-cell function, we analyzed insulin transcript abundance by qRT-PCR from the islets of poly I:C–treated LEW.1WR1 and LEW.1W rats.
We first observed that insulin transcript abundance in the islets of untreated LEW.1W and LEW1.WR1 rats was comparable (Fig. 5). On days 7 and 9, after poly I:C induction, however, LEW.1WR1 rats displayed significantly increased insulin transcript abundance, suggesting a change in β-cell function. By day 10 (when the first cases of T1D occurred), we observed a substantial reduction in insulin transcripts in LEW.1WR1 islets. In contrast, insulin mRNA levels in resistant LEW.1W rats showed little fluctuation over time, suggesting that poly I:C was not the cause of transcript variation in the susceptible rats.
Discussion
The present studies substantially extend our previous discovery that genome-encoded TCR sequences control genetic susceptibility to T1D. In the rat, this genome-encoded TCR element is Vβ13, the gene underlying the Iddm14 locus (9). Depletion of Vβ13a+ T cells prevents T1D in multiple rat strains (9), which, in conjunction with recent work in NOD mice, implies that TCR-based strategies could be useful in primary prevention of T1D in children (12). In addition, these data provide new insights into the earliest events that occur in islets in which β-cells are targeted for destruction.
We report here that Vβ13 transcripts become clonally focused in LEW.1WR1 islets and increase in abundance during diabetes progression. These events precede insulin transcript upregulation, which may represent a compensatory response to the autoimmune attack. We first observed that total T-cell abundance in LEW.1WR1 rats increases on days 7–9, while TCR transcripts of any isotype do not accumulate in islets of resistant rats (Fig. 1). The proportion of Vβ13+ TCR transcripts in LEW.1WR1 islets steadily increases during progression to diabetes (Fig. 2). This accumulation of T cells, the deletion of which prevents T1D (9), is consistent with a critical role for Vβ13+ T cells both early and throughout disease progression.
Although Vβ13+ T cells are required for diabetes, they are neither the only T cell present nor the majority of T cells in LEW.1WR1 islets. This is not surprising, given other features of T-cell populations that are related to diabetes. For example, CD8+ cytotoxic T lymphocytes are the most prevalent component of the immune infiltrate in diabetic human pancreatic tissue (30), whereas Vβ13a in the rat is preferentially expressed on CD4+ T cells (8). After in vitro expansion of islet T cells with IL-2, which promotes the proliferation of activated (antigen-experienced) T cells (28), >20% of the CD4+ T cells from LEW.1WR1 islets are Vβ13a+ (9).
Vβ13a+ transcripts in LEW.1WR1 islets are oligoclonally expanded beginning 5 days after induction (Fig. 3), evidence that Vβ13a+ T cells recognize critical islet autoantigens. This antigen recognition depends on inherited elements of the TCR (CDR1 and CDR2, and not solely the hypervariable CDR3 regions) because WF T cells bearing the low-risk TCR-Vβ13b allele do not accumulate in islets and do not cause T1D (Fig. 1). Furthermore, numerous CDR3 sequences were observed among clonally expanded Vβ13+ T-cell populations, suggesting that Vβ13a is required for autoantigen recognition in conjunction with permissive, but not monoclonally restricted, CDR3s. This is consistent with the hypothesis that TCR germline CDR1 and CDR2 sequences are a driving factor for recognition of islet autoantigens.
There is precedence for germline TCR sequence recognition of autoantigen in the NOD diabetes model and in other autoimmune diseases. For example, in celiac disease, characterized by T-cell autoreactivity toward certain gluten peptides, recognition of HLA-DQ8-α-I-gliadin (pMHC) is biased toward TCR-Vβ9*01+ TCRs. Mapping of the TCR-pMHC interface and CDR mutational analysis identified CDR1 (Leu37β) and CDR2 (Tyr57β) residues critical for interacting with α-I-gliadin (26). Interestingly, CDR1-37β is polymorphic in rat TCR-Vβ13 (7), suggesting that autoantigen engagement by CDR1 and CDR2 residues may be a critical mechanism of self-reactivity in multiple autoimmune diseases.
