Background: More than 25% of Mtb proteins have unknown functions, limiting our understanding of Mtb physiology and pathogenesis.
Results: The hypothetical Mtb protein Rv2179c is a new DEDD-type exoribonuclease.
Conclusion: Rv2179c is the founding member of a new exoribonuclease family with broad phylogenetic distribution.
Significance: Assigning RNase function to Rv2179c provides annotation for hundreds of bacterial orthologs and provides a new view of RNA processing in Mtb.
Keywords: Crystal Structure, Microbiology, Mycobacterium tuberculosis, Pseudomonas, Ribonuclease, Annotation
Abstract
Ribonucleases (RNases) maintain the cellular RNA pool by RNA processing and degradation. In many bacteria, including the human pathogen Mycobacterium tuberculosis (Mtb), the enzymes mediating several central RNA processing functions are still unknown. Here, we identify the hypothetical Mtb protein Rv2179c as a highly divergent exoribonuclease. Although the primary sequence of Rv2179c has no detectable similarity to any known RNase, the Rv2179c crystal structure reveals an RNase fold. Active site residues are equivalent to those in the DEDD family of RNases, and Rv2179c has close structural homology to Escherichia coli RNase T. Consistent with the DEDD fold, Rv2179c has exoribonuclease activity, cleaving the 3′ single-strand overhangs of duplex RNA. Functional orthologs of Rv2179c are prevalent in actinobacteria and found in bacteria as phylogenetically distant as proteobacteria. Thus, Rv2179c is the founding member of a new, large RNase family with hundreds of members across the bacterial kingdom.
Introduction
Metabolism of cellular RNA requires processing of precursor RNA and degradation of unwanted RNA through RNases. These processes rely on endo- and exoribonucleases that work together to set the cellular RNA balance. Bacteria code for strikingly different and phylogenetically limited sets of RNases. For example, the γ-proteobacterium Escherichia coli produces eight exoribonucleases and eight endoribonucleases, whereas the major human pathogen Mycobacterium tuberculosis (Mtb)2 encodes orthologs of only half of E. coli RNases, suggesting that some RNA processing functions in Mtb are carried out by additional, divergent members that remain to be identified.
Whereas endoribonucleases and their substrates in Mtb and other organisms have been studied extensively, our understanding of exoribonucleases and their functions remains incomplete. One large superfamily of nucleases that comprise several exoribonucleases is the DEDD family, which includes RNase D, oligoribonuclease, and RNase T (1). The DEDD family of RNases is defined by four acidic residues that form a characteristic metal binding catalytic site (1, 2). Of the DEDD family RNases, Mtb only codes for orthologs of RNase D and oligoribonuclease, but not RNase T. Despite its diverse and essential functions in the processing of rRNA and tRNA, RNase T is only found in γ-proteobacteria. RNase T belongs to the DEDDh subgroup of DEDD ribonucleases, characterized by a conserved histidine residue. Other characteristics include homodimerization that is required for activity and DNA exonuclease activity (3). Despite its role in processing several stable RNAs (4–7), RNase T is not essential in E. coli, suggesting overlapping functions of other E. coli RNases.
In a chemical proteomics screen using an ATP-based activity probe, we previously identified nearly 600 ATP-binding proteins in the Mtb proteome, including approximately 120 hypothetical proteins with unknown function (8). One of these hypothetical proteins is Rv2179c, a protein with no detectable sequence homology to any functionally characterized protein. Here, we identify Rv2179c as a novel, highly divergent RNase. The crystal structure of Rv2179c reveals a typical RNase fold, with active site residues forming a magnesium catalytic center. Despite <17% sequence identity (ClustalW2), Rv2179c is a close structural homolog of the E. coli DEDD family RNase T. Consistent with this structural homology, Rv2179c has 3′ exoribonuclease activity, identifying Rv2179c as a functional RNase. Rv2179c has hundreds of orthologs in the sequenced genomes of actinobacteria and phylogenetically distant organisms such as proteobacteria. Thus, Rv2179c establishes a highly divergent, previously unrecognized RNase family.
EXPERIMENTAL PROCEDURES
Cloning, Protein Expression, and Activity-based Protein Profiling
The cloning sequences for Rv2179c and the Pseudomonas putida ortholog were amplified from genomic DNA, cloned into the pET28b expression vector, and transformed into BL21(DE3). Cells were grown to an A600 of 0.8 in terrific broth, induced with 100 μm isopropyl β-d-1-thiogalactopyranoside, and expressed overnight at 18 °C. Recombinant proteins were purified by metal affinity chromatography and size exclusion chromatography. Point mutations were generated by the QuikChange protocol and confirmed by DNA sequencing. For ATP-based activity probe (ATP-ABP) labeling, recombinant Rv2179c was treated with 20 μm ATP-ABP for 1 h at 37 °C, followed by treatment with Cy5.5-azide (36 μm), tris(2-carboxyethyl)phosphine (2.5 mm), Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (100 μm), and CuSO4 (0.5 mm). The samples were vortexed and incubated at room temperature in the dark for 1.5 h. For competition experiments, recombinant protein was incubated with inhibitor for 30 min prior to probe labeling.
