Abstract
The genome of Buzura suppressaria nucleopolyhedrovirus (BusuNPV) was sequenced by 454 pyrosequencing technology. The size of the genome is 120,420 bp with 36.8% G+C content. It contains 127 hypothetical open reading frames (ORFs) covering 90.7% of the genome and includes the 37 conserved baculovirus core genes, 84 genes found in other baculoviruses, and 6 unique ORFs. No typical baculoviral homologous repeats (hrs) were present but the genome contained a region of repeated sequences. Gene Parity Plots revealed a 28.8 kb region conserved among the alpha- and beta-baculoviruses. Overall comparisons of BusuNPV to other baculoviruses point to a distinct species in group II Alphabaculovirus.
Introduction
The Baculovirdae is an insect-specific family of viruses with double stranded circular DNA genomes of 80 kb –180 kb. Among the so far sequenced baculoviruses, Xestia c-nigrum granulovirus (XecnGV) has the largest genome (178,733 bp) with the smallest in the Neodiprion lecontei nucleopolyhedrovirus (NeleNPV, 81,755 bp) [1], [2]. With the exception of members of Gammabaculovirus, two distinct progeny phenotypes are produced, the budded virus (BV) that disseminates systemically and the occlusion derived virus (ODV) required for oral infectivity [3]. The occlusion bodies (OBs) afford the embedded virions a certain amount of protection against environmental inactivating conditions such as UV lights and rainwater. The number of predicted ORFs in a single baculovirus range from 89 (NeleNPV) to 183 (Pseudaletia unipuncta GV, PsunGV) [2]. Among all the baculovirus predicted ORFs, 37 have been identified as core genes that exist in all sequenced baculoviruses and are essential for the viral life cycle [4], [5].
The family Baculoviridae is classified into 4 genera: Alphabaculovirus (NPVs isolated from Lepidoptera); Betabaculovirus (GVs isolated from Lepidoptera); Gammabaculovirus (NPVs isolated from Hymenoptera) and Deltabaculovirus (NPVs isolated from Diptera) [6], [7]. The Alphabaculovirus are further clustered into groups I and II based on phylogenetic analyses and the presence or absence of the gp64 gene. Only group I contains gp64 gene while group II has a gene encoding fusion protein (F) [8]–[11].
Buzura suppressaria is a pest insect of tea, tung oil, citrus and metasequoia plants. The Buzura suppressaria NPV (BusuNPV) was first isolated from dead larva of B. suppressaria and subsequently used as an insecticide against this pest [12], [13]. The virus is a single nucleocapsid NPV with a genome size of approximately 120 kb. So far, only a few of the BusuNPV genes have been identified, including those encoding polyhedrin [12], [14], ecdysteroid UDP-glucosyltransferase (egt) [15], polyhedron envelope protein gene (pep), the conotoxin-like protein gene (ctl), the inhibitor of apoptosis (iap), superoxide dismutase (sod) [16], and P10 [17]. A physical map of viral DNA was determined [12] and about 43.5 kb dispersed regions of the genome have been sequenced showing a distinct gene arrangement of BusuNPV [13]. In this manuscript we report the complete genome of BusuNPV. Sequence analysis showed that BusuNPV is a group II Alphabaculovirus with a genome distinct from other so far sequenced baculoviruses.
Results and Discussion
Sequencing and Genome Characteristics
The genome of BusuNPV was sequenced using the Roche 454 GS FLX system with shotgun strategy. A total of 97,246 reads were obtained with the average length of 340 bp. The BusuNPV genome was assembled by Roche GS De Novo assembler software and assisted by the published restriction maps [13]; the genome was covered 217 times.