Recognition of insulin B:9-23 in NOD mice depends on critical CDR1 and CDR2 residues in TCR-Vα5D4 (21). Furthermore, the human homolog of mouse TCR-Vα5D4 (TCR-Vα13-1) promotes an anti-insulin response in retrogenic NOD mice, indicating a pan-species mechanism of autoantigen (e.g., insulin) recognition (21). We analyzed rat TCR-Vα5 transcripts, bearing CDR1 and CDR2 sequences that are highly homologous with NOD TCR-Vα5D4. We observed a transient accumulation of rat TCR-Vα5 transcripts in LEW.1WR1 islets (Supplementary Fig. 3) and islet-specific clonal focusing of Vα5+ transcripts preceding diabetes onset (Fig. 4). Based on the rat usage of TCR-Vα5, we speculate that insulin may be a critical autoantigen early in rat diabetogenesis, as is likely in NOD mice (21,31).
The discordant kinetics of Vβ13 and Vα5 transcript accumulation in LEW.1WR1 islets suggest that Vβ13 may recognize multiple antigens during the course of diabetes development. Vβ13 steadily increases toward diabetes onset, whereas the transient increase in Vα5 abundance wanes at diabetes onset. The temporal abundance of Vα5 is consistent with the finding that insulin B:9-23 is an initiating autoantigen in NOD mice but is not a primary target of islet T cells at diabetes onset (31). We previously showed that depletion of Vβ13+ T cells prevents poly I:C–induced diabetes and preserves healthy islet architecture in LEW.1WR1 rats (9). If Vβ13+ T cells are required throughout progression to diabetes and/or recognize numerous islet antigens during diabetes progression, then a TCR-targeted strategy may not only protect against diabetes induction but also progression from autoimmunity to overt diabetes.
We observe significant T-cell accumulation in LEW.1WR1 islets 7–9 days after poly I:C induction. There is a substantial increase in insulin transcript abundance at this time as well. This may indicate that T cell–mediated β-cell death leads to compensatory upregulation of insulin gene transcription from remaining β-cells. As expected, insulin transcripts are severely diminished at or near diabetes onset. Substantial islet T-cell infiltration is not apparent in resistant rats (LEW.1W and WF), and accordingly there is no fluctuation in insulin transcript abundance in these samples, confirming that if β-cell destruction is reflected by insulin transcript fluctuation, it is not related to poly I:C treatment but to T-cell infiltration. This suggests that there is an immediate functional consequence of T-cell infiltration in the islet and a progressive decline in β-cell mass/function that persists for multiple days before hyperglycemia.
Our data also indicate that the trimolecular complex alone is not sufficient for rat autoimmune diabetes. Similar to the situation in humans, in whom diabetes develops in a limited proportion of individuals with high-risk MHC susceptibility genes, we have demonstrated that, despite a genetically high-risk background, including diabetes risk alleles at RT1.B/Du, Vβ13a, and Vα5, most LEW.1W rats are relatively free of islet T-cell infiltration and maintain normal insulin production after poly I:C induction. Treated LEW.1W rats showed no islet infiltration initially, but, after a delay, modest infiltration of diabetogenic Vβ13a+ T cells occurred (Supplementary Fig. 1). Therefore, Iddm37, the only quantitative trait locus interval that distinguishes LEW.1W from LEW.1WR1 (32), modifies the course of disease in LEW.1W rats by delaying islet infiltration by Vβ13a+ T cells, dampening islet T-cell infiltration, and thus protecting LEW.1W rats from disease. These results demonstrate that Vβ13a+ T cells, while necessary for diabetes, are not sufficient for poly I:C–induced islet infiltration; Iddm37 susceptibility alleles are required to promote the early and robust infiltration of LEW.1WR1 islets.
The present data clearly specify the mechanism by which anti-Vβ13 deletional therapy substantially prevents disease. A highly focused but small population of Vβ13+ T cells appears very early in prediabetic islets and expands rapidly. Deletion of this genetically critical population early on is hence a logical strategy for primary prevention of T1D, whether it occurs spontaneously or in response to immune perturbation. Whether this kind of deletional therapy would be effective for secondary prevention (i.e., after islet autoimmunity commences) is an open question, but it is a plausible expectation, based on preliminary data showing that depletion of Vβ13a+ T cells prevents diabetes even when started after poly I:C treatment has been started (M.H. and R.A.E., unpublished observations).
Perhaps the most important implication of the present data is that a search for genomic T1D susceptibility loci in the human TCR regions (in populations stratified for high-risk MHC haplotype) would be a worthwhile endeavor (33). Newer structural analyses of the TCR-pMHC also support this view (26,34). Finally, the staging of events leading to autoimmune diabetes in the LEW.1WR1 rat, characterized here, and the identification of corollary events in human diabetes will allow us to target therapeutics to specific stages in diabetes progression.
