Abstract
The 7mG (7-methylguanosine cap) formed on mRNA is fundamental to eukaryotic gene expression. Protein complexes recruited to 7mG mediate key processing events throughout the lifetime of the transcript. One of the most important mediators of 7mG functions is CBC (cap-binding complex). CBC has a key role in several gene expression mechanisms, including transcription, splicing, transcript export and translation. Gene expression can be regulated by signalling pathways which influence CBC function. The aim of the present review is to discuss the mechanisms by which CBC mediates and co-ordinates multiple gene expression events.
Keywords: cap-binding complex (CBC), 7-methylguanosine cap (7mG), splicing, transcription, translation
Abbreviations: ABA, abscisic acid; Ars2, arsenite-resistance protein 2; CBC, cap-binding complex; Cbp, cap-binding protein; Cdk9, cyclin-dependent kinase 9; CF IA, cleavage factor IA; CTD, C-terminal domain; CTIF, CBC-dependent translation initiation factor; DSIF, DRB sensitivity-inducing factor; eIF, eukaryotic initiation factor; hnRNP, heterogeneous nuclear ribonucleoprotein; MIF4G, middle domain of eIF4G; 7mG, 7-methylguanosine cap; mRNP, messenger ribonucleoprotein; mTORC1, mammalian target of rapamycin complex 1; NELF, negative elongation factor; NMD, nonsense-mediated decay; PABP, poly(A)-binding protein; PARN, poly(A)-specific ribonuclease; PHAX, phosphorylated adaptor of RNA export; PTC, premature termination codon; P-TEFb, positive transcription elongation factor b; RAM, RNMT-activating mini-protein; RNA pol II, RNA polymerase II; RNMT, RNA (guanine-7-)methyltransferase; RRM, RNA-recognition motif; snRNP, small nuclear ribonucleoprotein; SRSF1, serine/arginine-rich splicing factor 1; TMG, 2,2,7-trimethylguanosine cap; TREX, transcription export complex
INTRODUCTION
In the eukaryotic cell, the maturation and translation of RNA pol II (RNA polymerase II) transcripts into proteins requires a co-ordinated series of effective and efficient processing events. The first mRNA processing event is the formation of the 7mG (7-methylguanosine cap) at the 5′ end of nascent transcripts. Subsequent processing and translation is largely dependent on this structure. Protein complexes including CBC (cap-binding complex) and the eIF4F (eukaryotic initiation factor 4F) bind to 7mG and recruit the enzymes and factors to the transcript which mediate further processing, export and translation.
7mG SYNTHESIS
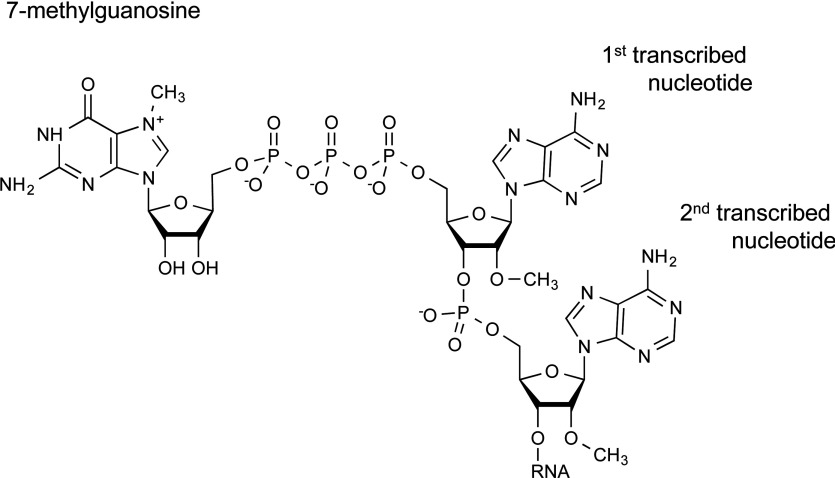
The first transcribed nucleotide of RNA pol II transcripts is modified by the addition of 7-methylguanosine during the early stages of transcription (Figure 1). All transcripts are synthesized with a 5′ triphosphate on the first nucleotide to which 7-methylguanosine is joined via a 5′–5′ triphosphate bridge, creating 7mG(5′)ppp(5′)X (X is the first nucleotide). Throughout the present review we use the abbreviation 7mG to refer to 7-methylguanosine in the cap structure. The 5′–5′ triphosphate linkage that joins 7-methylguanosine to the first nucleotide is thought to be found uniquely on RNA pol II transcripts. This unique structure enables certain processing factors to be recruited exclusively to RNA pol II transcripts [1–3]. Three enzymatic activities catalyse the addition of 7-methylguanosine: triphosphatase, guanylyltransferase and methyltransferase. The triphosphatase cleaves the terminal phosphate of the transcript and an RNA guanylyltransferase catalyses the addition of guanosine monophosphate to create G(5′)ppp(5′)X. Subsequently, a guanine-7-methyltransferase catalyses the addition of the methyl group to the N-7 position of the guanosine cap, creating 7mG(5′)ppp(5′)X [2,4,5]. In lower eukaryotes these three enzymatic activities reside in three distinct polypeptides; however, in metazoans the triphosphatase and guanylyltransferase reside in the same polypeptide, RNGTT [6]. In addition, in vertebrates the cap methyltransferase is a complex of two proteins; the catalytic subunit, RNMT [RNA (guanine-7-)methyltransferase], and the activating subunit, RAM (RNMT-activating mini-protein) [7].
Figure 1. 7mG structure.
The 7mG structure is depicted including the first and second transcribed nucleotides. Cap 2 structure is depicted, i.e. methylated on the O-2 position of ribose on the first and second transcribed nucleotides.
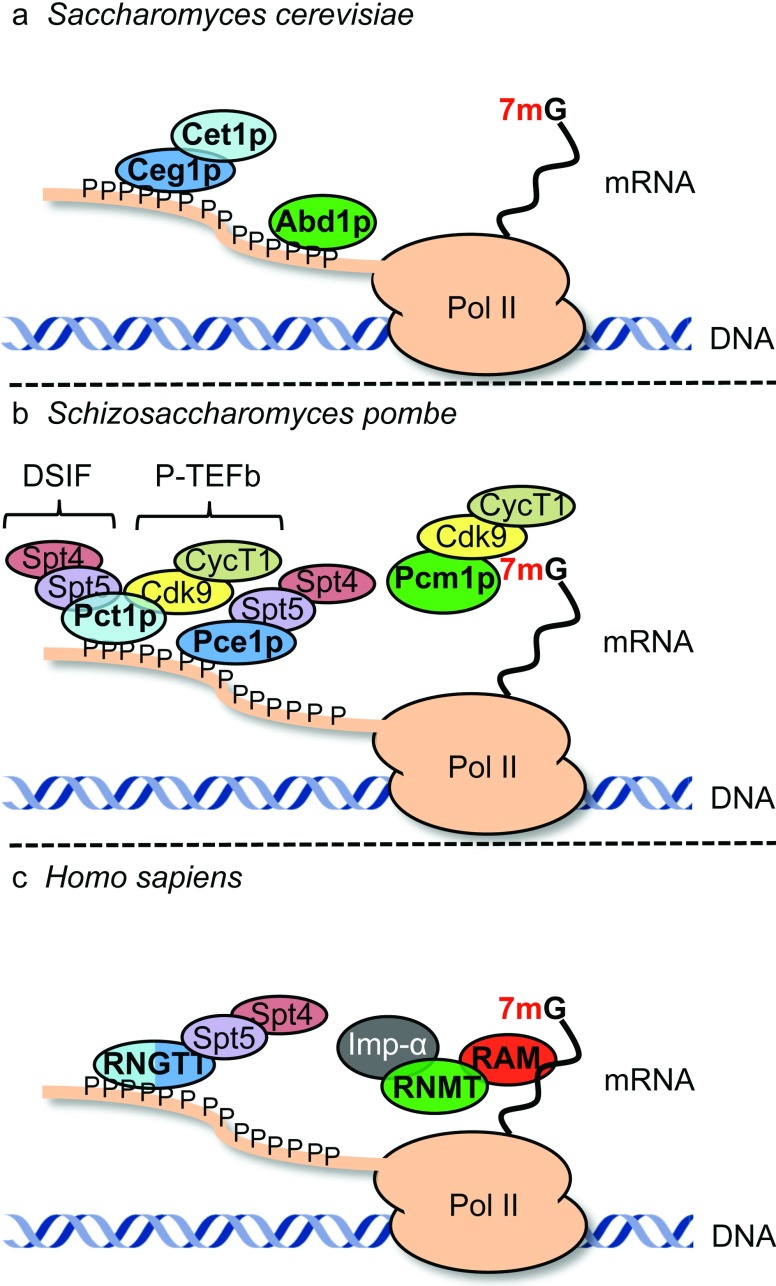
7mG is exclusively added to transcripts synthesized by RNA pol II since only this polymerase has a CTD (C-terminal domain), which when phosphorylated during the initial stages of transcription recruits the capping enzymes [8–10] (Figure 2). The capping enzymes have been demonstrated to promote transcription in lower and higher eukaryotes independently of their catalytic activities [11–13]. Although capping occurs predominantly during transcription, accumulating evidence suggests that cap-like structures can be also formed post-transcriptionally [14].
Figure 2. 7mG synthesis in S. cerevisiae, Schizosaccharomyces pombe and humans.
The RNA triphosphatase enzymes are depicted in light blue, the RNA guanylyltransferase in dark blue and the RNA guanine-7-methyltransferases in green. (a) In S. cerevisiae, the RNA guanylyltransferase (Ceg1p) and the RNA triphosphatase (Cet1p) interact with the Ser5-phosphorylated RNA pol II CTD via the Ceg1p subunit [168–172]. The RNA guanine-7-methyltransferase (Abd1p) is recruited to the transcription initiation site via an interaction with phosphorylated RNA pol II CTD [12,95,173]. (b) In S. pombe, the RNA triphosphatase (Pct1p) and the RNA guanylyltransferase (Pce1p) independently interact with phosphorylated RNA pol II CTD [171,174]. Pct1p interacts with DSIF (DRB sensitivity-inducing factor) (Spt4/Spt5) and P-TEFb (Cdk9/Cyclin T1) [175,176]. Pce1p interacts with DSIF [176,177]. The RNA guanine-7-methyltransferase (Pcm1p) functions at the transcription initiation site, but does not physically associate with RNA pol II. Pcm1p and P-TEFb are in a complex and Pcm1p is required for P-TEFb recruitment to chromatin and transcription elongation [11,178,179]. (c) In humans, the RNA triphosphatase and the RNA guanylyltransferase activities reside in the same polypeptide, RNGTT. RNGTT interacts with DSIF (Spt4/5), which stimulates its activity up to 5-fold [13,180]. RNGTT promotes transcription elongation independently of its catalytic activity by overcoming NELF-dependent transcriptional pausing [13]. RNGTT is recruited to transcription initiation sites via an interaction with Ser5-phosphorylated RNA pol II CTD, which stimulates guanylyltransferase activity [169,181,182]. The RNA guanine-7-methyltransferase complex (RNMT/RAM) interacts with RNGTT and indirectly with RNA pol II [5,183]. RNMT also interacts with importin-α (Imp-α), which stimulates cap methyltransferase activity [184].
The cap structure protects transcripts from exoribonucleolytic degradation [15,16]. Furthermore, it interacts with nuclear and cytoplasmic cap-binding proteins which mediate additional 7mG functions (Table 1 and Figure 3). CBC and eIF4F are the most thoroughly characterized cap-binding complexes, although other cap-binding proteins have been reported, including PARN [poly(A)-specific ribonuclease] deadenylase, PABPC1 [poly(A)-binding protein C1], PUM2 and Y14/Magoh [17–23].