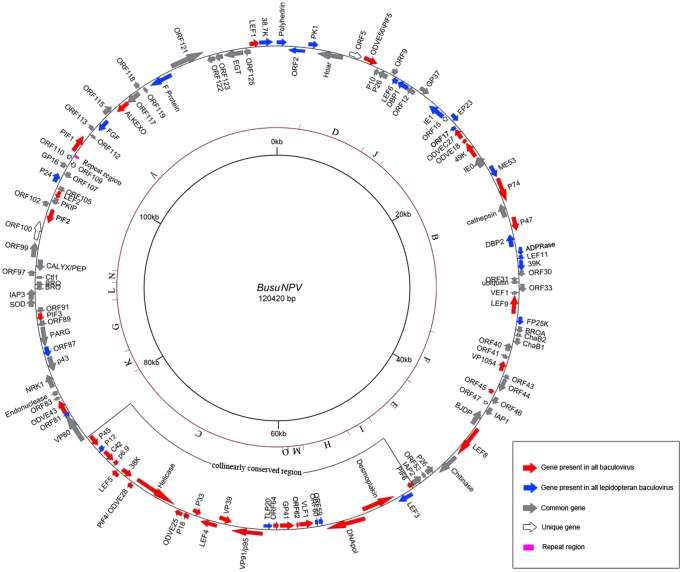
The size of the BusuNPV genome is 120,420 bp with a G+C content of 36.8% (Table S1) and 127 hypothetical ORFs of more than 150 bp. The polyhedrin gene was defined as the first ORF and the A of its initiation codon as the first nucleotide (nt) of the genome. So far, 78 baculoviral genomes have been completely sequenced including BusuNPV (Table S1). BusuNPV contains the 37 core genes conserved in all baculoviruses (shown as red in Fig. 1) and 25 other genes that are present in all sequenced lepidopteran baculovirus (shown as blue in Fig. 1). The genome also contains 59 additional genes commonly found in a variety of baculoviruses (shown as grey in Fig. 1) and also has 6 unique genes (shown as open arrows, Fig. 1). A restriction map of HindIII is presented in Fig. 1, which corroborates the previous physical map [13]. A region appears to be conserved in alpha- and beta-baculoviruses (see below) is also presented in this figure.
Figure 1. Genome map of BusuNPV.
ORFs are indicated by arrows with a displayed name. Arrows also signify transcription directions. Red arrows represent core genes, blue represent genes present in all lepidopteron baculoviruses, gray represent baculoviral common genes and open arrowers represent unique genes of BusuNPV. The pink square represent a repeat structure. The inner circle indicates genome scale position by 20HindIII restriction map is shown in the middle dark red circle. A region collinearly conserved in alpha- and betabaculoviruses is also shown.
Classification of BusuNPV
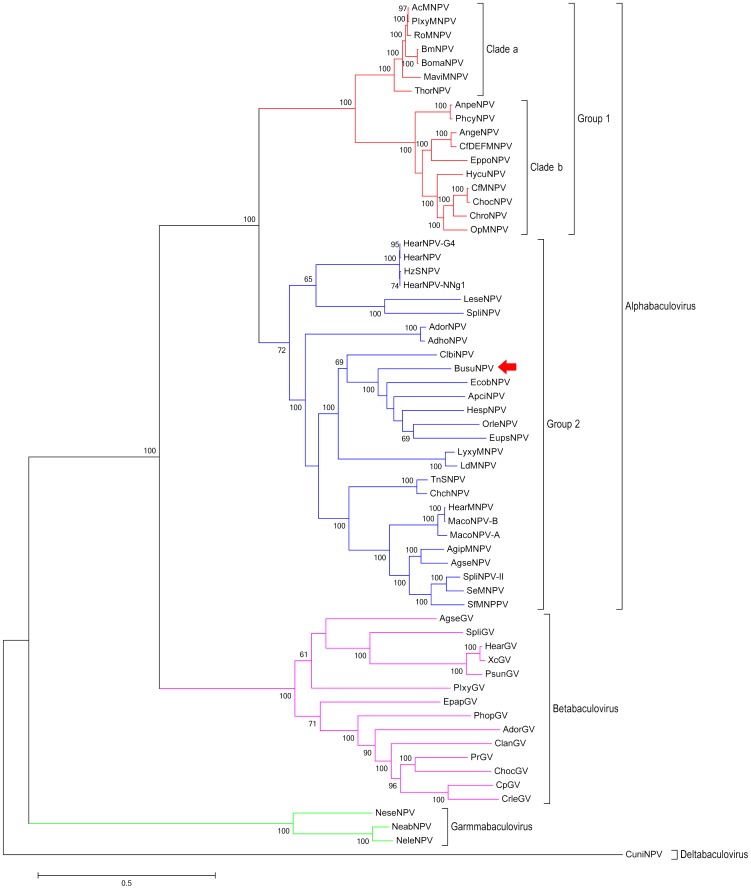