Supplementary Material
Article Information
Funding. This research was supported in part by American Diabetes Association (grants 7-11-BS-102 and 7-09-BS-18) and the National Institutes of Health (grants R01 AI092105-01 and R21 AI088480). This research made use of the Diabetes Endocrinology Research Center Morphology Core.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. R.A.E., J.P.M., and E.P.B. researched data; contributed to the discussion; and wrote, reviewed, and edited the manuscript. L.C. researched data and contributed to the discussion. M.H. researched data, contributed to the discussion, and reviewed and edited the manuscript. E.P.B. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db13-0462/-/DC1.
The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
References
- 1.Skyler JS, Ricordi C. Stopping type 1 diabetes: attempts to prevent or cure type 1 diabetes in man. Diabetes 2011;60:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mordes JP, Guberski DL, Leif JH, et al. LEW.1WR1 rats develop autoimmune diabetes spontaneously and in response to environmental perturbation. Diabetes 2005;54:2727–2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tirabassi RS, Guberski DL, Blankenhorn EP, et al. Infection with viruses from several families triggers autoimmune diabetes in LEW*1WR1 rats: prevention of diabetes by maternal immunization. Diabetes 2010;59:110–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mordes JP, Leif J, Novak S, DeScipio C, Greiner DL, Blankenhorn EP. The iddm4 locus segregates with diabetes susceptibility in congenic WF.iddm4 rats. Diabetes 2002;51:3254–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blankenhorn EP, Cort L, Greiner DL, Guberski DL, Mordes JP. Virus-induced autoimmune diabetes in the LEW.1WR1 rat requires Iddm14 and a genetic locus proximal to the major histocompatibility complex. Diabetes 2009;58:2930–2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blankenhorn EP, Rodemich L, Martin-Fernandez C, Leif J, Greiner DL, Mordes JP. The rat diabetes susceptibility locus Iddm4 and at least one additional gene are required for autoimmune diabetes induced by viral infection. Diabetes 2005;54:1233–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mordes JP, Cort L, Norowski E, et al. Analysis of the rat Iddm14 diabetes susceptibility locus in multiple rat strains: identification of a susceptibility haplotype in the Tcrb-V locus. Mamm Genome 2009;20:162–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stienekemeier M, Hofmann K, Gold R, Herrmann T. A polymorphism of the rat T-cell receptor beta-chain variable gene 13 (BV13S1) correlates with the frequency of BV13S1-positive CD4 cells. Immunogenetics 2000;51:296–305 [DOI] [PubMed] [Google Scholar]
- 9.Liu Z, Cort L, Eberwine R, et al. Prevention of type 1 diabetes in the rat with an allele-specific anti-T-cell receptor antibody: Vβ13 as a therapeutic target and biomarker. Diabetes 2012;61:1160–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang L, Stadinski BD, Michels A, Kappler JW, Eisenbarth GS. Immunization with an insulin peptide-MHC complex to prevent type 1 diabetes of NOD mice. Diabetes Metab Res Rev 2011;27:784–789 [DOI] [PubMed] [Google Scholar]
- 11.Michels AW, Ostrov DA, Zhang L, et al. Structure-based selection of small molecules to alter allele-specific MHC class II antigen presentation. J Immunol 2011;187:5921–5930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakayama M, Eisenbarth GS. Paradigm shift or shifting paradigm for type 1 diabetes. Diabetes 2012;61:976–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miyazaki T, Uno M, Uehira M, et al. Direct evidence for the contribution of the unique I-ANOD to the development of insulitis in non-obese diabetic mice. Nature 1990;345:722–724 [DOI] [PubMed] [Google Scholar]
- 14.Yokoi N, Hidaka S, Tanabe S, et al. Role of major histocompatibility complex class II in the development of autoimmune type 1 diabetes and thyroiditis in rats. Genes Immun 2012;13:139–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Todd JA, Bell JI, McDevitt HO. HLA-DQ beta gene contributes to susceptibility and resistance to insulin-dependent diabetes mellitus. Nature 1987;329:599–604 [DOI] [PubMed] [Google Scholar]