Table 1. CBC-interacting proteins (RNA-independent).
For each interacting protein, the CBC subunit mediating the interaction, the species in which the interaction was detected and the function of the interaction are presented. SLBP, stem–loop-binding protein; ZC3H18, zinc finger CCCH-type containing 18.
| Interacting protein | CBC subunit | Species detected | Function | Reference(s) |
|---|---|---|---|---|
| Importin-α | Cbp80 | H. sapiens, S. cerevisiae and X. laevis | Translocating CBC to nucleus, regulating the release of cargo from CBC and regulating CBC activity | [114,119,160] |
| Cdk9/Bur1p | Unclear | H. sapiens and S. cerevisiae | Transcription elongation and splicing | [63,64] |
| U4/U6.U5 tri-snRNP | Unclear | H. sapiens | Splicing | [34] |
| NELF-E | Unclear | H. sapiens | Histone RNA 3′ end processing | [32] |
| SLBP | Unclear | H. sapiens | Histone RNA 3′ end processing | [32] |
| Ars2 | Unclear | H. sapiens and D. melanogaster | Histone RNA 3′ end processing and miRNA biogenesis | [98,102,103] |
| PARN | Cbp80 | H. sapiens | mRNA stability | [108] |
| ZC3H18 | Unclear | H. sapiens | Unclear | [34,48] |
| PHAX | Unclear | Mammalian cells and X. laevis | U snRNA nuclear export | [115,116] |
| Aly component of TREX | Cbp80 | H. sapiens | mRNA nuclear export | [121,125] |
| hnRNP C | Cbp80 | H. sapiens | mRNA nuclear export | [126] |
| UPF1 | Cbp80 | Mammalian cells | NMD | [150,153] |
| eIF4G | Cbp80 | Mammalian cells and S. cerevisiae | CBC-dependent translation | [128,134,135] |
| CTIF | Cbp80 | Mammalian cells | CBC-dependent translation | [136,137] |
Figure 3. CBC functions.
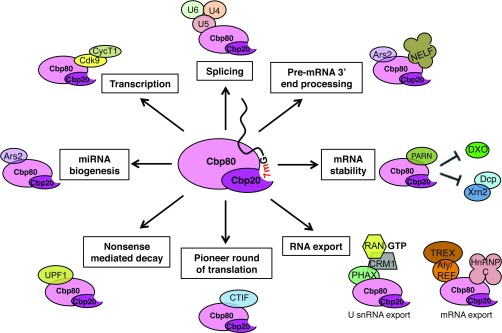
CBC, composed of Cbp20 and Cbp80 subunits, binds to 7mG located at the 5′ end of RNA pol II transcripts. CBC interacts with a spectrum of factors mediating RNA metabolism and translation mechanisms. CycT1, cyclin T1; Dcp, decapping mRNA; DXO, decapping exoribonuclease; Xm2, 5′–3′ exoribonuclease 2.
eIF4E is the cap-binding subunit of eIF4F, a complex required for cap-dependent translation initiation [24]. In the eIF4F complex, eIF4E binds to eIF4G, a scaffold protein to which other factors are recruited, including eIF4A, a DEAD-box RNA helicase required for 5′-UTR unwinding and eIF4G. The interaction of eIF4G with eIF4E is required for efficient 7mG binding [25,26]. Since there are many excellent reviews discussing eIF4F function [3,27,28], the present review will focus on the function and regulation of CBC. CBC is a multifaceted complex essential for gene expression, which integrates RNA processing events, transcript nuclear export and translation.
IDENTIFICATION OF CBC AS A NUCLEAR CAP-BINDING COMPLEX
CBC was first purified from nuclear extracts of HeLa cells on the basis of its affinity for 7mG [29,30]. CBC was demonstrated to consist of 20 and 80 kDa polypeptides, which were designated as Cbp20 (cap-binding protein 20) and Cbp80 respectively. It is likely that CBC is present in all eukaryotes, and its evolution correlates with the appearance of 7mG. Cbp20 is unlikely to be present in significant quantities as a monomer since it is unstable in the absence of Cbp80, both in mammals and yeast [31–34], and is undetectable in Cbp80-immunodepleted extracts [29,35]. Conversely, it is not clear whether Cbp80 can exist as a monomer and Cbp20 is not required for Cbp80 stability [33].
Cbp20 and Cbp80 bind to 7mG synergistically and neither subunit alone has significant affinity for the structure [29,30,36]. The crystal structure of CBC revealed that the 7mG-binding pocket resides in Cbp20, and this was validated by mutational analysis [37]. On binding to CBC, 7mG is positioned between two conserved tyrosine residues (Tyr20 and Tyr43) in the Cbp20 subunit [38,39], and biophysical studies indicated that these residues are essential for the interaction [38,40]. Cbp80 causes a conformational change in Cbp20, which is required for CBC to bind to 7mG with high affinity. The Cbp80 structure is highly ordered and composed of three helical domains connected by two linkers. The Cbp80 N-terminal helical domain is structurally similar to MIF4G (middle domain of eIF4G), and is required for cap-dependent translation [37,38,41]. The MIF4G and intermediate helical domains of Cbp80 mediate interaction with Cbp20.
In addition to binding 7mG, CBC binds directly to RNA via both subunits. Cbp20 contains a classical RRM (RNA-recognition motif). A splice variant of Cbp20 that does not bind to Cbp80 or 7mG, but does contain a portion of the RRM, retains RNA-binding activity, albeit reduced [42]. Cbp80 also binds to RNA [43,44]. As described throughout the present review, CBC often has gene-specific effects. It is possible that the RNA-binding domains of CBC may have an enhanced affinity for specific RNA sequences or motifs and thus have a role in mediating these gene-specific effects.
CBC interacts with transcripts shortly after transcription in the nucleus. Although one of its functions is to accompany the transcript through the nuclear pore (described in detail later), it is a predominantly nuclear complex. Cbp80 contains a nuclear localization signal at the N-terminus, which is required for its nuclear localization [45,46], and Cbp20 is likely to be co-imported into the nucleus with Cbp80 [47].
CBC recruits several factors to 7mG-modified transcripts which mediate processing events [34,48,49]. The contribution of CBC to gene expression has been addressed in yeast, plants and mammalian cells by depleting and reducing the expression of the subunits. CBP80 or CBP20 deletion in Saccharomyces cerevisiae results in significant changes in gene expression, with many genes exhibiting a change of 2-fold or more [50,51]. Although in S. cerevisiae CBP80 and CBP20 are not essential for cell viability [31,52,53], they are required for cell growth and proliferation [31,54]. Disruption of CBC genes in Arabidopsis thaliana is not lethal, but results in developmental delays, reduced stature and ABA (abscisic acid) hypersensitivity owing to a down-regulation of transcripts involved in ABA signalling [55,56]. In mammalian cells, siRNA-mediated depletion of Cbp80 results in deregulation of approximately 400 genes and a significant reduction in the cell proliferation rate [32]. To our knowledge there are no reports of Cbp80 or Cbp20 gene deletion in mammalian systems, and therefore it is not clear whether CBC is required for embryonic development or mammalian cell viability.
CBC AND TRANSCRIPTION
7mG formation is a co-transcriptional process and CBC is rapidly recruited to this structure during transcript synthesis. Using ChIP assays, Cbp20 and Cbp80 subunits were detected at the 5′ end of genes as well as within the gene bodies, suggesting that CBC may track with the elongating polymerase [32,57–61]. CBC is likely to be recruited to chromatin via an interaction with 7mG since monomeric Cbp20 and Cbp80 fail to be recruited [33,60], recruitment does not occur in Abd1p (cap methyltransferase)-deficient S. cerevisiae [33], and recruitment is mediated by RNA [62].
CBC can have a reciprocal relationship with transcription. The complex directly and indirectly recruits several transcriptional factors to promoters and for a subset of genes has an active role in transcription regulation [33,60,61,63,64]. In S. cerevisiae, CBC directly recruits Mot1p to a subset of gene promoters. Mot1p is a regulator of transcription that positively or negatively regulates the expression of RNA pol II-transcribed genes in a gene-specific manner [60]. On genes that Mot1p activates, CBC-dependent Mot1p recruitment results in the recruitment of general transcription factors and RNA pol II to promoters, stimulating transcription initiation. On genes that Mot1p represses, CBC-dependent Mot1p recruitment results in repression of RNA pol II recruitment [60]. It is currently unclear whether the mammalian homologue of Mot1p, BTAF1, has similar CBC-dependent functions. This CBC function is less likely to be prominent in mammals, since in these species the rate-limiting step in transcription, in general, is not recruitment of RNA pol II, but rather escape of promoter-proximal paused RNA pol II. CBC also indirectly interacts with Npl3p, an hnRNP (heterogeneous nuclear ribonucleoprotein)-like protein essential for growth in yeast [65]. CBC and Npl3p act synergistically to suppress transcription termination at weak polyadenylation sites by repressing the recruitment of the termination complex CF IA (cleavage factor IA) [33]. CBC also recruits Bur1p and Ctk1p, kinases that phosphorylate RNA pol II CTD and transcription factors, promoting transcriptional elongation [66,67]. CBC-dependent recruitment of Bur1p and Ctk1p stimulates RNA pol II CTD Ser2 phosphorylation and results in recruitment of histone methyltransferases and induction of H3K36me3 (histone H3 trimethylated on Lys36), a mark of active transcription [61,64].
In mammals, CBC also stimulates transcription elongation of a subset of genes via the recruitment of P-TEFb (positive transcription elongation factor b), which contains Cdk9 (cyclin-dependent kinase 9; the mammalian homologue of Bur1p), to promoter-proximal paused RNA pol II (Figure 4) [63]. Depletion of CBC results in decreased RNA pol II CTD Ser2 phosphorylation, accompanied by reduced RNA pol II in the body of a subset of genes [63]. It is currently unclear whether CBC influences transcription of a subset of genes or acts genome- wide.
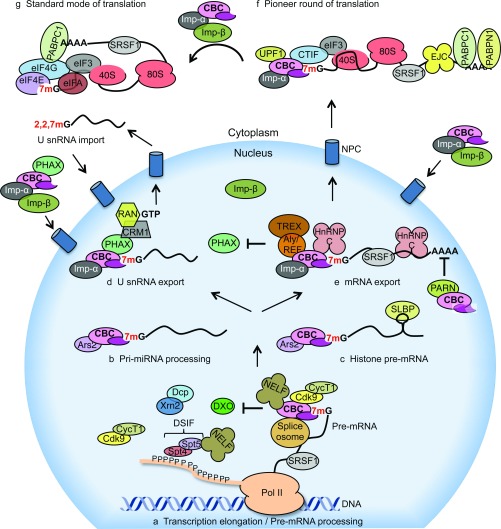
Figure 4. CBC functions in eukaryotic gene expression.
