Abstract
We have shown previously that liquid sample desorption electrospray ionization-mass spectrometry (DESI-MS) is able to measure large proteins and noncovalently-bound protein complexes (to 150 kDa) (Ferguson et al., Anal. Chem. 2011, 83, 6468-6473). In this study, we further investigate the application of liquid sample DESI-MS to probe protein-ligand interactions. Liquid sample DESI allows the direct formation of intact protein-ligand complex ions by spraying ligands toward separate protein sample solutions. This type of “reactive” DESI methodology can provide rapid information on binding stiochiometry, selectivity, and kinetics, as demonstrated by the binding of ribonuclease A (RNaseA, 13.7 kDa) with cytidine nucleotide ligands and the binding of lysozyme (14.3 kDa) with acetyl chitose ligands. A higher throughput method for ligand screening by liquid sample DESI was demonstrated, in which different ligands were sequentially injected as a segmented flow for DESI ionization. Furthermore, supercharging to enhance analyte charge can be integrated with liquid sample DESI-MS without interfering with the formation of protein-ligand complexes.
Keywords: Desorption electrospray ionization, mass spectrometry, noncovalent protein-ligand complexes, ligand screening, supercharging
INTRODUCTION
Protein-protein and protein-ligand interactions provide the basis for the formation of molecular assemblies and machines that perform a vast array of biological functions. Because of the biological significance of protein complexes, the ability to probe and measure protein complexes held together by non-covalent interactions in a sensitive and selective manner would have great utility. To this end, mass spectrometry (MS) coupled with electrospray ionization (ESI)1 has demonstrated its applicability for measuring non-covalent protein complexes. ESI-MS measurements of biomolecular assemblies provide information on binding partners, binding stoichiometries, and solution binding affinities.2-7 The conceptual simplicity of this method, along with its speed, sensitivity, and low sample consumption makes ESI-MS an attractive tool relative to other spectroscopic techniques for monitoring binding.
The recent development of ambient mass spectrometry techniques, such as desorption electrospray ionization (DESI)8 and direct analysis in real time (DART),9 has provided new capabilities and the means to provide direct ionization of analytes with little-to-no sample preparation required. In particular, DESI has been successfully used for the rapid analysis of a variety of different analytes, ranging from pharmaceuticals to tissue samples.8,10-15 For proteins, however, previous investigations of DESI from dried samples on surfaces have been limited to the small proteins (≤ 27 kDa)12 and small protein-substrate complexes (≤ 15 kDa, e.g., lysozyme with its natural substrate hexa-N-acetyl chitohexaose).8 Recently, DESI has been extended to allow for the direct analysis of liquid samples;16-19 ionization of the liquid sample occurs via interactions of the sample with charged microdroplets generated in a pneumatically assisted DESI spray and subsequent desolvation of the resulting secondary microdroplets containing the sample analyte. Several useful features of liquid sample DESI were uncovered in our previous studies. For instance, its increased tolerance to salt allows direct measurement of raw urine samples. Electrolyzed samples from an electrochemical cell can be directly desorbed into the gas phase for MS detection, enabling convenient on-line coupling of MS with electrochemistry.18,20-27 Very small sample volume samples (e.g., nL size),19,28 or at extreme solution pHs,29 as well as the effluent from high flow-rate liquid chromatography columns21,30 can be measured by liquid sample DESI. It also allows for the development of submillisecond time-resolved mass spectrometry for the study of fast reaction kinetics.31 Results from protein analysis by Julian’s group32 and our lab33 suggests that protein measurements by DESI can be enhanced by the liquid sample DESI option, especially for the analysis of native proteins because an acidic spray solvent can be used without denaturation of the protein structure.
Integration of a chemical reaction with DESI ionization, known as reactive DESI,10,18,19,34-39 is a further development that exploits the potential for coupling specific ion/molecule reactions40,41 with the ionization event and so greatly improves the selectivity and efficiency with which compounds with specific functionalities can be detected. It involves the use of a spray solution containing specific reagents intended to allow specific ionic and molecular reactions to proceed during the sampling process. An analytical advantage of reactive DESI, in contrast to traditional protocols10 employing solution phase derivatization followed by ESI, is that on-line derivation during reactive DESI is much faster, thereby speeding up the analytical process. In addition, by taking advantage of the short time scale during DESI ionization (<2 ms),42 transient reaction intermediates can be intercepted and detected in reactive DESI experiments.39,43
The focus of the present study is to exploit the special advantages of liquid sample DESI and reactive DESI for the mass spectrometric characterization of protein-ligand complexes. Monitoring protein-ligand binding reactions by reactive DESI could be of value as a high throughput method for screening of potent protein-binding ligands, a process that is important for drug discovery research. Previously, we demonstrated that liquid sample DESI’s capabilities to directly desorb/ionize non-covalent complexes such as the protein dimers of manganese superoxide dismutase and 93 kDa enolase, the 65 kDa hemoglobin (Hb) tetramer, and large proteins such as 150 kDa monoclonal antibodies.33 The results are consistent with the soft ionization nature of DESI and suggests the possibility to examine protein-ligand interaction events by DESI.