Crystallization, Structure Solution, and Analysis
Rv2179c protein was concentrated to 12.8 mg/ml. Crystals were grown by sitting drop vapor diffusion in 10% PEG 8000, 8% ethylene glycol, 100 mm HEPES, pH 7.5, with 2.5 mm AMPPNP and MgCl2 at 290 K. Crystals were cryoprotected in reservoir solution containing 20% ethylene glycol and vitrified by submersion in liquid nitrogen. Data were collected at 100 K on a Rigaku FR-E+ Superbright rotating anode diffraction source and a Rigaku Saturn 944+ CCD detector. To obtain phases, a crystal was soaked in reservoir solution with 20% ethylene glycol and 750 mm NaI. All data sets were reduced with the XDS suite (9). The structure was solved using iodide single anomalous dispersion. Seven anomalous sites were found by PHENIX.HYSS (10). PHASER_EP (11) was used to extend the set of anomalous sites to 11 and to calculate initial phases (figure of merit = 0.34). Phases were improved with PARROT to figure of merit = 0.70 (12). An initial model was built with BUCCANEER (13). The initial model was then refined against the native data using REFMAC5 (14) in iterative cycles with manual model building using COOT (15). For the complex structure, crystals were grown as described above with the following modifications: 25% Jeffamine ED2003, pH 7.0, 100 mm HEPES, pH 7.0, and protein at 25.6 mg/ml. Crystals were soaked in 35% Jeffamine ED2003, 100 mm HEPES, pH 7.0, 2.5 m AMP, and 2.5 mm MgCl2 for 1 h before vitrifying by submersion in liquid nitrogen. All structures were isomorphous. Figures were generated in PyMOL, CCP4mg (surface electrostatics) (16), and ConSurf (17). Structure validation was carried out using Molprobity (18). The coordinates and structure factors for Rv2179c and Rv2179c bound to AMP have been submitted to the Protein Data Bank under accession numbers 4HEC and 4HVJ, respectively.
RNase and DNase Assays
The following self-annealing RNA oligonucleotides were hybridized in 20 mm Tris-HCl, pH 7.5, 500 mm NaCl, and 1 mm EDTA (self-annealing sequences are underlined): 3′ overhang, 5′-GAGUGCGCACUCACUACAUGUACA-3′, and 5′ overhang, 5′-ACAUGUACAUCAGAGUGCGCACUC-3′. 5 μm hybridized overhang RNA was incubated with 5 μm purified Mtb Rv2179c, the P. putida ortholog, or the Rv2179c D145A mutant in a reaction containing 20 mm sodium phosphate, pH 7.4, 20 mm NaCl, 10 mm MgCl2 at 37 °C for the indicated times. DNA substrates were generated as described above for RNA, using the following oligonucleotides: 3′ overhang, 5′- GAGTGCGCACTCGAATGTACATCA-3′; 5′ overhang, 5′-ACATGTACATCAGAGTGCGCACTC-3′; blunt-ended DNA, 5′-GAATGTACATCATGATGTACATTC-3′. The pentaprobes are a set of RNA substrates that cover every combination of 5 bases in tandem on six ∼150-nucleotide-long RNAs (19). The complete sequence was cloned in six overlapping fragments, and the ∼150-base-long pentaprobe inserts were amplified by PCR to allow for in vitro transcription of 12 single-strand RNA segments (six in the forward and six in the reverse complementary direction). RNA was purified by sodium acetate and ethanol precipitation. One μg of each pentaprobe RNA was incubated with 1 μg of purified Rv2179c in 20 mm sodium phosphate, pH 7.4, 20 mm NaCl, 10 mm MgCl2 at 37 °C for the indicated times. RNase reactions were stopped by the addition of 2× formamide stop solution (80% (v/v) formamide, 5 mm EDTA, 0.1% (w/v) bromphenol blue, 0.1% (w/v) xylene cyanol FF) and heat inactivation at 70 °C for 5 min. All reactions were analyzed on 20% urea denaturing 1× TBE polyacrylamide gels. The results were visualized by SYBR Green II RNA staining.
RESULTS
Rv2179c Is an ATP-binding Protein
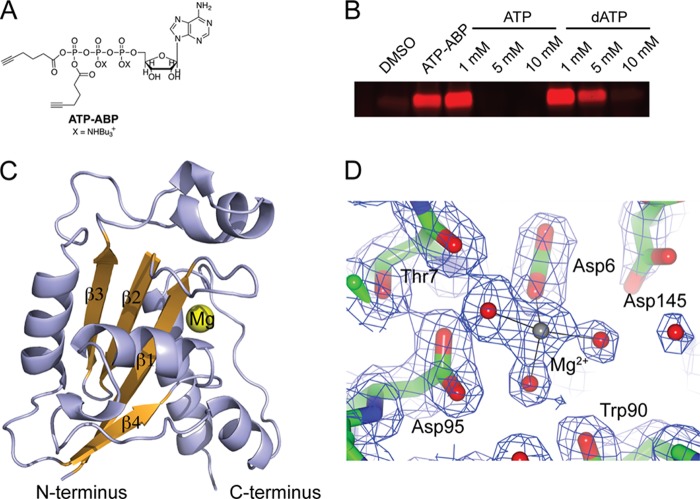
In a recent chemical biology screen using ATP-ABP (Fig. 1A and Ref. 20), we identified several hypothetical Mtb proteins as ATP-binding proteins (8). Using a quantitative mass spectrometry readout after ATP-ABP labeling and enrichment, the hypothetical protein Rv2179c showed 10-fold higher ATP-ABP binding than the control samples. To further probe the function of this ATP binder, we expressed Rv2179c recombinantly for biochemical analysis. To confirm ATP binding, we tested recombinant Rv2179c for binding to the ATP-ABP. Rv2179c readily bound the ATP-ABP, and binding of the ATP-ABP was reduced by ATP in a concentration-dependent manner (Fig. 1B), confirming that Rv2179c is an ATP-binding protein. Most ATP-binding proteins bind ATP but not dATP. However, we noted previously that many DNA- and RNA binding proteins also reacted with the ATP-ABP probe (8). To test whether Rv2179c might bind the probe due to DNA binding activity, we tested the ability of dATP to reduce ATP-ABP binding to Rv2179c. Unexpectedly, Rv2179c also bound to dATP (Fig. 1B). Thus, Rv2179c is an unusual ATP-binding protein that binds ATP and dATP.