(a) CBC is required for pre-mRNA processing. The co-transcriptional binding of CBC to 7mG prevents the decapping activities of pre-mRNA degradation complexes [DXO (decapping exoribonuclease) and Dcp (decapping mRNA) Xrn2 (5′–3′ exoribonuclease 2)] and promotes pre-mRNA processing. CBC recruits P-TEFb [Cdk9/Cyclin T1 (CycT1)] to transcription initiation sites of specific genes promoting phosphorylation of the RNA pol II CTD at Ser2 residues. This results in the recruitment of splicing factors including SRSF1, which regulates both constitutive and alternative splicing events. Furthermore, CBC interacts with splicing machinery components that results in the spliceosomal assembly. CBC interacts with NELF and promotes pre-mRNA processing of replication-dependent histone transcripts. (b) CBC forms a complex with Ars2 and promotes miRNA biogenesis by mediating pri-miRNA processing. (c) CBC/Ars2 promotes pre-mRNA processing of replication-dependent histone transcripts. (d) CBC promotes export of U snRNA. CBC interacts with PHAX, which recruits export factors including CRM1 and RAN·GTP. (e) CBC promotes export of mRNA. For export of transcripts over 300 nucleotides, hnRNP C interacts with CBC and inhibits the interaction between CBC and PHAX, allowing the CBC to interact with TREX and the transcript to be translocated to the cytoplasm. CBC interacts with the PARN deadenylase and inhibits its activity, protecting mRNAs from degradation. (f) CBC mediates the pioneer round of translation. Cbp80 interacts with CTIF, which recruits the 40S ribosomal subunit via eIF3 to the 5′ end of the mRNA for translation initiation. Upon binding of importin-β (Imp-β) to importin-α (Imp-α), mRNA is released from CBC and binds to eIF4E for the initiation of the standard mode of translation. CBC-bound mRNP components not found in eIF4E-bound mRNPs are CTIF, exon junction complex (EJC) and PABPN1. (g) The standard mode of translation is mediated by eIF4E cap-binding protein. eIF4E is a component of the eIF4F complex which promotes translation initiation.
CBC AND PRE-mRNA SPLICING
Eukaryotic pre-mRNA transcripts are synthesized as precursors containing an alternating series of exons and introns. The process of splicing excises introns and joins exons together to form the mature transcript [68]. The molecular machinery that catalyses splicing is called the spliceosome and is composed of five small nuclear ribonucleoprotein particles (U snRNPs) associated with a large number of additional proteins [69,70]. 7mG and CBC are required for efficient pre-mRNA splicing.
Incubation of uncapped or 7mG-capped transcripts with total or nuclear HeLa cell extracts initially revealed that the 7mG moiety is required for efficient splicing in mammalian systems [29,71–74]. Depletion of CBC from HeLa cell extracts resulted in inhibition of pre-mRNA splicing and reduced recruitment of U1 snRNP to the 5′ splice site of the 5′ proximal intron [29,35]. The effect of 7mG on splicing in vivo was initially observed in Xenopus laevis oocytes. Microinjected transcripts were only efficiently spliced if they were capped, and splicing of the 5′ proximal intron required 7mG [75]. Microinjection of X. laevis oocytes with antibodies raised against Cbp20 significantly decreased the splicing efficiency of microinjected transcripts [30].
In S. cerevisiae, only 3% of genes contain introns and are spliced [76]. Inactivation of the capping enzymes (Ceg1p or Abd1p) with temperature-sensitive mutants revealed that 7mG enhances splicing of a subset of these genes [77–80]. CBC is part of the splicing commitment complex [81], and depletion of CBC from cell extracts resulted in reduced in vitro splicing of pre-mRNA due to inhibition of spliceosome assembly [82,83]. The biological significance of the relationship between CBC and splicing is reflected in the observation that mutation of splicing components is synthetically lethal in CBC-deleted strains [31,84]. Furthermore, a Cbp20p mutant that cannot bind 7mG is synthetic lethal with the deletion of genes that encode for factors involved in the spliceosome assembly [85]. Notably, CBC has been demonstrated to couple splicing to transcription. Deletion of CBC results in a reduction in the recruitment of several splicing factors to the nascent transcript resulting in inhibition of co-transcriptional spliceosome assembly [86]. The dependency of splicing on CBC has also been observed in A. thaliana [87].
The mechanism by which CBC mediates splicing in mammalian cells has recently been investigated. A direct interaction of CBC with the protein components of the U4/U6.U5 tri-snRNP was observed, and these interactions were required for efficient co-transcriptional spliceosomal assembly [34]. Recent studies in mammalian cells have demonstrated that 7mG and CBC are required not only for the removal of 5′ proximal introns, but also for the removal of downstream introns [34,88]. It is currently unclear to what extent this mechanism is utilized throughout the genome.
Alternative splicing is the process by which the exons of a pre-mRNA are selected in different arrangements, producing multiple mRNAs, some of which produce distinct protein variants [89]. Previously, CBC was demonstrated to regulate alternative splicing in mammalian cells and in A. thaliana [63,90]. The model proposed is that CBC acts as a platform for the recruitment of splicing factors such as SRSF1 (serine/arginine-rich splicing factor 1) to elongating RNA pol II [63]. SRSF1 regulates constitutive and alternative splicing and has also been demonstrated to regulate cap-dependent translation initiation in the cytoplasm [91] (Figure 4).
CBC AND PRE-mRNA 3′ END PROCESSING
Most mammalian mRNAs have a poly(A)-tail consisting of 200–250 adenosines at the 3′ end which protects transcripts from degradation and stimulates their translation. Formation of the poly(A)-tail or polyadenylation is a two-step process. Initially the pre-mRNA is cleaved at the poly(A) site followed by polyadenylation of the 3′ end [5]. In vitro studies demonstrated that m7G-capped pre-mRNAs were cleaved more efficiently at the poly(A) sites than uncapped pre-mRNA [92,93], and the same observation was made with transcripts microinjected into X. laevis oocytes [94]. In mammalian cells, m7G-capped transcripts were demonstrated to be more efficiently cleaved at the poly(A) site than incompletely capped or uncapped transcripts [88,95]. This effect was mediated by CBC [96]. Although the exact molecular mechanism by which CBC promotes pre-mRNA cleavage at the poly(A)-site remains to be determined, it is likely to involve interactions of CBC with the polyadenylation machinery [96].
CBC also has a role in other 3′-end processing events. Replication-dependent histone mRNAs, unlike most other mRNAs, do not possess a poly(A)-tail, but have instead a conserved 3′ end stem–loop structure [97]. In mammalian cells, depletion of CBC causes aberrant production of poly(A)-tailed histone mRNAs [32,98]. CBC interactions with NELF (negative elongation factor) and Ars2 (arsenite-resistance protein 2) are required for efficient histone mRNA 3′-end processing (Figure 4) [32,98]. Recent evidence suggests that CBC can also determine the fate of replication-dependent histone mRNAs towards degradation or translation [99].
The effect of CBC on 3′-end processing is species-specific. In S. cerevisiae, 7mG appears to have minimal effect on pre-mRNA 3′-end processing. Inactivation of the RNA guanylyltransferase Ceg1p exhibits no overt effects on polyadenylation levels [77]. This suggests that the effect on mRNA 3′-end formation is a CBC function that evolved later [47]. However, as described above, CBC can suppress aberrant 3′-end processing in S. cerevisiae. CBC acts in synergy with Npl3p to repress the recruitment of the termination complex CF IA to weak polyadenylation sites [33].
CBC AND miRNA BIOGENESIS
miRNAs are endogenous transcripts that post-transcriptionally regulate gene expression in animals and plants. They are transcribed by RNA pol II as pri-miRNAs, thus carry 7mG and the poly(A)-tail [100]. During nuclear and cytoplasmic processing events, the pri-miRNA loses the 7mG and the poly(A)-tail and the mature, 21–23 nucleotide-long, miRNA is incorporated into RISC (RNA-induced silencing complex) to guide RNA silencing [101]. Ars2 and CBC have been demonstrated to be required for the biogenesis of a subset of miRNAs (Figure 4) [98,102,103]. Ars2 forms a complex with CBC and 7mG in mammalian cells [102]. Depletion of Ars2 with Cbp20 and Cbp80 from mammalian cells resulted in a decrease in miRNA biogenesis in a transcript-specific manner [98,102]. The same phenomenon was also observed in Drosophila melanogaster and A. thaliana [87,103,104].
CBC AND RNA STABILITY
Removal of 7mG by decapping enzymes leads to RNA degradation. This process can be regulated in a gene-specific manner by signalling pathways resulting in a biological response [105]. Electroporation of in vitro transcribed RNAs containing 7mG or incompletely capped structures into mammalian cells revealed that 7mG is required to stabilize mRNA [106]. The stability is, in part, mediated by cap-binding proteins, such as eIF4E, PABPC1 and CBC, that compete with the decapping enzymes for the 7mG structure [21,88,106,107]. Furthermore, CBC also interacts with and inhibits PARN deadenylase, which catalyses poly(A)-tail removal, an initial step in mRNA degradation (Figure 4) [108].
Recently, decapping and transcript degradation was demonstrated to occur co-transcriptionally and to limit bidirectional RNA pol II elongation and the production of aberrantly processed pre-mRNAs [88,109,110]. This suggests that the competition between CBC and the decapping complexes could regulate the balance between transcription elongation and degradation of the nascent transcript (Figure 4). CBC was also identified to interact with the NEXT (nuclear exosome targeting) complex, although the biological significance of this interaction remains unclear [34,111].
CBC AND RNA POL II TRANSCRIPT EXPORT: U snRNAs
In eukaryotic cells, RNA pol II transcripts are synthesized in the nucleus, but undergo essential processing and/or translation in the cytoplasm. Therefore transcript nuclear export is a key step in gene expression. U snRNAs, although synthesized and functioning in the nucleus, are processed and receive their protein partners in the cytoplasm. Efficient nuclear export of U snRNAs was found to be 7mG-dependent in X. laevis oocytes and requires CBC [30,112–114]. In order for CBC to promote U snRNA nuclear export, it interacts with PHAX (phosphorylated adaptor of RNA export) [115,116]. Upon phosphorylation by CK2 kinase, PHAX stimulates the nuclear export of U snRNAs in a process mediated by CRM1 in a RAN·GTP-dependent manner [115,117]. In the cytosol, PHAX dephosphorylation and importin-β are required for U snRNA release and recycling of CBC/PHAX back to the nucleus [115,117].
In S. cerevisiae, 30% of importin-α is isolated in a complex with CBC [114]. This interaction has also been observed in X. laevis oocytes and mammalian cells [114,118], and validated by co-crystallization of CBC with importin-α [119]. In the nucleus, importin-α/CBC binds to 7mG–U snRNA and the complex is exported. In the cytosol, importin-β interacts with importin-α and releases the U snRNA for processing including the formation of the 5′ TMG (2,2,7-trimethylguanosine cap) [114,119]. Subsequently, the importin-α/importin-β/CBC complex is re-imported into the nucleus. High levels of RAN·GTP in the nucleus promote dissociation of importin-β from the complex, and a new cycle begins (Figure 4). Of note, after nuclear re-entry of U snRNAs, the presence of the TMG prevents efficient binding by CBC [120].
CBC AND RNA POL II TRANSCRIPT EXPORT: mRNA
In higher eukaryotes, CBC stimulates mRNA nuclear export via an interaction with Aly/REF [121,122]. Aly/REF is a component of TREX (transcription export complex), a multi-subunit protein complex that links pre-mRNA processing with mRNA nuclear export in mammalian cells, required for the export of spliced and unspliced mRNAs [123]. TREX interacts with TAP, the mRNA export receptor [124]. Thus CBC promotes mRNA nuclear export by facilitating the recruitment of nuclear export machinery to the transcript (Figure 4) [121,122,125].
As described above, U snRNA and mRNA are exported by distinct mechanisms. The factor that distinguishes between these mechanisms is transcript length. The default export pathway utilizes PHAX. However, when CBC is bound to transcripts longer than 300 nucleotides, it interacts with hnRNP C, which abolishes the interaction between CBC and PHAX, thus selecting the TREX export pathway (Figure 4) [126].
In S. cerevisiae, both CBC and 7mG are dispensable for the majority if not all mRNA export [77,127].
