Abstract
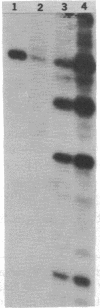
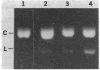
M13 recombinant DNA clones containing a 350-base sequence derived from the EcoRI fragment of two tandemly linked Rous-associated virus 2 (RAV-2) long terminal repeat (LTR) sequences have been used to map reverse transcriptase-associated endonuclease (RT-endonuclease) cleavage sites by primer extension studies. Under appropriate conditions, the alpha beta form of RT-endonuclease (composed of both the alpha and beta subunits) purified from Avian sarcoma virus (Pr-C and B-77 strains) introduces a specific break in the inverted complementary repeat sequence found at the junction of the LTRs. The cleavage sites occur in the same nucleotide sequence in (-) and (+) DNA strands; together they have the potential of generating a 6-base-pair staggered overlap that spans the junction. This supports the notion that the enzyme is involved in viral DNA integration. Other RT-endonuclease sites were analyzed. A second site, which occurs in the lac region of the M13 vector DNA upstream from the unique EcoRI cloning site, bears no apparent sequence homology to the site at the junction of the LTRs. However, it also lies within an inverted complementary repeat and, as is the case for the site in the LTR, the break occurs to the 5' side of the axis of symmetry. Cleavage at this second site is suppressed when the vector contains the RAV-2 LTR insert. Thus, the viral LTR appears to exert a cis effect that can influence a region over 300 base pairs away.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barnes W. M., Bevan M. Kilo-sequencing: an ordered strategy for rapid DNA sequence data acquisition. Nucleic Acids Res. 1983 Jan 25;11(2):349–368. doi: 10.1093/nar/11.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakesley R. W., Wells R. D. 'Single-stranded' DNA from phiX174 and M13 is cleaved by certain restriction endonucleases. Nature. 1975 Oct 2;257(5525):421–422. doi: 10.1038/257421a0. [DOI] [PubMed] [Google Scholar]
- Ehrlich S. D., Bertazzoni U., Bernardi G. The specificity of pancreatic deoxyribonuclease. Eur J Biochem. 1973 Dec 3;40(1):143–147. doi: 10.1111/j.1432-1033.1973.tb03178.x. [DOI] [PubMed] [Google Scholar]
- Gibson W., Verma I. M. Studies on the reverse transcriptase of RNA tumor viruses. Structural relatedness of two subunits of avian RNA tumor viruses. Proc Natl Acad Sci U S A. 1974 Dec;71(12):4991–4994. doi: 10.1073/pnas.71.12.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godson G. N., Roberts R. J. dna, single stranded/*metab. Virology. 1976 Sep;73(2):561–567. doi: 10.1016/0042-6822(76)90421-9. [DOI] [PubMed] [Google Scholar]
- Golomb M., Grandgenett D. P. Endonuclease activity of purified RNA-directed DNA polymerase from avian myeloblastosis virus. J Biol Chem. 1979 Mar 10;254(5):1606–1613. [PubMed] [Google Scholar]
- Hishinuma F., DeBona P. J., Astrin S., Skalka A. M. Nucleotide sequence of acceptor site and termini of integrated avian endogenous provirus ev1: integration creates a 6 bp repeat of host DNA. Cell. 1981 Jan;23(1):155–164. doi: 10.1016/0092-8674(81)90280-4. [DOI] [PubMed] [Google Scholar]
- Iida S., Marcoli R., Bickle T. A. Variant insertion element IS1 generates 8-base pair duplications of the target sequence. Nature. 1981 Nov 26;294(5839):374–376. doi: 10.1038/294374a0. [DOI] [PubMed] [Google Scholar]
- Ju G., Skalka A. M. Nucleotide sequence analysis of the long terminal repeat (LTR) of avian retroviruses: structural similarities with transposable elements. Cell. 1980 Nov;22(2 Pt 2):379–386. doi: 10.1016/0092-8674(80)90348-7. [DOI] [PubMed] [Google Scholar]
