Abstract
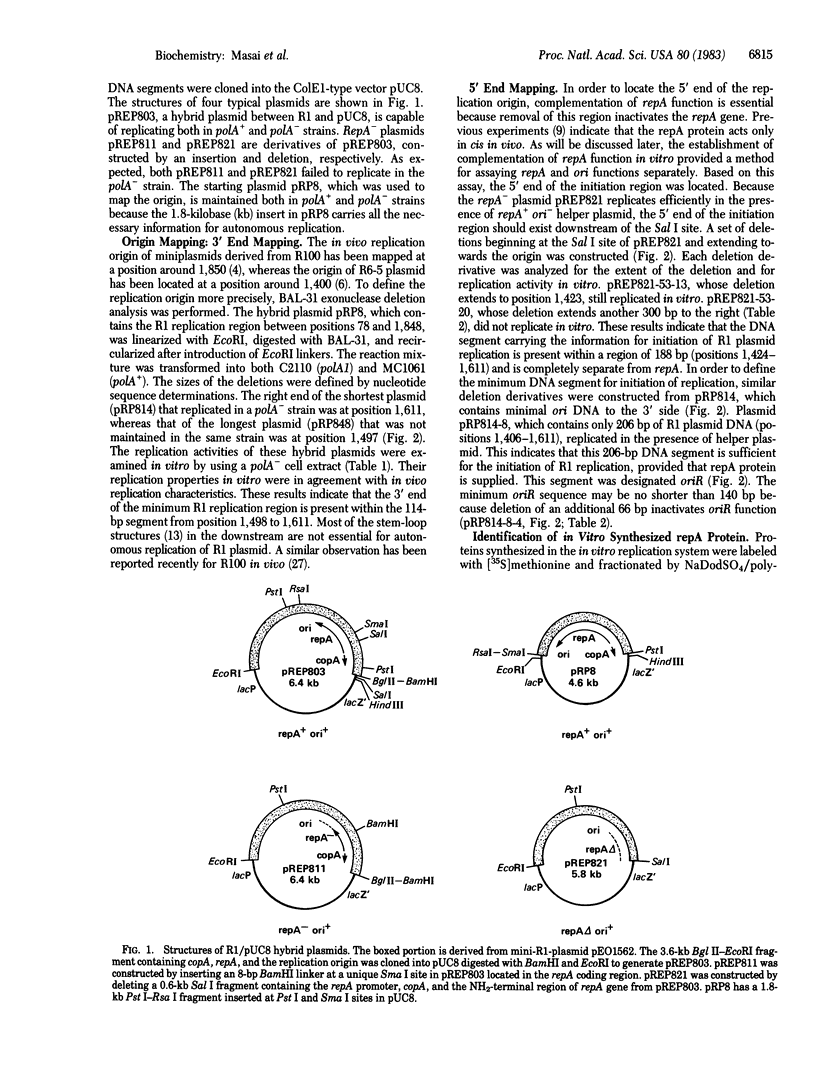
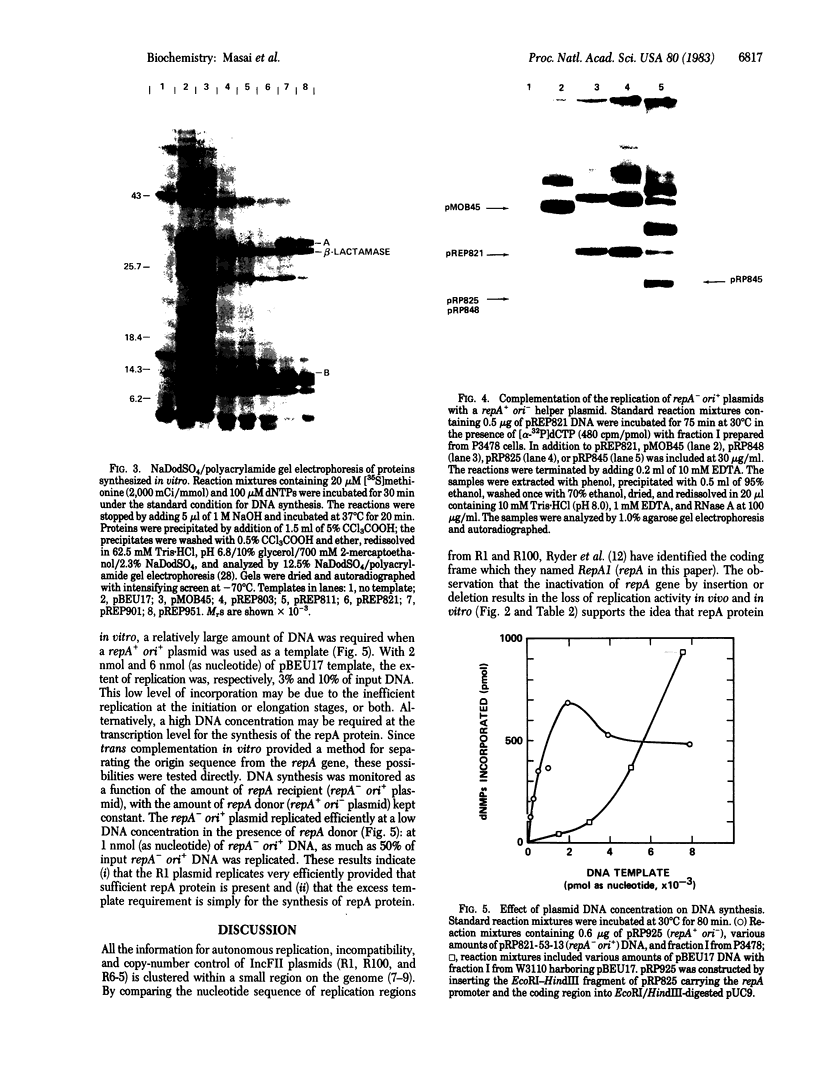
The 3.6-kilobase Bgl II-EcoRI fragment from R1 plasmid containing copA, repA, and the replication origin (ori) was inserted into the ColE1-type plasmid pUC8. The resulting hybrid plasmid replicates in extracts prepared from both polA- and polA+ cells, whereas pUC8 replicates only in a polA+ extract. This characteristic provides a method for assaying the repA and ori functions. Hybrid plasmids that were either repA- or ori- were unable to replicate in a polA- cell extract. Replication of the repA- ori+ plasmid was restored by complementation of the repA defect by a repA+ ori- plasmid in vitro. Successful complementation of the repA function in vitro provides a method for assaying the repA protein. In order to define the minimum DNA segment with origin function (oriR), deletions were introduced starting from either side of the insert, and the replication properties of the plasmids carrying these deletions were examined in a polA- cell extract. The right end of oriR was located at position 1,611 in the nucleotide coordinates defined previously [Ryder, T., Rosen, J., Armstrong, K., Davidson, D. & Ohtsubo, E. (1981) in The Initiation of DNA Replication: ICN-UCLA Symposia on Molecular and Cellular Biology, ed. Ray, D.S. (Academic, New York), Vol. 22, 91-111]. By complementing repA- ori+ plasmids with the repA+ ori- plasmid, the left end of oriR was localized at position 1,424. Therefore, the oriR sequence, localized within a region of 188 base pairs, is separate from the repA gene. A hybrid plasmid carrying the 206-base-pair segment between positions 1,406 and 1,611 also replicates in a polA- cell extract when the repA function is supplied in trans. Removal of an additional 66 base pairs (positions 1,406-1,471) inactivates the function of the minimal oriR segment.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bittner M., Vapnek D. Versatile cloning vectors derived from the runaway-replication plasmid pKN402. Gene. 1981 Dec;15(4):319–329. doi: 10.1016/0378-1119(81)90175-x. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980 Apr;138(2):179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Cohen S. N., Miller C. A. Non-chromosomal antibiotic resistance in bacteria. II. Molecular nature of R-factors isolated from Proteus mirabilis and Escherichia coli. J Mol Biol. 1970 Jun 28;50(3):671–687. doi: 10.1016/0022-2836(70)90092-6. [DOI] [PubMed] [Google Scholar]
- Cohen S. N., Silver R. P., Sharp P. A., McCoubrey A. E. The problems of drug-resistant pathogenic bacteria. Studies on the molecular nature of R factors. Ann N Y Acad Sci. 1971 Jun 11;182:172–187. doi: 10.1111/j.1749-6632.1971.tb30655.x. [DOI] [PubMed] [Google Scholar]