Reactive liquid sample DESI was used to measure the non-covalent complexes formed between ribonuclease A (RNaseA) and cytidine nucleotide ligands and between lysozyme andN-acetylglucosamine ligands. The protein target and the ligands were combined by either having the protein or the ligand in the DESI spray (and the other component resided in the “liquid sample” droplet), and the non-covalent complexes were formed during the brief time the components of the DESI spray mixed with the analyte droplet prior to DESI desorption/ionization. Liquid sample DESI-MS measurements of solution phase binding constants were compared to previously reported values using other biophysical methods. The effects of increasing analyte charge by ESI “supercharging” on measuring protein-ligand complexes were investigated as well.
EXPERIMENTAL SECTION
Materials
Ribonuclease A (bovine pancreas), lysozyme (chicken egg white), cytidine 2′-monophosphate (2′-CMP), cytidine 5′-diphosphate disodium salt (CDP), cytidine 5′-triphosphate disodium salt (CTP), sulfolane, formic acid (FA), ammonium acetate (NH4OAc), and acetonitrile (ACN) were all purchased from Sigma-Aldrich (St. Louis, MO). N,N’,N’’-triacetylchitotriose (NAG3) and N,N’,N’’,N’’’,N’’’’,N’’’’’-hexaacetylchitohexaose (NAG6) were purchased from Santa Cruz Biotechnology (Dallas, TX).
Apparatus
The liquid sample DESI source, in which a beam of charged microdroplets from the DESI spray probe is directed at the outlet of a sample introduction capillary for desorption and ionization, has been described previously.18,33 The sample introduction capillary outlet was positioned between the DESI spray probe and the mass spectrometer interface. A quadrupole-ion mobility-time-of-flight (TOF) instrument (Synapt HDMS G1, Waters Corporation, Milford, MA) was used for the MS measurements. The standard nanoelectrospray source was removed to accommodate the liquid sample DESI source. The DESI spray probe was supplied with a voltage of +5 kV and high-pressure nebulizing nitrogen gas (120 psi) to generate the charged microdroplet beam. The Synapt HDMS parameters were: sampling cone voltage 80 V, extraction cone voltage 5 V, source temperature 80°C, trap collisional energy 6, and transfer collisional energy 4. Data were analyzed using MassLynx 4.1 and Driftscope v2.0 (Waters Corporation).
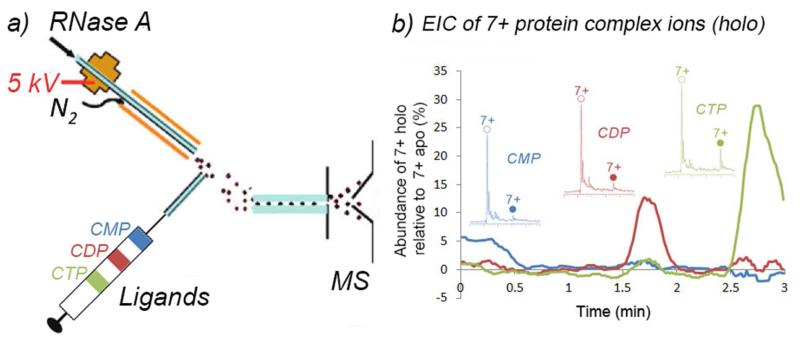
Mixing of the proteins and the ligands to form the protein-ligand complexes were accomplished in either of two modes of the liquid sample DESI source. One mode was to introduce the ligand solution (in water with 20 mM NH4OAc, pH 6.5) into the DESI spray probe at a flow rate of 5 μL min-1. The protein solutions were introduced through the sample introduction capillary at a flow rate of 5 μL min-1, as shown in Figure 1. The other mode was to introduce the protein solution into the DESI spray probe, and the ligand solution was pumped through the sample introduction capillary.
Figure 1.
Schematic showing the reactive liquid sample DESI source for MS of protein-ligand complexes. As depicted, a protein solution is infused through a sample transfer capillary, and the emerging droplet containing the protein is exposed to the charged droplets containing ligand molecules from the DESI probe. The sampling can be reversed, with the ligand solution introduced via the sample transfer capillary and the protein sprayed through the DESI probe.
RESULTS AND DISCUSSION
Probing Protein-Ligand Interactions Using Reactive DESI-MS
Figure 1 shows the scheme for the reactive liquid sample DESI-MS platform. The solutions containing the protein and the ligand(s) are infused from the sample introduction capillary and DESI probe, respectively (or vice versa). Desorption/ionization occurs immediately after the solution mixing and complex formation may occur during the initial mixing and/or after the microdroplets leave the surface.