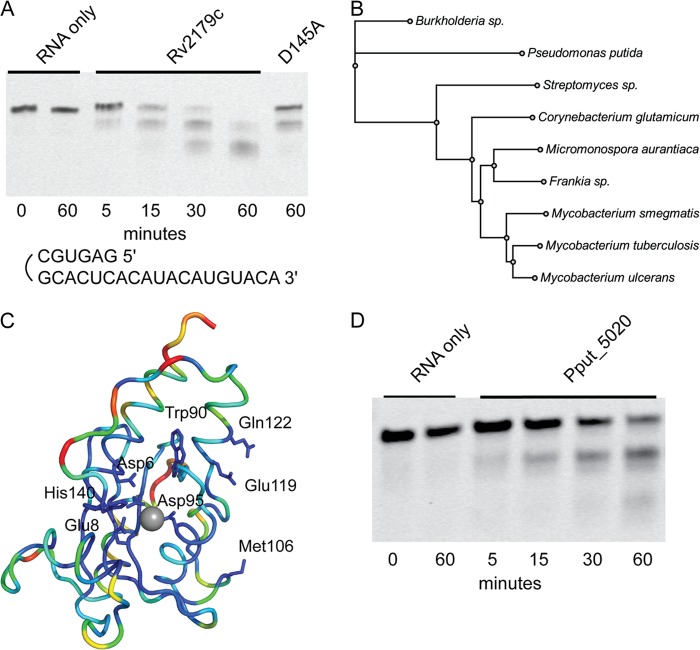
FIGURE 1.
Rv2179c has an RNase fold. A, ATP-ABP activity probe used for the identification of Rv2179c. B, Rv2179c binds ATP-ABP, and binding is inhibited by ATP and dATP. C, overall structure of Rv2179c shows an RNase fold. D, 2Fo − Fc electron density map shows active site residues and the magnesium ion contoured at 1σ. Mg2+ is coordinated by three water molecules and the carboxylate group of Asp-6.
Sequence Similarity Does Not Reveal Function
Rv2179c is annotated as a hypothetical protein. A search of the TB Database, Tuberculist, Pfam, PATRIC, and the NCBI Conserved Domain server did not reveal any similarity to known proteins or conserved domains. Thus, sequence analysis did not provide any functional predictions. In Mtb, Rv2179c is found in an operon with the probable membrane protein Rv2180c, also with unknown function. A BLAST search identified hundreds of sequences homologous to Rv2179c, mostly among the actinobacteria, but also among distant members including proteobacteria. Within the mycobacteria, Rv2179c orthologs are found in all sequenced species, including the nonpathogenic, fast growing Mycobacterium smegmatis. These data indicate that Rv2179c belongs to a large, phylogenetically broadly distributed protein family.
Rv2179c Has a Conserved RNase Fold
To understand the structural basis for the unusual nucleotide binding properties of Rv2179c, to test the idea that Rv2179c is a DNA or RNA binding protein, and to gain further insight into its function, we solved the crystal structure of Rv2179c (Fig. 1C). Full-length Rv2179c crystallized in space group P212121 with two copies of the protomer in the asymmetric unit. Crystals diffracted to 1.8 Å, and the structure was solved by single anomalous dispersion using a crystal soaked in iodide (21). The structure was refined against native data to an Rwork = 18.8% and an Rfree = 22.8% (Table 1). Of the 168 residues of Rv2179c, the crystal structure comprises residues 1–159 of chain A, 1–162 of chain B, 121 water molecules, and one magnesium ion per protomer. Interpretable electron density was missing for a loop in chain A from residue 136 to 141. Rv2179c adopts a compact α/β-fold with a central, four-stranded β-sheet (Fig. 1C). The central sheet consists of three antiparallel strands and one parallel strand. The α-helices arrange around this central sheet. The two protomers in the asymmetric unit are similar to within a Cα root mean square deviation of 0.3 Å and form a dimer through an extensive interface. Both protomers in the asymmetric unit bind a magnesium ion through Asp-6 (Fig. 1D).
TABLE 1.