Phyogenetic analysis on the 37 core proteins from the 62 representing baculoviruses placed BusuNPV in group II of the genus Alphabaculovirus (Fig. 2), which is consistent with the previous reports [13], [16]. It formed a subclade with other six NPVs including Ectropis obliqua NPV (EcobNPV), Apocheima cinerarium NPV (ApciNPV), Euproctis pseudoconspersa NPV (EupsNPV), Hemileuca sp. NPV (HespNPV), Clanis bilineata NPV (ClbiNPV) and Orgyia leucostigma NPV (OrleNPV) [18], [19].
Figure 2. Phylogenetic tree using 37 core proteins of 62 sequenced baculoviruses based on Maximum Likelihood method.
It tested by Bootstrap method with a value of 1000. The bootstrap values greater than 50% are showed in front of every nodes. Arrow points to BusuNPV.
Comparison to other Baculoviruses
The nucleotide identities between the ORFs of BusuNPV and other representative baculoviruses are shown in Table S2. The overall genomic nucleic acid identity to EcobNPV, EupsNPV, OrleNPV, HespNPV, ClbiNPV and ApciNPV was about 27.2%, 27.0%, 26.7% 22.0%, 24.2% and 27.4%, respectively. The observed low identities imply that BusuNPV is evolutionarily quite divergent from the fully sequenced baculoviruses.
Gene-parity plots of BusuNPV against the other 6 viruses in the same subclade demonstrated colinearity with some inversions over the whole genome (Fig. 3a). Some colinearity was also found with representatives of group I alphabaculoviruses and betabaculoviruses, but almost no colinearity with those from gamma- and deltabaculoviruses (Fig. 3b). Interestingly, a 28.8 kb region from Busu55 to Busu79 is almost totally collinearly conserved in alpha- and betabaculoviruses (Table 1, Fig. 1). This region contains 25 ORFs in BusuNPV, 20 of which are conserved in all baculoviruses (Table 1, Fig. 1). It is likely that this region existed in the common ancestor of alpha- and betabaculoviruses.
Figure 3. Gene-parity plot analysis.