To test the performance of reactive liquid sample DESI for measuring protein-ligand complexes formed in this manner, the binding of RNaseA with cytidine nucleotide ligands was investigated. RNaseA is an enzyme that catalyzes the cleavage of the P-O 5′-bond of single-stranded ribonucleic acids on the 3′-side of cytidine and uridine residues. The complexation of mono- and oligonucleotide ligands with ribonuclease A has been measured using ESI-MS by many laboratories.44-48
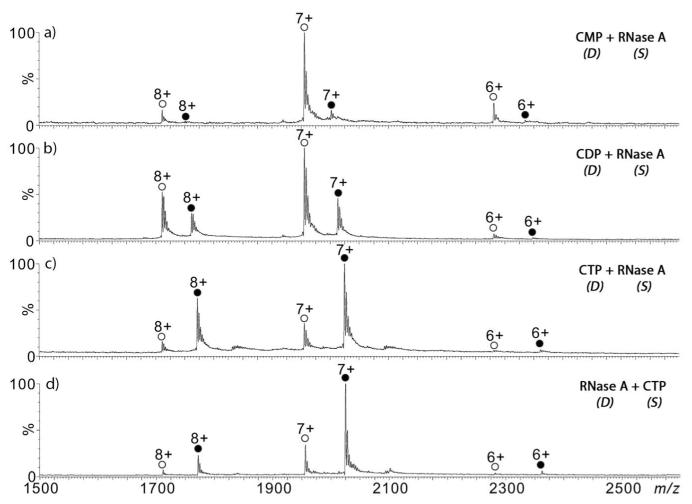
RNaseA (13,682 Da, 5 μM) was prepared in 20 mM NH4OAc aqueous solution and the cytidine nucleotide ligands (2′-CMP, CDP, and CTP) were dissolved in 20 mM NH4OAc (aq) and infused into the DESI spray probe. Multiply charged ions for both the free protein (apo-form) and the ligand-bound protein (holo-form) were detected (Figure 2a-c), but with [holo]/[apo] ratios that were dependent on the nature of the nucleotide ligand. As expected from previous studies, a protein-ligand stoichiometry of 1:1 was the dominant form of the complex measured.44 The total ion chromatograms (TICs) and extracted ion chromatograms (EICs) for the 7+ charge apo- and holo-ions (Figures S6-8) showed very good stability and reflecting the reproducibility of this method. Moreover, the relative abundance of the holo-complex was higher with increasing number of phosphate groups for the same concentration of ligand; with 5 μM protein and 50 μM ligand, the measured [holo]/[apo] ratio was 0.13±0.017, 0.37±0.04, and 3.21±0.22 (based on 5 replicate measurements) for 2′-CMP, CDP, and CTP, respectively, which agrees well with their solution phase binding trends.44
Figure 2.
Reactive liquid sample DESI-MS spectra of RNaseA (5 μM in 20 mM NH4OAc aqueous solution, pH 6.5) in the sample transfer capillary (designated as “S” in the figure) with the DESI probe (designated as “D” in the figure) spraying reagents containing (a) 50 μM 2′-CMP; (b) 50 μM CDP; (c) 50 μM CTP in 20 mM NH4OAc. (d) Reactive DESI-MS spectrum of CTP (50 μM in 20 mM NH4OAc aqueous solution) in the sample transfer capillary (“S“) with the DESI probe (“D”) spraying 5 μM RNaseA (20 mM NH4OAc). Unfilled circles represent free protein ions (apo) and solid circles represent protein-ligand complex ions (holo).
Similar results were obtained when the RNaseA solution was infused from the DESI probe and the ligand solution was injected through the sample introduction capillary. For the CTP ligand, the measured [holo] [apo] ratio was 3.12 (Figure 2d), which is similar to the value measured by introducing the ligand solution through the DESI spray probe (Figure 2c).
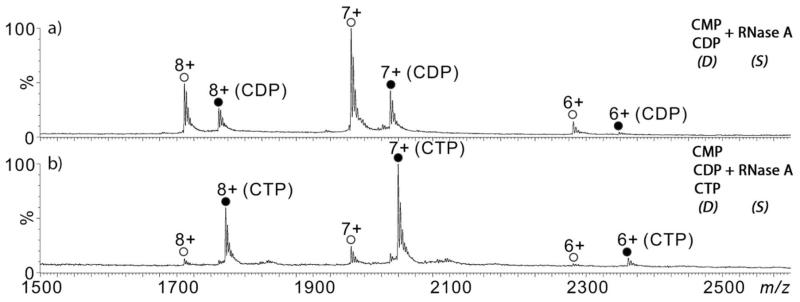
Their relative solution binding affinities can be tested directly also using liquid sample DESI by performing a competitive binding experiment49 in which equimolar mixtures of ligands are presented to the protein target. The rank order of ligand binding can be determined if the total ligand concentration is higher than the total protein concentration. A non-competitive binding experiment in which the protein concentration is greater than the total ligand concentration showed ions for the protein-ligand complexes for each of the ligands (data not shown). Figure 3a shows the mass spectrum resulting from a mixture of 50 μM CDP and 50 μM 2′-CMP sprayed towards a RNaseA (5 μM) sample solution. Only ions for the RNaseA-CDP complex (and the apo-protein) could be measured, which is consistent with the data collected from the individual ligands (Figure 2) that suggests that CDP has a higher affinity to RNase-A than does 2′-CMP. Similarly, only RNaseA-CTP complex ions were observed when a mixture of 50 μM each of CMP, CDP, and CTP mixture was sprayed at the protein (Figure 3b). Thus, the rank order of binding affinities to RNaseA is CTP > CDP > 2′-CMP.
Figure 3.