Crystallographic and refinement statistics for Rv2179c and Rv2179c in complex with AMP
| Data set | Iodide soak | Native | AMP soak |
|---|---|---|---|
| Data collection | |||
| Space group | P212121 | P212121 | P212121 |
| Unit cell paramters (Å) | a = 42.9 Å, b = 76.6, c = 105.4 | a = 42.8, b = 76.7, c = 106.0 | a = 42.5, b = 76.3, c = 105.6 |
| Resolution (Å) | 50–2.40 (2.46–2.40) | 50–1.80 (1.85–1.80) | 50–2.10 (2.15–2.10) |
| Mean I/σ(I) | 19.2 (6.1)a | 27.3 (2.4) | 20.9 (4.7) |
| Rmerge | 0.115 (0.537)a | 0.036 (0.434) | 0.079 (0.398) |
| Completeness | 99.9% (100%)a | 99.1% (96.4%) | 99.3 (94.1) |
| Multiplicity | 15.1 (15.1)a | 4.7 (2.9) | 6.9 (5.4) |
| No. of unique reflections | 26,188 (1923)a | 32.926 (2329) | 20.675 (1427) |
| SigAno | 1.26 (0.84) | NAb | NA |
| Refinement | |||
| Rwork | 0.188 | 0.177 | |
| Rfree | 0.228 | 0.227 | |
| r.m.s.d.c bonds (Å) | 0.014 | 0.011 | |
| r.m.s.d. angles (°) | 1.65 | 1.52 | |
| Molprobity score | 1.02 (100th %) | 1.19 (100th %) | |
| PDB entry | 4HEC | 4HVJ | |
a For the iodide data set, Friedel mates were kept separately. The iodide structure was not refined and deposited.
b NA, not applicable.
c r.m.s.d., root mean square deviation.
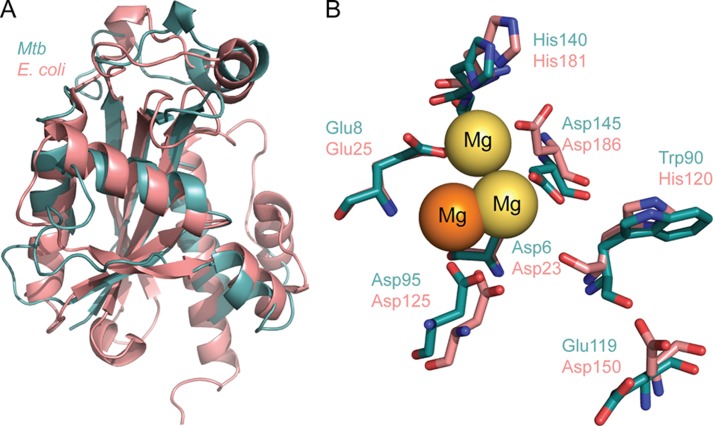
A search for the closest structural homolog using PDBeFold and the DALI server produced the E. coli RNase T (Protein Data Bank IC code 3V9W (22)) as the closest structural match in the Protein Data Bank. The agreement between the Cα carbons of the two structures is 2.2 Å (Fig. 2A). Whereas secondary structure elements share the same overall topology and superpose well, most loop regions differ significantly from E. coli RNase T. The loop that connects strands 2 and 3 in particular contains 9 additional residues compared with the E. coli enzyme, and all other loops have largely different conformations. The overall similarity in topology and fold, however, clearly identifies Rv2179c as a structural RNase T homolog.
FIGURE 2.
Rv2179c is a structural ortholog of E. coli RNase T. A, overlay of Rv2179c and E. coli RNase T shows close similarity of topology and secondary structure elements. B, overlay of the DEDD family active site residues identifies Rv2179c as a DEDDh family member. Orange magnesium ion, Rv2179c. Yellow magnesium ions, RNase T.
Rv2179c Is Similar to DEDD-type RNases
DEDD family nucleases such as RNase T are defined by four acidic catalytic residues within three sequence motifs, ExoI through ExoIII (1). The DEDD residues coordinate a divalent metal ion that forms the catalytic center. The superposition of Rv2179c with E. coli RNase T reveals the presence of all four DEDD residues and a striking agreement of their position between Rv2179c and RNase T. The E. coli DEDD residues Asp-23, Glu-25, Asp-125, and Asp-186 superpose closely with Rv2179c Asp-6, Glu-8, Asp-95, and Asp-145 (Fig. 2B). In addition, other residues that are highly conserved or essential for efficient catalysis in RNase T such as Asp-150 have structural equivalents in Rv2179c (Glu-119). Also similar to RNase T, Rv2179c has a His in the ExoIII motif (His-140), grouping it with the DEDDh subfamily. The position of another His in RNase T with a role in substrate binding, His-120, is taken by Trp-90 in Rv2179c. Interestingly, the His and Trp superpose to align the imidazole and indole nitrogen atoms, suggesting that Trp-90 is a functional His-120 equivalent. The magnesium ions in the catalytic sites are within 4.2 Å of each other in the Rv2179c and E. coli RNase T structures and are coordinated by Asp-6 and Asp-23, respectively (Fig. 2B).