CBC AND THE PIONEER ROUND OF TRANSLATION
The majority of translation in eukaryotes is dependent on eIF4E binding to 7mG, which drives translation initiation by recruiting the 40S ribosomal subunit [3,24]. CBC has an analogous function to eIF4F in translation with the exception that it drives the first round or rounds of translation known as the pioneer round of translation. The pioneer round of translation was originally identified by a screen performed in S. cerevisiae in which a Cbp80p mutant was found to be synthetic lethal with an eIF4G mutant deficient for eIF4E and Pab1p binding [128]. Although the pioneer round of translation was initially believed only to have a quality control role in orchestrating NMD (nonsense-mediated decay) [129,130], evidence suggests that CBC-bound transcripts can undergo multiple rounds of translation generating functional products [131–133]. However, in contrast to the standard mode of translation mediated by eIF4F, the pioneer round of translation does not generate abundant amounts of protein.
The mechanism of CBC-mediated translation initiation is not well characterized and is somewhat controversial. CBC has been demonstrated to interact with eIF4G in yeast and mammalian cells [128,134,135]. However, a novel protein with similarity to eIF4G designated as CTIF (CBC-dependent translation initiation factor) was demonstrated to interact with Cbp80 [136]. CTIF is required during the initiation of the pioneer round of translation, but is not involved in the standard mode of translation [136,137]. CTIF interacts with eIF3g, a component of the eIF3 complex, and recruits the 40S ribosomal subunit to CBC-bound mRNA for the pioneer round of translation (Figure 4) [137]. Depletion of CTIF causes redistribution of CBC from polysomes to subpolysomal fractions, suggesting that CTIF is an essential component of the pioneer round of translation that functions in a manner similar to eIF4G in the standard mode of translation [136,137]. In contrast with the eIF4F-dependent translation where the eIF4E-bound mRNA forms a circular structure [138,139], similar circularization has not been observed in CBC-dependent translation [140].
The transition from the pioneer round of translation to the standard mode of translation and the exchange of CBC for eIF4F at 7mG is regulated by importins [118]. Importin-α, as described previously, interacts with Cbp80 [114,119]. In the cytosol, importin-β interacts with importin-α and promotes the dissociation of the mRNA from CBC. Subsequently, eIF4E interacts with 7mG promoting eIF4E-dependent translation initiation (Figure 4) [118].
Fundamental questions remain concerning the mechanism and biological significance of the pioneer round of translation. It is currently unclear what proportion of transcripts utilize the pioneer round of translation, to what extent and under which growth conditions.
CBC AND NONSENSE-MEDIATED DECAY
NMD is a mRNA surveillance pathway that predisposes aberrant mRNAs containing a PTC (premature termination codon) for degradation [141]. NMD can play an important role in regulating the steady-state levels of a subset of mRNAs, modulating several biological responses [142]. The major facilitators of NMD are the UPF proteins (UPF1, UPF2 and UPF3), which are required to detect PTCs [143].
NMD is a translation-dependent process and transcripts engaged in CBC-, eIF4E- or IRES (internal ribosome entry site)-mediated translation are all subject to NMD [140,144–149]. UPF1 can be co-purified with CBC- and eIF4E-bound transcripts in an RNA-dependent manner [140,144,145,149]. Therefore NMD can act at any round of translation and is independent of a specific translation initiation mechanism [146,148,149].
In mammalian cells, siRNA-mediated depletion of Cbp80 results in partial inhibition of NMD [136,150]. However, in S. cerevisiae and A. thaliana CBC is dispensable for NMD [147,151,152]. In mammalian cells, CBC was demonstrated to interact physically with UPF1 via Cbp80 (Figure 4) [150,153], and inhibition of this interaction abolished NMD [153]. Although the exact molecular details are unclear, CBC interaction with UPF1 promotes NMD machinery assembly on the aberrant mRNA, promoting decay [150,153]. However, there is controversy regarding the direct interaction between CBC and UPF1 since several studies have failed to detect it after RNase treatment [140,154].
REGULATION OF CBC
Gene expression is a heavily regulated process and examples are emerging of the regulation of CBC function. Growth factors and mTORC1 (mammalian target of rapamycin complex 1) kinase have been demonstrated to regulate CBC affinity for 7mG [119,155–157]. Nutrient availability and growth factors activate mTORC1 kinase, which phosphorylates S6 kinase [158]. S6 kinase phosphorylates Cbp80, stimulating the affinity of CBC for 7mG in vitro [156]. In vivo, Cbp80 phosphorylation was observed to be stimulated by growth factors and could be inhibited by rapamycin, an mTORC1 inhibitor. Increased Cbp80 phosphorylation was observed to correlate with increased cap-dependent splicing activity [156], and the recruitment of the S6 kinase to CBC-bound mRNPs (messenger ribonucleoproteins) stimulated translation efficiency [159]. Interestingly, binding of importin-α and importin-β to CBC was essential for growth-factor-mediated stimulation of 7mG binding [119]. Furthermore, overexpression of constitutively active RAN within cells, which abolishes the interaction between importin-β and the CBC–importin-α complex, stimulates CBC cap-binding activity [160].
SPECIES-SPECIFIC EFFECTS OF CBC
As discussed throughout the present review, certain CBC functions are observed in all species that express the complex, whereas other functions have only been observed in a subset of species. During evolution, selective pressure has resulted in divergence of gene expression mechanisms, including those involving CBC [161]. Cbp20 is primarily responsible for 7mG binding and is highly conserved. However, Cbp80 is a platform for recruiting proteins to the transcript and appears to have evolved to co-ordinate the increasing complexity of gene expression mechanisms in mammals [37,39,114]. CBC binds to NELF-E, Ars2 and CTIF to mediate histone pre-mRNA processing, miRNA biogenesis and the pioneer round of translation respectively [32,98,102,103,136]. Homologues of NELF-E, Ars2 and CTIF have not been identified in yeast and, to our knowledge, in S. cerevisae these events are not mediated by CBC [50,77].
Splicing is another gene regulation mechanism which has changed dramatically during evolution. In S. cerevisiae, for example, only 3% of genes contain introns and therefore splicing is not prevalent, whereas conversely the majority of metazoan genes contain multiple introns [76]. In metazoans, several gene expression steps have evolved to become coupled to splicing which is obviously not the case in S. cerevisiae [162]. This may explain why CBC is dispensable for mRNA export and NMD in S. cerevisiae, since it interacts with the splicing machinery during these processes.
However, when discussing species specificity it must be stressed that most information about CBC function (and all gene expression mechanisms) comes from a limited number of organisms, and therefore the true extent of conservation of CBC function is not clear. Furthermore, different model organisms are amenable to distinct forms of experimentation, which may result in the appearance of differences in CBC function. For example, in S. cerevisiae, temperature-sensitive mutants allow inhibition of protein function and/or expression within minutes, whereas in human cells siRNA-mediated suppression of CBC leads to loss of protein expression over hours and days, which may lead to indirect effects being observed. Research into the function of CBC may benefit from an inhibitor of the Cbp20–Cbp80 interaction, which would rapidly inhibit CBC function, thus reducing the observation of indirect effects.
FUTURE PERSPECTIVES
There has been intense research into CBC function over the last decade, which has raised many intriguing questions about how this complex influences gene expression.
Do Cbp20 and Cbp80 have CBC-independent functions? The majority of Cbp20 and Cbp80 functions are as a heterodimer. However, there is evidence that these subunits may also function independently. Cbp80, but not Cbp20, was co-purified with eIF4E-bound transcripts, indicating that a pool of Cbp80 exists independently of Cbp20 [140]. Furthermore, gene expression profiling of CBP20- and CBP80-deletion strains in S. cerevisiae revealed that only 15% of regulated transcripts were common to both strains, suggesting that these subunits have independent functions [50]. The question of whether Cbp80 has an independent role is experimentally important since the function of CBC is often inferred from using reagents that only target Cbp80 expression or interactions. An alternatively spliced isoform of Cbp20, Cbp20s, was identified which is likely to function independently of Cbp80, since it does not interact with either Cbp80 or 7mG [42]. The novel Cbp20 isoform was observed to bind to mRNA, presumably via a conserved RRM domain, and was recruited to active transcription sites. It will be interesting to determine the Cbp80 and m7G-independent functions of Cbp20s, and whether Cbp20 also has these functions in the CBC complex.
Does CBC have enhanced affinity for certain transcripts? Many of the studies described in the present review have provided evidence that CBC exhibits transcript specificity. Both Cbp20 and Cbp80 can bind to transcripts independently of 7mG and therefore it is plausible that these subunits could add specificity to the interaction with RNA [42–44].
What is the role of CBC in long non-coding RNA processing? Over recent years there has been explosion of knowledge and interest in lncRNAs (long non-coding RNAs) and other pervasive 7mG-containing transcripts [163,164]. It seems likely that CBC will bind to these transcripts and therefore an obvious question is whether CBC has effects on their metabolism and function.
Can CBC be targeted therapeutically? Although there is no direct evidence to suggest that Cbp20 or Cbp80 are proto-oncogenes, mRNA expression profiling has detected elevated expression of Cbp20 in a subset of tumours [165]. Moreover, as described previously, CBC function is regulated by the mTOR signalling pathway, which when deregulated causes cell transformation [166,167]. Considering that a common feature of tumour cells is enhanced gene expression rates, tumour cells may be predicted to be more sensitive than untransformed cells to inhibition of CBC function. Inhibitors of the Cbp20–Cbp80 or Cbp20–7mG interactions would be not only useful experimentally to determine the direct functions of CBC, but could also be used to determine whether this interface should be pursued as a therapeutic target.
CONCLUSION
CBC plays a pivotal role in post-transcriptional processing events. CBC binds to RNA pol II transcripts during transcription and accompanies them through much of their lifetime, facilitating processing events, including transcription regulation, pre-mRNA processing, mRNA export and the pioneer round of translation (Table 1 and Figure 3). With the first discoveries of regulation of CBC function, it is an exciting possibility that CBC is a complex through which cellular signalling pathways can change the gene expression landscape and cellular physiology.
ACKNOWLEDGEMENTS
We thank Michael Aregger, Nicola Phillips and the Cowling laboratory for advice, and Marianne Schimpl for assistance with the Figures. We apologize to the colleagues whose work could not be cited due to space constraints.
FUNDING
This review was supported by a UK Medical Research Council Career Development Award (to V.H.C.) and a Wellcome Trust strategic award [number WT 097818/Z/11/A (to T.G.P.)].