- Diaz R., Nordström K., Staudenbauer W. L. Plasmid R1 DNA replication dependent on protein synthesis in cell-free extracts of E. coli. Nature. 1981 Jan 22;289(5795):326–328. doi: 10.1038/289326a0. [DOI] [PubMed] [Google Scholar]
- Diaz R., Staudenbauer W. L. Origin and direction of mini-R1 plasmid DNA replication in cell extracts of Escherichia coli. J Bacteriol. 1982 Jun;150(3):1077–1084. doi: 10.1128/jb.150.3.1077-1084.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollek R., Oertel W., Goebel W. Isolation and characterization of the minimal fragment required for autonomous replication ("basic replicon") of a copy mutant (pKN102) of the antibiotic resistance factor R1. Mol Gen Genet. 1978 Jun 1;162(1):51–57. doi: 10.1007/BF00333850. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Light J., Molin S. Replication control functions of plasmid R1 act as inhibitors of expression of a gene required for replication. Mol Gen Genet. 1981;184(1):56–61. doi: 10.1007/BF00271195. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Meynell E., Meynell G. G., Datta N. Phylogenetic relationships of drug-resistance factors and other transmissible bacterial plasmids. Bacteriol Rev. 1968 Mar;32(1):55–83. doi: 10.1128/br.32.1.55-83.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T., Easton A. M., Rownd R. H. Cloning of replication, incompatibility, and stability functions of R plasmid NR1. J Bacteriol. 1980 Jan;141(1):87–99. doi: 10.1128/jb.141.1.87-99.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molin S., Stougaard P., Light J., Nordström M., Nordström K. Isolation and characterization of new copy mutants of plasmid R1, and identification of a polypeptide involved in copy number control. Mol Gen Genet. 1981;181(1):123–130. doi: 10.1007/BF00339015. [DOI] [PubMed] [Google Scholar]
- Molin S., Stougaard P., Uhlin B. E., Gustafsson P., Nordström K. Clustering of genes involved in replication, copy number control, incompatibility, and stable maintenance of the resistance plasmid R1drd-19. J Bacteriol. 1979 Apr;138(1):70–79. doi: 10.1128/jb.138.1.70-79.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo E., Feingold J., Ohtsubo H., Mickel S., Bauer W. Unidirectional replication in Escherichia coli of three small plasmids derived from R factor R12. Plasmid. 1977 Nov;1(1):8–18. doi: 10.1016/0147-619x(77)90004-x. [DOI] [PubMed] [Google Scholar]
- Ohtsubo H., Vassino B., Ryder T., Ohtsubo E. A simple method for shortening a plasmid genome using a system of plasmid cointegration mediated by a Tn3 mutant. Gene. 1982 Dec;20(2):245–254. doi: 10.1016/0378-1119(82)90043-9. [DOI] [PubMed] [Google Scholar]