Rv2179c Forms a Dimer
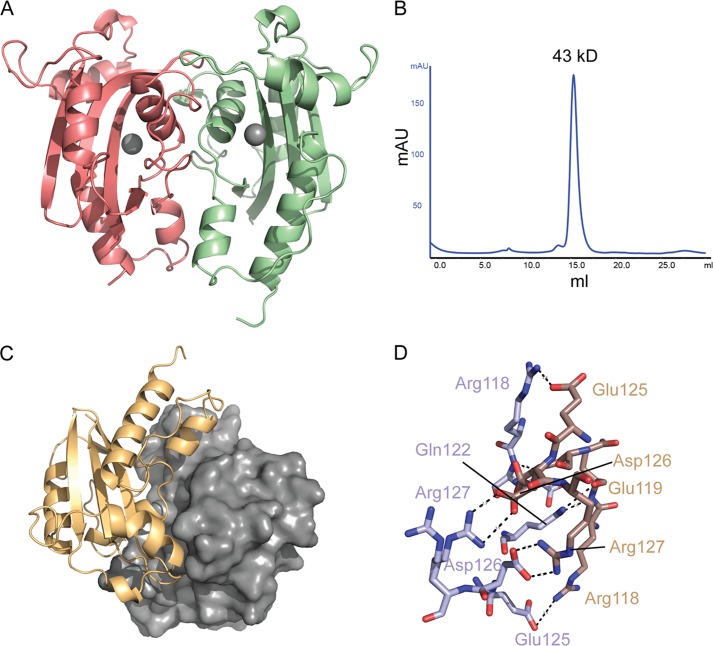
Rv2179c crystallized as a dimer with an extensive dimer interface (Fig. 3A), suggesting that the dimer is functional and also forms in solution. Size exclusion chromatography confirmed dimerization in solution (Fig. 3B). Interestingly, E. coli RNase T also forms a dimer and requires dimerization for substrate recognition (3). Closer inspection of the crystal contacts showed an extensive interface, involving 10 hydrogen bonds and 145 nonbonded contacts covering an area of ∼1200 Å2 (Fig. 3C). As in the RNase T structure (22), the protomers of Rv2179c are approximately related by a 180° rotation. Hydrogen bonds between the protomers are found along the entire interface, with Arg-127, Gln-122, and Glu-119 making two contacts each to Asp-126, Glu-119, and Gln-122, respectively (Fig. 3D). The orientation of the protomers toward each other is slightly different from that found in the E. coli structure (Fig. 4, A and B), suggesting that Rv2179c might recognize other or additional substrates than E. coli RNase T.
FIGURE 3.
Rv2179c forms a dimer in the crystal and in solution. A, overall orientation of the two Rv2179c protomers in the dimer. B, size exclusion chromatography showing elution of recombinant Rv2179c (molecular mass ∼20 kDa) at a molecular mass consistent with a dimer. C, dimer interface showing large complementary surfaces, together creating a 1200-Å interface. D, network of hydrogen bonds stabilizing the dimer interface.
FIGURE 4.
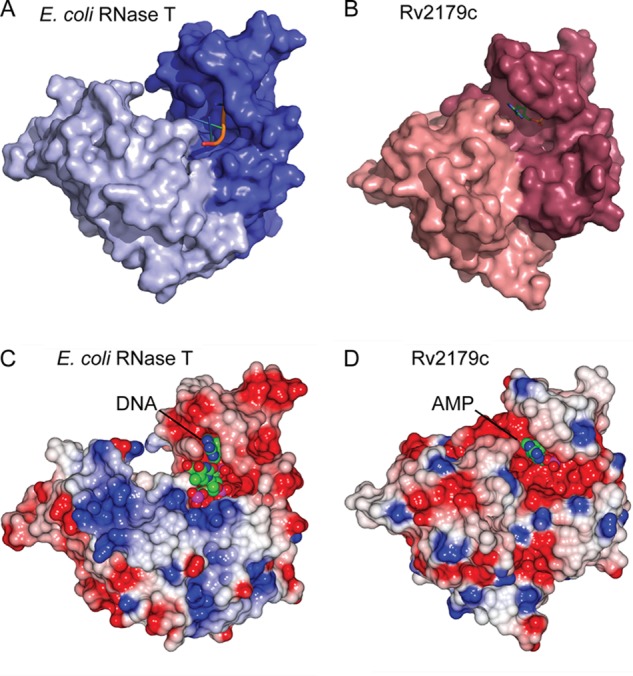
The Rv2179c dimer is oriented similarly to that of RNase T. A, surface representation of the RNase T protomers. B, Rv2179c protomers. C, electrostatic surface potential of RNase T. D, Rv2179c showing a more extensive electropositive binding groove in RNase T.
The Rv2179c Substrate Binding Site Is Distinct
In E. coli RNase T, the substrate binding site from one protomer of the dimer and the catalytic site from the other together form the functional RNase (23). The overall orientation of the protomers is similar in Rv2179c, suggesting a similar substrate binding mode involving both protomers (Fig. 4, A and B). The substrate binding site of RNase T is formed by three distinct regions (1). The resulting surface is electropositive and neutralizes the phosphate backbone charge of substrate RNA (22). In Rv2179c, the general charge composition of the equivalent region is similar. Arg-115, Arg-121, and Arg-118 from the other protomer form an electropositive area leading toward the active site cavity. This electropositive area does not extend as far as it does on RNase T and comprises fewer basic residues than in RNase T, suggesting that Rv2179c might bind shorter substrates (Fig. 4, C and D). The nucleotide-binding residues of RNase T are not conserved in Rv2179c, and nucleotide binding site I is missing.