References
- 1.Shatkin A. J. Capping of eucaryotic mRNAs. Cell. 1976;9:645–653. doi: 10.1016/0092-8674(76)90128-8. [DOI] [PubMed] [Google Scholar]
- 2.Furuichi Y., Shatkin A. J. Viral and cellular mRNA capping: past and prospects. Adv. Virus Res. 2000;55:135–184. doi: 10.1016/S0065-3527(00)55003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Topisirovic I., Svitkin Y. V, Sonenberg N., Shatkin A. J. Cap and cap-binding proteins in the control of gene expression. Wiley Interdiscip. Rev. RNA. 2011;2:277–298. doi: 10.1002/wrna.52. [DOI] [PubMed] [Google Scholar]
- 4.Cowling V. H. Regulation of mRNA cap methylation. Biochem. J. 2010;425:295–302. doi: 10.1042/BJ20091352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shatkin A. J., Manley J. L. The ends of the affair: capping and polyadenylation. Nat. Struct. Biol. 2000;7:838–842. doi: 10.1038/79583. [DOI] [PubMed] [Google Scholar]
- 6.Shuman S. What messenger RNA capping tells us about eukaryotic evolution. Nat. Rev. Mol. Cell Biol. 2002;3:619–625. doi: 10.1038/nrm880. [DOI] [PubMed] [Google Scholar]
- 7.Gonatopoulos-Pournatzis T., Dunn S., Bounds R., Cowling V. H. RAM/Fam103a1 is required for mRNA cap methylation. Mol. Cell. 2011;44:585–596. doi: 10.1016/j.molcel.2011.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol. Cell. 2009;36:541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perales R., Bentley D. “Cotranscriptionality”: the transcription elongation complex as a nexus for nuclear transactions. Mol. Cell. 2009;36:178–191. doi: 10.1016/j.molcel.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heidemann M., Hintermair C., Voß K., Eick D. Dynamic phosphorylation patterns of RNA polymerase II CTD during transcription. Biochim. Biophys. Acta. 2013;1829:55–62. doi: 10.1016/j.bbagrm.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 11.Guiguen A., Soutourina J., Dewez M., Tafforeau L., Dieu M., Raes M., Vandenhaute J., Werner M., Hermand D. Recruitment of P-TEFb (Cdk9-Pch1) to chromatin by the cap-methyl transferase Pcm1 in fission yeast. EMBO J. 2007;26:1552–1559. doi: 10.1038/sj.emboj.7601627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schroeder S. C., Zorio D. A., Schwer B., Shuman S., Bentley D. A function of yeast mRNA cap methyltransferase, Abd1, in transcription by RNA polymerase II. Mol. Cell. 2004;13:377–387. doi: 10.1016/s1097-2765(04)00007-3. [DOI] [PubMed] [Google Scholar]
- 13.Mandal S. S., Chu C., Wada T., Handa H., Shatkin A. J., Reinberg D. Functional interactions of RNA-capping enzyme with factors that positively and negatively regulate promoter escape by RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 2004;101:7572–7577. doi: 10.1073/pnas.0401493101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schoenberg D. R., Maquat L. E. Re-capping the message. Trends Biochem. Sci. 2009;34:435–442. doi: 10.1016/j.tibs.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furuichi Y., LaFiandra A., Shatkin A. J. 5′-Terminal structure and mRNA stability. Nature. 1977;266:235–239. doi: 10.1038/266235a0. [DOI] [PubMed] [Google Scholar]
- 16.Green M. R., Maniatis T., Melton D. A. Human β-globin pre-mRNA synthesized in vitro is accurately spliced in Xenopus oocyte nuclei. Cell. 1983;32:681–694. doi: 10.1016/0092-8674(83)90054-5. [DOI] [PubMed] [Google Scholar]
- 17.Gao M., Fritz D. T., Ford L. P., Wilusz J. Interaction between a poly(A)-specific ribonuclease and the 5′ cap influences mRNA deadenylation rates in vitro. Mol. Cell. 2000;5:479–488. doi: 10.1016/s1097-2765(00)80442-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dehlin E., Wormington M., Körner C. G., Wahle E. Cap-dependent deadenylation of mRNA. EMBO J. 2000;19:1079–1086. doi: 10.1093/emboj/19.5.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez J., Ren Y. G., Thuresson A. C., Hellman U., Astrom J., Virtanen A. A 54-kDa fragment of the poly(A)-specific ribonuclease is an oligomeric, processive, and cap-interacting poly(A)-specific 3′ exonuclease. J. Biol. Chem. 2000;275:24222–24230. doi: 10.1074/jbc.M001705200. [DOI] [PubMed] [Google Scholar]
- 20.Wu M., Nilsson P., Henriksson N., Niedzwiecka A., Lim M. K., Cheng Z., Kokkoris K., Virtanen A., Song H. Structural basis of m(7)GpppG binding to poly(A)-specific ribonuclease. Structure. 2009;17:276–286. doi: 10.1016/j.str.2008.11.012. [DOI] [PubMed] [Google Scholar]
- 21.Khanna R., Kiledjian M. Poly(A)-binding-protein-mediated regulation of hDcp2 decapping in vitro. EMBO J. 2004;23:1968–1976. doi: 10.1038/sj.emboj.7600213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cao Q., Padmanabhan K., Richter J. D. Pumilio 2 controls translation by competing with eIF4E for 7-methyl guanosine cap recognition. RNA. 2010;16:221–227. doi: 10.1261/rna.1884610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chuang T.-W., Chang W.-L., Lee K.-M., Tarn W.-Y. The RNA-binding protein Y14 inhibits mRNA decapping and modulates processing body formation. Mol. Biol. Cell. 2013;24:1–13. doi: 10.1091/mbc.E12-03-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sonenberg N., Hinnebusch A. G. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haghighat A., Sonenberg N. eIF4G dramatically enhances the binding of eIF4E to the mRNA 5′-cap structure. J. Biol. Chem. 1997;272:21677–21680. doi: 10.1074/jbc.272.35.21677. [DOI] [PubMed] [Google Scholar]
- 26.Gross J. D., Moerke N. J., von der Haar T., Lugovskoy A. A., Sachs A. B., McCarthy J. E. G., Wagner G. Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell. 2003;115:739–750. doi: 10.1016/s0092-8674(03)00975-9. [DOI] [PubMed] [Google Scholar]
- 27.Raught B., Gingras A. C. eIF4E activity is regulated at multiple levels. Int. J. Biochem. Cell Biol. 1999;31:43–57. doi: 10.1016/s1357-2725(98)00131-9. [DOI] [PubMed] [Google Scholar]
- 28.Sonenberg N. eIF4E, the mRNA cap-binding protein: from basic discovery to translational research. Biochem. Cell Biol. 2008;86:178–183. doi: 10.1139/O08-034. [DOI] [PubMed] [Google Scholar]
- 29.Izaurralde E., Lewis J., McGuigan C., Jankowska M., Darzynkiewicz E., Mattaj I. W. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell. 1994;78:657–668. doi: 10.1016/0092-8674(94)90530-4. [DOI] [PubMed] [Google Scholar]
- 30.Izaurralde E., Lewis J., Gamberi C., Jarmolowski A., McGuigan C., Mattaj I. W. A cap-binding protein complex mediating U snRNA export. Nature. 1995;376:709–712. doi: 10.1038/376709a0. [DOI] [PubMed] [Google Scholar]
- 31.Fortes P., Kufel J., Fornerod M., Polycarpou-Schwarz M., Lafontaine D., Tollervey D., Mattaj I. W. Genetic and physical interactions involving the yeast nuclear cap-binding complex. Mol. Cell. Biol. 1999;19:6543–6553. doi: 10.1128/mcb.19.10.6543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Narita T., Yung T. M. C., Yamamoto J., Tsuboi Y., Tanabe H., Tanaka K., Yamaguchi Y., Handa H. NELF interacts with CBC and participates in 3′ end processing of replication-dependent histone mRNAs. Mol. Cell. 2007;26:349–365. doi: 10.1016/j.molcel.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 33.Wong C.-M., Qiu H., Hu C., Dong J., Hinnebusch A. G. Yeast cap binding complex impedes recruitment of cleavage factor IA to weak termination sites. Mol. Cell. Biol. 2007;27:6520–6531. doi: 10.1128/MCB.00733-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pabis M., Neufeld N., Steiner M. C., Bojic T., Shav-Tal Y., Neugebauer K. M. The nuclear cap-binding complex interacts with the U4/U6·U5 tri-snRNP and promotes spliceosome assembly in mammalian cells. RNA. 2013;19:1054–1063. doi: 10.1261/rna.037069.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewis J. D., Izaurralde E., Jarmolowski A., McGuigan C., Mattaj I. W. A nuclear cap-binding complex facilitates association of U1 snRNP with the cap-proximal 5′ splice site. Genes Dev. 1996;10:1683–1698. doi: 10.1101/gad.10.13.1683. [DOI] [PubMed] [Google Scholar]
- 36.Kataoka N., Ohno M., Moda I., Shimura Y. Identification of the factors that interact with NCBP, an 80 kDa nuclear cap binding protein. Nucleic Acids Res. 1995;23:3638–3641. doi: 10.1093/nar/23.18.3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mazza C., Ohno M., Segref A., Mattaj I. W., Cusack S. Crystal structure of the human nuclear cap binding complex. Mol. Cell. 2001;8:383–936. doi: 10.1016/s1097-2765(01)00299-4. [DOI] [PubMed] [Google Scholar]
- 38.Calero G., Wilson K. F., Ly T., Rios-Steiner J. L., Clardy J. C., Cerione R. A. Structural basis of m7GpppG binding to the nuclear cap-binding protein complex. Nat. Struct. Biol. 2002;9:912–917. doi: 10.1038/nsb874. [DOI] [PubMed] [Google Scholar]
- 39.Mazza C., Segref A., Mattaj I. W., Cusack S. Large-scale induced fit recognition of an m(7)GpppG cap analogue by the human nuclear cap-binding complex. EMBO J. 2002;21:5548–5557. doi: 10.1093/emboj/cdf538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Worch R., Jankowska-Anyszka M., Niedzwiecka A., Stepinski J., Mazza C., Darzynkiewicz E., Cusack S., Stolarski R. Diverse role of three tyrosines in binding of the RNA 5′ cap to the human nuclear cap binding complex. J. Mol. Biol. 2009;385:618–627. doi: 10.1016/j.jmb.2008.10.092. [DOI] [PubMed] [Google Scholar]
- 41.Marintchev A., Wagner G. eIF4G and CBP80 share a common origin and similar domain organization: implications for the structure and function of eIF4G. Biochemistry. 2005;44:12265–12272. doi: 10.1021/bi051271v. [DOI] [PubMed] [Google Scholar]
- 42.Pabis M., Neufeld N., Shav-Tal Y., Neugebauer K. M. Binding properties and dynamic localization of an alternative isoform of the cap-binding complex subunit CBP20. Nucleus. 2010;1:412–421. doi: 10.4161/nucl.1.5.12839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Castello A., Fischer B., Eichelbaum K., Horos R., Beckmann B. M., Strein C., Davey N. E., Humphreys D. T., Preiss T., Steinmetz L. M., et al. Insights into RNA biology from an atlas of mammalian mRNA-binding. proteins. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 44.Baltz A. G., Munschauer M., Schwanhäusser B., Vasile A., Murakawa Y., Schueler M., Youngs N., Penfold-Brown D., Drew K., Milek M., et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell. 2012;46:674–690. doi: 10.1016/j.molcel.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 45.Izaurralde E., McGuigan C., Mattaj I. W. Nuclear localization of a cap-binding protein complex. Cold Spring Harb. Symp. Quant. Biol. 1995;60:669–675. doi: 10.1101/sqb.1995.060.01.072. [DOI] [PubMed] [Google Scholar]
- 46.Visa N., Izaurralde E., Ferreira J., Daneholt B., Mattaj I. W. A nuclear cap-binding complex binds Balbiani ring pre-mRNA cotranscriptionally and accompanies the ribonucleoprotein particle during nuclear export. J. Cell Biol. 1996;133:5–14. doi: 10.1083/jcb.133.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lewis J. D., Izaurralde E. The role of the cap structure in RNA processing and nuclear export. Eur. J. Biochem. 1997;247:461–469. doi: 10.1111/j.1432-1033.1997.00461.x. [DOI] [PubMed] [Google Scholar]