Reactive DESI-MS spectra of RNaseA (5 μM in 20 mM NH4OAc aqueous solution) in the sample transfer capillary (“S” ) and the DESI probe (“D”) spraying a mixture of (a) 50 μM CDP and 50 μM 2′-CMP (20 mM NH4OAc), and (b) 50 μM CTP, 50 μM CDP, and 50 μM 2′-CMP (20 mM NH4OAc). (apo – open circles; holo – filled circles)
The solution phase dissociation constant (Kd) of the protein-ligand binding reaction can be measured using the liquid sample DESI apparatus by monitoring the formation of the complex as a function of ligand concentration. A solution of 5 μM RNaseA (20 mM NH4OAc) was infused through the sample transfer capillary at a flow rate of 5 μL min-1. Solutions of CTP ligand with different concentrations (from 1-15 μM in 20 mM NH4OAc) were sprayed at a flow rate of 5 μL min-1 by the DESI probe. The liquid sample DESI-based Kd was determined based on the relative abundance of the holo- and apo-forms of the protein according to the method previously described by Daniel et al.50 The measured Kd was 1.91 μM (see Supporting Information), which is slightly higher than a value of 0.72 μM reported in an earlier ESI-MS study48 and 1.6 μM from a more recent study,51 but it is also consistent with other values using other biophysical methods.52,53
Liquid Sample DESI-MS of Lysozyme-Acetyl Chitose Complexes
Our reactive liquid sample DESI-MS platform was used to measure another protein-ligand system previously studied by ESI-MS, lysozyme binding with oligoacetyl chitose ligands.54-56 An aqueous solution of lysozyme (5 μM in 20 mM NH4OAc) was infused through the sample transfer capillary at a flow rate of 5 μL min-1. The ligands, N,N’,N’’,N’’’,N’’’’,N’’’’’-hexaacetylchitohexaose (NAG6) and N,N’,N’’-triacetylchitotriose (NAG3), were dissolved in 20 mM NH4OAc (aq) at a concentration of 200 μM and were infused through the DESI spray capillary at 5 μL min-1. A protein-ligand stoichiometry of 1:1 was measured by reactive liquid sample DESI-MS (Figure 4). The [holo]/[apo] ratios for NAG3 and NAG6 were 0.28 and 0.31, suggesting that their binding affinities are similar and consistent with earlier work.57 The calculated Kd of the lysozyme-NAG3 system is 169.1 μM, which is also larger than the ESI-MS measured value (59.8 μM) reported before.56 This may be due to relatively poor mixing efficiency between the protein solution and ligand-containing droplets and/or weaker gas phase avidity compared to the RNaseA-nucleotide complexes (see Supporting Information).
Figure 4.
Reactive DESI-MS spectra of lysozyme in the sample transfer capillary (“S”) and the DESI probe (“D”) spraying (a) 200 μM NAG3, and (b) 200 μM NAG6 in 20 mM NH4OAc aqueous solution. (apo – open circles; holo – filled circles)
High Throughput Ligand Screening Using Segment Flow Infusion
Potentially, liquid sample DESI could be an effective method for high throughput monitoring of protein-ligand binding by rapidly switching the ligand composition. To test this, we employed a segmented flow technique58-60 coupled to liquid sample DESI. RNaseA (5 μM) in 20 mM NH4OAc aqueous solution was sprayed through the DESI probe (5 μL min-1). The 3 different nucleotide ligands (20 μM in 20 mM NH4OAc aqueous solution) were packed in a syringe as segmented plugs separated by air (Figure 5a). The packed segmented ligands were infused using the syringe at a flow rate of 10 μL min-1 and the resulting MS spectra were recorded continuously. The extracted ion chromatograms (EIC) of the 7+-charged protein-ligand complex ions are shown in Figure 5b, with the y-axis representing the relative abundance of the holo-complex compared to that of the apo-protein. In this example, a sample plug of 2′-CMP was eluted first, followed by CDP and CTP (Figure 5a). From the EIC, the different complexes with different ligands were observed in the order of their packing in the syringe. Consistent with the previously discussed measurements, CTP has the greatest affinity towards RNaseA of the 3 ligands tested. In this example, the total screening time for the three ligands was only 3 min, but one could easily envision segmenting many more ligands over a similar time period.
Figure 5.
(a) Scheme showing the segmented flow experiment apparatus and (b) EICs of the 7+ complex ions. The three different traces represent the three different complexes (blue – 2′- CMP; red – CDP; green – CTP). The recorded mass spectra of the different complexes are shown in the inset. (apo – open circles; holo – filled circles)
Supercharging and Reactive Liquid Sample DESI
For protein and peptide analysis by MS, there is value to increase analyte multiple charging:61 (a) structural analysis via top-down MS approaches using electron-based ion activation/dissociation techniques, such as electron capture dissociation (ECD)62 and electron transfer dissociation (ETD)63 is more effective with higher charging; (b) signal-to-noise (S/N) ratio, mass resolution, and mass accuracy are enhanced at lower mass-to-charge (m/z) ratio and higher z for most mass analyzers, including orbitrap64 and ion cyclotron resonance65,66 systems.