- 16.Carrasco-Marin E, Shimizu J, Kanagawa O, Unanue ER. The class II MHC I-Ag7 molecules from non-obese diabetic mice are poor peptide binders. J Immunol 1996;156:450–458 [PubMed] [Google Scholar]
- 17.Kanagawa O, Martin SM, Vaupel BA, Carrasco-Marin E, Unanue ER. Autoreactivity of T cells from nonobese diabetic mice: an I-Ag7-dependent reaction. Proc Natl Acad Sci USA 1998;95:1721–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Latek RR, Suri A, Petzold SJ, et al. Structural basis of peptide binding and presentation by the type I diabetes-associated MHC class II molecule of NOD mice. Immunity 2000;12:699–710 [DOI] [PubMed] [Google Scholar]
- 19.Sosinowski T, Eisenbarth GS. Type 1 diabetes: primary antigen/peptide/register/trimolecular complex. Immunol Res 2013;55:270–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakayama M. Insulin as a key autoantigen in the development of type 1 diabetes. Diabetes Metab Res Rev 2011;27:773–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakayama M, Castoe T, Sosinowski T, et al. Germline TRAV5D-4 T-cell receptor sequence targets a primary insulin peptide of NOD mice. Diabetes 2012;61:857–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levisetti MG, Suri A, Petzold SJ, Unanue ER. The insulin-specific T cells of nonobese diabetic mice recognize a weak MHC-binding segment in more than one form. J Immunol 2007;178:6051–6057 [DOI] [PubMed] [Google Scholar]
- 23.Stadinski BD, Zhang L, Crawford F, Marrack P, Eisenbarth GS, Kappler JW. Diabetogenic T cells recognize insulin bound to IAg7 in an unexpected, weakly binding register. Proc Natl Acad Sci USA 2010;107:10978–10983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu B, Gauthier L, Hausmann DH, Wucherpfennig KW. Binding of conserved islet peptides by human and murine MHC class II molecules associated with susceptibility to type I diabetes. Eur J Immunol 2000;30:2497–2506 [DOI] [PubMed] [Google Scholar]
- 25.Yin L, Dai S, Clayton G, et al. Recognition of self and altered self by T cells in autoimmunity and allergy. Protein Cell 2013;4:8–16 [DOI] [PMC free article] [PubMed]
- 26.Broughton SE, Petersen J, Theodossis A, et al. Biased T cell receptor usage directed against human leukocyte antigen DQ8-restricted gliadin peptides is associated with celiac disease. Immunity 2012;37:611–621 [DOI] [PubMed] [Google Scholar]
- 27.Ellerman KE, Like AA. Susceptibility to diabetes is widely distributed in normal class IIu haplotype rats. Diabetologia 2000;43:890–898 [DOI] [PubMed] [Google Scholar]
- 28.Jarchum I, Takaki T, DiLorenzo TP. Efficient culture of CD8(+) T cells from the islets of NOD mice and their use for the study of autoreactive specificities. J Immunol Methods 2008;339:66–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simone E, Daniel D, Schloot N, et al. T cell receptor restriction of diabetogenic autoimmune NOD T cells. Proc Natl Acad Sci USA 1997;94:2518–2521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Willcox A, Richardson SJ, Bone AJ, Foulis AK, Morgan NG. Analysis of islet inflammation in human type 1 diabetes. Clin Exp Immunol 2009;155:173–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prasad S, Kohm AP, McMahon JS, Luo X, Miller SD. Pathogenesis of NOD diabetes is initiated by reactivity to the insulin B chain 9-23 epitope and involves functional epitope spreading. J Autoimmun 2012;39:347–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kohoutavá M, Günther E, Stark O. Genetic definition of a further gene region and identification of at least three different histocompatibility genes in the rat major histocompatibility system. Immunogenetics 1980;11:483–490 [DOI] [PubMed] [Google Scholar]
- 33.Pierce BG, Eberwine R, Noble JA, et al. The missing heritability in T1D and potential new targets for prevention. J Diabetes Res 2013;2013:737485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sethi DK, Schubert DA, Anders AK, et al. A highly tilted binding mode by a self-reactive T cell receptor results in altered engagement of peptide and MHC. J Exp Med 2011;208:91–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shirwan H, Barwari L, Fuss I, Makowka L, Cramer DV. Structure and repertoire usage of rat TCR alpha-chain genes in T cells infiltrating heart allografts. J Immunol 1995;154:1964–1972 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.