To explore substrate binding further, we solved the crystal structure of Rv2179c in complex with AMP to a resolution of 2.1 Å. The structure was solved by molecular replacement using the apo structure as search model and refined to an Rwork of 18% and an Rfree of 22.7% (Table 1). The apo and complex structures were nearly identical, with a Cα root mean square deviation of 0.15 Å. Clear electron density was visible for AMP (Fig. 5A). A superposition of Rv2179c-AMP with E. coli RNase T bound to a single-strand DNA (22) showed that AMP bound in a position that is occupied by an adenosine nucleotide at the 3′ end of the bound DNA in the E. coli structure (Fig. 5B). The nucleobase of AMP bound to Rv2179c forms an aromatic stack with Trp-46 whereas in the E. coli RNase T structure, the 3′ adenosine nucleobase forms an aromatic stack with Phe-76. The sugar moieties of the adenosine nucleotides in the E. coli RNase T-DNA and Rv2179c-AMP complexes are positioned similarly, forming a hydrogen bond with a backbone amide and an equivalent interaction with the side chain of a glutamate (Glu-8 in Rv2179c and Glu-25 in E. coli). The phosphate of AMP bound to Rv2179c interacts with a magnesium ion positioned off of Asp-6, which is equivalent to the interaction between the phosphate of the 3′ nucleotide and a magnesium ion positioned off Asp-23 in the E. coli RNase T structure. These data show that Rv2179c has a nucleotide binding site equivalent to RNase T.
FIGURE 5.
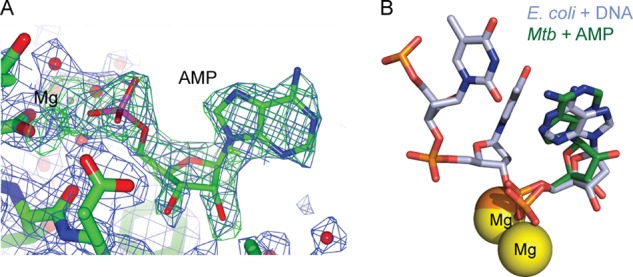
Rv2179c binds AMP in the active site. A, omit density map for AMP in the AMP-bound structure. Both AMP and magnesium were omitted for the calculation of the maps. The σA-weighted 2Fo − Fc map contoured at 1σ is shown in blue, the σA-weighted Fo − Fc map contoured at 3σ is shown in green. B, Rv2179c structure in complex with AMP showing AMP binding consistent with substrate binding in E. coli RNase T.
Rv2179c Has 3′ Exoribonuclease Activity
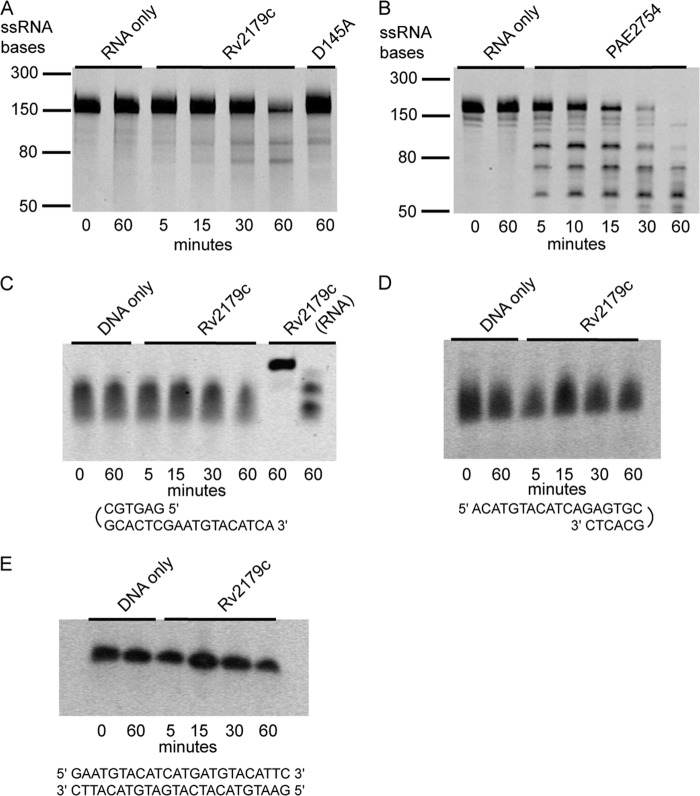
The Rv2179c crystal structure suggested RNase activity. To test for endoribonuclease activity, we used a pentaprobe RNA library that contains all possible combinations of RNA pentanucleotide sequences (23). Rv2179c showed limited cleavage of few sites, but overall activity was negligible compared with that of a known endoribonuclease under the same conditions (Fig. 6, A and B). Because several DEDD-type ribonucleases also have activity on DNA substrates, we next tested the activity of Rv2179c on single-strand DNA with 3′ and 5′ overhangs as well as on blunt-end DNA. Rv2179c did not show activity on any DNA substrate (Fig. 6, C–E).
FIGURE 6.
Rv2179c does not have endonuclease or DNase activity. A and B, endoribonuclease assay using pentaprobes representing all possible 5-nucleotide combinations shows limited activity of Rv2179c compared with the endoribonuclease PAE2754 from Pyrobaculum aerophilum (B, Ref. 24). C, activity of Rv2179c on 3′ DNA overhangs. D and E, activity of Rv2179c on 5′ DNA overhangs (D) and on blunt-end DNA (E).
To test for exoribonuclease activity, we assayed the activity of Rv2179c on defined RNA oligonucleotides that present 3′ or 5′ single-strand overhangs. Rv2179c had no activity on 5′ single-strand overhangs, but readily cleaved 3′ overhangs in a time-dependent manner (Fig. 7A). We next tested whether the Rv2179c active site residues correspond to the canonical DEDD residues. Similar to E. coli RNase T, mutation of a magnesium-coordinating DEDD residue, D145A, or addition of the metal chelator EDTA reduced Rv2179c activity, confirming that the activity resides in the magnesium center (Fig. 7A).
FIGURE 7.