- 48.Merz C., Urlaub H., Will C. L., Lührmann R. Protein composition of human mRNPs spliced in vitro and differential requirements for mRNP protein recruitment. RNA. 2007;13:116–128. doi: 10.1261/rna.336807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Müller-McNicoll M., Neugebauer K. M. How cells get the message: dynamic assembly and function of mRNA-protein complexes. Nat. Rev. Genet. 2013;14:275–287. doi: 10.1038/nrg3434. [DOI] [PubMed] [Google Scholar]
- 50.Baron-Benhamou J., Fortes P., Inada T., Preiss T., Hentze M. W. The interaction of the cap-binding complex (CBC) with eIF4G is dispensable for translation in yeast. RNA. 2003;9:654–662. doi: 10.1261/rna.5100903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hossain M. A., Claggett J. M., Nguyen T., Johnson T. L. The cap binding complex influences H2B ubiquitination by facilitating splicing of the SUS1 pre-mRNA. RNA. 2009;15:1515–1527. doi: 10.1261/rna.1540409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Uemura H., Jigami Y. GCR3 encodes an acidic protein that is required for expression of glycolytic genes in Saccharomyces cerevisiae. J. Bacteriol. 1992;174:5526–5532. doi: 10.1128/jb.174.17.5526-5532.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abovich N., Liao X. C., Rosbash M. The yeast MUD2 protein: an interaction with PRP11 defines a bridge between commitment complexes and U2 snRNP addition. Genes Dev. 1994;8:843–854. doi: 10.1101/gad.8.7.843. [DOI] [PubMed] [Google Scholar]
- 54.Das B., Guo Z., Russo P., Chartrand P., Sherman F. The role of nuclear cap binding protein Cbc1p of yeast in mRNA termination and degradation. Mol. Cell. Biol. 2000;20:2827–2838. doi: 10.1128/mcb.20.8.2827-2838.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hugouvieux V., Kwak J. M., Schroeder J. I. An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell. 2001;106:477–487. doi: 10.1016/s0092-8674(01)00460-3. [DOI] [PubMed] [Google Scholar]
- 56.Papp I., Mur L. A., Dalmadi A., Dulai S., Koncz C. A mutation in the cap binding protein 20 gene confers drought tolerance to Arabidopsis. Plant Mol. Biol. 2004;55:679–686. doi: 10.1007/s11103-004-1680-2. [DOI] [PubMed] [Google Scholar]
- 57.Listerman I., Sapra A. K., Neugebauer K. M. Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat. Struct. Mol. Biol. 2006;13:815–822. doi: 10.1038/nsmb1135. [DOI] [PubMed] [Google Scholar]
- 58.Glover-Cutter K., Kim S., Espinosa J., Bentley D. L. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat. Struct. Mol. Biol. 2008;15:71–78. doi: 10.1038/nsmb1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zenklusen D., Vinciguerra P., Wyss J.-C., Stutz F. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol. Cell. Biol. 2002;22:8241–8253. doi: 10.1128/MCB.22.23.8241-8253.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lahudkar S., Shukla A., Bajwa P., Durairaj G., Stanojevic N., Bhaumik S. R. The mRNA cap-binding complex stimulates the formation of pre-initiation complex at the promoter via its interaction with Mot1p in vivo. Nucleic Acids Res. 2011;39:2188–2209. doi: 10.1093/nar/gkq1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lidschreiber M., Leike K., Cramer P. Cap completion and CTD kinase recruitment underlie the initiation-elongation transition of RNA polymerase II. Mol. Cell. Biol. 2013;33:3805–3816. doi: 10.1128/MCB.00361-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schroder P. A., Moore M. J. Association of ribosomal proteins with nascent transcripts in S. cerevisiae. RNA. 2005;11:1521–1529. doi: 10.1261/rna.2134305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lenasi T., Peterlin B., Barboric M. Cap-binding protein complex links pre-mRNA capping to transcription elongation and alternative splicing through positive transcription elongation factor b (P-TEFb) J. Biol. Chem. 2011;286:22758–22768. doi: 10.1074/jbc.M111.235077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hossain M. A., Chung C., Pradhan S. K., Johnson T. L. The yeast cap binding complex modulates transcription factor recruitment and establishes proper histone H3K36 trimethylation during active transcription. Mol. Cell. Biol. 2013;33:785–799. doi: 10.1128/MCB.00947-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shen E. C., Stage-Zimmermann T., Chui P., Silver P. The yeast mRNA-binding protein Npl3p interacts with the cap-binding complex. J. Biol. Chem. 2000;275:23718–23724. doi: 10.1074/jbc.M002312200. [DOI] [PubMed] [Google Scholar]
- 66.Peterlin B. M., Price D. H. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 67.Bartkowiak B., Liu P., Phatnani H. P., Fuda N. J., Cooper J. J., Price D. H., Adelman K., Lis J. T., Greenleaf A. L. CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev. 2010;24:2303–2316. doi: 10.1101/gad.1968210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tollervey D., Caceres J. F. RNA processing marches on. Cell. 2000;103:703–709. doi: 10.1016/s0092-8674(00)00174-4. [DOI] [PubMed] [Google Scholar]
- 69.Sanford J. R., Caceres J. F. Pre-mRNA splicing: life at the centre of the central dogma. J. Cell Sci. 2004;117:6261–6263. doi: 10.1242/jcs.01513. [DOI] [PubMed] [Google Scholar]
- 70.Wahl M. C., Will C. L., Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 71.Konarska M. M., Padgett R. A., Sharp P. A. Recognition of cap structure in splicing in vitro of mRNA precursors. Cell. 1984;38:731–736. doi: 10.1016/0092-8674(84)90268-x. [DOI] [PubMed] [Google Scholar]
- 72.Edery I., Sonenberg N. Cap-dependent RNA splicing in a HeLa nuclear extract. Proc. Natl. Acad. Sci. U.S.A. 1985;82:7590–7594. doi: 10.1073/pnas.82.22.7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ohno M., Sakamoto H., Shimura Y. Preferential excision of the 5′ proximal intron from mRNA precursors with two introns as mediated by the cap structure. Proc. Natl. Acad. Sci. U.S.A. 1987;84:5187–5191. doi: 10.1073/pnas.84.15.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Patzelt E., Thalmann E., Hartmuth K., Blaas D., Kuechler E. Assembly of pre-mRNA splicing complex is cap dependent. Nucleic Acids Res. 1987;15:1387–1399. doi: 10.1093/nar/15.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Inoue K., Ohno M., Sakamoto H., Shimura Y. Effect of the cap structure on pre-mRNA splicing in Xenopus oocyte nuclei. Genes Dev. 1989;3:1472–1479. doi: 10.1101/gad.3.9.1472. [DOI] [PubMed] [Google Scholar]
- 76.Ast G. How did alternative splicing evolve? Nat. Rev. Genet. 2004;5:773–782. doi: 10.1038/nrg1451. [DOI] [PubMed] [Google Scholar]
- 77.Fresco L. D., Buratowski S. Conditional mutants of the yeast mRNA capping enzyme show that the cap enhances, but is not required for, mRNA splicing. RNA. 1996;2:584–596. [PMC free article] [PubMed] [Google Scholar]
- 78.Schwer B., Shuman S. Conditional inactivation of mRNA capping enzyme affects yeast pre-mRNA splicing in vivo. RNA. 1996;2:574–583. [PMC free article] [PubMed] [Google Scholar]
- 79.Schwer B., Mao X., Shuman S. Accelerated mRNA decay in conditional mutants of yeast mRNA capping enzyme. Nucleic Acids Res. 1998;26:2050–2057. doi: 10.1093/nar/26.9.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schwer B., Saha N., Mao X., Chen H. W., Shuman S. Structure-function analysis of yeast mRNA cap methyltransferase and high-copy suppression of conditional mutants by AdoMet synthase and the ubiquitin conjugating enzyme Cdc34p. Genetics. 2000;155:1561–1576. doi: 10.1093/genetics/155.4.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang D., Rosbash M. Identification of eight proteins that cross-link to pre-mRNA in the yeast commitment complex. Genes Dev. 1999;13:581–592. doi: 10.1101/gad.13.5.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Colot H. V, Stutz F., Rosbash M. The yeast splicing factor Mud13p is a commitment complex component and corresponds to CBP20, the small subunit of the nuclear cap-binding complex. Genes Dev. 1996;10:1699–1708. doi: 10.1101/gad.10.13.1699. [DOI] [PubMed] [Google Scholar]
- 83.Lewis J. D., Görlich D., Mattaj I. W. A yeast cap binding protein complex (yCBC) acts at an early step in pre-mRNA splicing. Nucleic Acids Res. 1996;24:3332–3336. doi: 10.1093/nar/24.17.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fortes P., Bilbao-Cortés D., Fornerod M., Rigaut G., Raymond W., Séraphin B., Mattaj I. W. Luc7p, a novel yeast U1 snRNP protein with a role in 5′ splice site recognition. Genes Dev. 1999;13:2425–2438. doi: 10.1101/gad.13.18.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Qiu Z. R., Chico L., Chang J., Shuman S., Schwer B. Genetic interactions of hypomorphic mutations in the m7G cap-binding pocket of yeast nuclear cap binding complex: an essential role for Cbc2 in meiosis via splicing of MER3 pre-mRNA. RNA. 2012;18:1996–2011. doi: 10.1261/rna.033746.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Görnemann J., Kotovic K. M., Hujer K., Neugebauer K. M. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol. Cell. 2005;19:53–63. doi: 10.1016/j.molcel.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 87.Laubinger S., Sachsenberg T., Zeller G., Busch W., Lohmann J. U., Rätsch G., Weigel D. Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 2008;105:8795–8800. doi: 10.1073/pnas.0802493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jiao X., Chang J. H., Kilic T., Tong L., Kiledjian M. A mammalian pre-mRNA 5′ end capping quality control mechanism and an unexpected link of capping to pre-mRNA processing. Mol. Cell. 2013;50:104–115. doi: 10.1016/j.molcel.2013.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Blencowe B. J. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 90.Raczynska K. D., Simpson C. G., Ciesiolka A., Szewc L., Lewandowska D., McNicol J., Szweykowska-Kulinska Z., Brown J. W. S., Jarmolowski A. Involvement of the nuclear cap-binding protein complex in alternative splicing in Arabidopsis thaliana. Nucleic Acids Res. 2010;38:265–278. doi: 10.1093/nar/gkp869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Michlewski G., Sanford J. R., Cáceres J. F. The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E-BP1. Mol. Cell. 2008;30:179–189. doi: 10.1016/j.molcel.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 92.Gilmartin G. M., McDevitt M. A., Nevins J. R. Multiple factors are required for specific RNA cleavage at a poly(A) addition site. Genes Dev. 1988;2:578–587. doi: 10.1101/gad.2.5.578. [DOI] [PubMed] [Google Scholar]