- Klaer R., Kühn S., Fritz H. J., Tillmann E., Saint-Girons I., Habermann P., Pfeifer D., Starlinger P. Studies on transposition mechanisms and specificity of IS4. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):215–224. doi: 10.1101/sqb.1981.045.01.034. [DOI] [PubMed] [Google Scholar]
- Klempnauer K. H., Gonda T. J., Bishop J. M. Nucleotide sequence of the retroviral leukemia gene v-myb and its cellular progenitor c-myb: the architecture of a transduced oncogene. Cell. 1982 Dec;31(2 Pt 1):453–463. doi: 10.1016/0092-8674(82)90138-6. [DOI] [PubMed] [Google Scholar]
- Leis J., Duyk G., Johnson S., Longiaru M., Skalka A. Mechanism of action of the endonuclease associated with the alpha beta and beta beta forms of avian RNA tumor virus reverse transcriptase. J Virol. 1983 Feb;45(2):727–739. doi: 10.1128/jvi.45.2.727-739.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Gronenborn B., Müller-Hill B., Hans Hopschneider P. Filamentous coliphage M13 as a cloning vehicle: insertion of a HindII fragment of the lac regulatory region in M13 replicative form in vitro. Proc Natl Acad Sci U S A. 1977 Sep;74(9):3642–3646. doi: 10.1073/pnas.74.9.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra T. K., Grandgenett D. P., Parsons J. T. Avian retrovirus pp32 DNA-binding protein. I. Recognition of specific sequences on retrovirus DNA terminal repeats. J Virol. 1982 Oct;44(1):330–343. doi: 10.1128/jvi.44.1.330-343.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rho H. M., Grandgenett D. P., Green M. Sequence relatedness between the subunits of avian myeloblastosis virus reverse transcriptase. J Biol Chem. 1975 Jul 10;250(13):5278–5280. [PubMed] [Google Scholar]
- Rice C. M., Strauss J. H. Synthesis, cleavage and sequence analysis of DNA complementary to the 26 S messenger RNA of Sindbis virus. J Mol Biol. 1981 Aug 15;150(3):315–340. doi: 10.1016/0022-2836(81)90550-7. [DOI] [PubMed] [Google Scholar]
- Rushlow K. E., Lautenberger J. A., Reddy E. P., Souza L. M., Baluda M. A., Chirikjian J. G., Papas T. S. Nucleotide sequence analysis of the long terminal repeat of avian myeloblastosis virus and adjacent host sequences. J Virol. 1982 Jun;42(3):840–846. doi: 10.1128/jvi.42.3.840-846.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Schiff R. D., Grandgenett D. P. Virus-coded origin of a 32,000-dalton protein from avian retrovirus cores: structural relatedness of p32 and the beta polypeptide of the avian retrovirus DNA polymerase. J Virol. 1978 Oct;28(1):279–291. doi: 10.1128/jvi.28.1.279-291.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker C., Goff S., Gilboa E., Paskind M., Mitra S. W., Baltimore D. Structure of a cloned circular Moloney murine leukemia virus DNA molecule containing an inverted segment: implications for retrovirus integration. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3932–3936. doi: 10.1073/pnas.77.7.3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varmus H. E. Form and function of retroviral proviruses. Science. 1982 May 21;216(4548):812–820. doi: 10.1126/science.6177038. [DOI] [PubMed] [Google Scholar]
- Wells R. D., Goodman T. C., Hillen W., Horn G. T., Klein R. D., Larson J. E., Müller U. R., Neuendorf S. K., Panayotatos N., Stirdivant S. M. DNA structure and gene regulation. Prog Nucleic Acid Res Mol Biol. 1980;24:167–267. doi: 10.1016/s0079-6603(08)60674-1. [DOI] [PubMed] [Google Scholar]