Variables such as solution pH and solvent composition affect ESI-based multiple charging of peptides and proteins. Another approach to enhance multiple charging is by addition of so-called “supercharging” reagents, such as m-nitrobenzyl alcohol (m-NBA), which has been systematically studied by Williams’ group.67-70 Our lab has introduced new supercharging reagents, including sulfolane, and we have studied the supercharging effects on proteins (and protein complexes) under native and denaturing solution environments.33,71,72
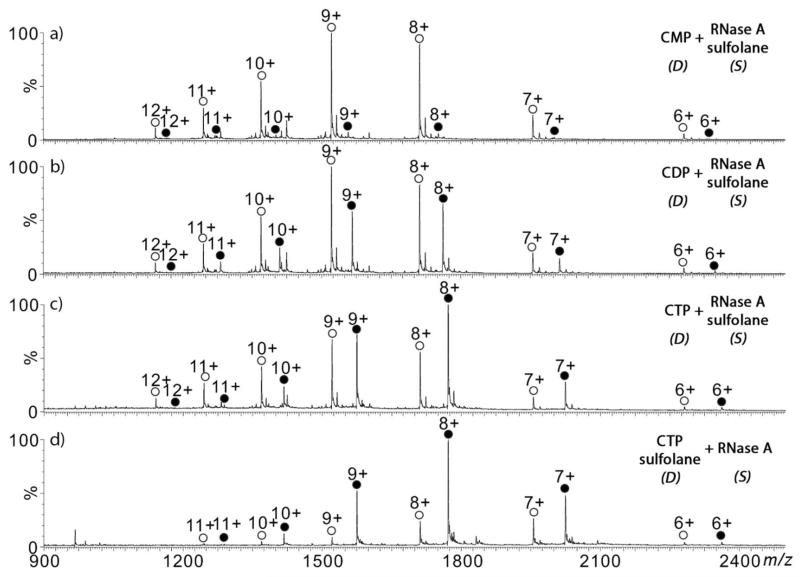
Previously, we demonstrated the ability to incorporate supercharging with electrospray laser desorption ionization (ELDI)73 and liquid sample DESI;33 for DESI, increased multiple charging was observed for proteins and protein complexes ions when the supercharging reagent was sprayed from the DESI probe. In the present study, RNaseA was mixed with 200 mM sulfolane and infused into the sample introduction capillary. The DESI spray solution was composed of the nucleotide ligand in NH4OAc aqueous solution (Figure 6a-c). With the supercharging reagent doped into the protein solution, multiple charging increased from 8+ to 12+ for both the apo- and holo-protein states. The measured [holo]/[apo] ratios of the complex ions for 2′-CMP and CDP were 0.06 and 0.58, respectively, and similar to the data generated without the presence of supercharging reagents (Figure 2).
Figure 6.
Reactive DESI-MS spectra of mixture composed of 5 μM RNase A and 200 mM sulfolane (in 20 mM NH4OAc aqueous solution; 5 μL min-1) in the sample transfer capillary (“S”) and the DESI probe (“D”) spraying (5 μL min-1) 50 μM of (a) CMP, (b) CDP, and (c) CTP in 20 mM NH4OAc aqueous solution. (d) Sulfolane (200 mM) was mixed with CTP and sprayed through the DESI probe. (apo – open circles; holo – filled circles)
The supercharging agent can be introduced by pre-mixing with the small molecule ligand instead of with the protein. Figure 6d also shows the result of this method for RNaseA binding to CTP. Sulfolane and CTP were mixed together and sprayed through the DESI spray probe towards the RNaseA solution, which was infused from sample introduction capillary. The measured [holo]/[apo] ratio of 3.37 is similar to the 3.21 value measured without supercharging (Figure 2). However we note that the [holo]/[apo] ratio in Figure 6c is smaller. We suspect that some change in the DESI probe position or sample capillary position occurred, and this has some effect on the [holo]/[apo] ratio. Ideally, both DESI probe and sample capillary positions should be well controlled for each experiment.
Similar results with the addition of sulfolane supercharging agent were observed for the lysozyme-NAG complex system. Ions for the complex were shifted to higher charge state, and the measured [holo]/[apo] ratio was similar to results without the addition of supercharging agent (see Supporting Information).
The results are consistent with our previous data that showed that the addition of supercharging agents at the concentrations used allowed the protein to retain a structure that is sufficient for recognizing and binding to small molecule ligands,71,72,74 and the measured affinities are similar to previous measurement without the addition of supercharging agents. Even for the example in which sulfolane is added to the protein solution and the supercharged protein reacts with the ligand (Figure 6a-c), no alteration of binding affinities were noted.
Mechanism of Liquid Sample DESI as it Relates to Protein-Ligand Complex Measurements
The results described in this study are consistent with the previously proposed droplet pick-up mechanism for DESI.75 As the protein (or ligand) is sprayed out of the DESI probe, the charged microdroplet carrying the protein (or ligand) will pick up the sample containing its binding partner.
In principle, there are several factors that could preclude the utility of this method for measuring solution phase binding constants. We assumed that the efficiency for mixing of the DESI probe solution and sample introduction capillary solution is high under the experimental conditions explored for this study. However, the effect of gas and solution flow, temperature, spray voltage, and geometry may play a role in the mixing efficiency (and will be the subject of a future study).