Rv2179c has 3′ exoribonuclease activity and establishes a new RNase family. A, exoribonuclease assay using RNA oligonucleotides with 3′ overhangs shows degradation of single strand 3′ ends. B, unrooted phylogenetic tree of nine sequences showing distribution of Rv2179c orthologs among bacterial species. C, sequence conservation in >400 Rv2179c orthologs mapped on the Rv2179c structure using ConSurf. Blue represents high conservation; red, low conservation. D, 3′ exoribonuclease activity of the distant Rv2179c ortholog Pput_5020 from P. putida with the same substrate shown in A.
Rv2179c Defines a Novel RNase Family
A BLAST search for Rv2179c orthologs identified hundreds of hypothetical proteins with broad phylogenetic distribution. In the genera Gordonia, Nocardia, Rhodococcus, and Corynebacterium, sequence identity to Rv2179c was >60%, and sequence similarity was >70%, suggesting functional similarity. More distant relatives include members in Gram-negative species such as Burkholderia spp. and Pseudomonas spp. (Fig. 7B), although their presence in Gram-negative bacteria is scattered and associated with regions showing evidence of horizontal gene transfer. A multiple sequence alignment showed almost absolute conservation of the ExoI–III motifs and high similarity in other regions in all orthologs. The orthologs in Gram-negative members still showed sequence identity of 28% and similarity of 40% (P. putida). All members of the family had a His residue in the ExoIII motif, grouping them all with the DEDDh family.
To understand sequence conservation in the context of the Rv2179c structure, we mapped sequence conservation of >400 orthologs onto the Rv2179c structure using ConSurf (Fig. 7C). The DEDD residues were absolutely conserved, and most residues of the central β-sheet were highly conserved. Sequence variability mapped primarily to helices 3 and 5 and the loop between helix 6 and 7. The residues providing Rv2179c dimer interactions were also highly conserved, especially residues Met-106 and Glu-119 that both form hydrogen bonds with the other protomer. These data suggest that dimerization is conserved across the family. To test whether phylogenetically distant members of the Rv2179c family are also functional exoribonucleases, we expressed the ortholog from the proteobacterium P. putida. The P. putida ortholog showed activity similar to that of Rv2179c on 3′ ssRNA (Fig. 7D), indicating conservation of function across the entire family, including the most distant members.
DISCUSSION
The functional annotation of proteins remains a major challenge and has not kept up with the rapid increase in genomic information. Whereas sequence-based annotation can provide some prediction of function, the majority of proteins in bacterial genomes are often unknown or hypothetical proteins. Due to the absence of orthologs in better characterized model organisms, species-specific proteins with specialized functions are especially challenging to annotate based on sequence. In Mtb, for example, >25% of proteins are hypothetical with no predicted function. ABPP is a unique approach that allows for global functional annotation when sequence divergence masks function. Using ABPP, we previously annotated nearly 600 Mtb proteins as ATP-binding proteins, providing the first biochemical information for ∼120 hypothetical Mtb proteins. Here, we combine ABPP and structural analysis to define the biochemical function of the hypothetical Mtb protein Rv2179c, leading to the identification of a novel family of RNases with hundreds of members and wide phylogenomic distribution. This study exemplifies how ABPP and structural analysis can probe divergent enzyme space and drive functional annotation independent of sequence.
Rv2179c is a hypothetical protein for which computational methods failed to produce functional predictions. Rv2179c showed unusual nucleotide binding: in an uncharacteristic departure from most ATP-binding proteins, Rv2179c also bound dATP, which is not recognized by canonical ATP cofactor binding sites. These characteristics suggested a function different from common ATP-binding proteins. Unexpectedly, the Rv2179c crystal structure revealed a typical RNase fold. The active site similarity with RNase T further identified Rv2179c as a DEDD-type RNase. The identification of Rv2179c as an RNase explains its nontypical ATP binding properties. DEDD family RNases are related to the proofreading domains of DNA polymerase III and commonly have DNA binding and cleavage activity in addition to RNase activity. Thus, Rv2179c recognizes ATP and dATP by virtue of its RNA and DNA binding properties. These data explain the identification of large numbers of RNA- and DNA-binding proteins in our and other ABPP screens using an acylphosphate ATP-ABP (20, 24). Whereas some of these enzymes may bind ATP as a cofactor outside of the RNA binding site similar to, for example, DNA gyrase, it is likely that most recognize ATP in the RNA binding site. These results identify an unexpected property of the commonly used ATP-ABP and highlight the complexity of binding of even well defined chemical entities in complex proteomes.
The cellular substrates of Rv2179c remain to be defined. Our functional and structural characterization of Rv2179c revealed several similarities to RNase T, but also to oligoribonuclease, a salvage enzyme that degrades short RNAs. However, Mtb codes separately for an oligoribonuclease ortholog, Rv2511, and the E. coli oligoribonuclease structure is less similar to Rv2179c than the RNase T structure. In addition, oligoribonuclease is specific for 2–5-nucleotide-long substrates, unlike the activity on longer substrates seen for Rv2179c. The similarity to RNase T suggests that Rv2179c might also process structural RNAs such as rRNA and tRNA. Not only the overall fold and 3′ exoribonuclease activity are shared between the two enzymes. Both enzymes also dimerize, a critical factor in substrate recognition by RNase T. The RNA binding sites of RNase T and Rv2179c, however, show several differences that suggest different substrates. Thus, Rv2179c may have a cellular function that is unique to Mtb and related bacteria.