- 93.Cooke C., Alwine J. C. The cap and the 3′ splice site similarly affect polyadenylation efficiency. Mol. Cell. Biol. 1996;16:2579–2584. doi: 10.1128/mcb.16.6.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Georgiev O., Mous J., Birnstiel M. L. Processing and nucleo-cytoplasmic transport of histone gene transcripts. Nucleic Acids Res. 1984;12:8539–8551. doi: 10.1093/nar/12.22.8539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.McCracken S., Fong N., Rosonina E., Yankulov K., Brothers G., Siderovski D., Hessel A., Foster S., Program A. E., Shuman S., et al. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 1997;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Flaherty S. M., Fortes P., Izaurralde E., Mattaj I. W., Gilmartin G. M. Participation of the nuclear cap binding complex in pre-mRNA 3′ processing. Proc. Natl. Acad. Sci. U.S.A. 1997;94:11893–11898. doi: 10.1073/pnas.94.22.11893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Marzluff W. F., Wagner E. J., Duronio R. J. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat. Rev. Genet. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gruber J. J., Olejniczak S. H., Yong J., La Rocca G., Dreyfuss G., Thompson C. B. Ars2 promotes proper replication-dependent histone mRNA 3′ end formation. Mol. Cell. 2012;45:87–98. doi: 10.1016/j.molcel.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Choe J., Kim K. M., Park S., Lee Y. K., Song O.-K., Kim M. K., Lee B.-G., Song H. K., Kim Y. K. Rapid degradation of replication-dependent histone mRNAs largely occurs on mRNAs bound by nuclear cap-binding proteins 80 and 20. Nucleic Acids Res. 2012;41:1307–1318. doi: 10.1093/nar/gks1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bartel D. P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Macias S., Cordiner R. A., Cáceres J. F. Cellular functions of the microprocessor. Biochem. Soc. Trans. 2013;41:838–843. doi: 10.1042/BST20130011. [DOI] [PubMed] [Google Scholar]
- 102.Gruber J. J., Zatechka D. S., Sabin L. R., Yong J., Lum J. J., Kong M., Zong W.-X., Zhang Z., Lau C.-K., Rawlings J., et al. Ars2 links the nuclear cap-binding complex to RNA interference and cell proliferation. Cell. 2009;138:328–339. doi: 10.1016/j.cell.2009.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sabin L. R., Zhou R., Gruber J. J., Lukinova N., Bambina S., Berman A., Lau C.-K., Thompson C. B., Cherry S. Ars2 regulates both miRNA- and siRNA- dependent silencing and suppresses RNA virus infection in Drosophila. Cell. 2009;138:340–351. doi: 10.1016/j.cell.2009.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kim S., Yang J.-Y., Xu J., Jang I.-C., Prigge M. J., Chua N.-H. Two cap-binding proteins CBP20 and CBP80 are involved in processing primary microRNAs. Plant Cell Physiol. 2008;49:1634–1644. doi: 10.1093/pcp/pcn146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li Y., Kiledjian M. Regulation of mRNA decapping. Wiley Interdiscip. Rev. RNA. 2010;1:253–65. doi: 10.1002/wrna.15. [DOI] [PubMed] [Google Scholar]
- 106.Grudzien E., Kalek M., Jemielity J., Darzynkiewicz E., Rhoads R. E. Differential inhibition of mRNA degradation pathways by novel cap analogs. J. Biol. Chem. 2006;281:1857–1867. doi: 10.1074/jbc.M509121200. [DOI] [PubMed] [Google Scholar]
- 107.Schwartz D. C., Parker R. mRNA decapping in yeast requires dissociation of the cap binding protein, eukaryotic translation initiation factor 4E. Mol. Cell. Biol. 2000;20:7933–7942. doi: 10.1128/mcb.20.21.7933-7942.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Balatsos N., Nilsson P., Mazza C., Cusack S., Virtanen A. Inhibition of mRNA deadenylation by the nuclear cap binding complex (CBC) J. Biol. Chem. 2006;281:4517–4522. doi: 10.1074/jbc.M508590200. [DOI] [PubMed] [Google Scholar]
- 109.Brannan K., Kim H., Erickson B., Glover-Cutter K., Kim S., Fong N., Kiemele L., Hansen K., Davis R., Lykke-Andersen J., et al. mRNA decapping factors and the exonuclease Xrn2 function in widespread premature termination of RNA polymerase II transcription. Mol. Cell. 2012;46:311–324. doi: 10.1016/j.molcel.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Davidson L., Kerr A., West S. Co-transcriptional degradation of aberrant pre-mRNA by Xrn2. EMBO J. 2012;31:2566–2578. doi: 10.1038/emboj.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lubas M., Christensen M. S., Kristiansen M. S., Domanski M., Falkenby L. G., Lykke-Andersen S., Andersen J. S., Dziembowski A., Jensen T. H. Interaction profiling identifies the human nuclear exosome targeting complex. Mol. Cell. 2011;43:624–637. doi: 10.1016/j.molcel.2011.06.028. [DOI] [PubMed] [Google Scholar]
- 112.Hamm J., Mattaj I. W. Monomethylated cap structures facilitate RNA export from the nucleus. Cell. 1990;63:109–118. doi: 10.1016/0092-8674(90)90292-m. [DOI] [PubMed] [Google Scholar]
- 113.Izaurralde E., Stepinski J., Darzynkiewicz E., Mattaj I. W. A cap binding protein that may mediate nuclear export of RNA polymerase II-transcribed RNAs. J. Cell Biol. 1992;118:1287–1295. doi: 10.1083/jcb.118.6.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Görlich D., Kraft R., Kostka S., Vogel F., Hartmann E., Laskey R., Mattaj I. W., Izaurralde E. Importin provides a link between nuclear protein import and U snRNA export. Cell. 1996;87:21–32. doi: 10.1016/s0092-8674(00)81319-7. [DOI] [PubMed] [Google Scholar]
- 115.Ohno M., Segref A., Bachi A., Wilm M., Mattaj I. W. PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell. 2000;101:187–198. doi: 10.1016/S0092-8674(00)80829-6. [DOI] [PubMed] [Google Scholar]
- 116.Segref A., Mattaj I. W., Ohno M. The evolutionarily conserved region of the U snRNA export mediator PHAX is a novel RNA-binding domain that is essential for U snRNA export. RNA. 2001;7:351–360. doi: 10.1017/s1355838201002278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kitao S., Segref A., Kast J., Wilm M., Mattaj I. W., Ohno M. A compartmentalized phosphorylation/dephosphorylation system that regulates U snRNA export from the nucleus. Mol. Cell. Biol. 2008;28:487–497. doi: 10.1128/MCB.01189-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sato H., Maquat L. E. Remodeling of the pioneer translation initiation complex involves translation and the karyopherin importin β. Genes Dev. 2009;23:2537–2550. doi: 10.1101/gad.1817109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dias S. M. G., Wilson K. F., Rojas K. S., Ambrosio A. L. B., Cerione R. A. The molecular basis for the regulation of the cap-binding complex by the importins. Nat. Struct. Mol. Biol. 2009;16:930–937. doi: 10.1038/nsmb.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Schwer B., Erdjument-Bromage H., Shuman S. Composition of yeast snRNPs and snoRNPs in the absence of trimethylguanosine caps reveals nuclear cap binding protein as a gained U1 component implicated in the cold-sensitivity of tgs1Δ cells. Nucleic Acids Res. 2011;39:6715–6728. doi: 10.1093/nar/gkr279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cheng H., Dufu K., Lee C.-S., Hsu J. L., Dias A., Reed R. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell. 2006;127:1389–1400. doi: 10.1016/j.cell.2006.10.044. [DOI] [PubMed] [Google Scholar]
- 122.Nojima T., Hirose T., Kimura H., Hagiwara M. The interaction between cap-binding complex and RNA export factor is required for intronless mRNA export. J. Biol. Chem. 2007;282:15645–15651. doi: 10.1074/jbc.M700629200. [DOI] [PubMed] [Google Scholar]
- 123.Rodrigues J. P., Rode M., Gatfield D., Blencowe B. J., Carmo-Fonseca M., Izaurralde E. REF proteins mediate the export of spliced and unspliced mRNAs from the nucleus. Proc. Natl. Acad. Sci. U.S.A. 2001;98:1030–1035. doi: 10.1073/pnas.031586198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Köhler A., Hurt E. Exporting RNA from the nucleus to the cytoplasm. Nat. Rev. Mol. Cell Biol. 2007;8:761–773. doi: 10.1038/nrm2255. [DOI] [PubMed] [Google Scholar]
- 125.Chi B., Wang Q., Wu G., Tan M., Wang L., Shi M., Chang X., Cheng H. Aly and THO are required for assembly of the human TREX complex and association of TREX components with the spliced mRNA. Nucleic Acids Res. 2012;41:1294–1306. doi: 10.1093/nar/gks1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.McCloskey A., Taniguchi I., Shinmyozu K., Ohno M. hnRNP C tetramer measures RNA length to classify RNA polymerase II transcripts for export. Science. 2012;335:1643–1646. doi: 10.1126/science.1218469. [DOI] [PubMed] [Google Scholar]
- 127.Izaurralde E., Adam S. Transport of macromolecules between the nucleus and the cytoplasm. RNA. 1998;4:351–364. [PMC free article] [PubMed] [Google Scholar]
- 128.Fortes P., Inada T., Preiss T., Hentze M., Mattaj I. The yeast nuclear cap binding complex can interact with translation factor eIF4G and mediate translation initiation. Mol. Cell. 2000;6:191–196. [PubMed] [Google Scholar]
- 129.Chiu S.-Y., Lejeune F., Ranganathan A. C., Maquat L. E. The pioneer translation initiation complex is functionally distinct from but structurally overlaps with the steady-state translation initiation complex. Genes Dev. 2004;18:745–754. doi: 10.1101/gad.1170204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Maquat L. E., Tarn W.-Y., Isken O. The pioneer round of translation: features and functions. Cell. 2010;142:368–374. doi: 10.1016/j.cell.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sharma A., Yilmaz A., Marsh K., Cochrane A., Boris-Lawrie K. Thriving under stress: selective translation of HIV-1 structural protein mRNA during Vpr-mediated impairment of eIF4E translation activity. PLoS Pathog. 2012;8:e1002612. doi: 10.1371/journal.ppat.1002612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Apcher S., Daskalogianni C., Lejeune F., Manoury B., Imhoos G., Heslop L., Fåhraeus R. Major source of antigenic peptides for the MHC class I pathway is produced during the pioneer round of mRNA translation. Proc. Natl. Acad. Sci. U.S.A. 2011;108:11572–7. doi: 10.1073/pnas.1104104108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Garre E., Romero-Santacreu L., De Clercq N., Blasco-Angulo N., Sunnerhagen P., Alepuz P. Yeast mRNA cap-binding protein Cbc1/Sto1 is necessary for the rapid reprogramming of translation after hyperosmotic shock. Mol. Biol. Cell. 2011;23:137–150. doi: 10.1091/mbc.E11-05-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.McKendrick L., Thompson E., Ferreira J., Morley S. J., Lewis J. D. Interaction of eukaryotic translation initiation factor 4G with the nuclear cap-binding complex provides a link between nuclear and cytoplasmic functions of the m(7) guanosine cap. Mol. Cell. Biol. 2001;21:3632–3641. doi: 10.1128/MCB.21.11.3632-3641.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lejeune F., Ranganathan A. C., Maquat L. E. eIF4G is required for the pioneer round of translation in mammalian cells. Nat. Struct. Mol. Biol. 2004;11:992–1000. doi: 10.1038/nsmb824. [DOI] [PubMed] [Google Scholar]
- 136.Kim K. M., Cho H., Choi K., Kim J., Kim B.-W., Ko Y.-G., Jang S. K., Kim Y. K. A new MIF4G domain-containing protein, CTIF, directs nuclear cap-binding protein CBP80/20-dependent translation. Genes Dev. 2009;23:2033–2045. doi: 10.1101/gad.1823409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Choe J., Oh N., Park S., Lee Y. K., Song O.-K., Locker N., Chi S.-G., Kim Y. K. Translation initiation on mRNAs bound by nuclear cap-binding protein complex CBP80/20 requires interaction between CBP80/20-dependent translation initiation factor and eukaryotic translation initiation factor 3g. J. Biol. Chem. 2012;287:18500–18509. doi: 10.1074/jbc.M111.327528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Imataka H., Gradi A., Sonenberg N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 1998;17:7480–7489. doi: 10.1093/emboj/17.24.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wells S. E., Hillner P. E., Vale R. D., Sachs A. B. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell. 1998;2:135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- 140.Rufener S. C., Mühlemann O. eIF4E-bound mRNPs are substrates for nonsense-mediated mRNA decay in mammalian cells. Nat. Struct. Mol. Biol. 2013;20:710–717. doi: 10.1038/nsmb.2576. [DOI] [PubMed] [Google Scholar]