The kinetics of protein-ligand association and dissociation may govern the type of complexes that can be probed by our method. As the DESI ionization time scale is around 1-2 ms, the on-rate for formation of the RNaseA and lysozyme complexes should fall in this time regime. However, the reaction rate for complex formation may be driven further from a concentration effect that might occur as the final droplet containing the complex is evaporating, potentially raising the concentration of the reactants prior to desorption/ionization into the gas phase complex. Previous work with ELDI-MS showed that reduction of disulfide bonds can occur in the millisecond timescale for reactions that take many minutes in solution.73
CONCLUSIONS
Liquid sample DESI was used as a method to separately introduce reactants and mix to form noncovalent protein-ligand complexes to be measured by mass spectrometry. For RNaseA binding to cytidine nucleotides and lysozyme binding to saccharides, the measured binding affinities using this ambient ionization method with MS were consistent with other measurements using conventional MS and biophysical tools.
Liquid sample DESI-MS offers some sample handling convenience, as the protein does not need to be mixed with the ligand (or ligand mixture) prior to the measurement. Further, a potentially higher throughput method of sample introduction was tested by using a segmented flow arrangement. A library of ligand compounds can be introduced in a single flow and each compound can be tested for binding and measured sequentially using MS detection. The results described in this study agree well with the previously proposed droplet pick-up mechanism for DESI.75 The fast mixing of two partners of a protein/ligand pair in liquid sample DESI may allow for the monitoring of the early stages of a binding event in solution.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Science Foundation (Grant CHE-0911160), NSF Career (CHE-1149367), the ASMS Research Award (Thermo Scientific), and the National Natural Science Foundation of China (NNSFC No. 21328502) to H.C., the Ruth L. Kirschstein National Research Service Award (Grant GM007185, UCLA Cellular and Molecular Biology Training Grant, for C.N.F.), and the National Institutes of Health (Grant R01 GM103479 to J.A.L.).
Footnotes
ASSOCIATED CONTENT
Supporting Information
Supercharging spectra of lysozyme-NAGn complex data and the Kd calculations are provided in supporting information. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- (1).Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Science. 1989;246:64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- (2).Loo JA. Mass Spectrom. Rev. 1997;16:1–23. doi: 10.1002/(SICI)1098-2787(1997)16:1<1::AID-MAS1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- (3).Loo JA. Int. J. Mass Spectrom. 2000;200:175–186. [Google Scholar]
- (4).Loo JA. Int. J. Mass Spectrom. 2001;204:113–123. [Google Scholar]
- (5).van den Heuvel RH, Heck AJR. Curr. Opin. Chem. Biol. 2004;8:519–526. doi: 10.1016/j.cbpa.2004.08.006. [DOI] [PubMed] [Google Scholar]
- (6).Benesch JLP, Ruotolo BT, Simmons DA, Robinson CV. Chem. Rev. 2007;107:3544–3567. doi: 10.1021/cr068289b. [DOI] [PubMed] [Google Scholar]
- (7).Li JX, Shefcheck K, Callahan J, Fenselau C. Int. J. Mass Spectrom. 2008;278:109–113. doi: 10.1016/j.ijms.2008.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Takats Z, Wiseman JM, Gologan B, Cooks RG. Science. 2004;306:471–473. doi: 10.1126/science.1104404. [DOI] [PubMed] [Google Scholar]
- (9).Cody RB, Laramee JA, Durst HD. Anal. Chem. 2005;77:2297–2302. doi: 10.1021/ac050162j. [DOI] [PubMed] [Google Scholar]
- (10).Cotte-Rodriguez I, Chen H, Cooks RG. Chem. Commun. 2006:953–955. doi: 10.1039/b515122h. [DOI] [PubMed] [Google Scholar]
- (11).Kauppila TJ, Wiseman JM, Ketola RA, Kotiaho T, Cooks RG, Kostiainen R. Rapid Commun. Mass Spectrom. 2006;20:387–392. doi: 10.1002/rcm.2304. [DOI] [PubMed] [Google Scholar]
- (12).Shin YS, Drolet B, Mayer R, Dolence K, Basile F. Anal. Chem. 2007;79:3514–3518. doi: 10.1021/ac062451t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Denes J, Katona M, Hosszu A, Czuczy N, Takats Z. Anal. Chem. 2009;81:1669–1675. doi: 10.1021/ac8024812. [DOI] [PubMed] [Google Scholar]