This study exemplifies the facility of ABPP for protein functional annotation. ABPP can probe divergent enzymes and provide mechanistic insight into proteins that are intractable by sequence-based approaches. Whereas the conservation of sequence between bacterial RNases is usually high, the Rv2179c sequence is highly divergent and has only <17% identity to its closest functionally characterized ortholog. Yet, the identical topology of Rv2179c and other DEDD family members indicates that Rv2179c diverged from a common ancestor. Many orthologs of Rv2179c are found in actinobacteria to proteobacteria. Thus, Rv2179c is the founding member of a new bacterial RNase family with hundreds of members.
Acknowledgments
We thank the entire Seattle Structural Genomics Center for Infectious Diseases (SSGCID) team and Aaron Wright for ABPP reagents and help.
This work was supported, in whole or in part, by National Institutes of Health Federal Contracts HHSN272200700057C and HHSN272201200025C from the NIAID.
The atomic coordinates and structure factors (codes 4HEC and 4HVJ) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- Mtb
- Mycobacterium tuberculosis
- AMPPNP
- adenosine 5′-(β,γ-imino)triphosphate
- ABPP
- activity-based protein profiling
- ATP-ABP
- ATP-based activity probe.
REFERENCES
- 1. Zuo Y., Deutscher M. P. (2001) Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Res. 29, 1017–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beese L. S., Steitz T. A. (1991) Structural basis for the 3′-5′ exonuclease activity of Escherichia coli DNA polymerase I: a two-metal ion mechanism. EMBO J. 10, 25–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li Z., Zhan L., Deutscher M. P. (1996) Escherichia coli RNase T functions in vivo as a dimer dependent on cysteine 168. J. Biol. Chem. 271, 1133–1137 [DOI] [PubMed] [Google Scholar]
- 4. Li Z., Deutscher M. P. (1995) The tRNA processing enzyme RNase T is essential for maturation of 5S RNA. Proc. Natl. Acad. Sci. U.S.A. 92, 6883–6886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li Z., Deutscher M. P. (1996) Maturation pathways for E. coli tRNA precursors: a random multienzyme process in vivo. Cell 86, 503–512 [DOI] [PubMed] [Google Scholar]
- 6. Li Z., Pandit S., Deutscher M. P. (1998) 3′ exoribonucleolytic trimming is a common feature of the maturation of small, stable RNAs in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 95, 2856–2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li Z., Pandit S., Deutscher M. P. (1999) Maturation of 23S ribosomal RNA requires the exoribonuclease RNase T. RNA 5, 139–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ansong C., Ortega C., Payne S. H., Haft D. H., Chauvignè-Hines L. M., Lewis M. P., Ollodart A. R., Purvine S. O., Shukla A. K., Fortuin S., Smith R. D., Adkins J. N., Grundner C., Wright A. T. (2013) Identification of widespread adenosine nucleotide binding in Mycobacterium tuberculosis. Chem. Biol. 20, 123–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Read R. J., McCoy A. J. (2011) Using SAD data in Phaser. Acta Crystallogr. D Biol. Crystallogr. 67, 338–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cowtan K. (2010) Recent developments in classical density modification. Acta Crystallogr. D Biol. Crystallogr. 66, 470–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cowtan K. (2006) The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 62, 1002–1011 [DOI] [PubMed] [Google Scholar]
- 14. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of COOT. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McNicholas S., Potterton E., Wilson K. S., Noble M. E. (2011) Presenting your structures: the CCP4mg molecular-graphics software. Acta Crystallogr. D Biol. Crystallogr. 67, 386–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Landau M., Mayrose I., Rosenberg Y., Glaser F., Martz E., Pupko T., Ben-Tal N. (2005) ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 33, W299–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kwan A. H., Czolij R., Mackay J. P., Crossley M. (2003) Pentaprobe: a comprehensive sequence for the one-step detection of DNA-binding activities. Nucleic Acids Res. 31, e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Patricelli M. P., Szardenings A. K., Liyanage M., Nomanbhoy T. K., Wu M., Weissig H., Aban A., Chun D., Tanner S., Kozarich J. W. (2007) Functional interrogation of the kinome using nucleotide acyl phosphates. Biochemistry 46, 350–358 [DOI] [PubMed] [Google Scholar]
- 21. Abendroth J., Gardberg A. S., Robinson J. I., Christensen J. S., Staker B. L., Myler P. J., Stewart L. J., Edwards T. E. (2011) SAD phasing using iodide ions in a high-throughput structural genomics environment. J. Struct. Funct. Genomics 12, 83–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zuo Y., Zheng H., Wang Y., Chruszcz M., Cymborowski M., Skarina T., Savchenko A., Malhotra A., Minor W. (2007) Crystal structure of RNase T, an exoribonuclease involved in tRNA maturation and end turnover. Structure 15, 417–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McKenzie J. L., Duyvestyn J. M., Smith T., Bendak K., Mackay J., Cursons R., Cook G. M., Arcus V. L. (2012) Determination of ribonuclease sequence-specificity using Pentaprobes and mass spectrometry. RNA 18, 1267–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Qiu H., Wang Y. (2007) Probing adenosine nucleotide-binding proteins with an affinity-labeled nucleotide probe and mass spectrometry. Anal. Chem. 79, 5547–5556 [DOI] [PMC free article] [PubMed] [Google Scholar]