- 141.Schoenberg D. R., Maquat L. E. Regulation of cytoplasmic mRNA decay. Nat. Rev. Genet. 2012;13:246–259. doi: 10.1038/nrg3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Stalder L., Mühlemann O. The meaning of nonsense. Trends Cell Biol. 2008;18:315–321. doi: 10.1016/j.tcb.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 143.Kervestin S., Jacobson A. NMD: a multifaceted response to premature translational termination. Nat. Rev. Mol. Cell Biol. 2012;13:700–712. doi: 10.1038/nrm3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ishigaki Y., Li X., Serin G., Maquat L. E. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell. 2001;106:607–617. doi: 10.1016/s0092-8674(01)00475-5. [DOI] [PubMed] [Google Scholar]
- 145.Lejeune F., Ishigaki Y., Li X., Maquat L. E. The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: dynamics of mRNP remodeling. EMBO J. 2002;21:3536–3545. doi: 10.1093/emboj/cdf345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Maderazo A. B., Belk J. P., He F., Jacobson A. Nonsense-containing mRNAs that accumulate in the absence of a functional nonsense-mediated mRNA decay pathway are destabilized rapidly upon its restitution. Mol. Cell. Biol. 2003;23:842–851. doi: 10.1128/MCB.23.3.842-851.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Gao Q., Das B., Sherman F., Maquat L. E. Cap-binding protein 1-mediated and eukaryotic translation initiation factor 4E-mediated pioneer rounds of translation in yeast. Proc. Natl. Acad. Sci. U.S.A. 2005;102:4258–4263. doi: 10.1073/pnas.0500684102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Holbrook J. A., Neu-Yilik G., Gehring N. H., Kulozik A. E., Hentze M. W. Internal ribosome entry sequence-mediated translation initiation triggers nonsense-mediated decay. EMBO Rep. 2006;7:722–726. doi: 10.1038/sj.embor.7400721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Durand S., Lykke-Andersen J. Nonsense-mediated mRNA decay occurs during eIF4F-dependent translation in human cells. Nat. Struct. Mol. Biol. 2013;20:702–709. doi: 10.1038/nsmb.2575. [DOI] [PubMed] [Google Scholar]
- 150.Hosoda N., Kim Y. K., Lejeune F., Maquat L. E. CBP80 promotes interaction of Upf1 with Upf2 during nonsense-mediated mRNA decay in mammalian cells. Nat. Struct. Mol. Biol. 2005;12:893–901. doi: 10.1038/nsmb995. [DOI] [PubMed] [Google Scholar]
- 151.Kuperwasser N., Brogna S., Dower K., Rosbash M. Nonsense-mediated decay does not occur within the yeast nucleus. RNA. 2004;10:1907–1915. doi: 10.1261/rna.7132504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Dzikiewicz-Krawczyk A., Piontek P., Szweykowska-Kulińska Z., Jarmołowski A. The nuclear cap-binding protein complex is not essential for nonsense-mediated mRNA decay (NMD) in plants. Acta Biochim. Pol. 2008;55:825–828. [PubMed] [Google Scholar]
- 153.Hwang J., Sato H., Tang Y., Matsuda D., Maquat L. E. UPF1 association with the cap-binding protein, CBP80, promotes nonsense-mediated mRNA decay at two distinct steps. Mol. Cell. 2010;39:396–409. doi: 10.1016/j.molcel.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Okada-Katsuhata Y., Yamashita A., Kutsuzawa K., Izumi N., Hirahara F., Ohno S. N- and C-terminal Upf1 phosphorylations create binding platforms for SMG-6 and SMG-5:SMG-7 during NMD. Nucleic Acids Res. 2012;40:1251–1266. doi: 10.1093/nar/gkr791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wilson K. F., Fortes P., Singh U. S., Ohno M., Mattaj I. W., Cerione R. A. The nuclear cap-binding complex is a novel target of growth factor receptor-coupled signal transduction. J. Biol. Chem. 1999;274:4166–4173. doi: 10.1074/jbc.274.7.4166. [DOI] [PubMed] [Google Scholar]
- 156.Wilson K. F., Wu W. J., Cerione R. A. Cdc42 stimulates RNA splicing via the S6 kinase and a novel S6 kinase target, the nuclear cap-binding complex. J. Biol. Chem. 2000;275:37307–37310. doi: 10.1074/jbc.C000482200. [DOI] [PubMed] [Google Scholar]
- 157.Dias S. M. G., Cerione R. A., Wilson K. F. Unloading RNAs in the cytoplasm. Nucleus. 2010;2:139–143. doi: 10.4161/nucl.1.2.10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Laplante M., Sabatini D. M. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ma X. M., Yoon S.-O., Richardson C. J., Jülich K., Blenis J. SKAR links pre-mRNA splicing to mTOR/S6K1-mediated enhanced translation efficiency of spliced mRNAs. Cell. 2008;133:303–313. doi: 10.1016/j.cell.2008.02.031. [DOI] [PubMed] [Google Scholar]
- 160.Ly T. K., Wang J., Pereira R., Rojas K. S., Peng X., Feng Q., Cerione R. A., Wilson K. F. Activation of the Ran GTPase is subject to growth factor regulation and can give rise to cellular transformation. J. Biol. Chem. 2010;285:5815–5826. doi: 10.1074/jbc.M109.071886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hedges S. B. The origin and evolution of model organisms. Nat. Rev. Genet. 2002;3:838–849. doi: 10.1038/nrg929. [DOI] [PubMed] [Google Scholar]
- 162.Reed R. Coupling transcription, splicing and mRNA export. Curr. Opin. Cell Biol. 2003;15:326–331. doi: 10.1016/s0955-0674(03)00048-6. [DOI] [PubMed] [Google Scholar]
- 163.Guttman M., Amit I., Garber M., French C., Lin M. F., Feldser D., Huarte M., Zuk O., Carey B. W., Cassady J. P., et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–237. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Preker P., Almvig K., Christensen M. S., Valen E., Mapendano C. K., Sandelin A., Jensen T. H. PROMoter uPstream Transcripts share characteristics with mRNAs and are produced upstream of all three major types of mammalian promoters. Nucleic Acids Res. 2011;39:7179–7193. doi: 10.1093/nar/gkr370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Rhodes D. R., Kalyana-Sundaram S., Mahavisno V., Varambally R., Yu J., Briggs B. B., Barrette T. R., Anstet M. J., Kincead-Beal C., Kulkarni P., et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Mamane Y., Petroulakis E., Rong L., Yoshida K., Ler L. W., Sonenberg N. eIF4E: from translation to transformation. Oncogene. 2004;23:3172–3179. doi: 10.1038/sj.onc.1207549. [DOI] [PubMed] [Google Scholar]
- 167.Ruggero D., Sonenberg N. The Akt of translational control. Oncogene. 2005;24:7426–7434. doi: 10.1038/sj.onc.1209098. [DOI] [PubMed] [Google Scholar]
- 168.Ho C., Schwer B., Shuman S. Genetic, physical, and functional interactions between the triphosphatase and guanylyltransferase components of the yeast mRNA capping apparatus. Mol. Cell. Biol. 1998;18:5189–5198. doi: 10.1128/mcb.18.9.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ho C. K., Sriskanda V., McCracken S., Bentley D., Schwer B., Shuman S. The guanylyltransferase domain of mammalian mRNA capping enzyme binds to the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 1998;273:9577–9585. doi: 10.1074/jbc.273.16.9577. [DOI] [PubMed] [Google Scholar]
- 170.Ho C. K., Shuman S. Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol. Cell. 1999;3:405–411. doi: 10.1016/s1097-2765(00)80468-2. [DOI] [PubMed] [Google Scholar]
- 171.Takagi T., Cho E.-J., Janoo R. T. K., Polodny V., Takase Y., Keogh M. C., Woo S.-A., Fresco-Cohen L. D., Hoffman C. S., Buratowski S. Divergent subunit interactions among fungal mRNA 5′-capping machineries. Eukaryot. Cell. 2002;1:448–457. doi: 10.1128/EC.1.3.448-457.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Gu M., Rajashankar K. R., Lima C. D. Structure of the Saccharomyces cerevisiae Cet1-Ceg1 mRNA capping apparatus. Structure. 2010;18:216–227. doi: 10.1016/j.str.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Schroeder S. C., Schwer B., Shuman S., Bentley D. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 2000;14:2435–2440. doi: 10.1101/gad.836300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Pei Y., Hausmann S., Ho C. K., Schwer B., Shuman S. The length, phosphorylation state, and primary structure of the RNA polymerase II carboxyl-terminal domain dictate interactions with mRNA capping enzymes. J. Biol. Chem. 2001;276:28075–28082. doi: 10.1074/jbc.M102170200. [DOI] [PubMed] [Google Scholar]
- 175.Pei Y., Schwer B., Shuman S. Interactions between fission yeast Cdk9, its cyclin partner Pch1, and mRNA capping enzyme Pct1 suggest an elongation checkpoint for mRNA quality control. J. Biol. Chem. 2003;278:7180–7188. doi: 10.1074/jbc.M211713200. [DOI] [PubMed] [Google Scholar]
- 176.Pei Y., Shuman S. Interactions between fission yeast mRNA capping enzymes and elongation factor Spt5. J. Biol. Chem. 2002;277:19639–19648. doi: 10.1074/jbc.M200015200. [DOI] [PubMed] [Google Scholar]
- 177.Schneider S., Pei Y., Shuman S., Schwer B. Separable functions of the fission yeast Spt5 carboxyl-terminal domain (CTD) in capping enzyme binding and transcription elongation overlap with those of the RNA polymerase II CTD. Mol. Cell. Biol. 2010;30:2353–2364. doi: 10.1128/MCB.00116-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Pei Y., Du H., Singer J., Stamour C., Granitto S., Shuman S., Fisher R. P. Cyclin-dependent kinase 9 (Cdk9) of fission yeast is activated by the CDK-activating kinase Csk1, overlaps functionally with the TFIIH-associated kinase Mcs6, and associates with the mRNA cap methyltransferase Pcm1 in vivo. Mol. Cell. Biol. 2006;26:777–788. doi: 10.1128/MCB.26.3.777-788.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Viladevall L., St Amour C. V, Rosebrock A., Schneider S., Zhang C., Allen J. J., Shokat K. M., Schwer B., Leatherwood J. K., Fisher R. P. TFIIH and P-TEFb coordinate transcription with capping enzyme recruitment at specific genes in fission yeast. Mol. Cell. 2009;33:738–751. doi: 10.1016/j.molcel.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Wen Y., Shatkin A. J. Transcription elongation factor hSPT5 stimulates mRNA capping. Genes Dev. 1999;13:1774–1779. doi: 10.1101/gad.13.14.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Yue Z., Maldonado E., Pillutla R., Cho H., Reinberg D., Shatkin A. J. Mammalian capping enzyme complements mutant Saccharomyces cerevisiae lacking mRNA guanylyltransferase and selectively binds the elongating form of RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 1997;94:12898–12903. doi: 10.1073/pnas.94.24.12898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Ghosh A., Shuman S., Lima C. D. Structural insights to how mammalian capping enzyme reads the CTD code. Mol. Cell. 2011;43:299–310. doi: 10.1016/j.molcel.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Pillutla R. C., Yue Z., Maldonado E., Shatkin A. J. Recombinant human mRNA cap methyltransferase binds capping enzyme/RNA polymerase IIo complexes. J. Biol. Chem. 1998;273:21443–21446. doi: 10.1074/jbc.273.34.21443. [DOI] [PubMed] [Google Scholar]
- 184.Wen Y., Shatkin A. J. Cap methyltransferase selective binding and methylation of GpppG-RNA are stimulated by importin-α. Genes Dev. 2000;14:2944–2949. doi: 10.1101/gad.848200. [DOI] [PMC free article] [PubMed] [Google Scholar]