- (14).Douglass KA, Venter AR. J. Mass Spectrom. 2013;48:553–560. doi: 10.1002/jms.3206. [DOI] [PubMed] [Google Scholar]
- (15).Harris GA, Galhena AS, Fernandez FM. Anal. Chem. 2011;83:4508–4538. doi: 10.1021/ac200918u. [DOI] [PubMed] [Google Scholar]
- (16).Chipuk JE, Brodbelt JS. J. Am. Soc. Mass Spectrom. 2008;19:1612–1620. doi: 10.1016/j.jasms.2008.07.002. [DOI] [PubMed] [Google Scholar]
- (17).Ma X, Zhao M, Lin Z, Zhang S, Yang C, Zhang X. Anal. Chem. 2008;80:6131–6136. doi: 10.1021/ac800803x. [DOI] [PubMed] [Google Scholar]
- (18).Miao ZX, Chen H. J. Am. Soc. Mass Spectrom. 2009;20:10–19. doi: 10.1016/j.jasms.2008.09.023. [DOI] [PubMed] [Google Scholar]
- (19).Zhang Y, Chen H. Int. J. Mass Spectrom. 2010;289:98–107. [Google Scholar]
- (20).Li JW, Dewald HD, Chen H. Anal. Chem. 2009;81:9716–9722. doi: 10.1021/ac901975j. [DOI] [PubMed] [Google Scholar]
- (21).Zhang Y, Yuan Z, Dewald HD, Chen H. Chem. Commun. 2011;47:4171–4173. doi: 10.1039/c0cc05736c. [DOI] [PubMed] [Google Scholar]
- (22).Zhang Y, Dewald HD, Chen H. J. Proteome Res. 2011;10:1293–1304. doi: 10.1021/pr101053q. [DOI] [PubMed] [Google Scholar]
- (23).Lu M, Wolff C, Cui W, Chen H. Anal. Bioanal. Chem. 2012;403:355–365. doi: 10.1007/s00216-011-5679-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Zhang Y, Cui W, Zhang H, Dewald HD, Chen H. Anal. Chem. 2012 doi: 10.1021/ac300106y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Liu P, Lanekoff IT, Laskin J, Dewald HD, Chen H. Anal. Chem. 2012;84:5737–5743. doi: 10.1021/ac300916k. [DOI] [PubMed] [Google Scholar]
- (26).Liu P, Lu M, Zheng Q, Dewald HD, Zhang Y, Chen H. Analyst. 2013;138:5519–5539. doi: 10.1039/c3an00709j. [DOI] [PubMed] [Google Scholar]
- (27).Zheng Q, Zhang H, Chen H. Int. J. Mass Spectrom. 2013 doi: 10.1016/j.ijms.2013.04.009. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Sun X, Miao Z, Yuan Z, Harrington P. d. B., Colla J, Chen H. Int. J. Mass Spectrom. 2011;301:102–108. [Google Scholar]
- (29).Pan N, Liu P, Chen H, Cui W, Tang B, Shi J. Analyst. 2013;138:1321–1324. doi: 10.1039/c3an36737a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Liu Y, Miao ZX, Lakshmanan R, Ogorzalek Loo RR, Loo JA, Chen H. Int. J. Mass Spectrom. 2012;325:161–166. doi: 10.1016/j.ijms.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Miao Z, Chen H, Liu P, Liu Y. Anal. Chem. 2011;83:3994–3997. doi: 10.1021/ac200842e. [DOI] [PubMed] [Google Scholar]
- (32).Moore BN, Hamdy O, Julian RR. Int. J. Mass Spectrom. 2012:330–332. 220–225. doi: 10.1016/j.ijms.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Ferguson CN, Benchaar SA, Miao ZX, Loo JA, Chen H. Anal. Chem. 2011;83:6468–6473. doi: 10.1021/ac201390w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Chen HW, Talaty NN, Takats Z, Cooks RG. Anal. Chem. 2005;77:6915–6927. doi: 10.1021/ac050989d. [DOI] [PubMed] [Google Scholar]
- (35).Chen H, Cotte-Rodriguez I, Cooks RG. Chem. Commun. 2006:597–599. doi: 10.1039/b516448f. [DOI] [PubMed] [Google Scholar]
- (36).Huang G, Chen H, Zhang X, Cooks RG, Ouyang Z. Anal. Chem. 2007;79:8327–8332. doi: 10.1021/ac0711079. [DOI] [PubMed] [Google Scholar]
- (37).Nyadong L, Green MD, De Jesus VR, Newton PN, Fernandez FM. Anal. Chem. 2007;79:2150–2157. doi: 10.1021/ac062205h. [DOI] [PubMed] [Google Scholar]
- (38).Cotte-Rodriguez I, Hernandez-Soto H, Chen H, Cooks RG. Anal. Chem. 2008;80:1512–1519. doi: 10.1021/ac7020085. [DOI] [PubMed] [Google Scholar]
- (39).Perry RH, Splendore M, Chien A, Davis NK, Zare RN. Angew. Chem. Int. Ed. 2011;50:250–254. doi: 10.1002/anie.201004861. [DOI] [PubMed] [Google Scholar]
- (40).O’Hair RA. J. Chem. Commun. 2006:1469–1481. doi: 10.1039/b516348j. [DOI] [PubMed] [Google Scholar]
- (41).Schröder D, Schwarz H. Proc. Natl. Acad. Sci. USA. 2008;105:18114–18119. doi: 10.1073/pnas.0801849105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Li YL, Su X, Stahl PD, Gross ML. Anal. Chem. 2007;79:1569–1574. doi: 10.1021/ac0615910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Miao ZX, Wu SY, Chen H. J. Am. Soc. Mass Spectrom. 2010;21:1730–1736. doi: 10.1016/j.jasms.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Yin S, Xie YM, Loo JA. J. Am. Soc. Mass Spectrom. 2008;19:1199–1208. doi: 10.1016/j.jasms.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Benkestock K, Sundqvist G, Edlund P-O, Roeraade J. J. Mass Spectrom. 2004;39:1059–1067. doi: 10.1002/jms.685. [DOI] [PubMed] [Google Scholar]