- Rosen J., Ryder T., Inokuchi H., Ohtsubo H., Ohtsubo E. Genes and sites involved in replication and incompatibility of an R100 plasmid derivative based on nucleotide sequence analysis. Mol Gen Genet. 1980;179(3):527–537. doi: 10.1007/BF00271742. [DOI] [PubMed] [Google Scholar]
- Rosen J., Ryder T., Ohtsubo H., Ohtsubo E. Role of RNA transcripts in replication incompatibility and copy number control in antibiotic resistance plasmid derivatives. Nature. 1981 Apr 30;290(5809):794–797. doi: 10.1038/290794a0. [DOI] [PubMed] [Google Scholar]
- Rubin C. M., Schmid C. W. Pyrimidine-specific chemical reactions useful for DNA sequencing. Nucleic Acids Res. 1980 Oct 24;8(20):4613–4619. doi: 10.1093/nar/8.20.4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver L., Chandler M., de la Tour E. B., Caro L. Origin and direction of replication of the drug resistance plasmid R100.1 and of a resistance transfer factor derivative in synchronized cultures. J Bacteriol. 1977 Sep;131(3):929–942. doi: 10.1128/jb.131.3.929-942.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudenbauer W. L. Replication of small plasmids in extracts of Escherichia coli. Mol Gen Genet. 1976 Jun 15;145(3):273–280. doi: 10.1007/BF00325823. [DOI] [PubMed] [Google Scholar]
- Stougaard P., Molin S., Nordström K. RNAs involved in copy-number control and incompatibility of plasmid R1. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6008–6012. doi: 10.1073/pnas.78.10.6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Synenki R. M., Nordheim A., Timmis K. N. Plasmid replication functions. III. Origin and direction of replication of a "mini" plasmid derived from R6-5. Mol Gen Genet. 1979 Jan 5;168(1):27–36. doi: 10.1007/BF00267930. [DOI] [PubMed] [Google Scholar]
- Taylor D. P., Cohen S. N. Structural and functional analysis of cloned DNA segments containing the replication and incompatibility regions of a miniplasmid derived from a copy number mutant of NR1. J Bacteriol. 1979 Jan;137(1):92–104. doi: 10.1128/jb.137.1.92-104.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa J., Itoh T. Plasmid ColE1 incompatibility determined by interaction of RNA I with primer transcript. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6096–6100. doi: 10.1073/pnas.78.10.6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlin B. E., Molin S., Gustafsson P., Nordström K. Plasmids with temperature-dependent copy number for amplification of cloned genes and their products. Gene. 1979 Jun;6(2):91–106. doi: 10.1016/0378-1119(79)90065-9. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Yoshikawa M. Identification and mapping of the replication genes of an R factor, R100-1, integrated into the chromosome of Escherichia coli K-12. J Bacteriol. 1974 Jun;118(3):1123–1131. doi: 10.1128/jb.118.3.1123-1131.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]