- (46).Camilleri P, Haskins N. J. Rapid Commun. Mass Spectrom. 1993;7:603–604. [Google Scholar]
- (47).Sundqvist G, Benkestock K, Roeraade J. Rapid Commun. Mass Spectrom. 2005;19:1011–1016. doi: 10.1002/rcm.1880. [DOI] [PubMed] [Google Scholar]
- (48).Zhang S, Van Pelt CK, Wilson DB. Anal. Chem. 2003;75:3010–3018. doi: 10.1021/ac034089d. [DOI] [PubMed] [Google Scholar]
- (49).Loo JA, Hu P, McConnell P, Mueller WT. J. Am. Soc. Mass Spectrom. 1997;8:234–243. [Google Scholar]
- (50).Daniel JM, McCombie G, Wendt S, Zenobi R. J. Am. Soc. Mass Spectrom. 2003;14:442–448. doi: 10.1016/S1044-0305(03)00132-6. [DOI] [PubMed] [Google Scholar]
- (51).Jaquillard L, Saab F, Schoentgen F, Cadene M. J. Am. Soc. Mass Spectrom. 2012;23:908–922. doi: 10.1007/s13361-011-0305-7. [DOI] [PubMed] [Google Scholar]
- (52).Cathou RE, Hammes GG. J. Am. Chem. Soc. 1965;87:4674–4680. doi: 10.1021/ja00949a003. [DOI] [PubMed] [Google Scholar]
- (53).Plotnikov V, Rochalski A, Brandts M, Brandts JF, Williston S, Frasca V, Lin L-N. Assay Drug Dev. Technol. 2002;1:83–90. doi: 10.1089/154065802761001338. [DOI] [PubMed] [Google Scholar]
- (54).Ganem B, Li YT, Henion JD. J. Am. Chem. Soc. 1991;113:7818–7819. [Google Scholar]
- (55).Cubrilovic D, Zenobi R. Anal. Chem. 2013;85:2724–2730. doi: 10.1021/ac303197p. [DOI] [PubMed] [Google Scholar]
- (56).Jecklin MC, Touboul D, Bovet C, Wortmann A, Zenobi R. J. Am. Soc. Mass Spectrom. 2008;19:332–343. doi: 10.1016/j.jasms.2007.11.007. [DOI] [PubMed] [Google Scholar]
- (57).Holler E, Rupley JA, Hess GP. Biochem. Biophys. Res. Commun. 1970;40:166–170. doi: 10.1016/0006-291x(70)91061-2. [DOI] [PubMed] [Google Scholar]
- (58).Gunther A, Jhunjhunwala M, Thalmann M, Schmidt MA, Jensen KF. Langmuir. 2005;21:1547–1555. doi: 10.1021/la0482406. [DOI] [PubMed] [Google Scholar]
- (59).Hatakeyama T, Chen DL, Ismagilov RF. J. Am. Chem. Soc. 2006;128:2518–2519. doi: 10.1021/ja057720w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Li Q, Pei J, Song P, Kennedy RT. Anal. Chem. 2010;82:5260–5267. doi: 10.1021/ac100669z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Valeja SG, Tipton JD, Emmett MR, Marshall AG. Anal. Chem. 2010;82:7515–7519. doi: 10.1021/ac1016858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Zubarev RA, Kelleher NL, McLafferty FW. J. Am. Chem. Soc. 1998;120:3265–3266. [Google Scholar]
- (63).Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Proc. Natl. Acad. Sci USA. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Hu QZ, Noll RJ, Li HY, Makarov A, Hardman M, Cooks RG. J. Mass Spectrom. 2005;40:430–443. doi: 10.1002/jms.856. [DOI] [PubMed] [Google Scholar]
- (65).Marshall AG, Hendrickson CL, Jackson GS. Mass Spectrom. Rev. 1998;17:1–35. doi: 10.1002/(SICI)1098-2787(1998)17:1<1::AID-MAS1>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- (66).Marshall AG, Hendrickson CL. Rapid Commun. Mass Spectrom. 2001;15:232–235. doi: 10.1002/(SICI)1097-0231(19990815)13:15<1639::AID-RCM691>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- (67).Iavarone AT, Jurchen JC, Williams ER. Anal. Chem. 2001;73:1455–1460. doi: 10.1021/ac001251t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Iavarone AT, Williams ER. Int. J. Mass Spectrom. 2002;219:63–72. [Google Scholar]
- (69).Iavarone AT, Williams ER. J. Am. Chem. Soc. 2003;125:2319–2327. doi: 10.1021/ja021202t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Iavarone AT, Williams ER. Anal. Chem. 2003;75:4525–4533. doi: 10.1021/ac034144i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Lomeli SH, Yin S, Ogorzalek Loo RR, Loo JA. J. Am. Soc. Mass Spectrom. 2009;20:593–596. doi: 10.1016/j.jasms.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Lomeli SH, Peng IX, Yin S, Ogorzalek Loo RR, Loo JA. J. Am. Soc. Mass Spectrom. 2010;21:127–131. doi: 10.1016/j.jasms.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Peng IX, Ogorzalek Loo RR, Shiea J, Loo JA. Anal. Chem. 2008;80:6995–7003. doi: 10.1021/ac800870c. [DOI] [PubMed] [Google Scholar]
- (74).Yin S, Loo JA. Int. J. Mass Spectrom. 2011;300:118–122. doi: 10.1016/j.ijms.2010.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Takats Z, Wiseman JM, Cooks RG. J. Mass Spectrom. 2005;40:1261–1275. doi: 10.1002/jms.922. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.