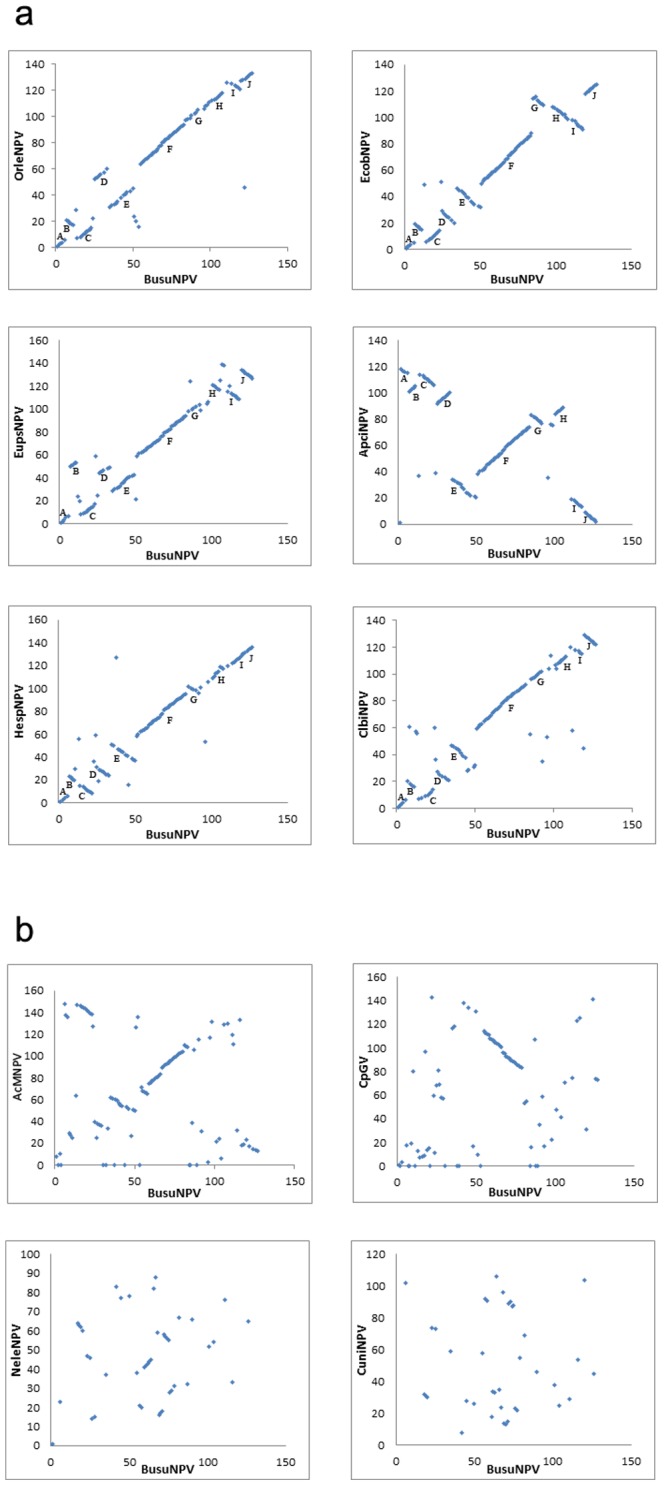
a. Gene-parity plots of BusuNPV with OrleNPV, EupsNPV, ApciNPV, HespNPV, ClbiNPV and EcboNPV based on ORF order. The gene cluster marked by alphabet sorted by their order in BusuNPV. b. Gene-parity plot of BusuNPV with AcMNPV, HearNPV G4, CpGV, NeleNPV and CuniNPV based on ORF order.
Table 1. Collinearly conserved region in alpha- and betabaculoviruses#.
| Gene name | ORF position | ||||
| BusuNPV | AcMNPV | HearNPV | LdMNPV | CpGV | |
| PIF-6* | 55 | 68 | 64 | 80 | 114 |
| LEF-3 | 56 | 67 | 65 | 81 | 113 |
| Desmoplakin* | 57 | 66 | 66 | 82 | 112 |
| DNA-pol* | 58 | 65 | 67 | 83 | 111 |
| ORF-59 | 59 | 75 | 69 | 84 | 108 |
| ORF-60 | 60 | 76 | 70 | 85 | 107 |
| VLF-1* | 61 | 77 | 71 | 86 | 106 |
| P78/83* | 62 | 78 | 72 | 87 | 105 |
| GP41* | 63 | 80 | 73 | 88 | 104 |
| AC81* | 64 | 81 | 74 | 89 | 103 |
| TLP-20$ | 65 | 82 | 75 | 90 | 102 |
| VP91/p95* | 66 | 83 | 76 | 91 | 101 |
| VP39* | 67 | 89 | 78 | 92 | 96 |
| LEF-4* | 68 | 90 | 79 | 93 | 95 |
| P33* | 69 | 92 | 80 | 94 | 93 |
| P18* | 70 | 93 | 81 | 95 | 92 |
| ODV-E25* | 71 | 94 | 82 | 96 | 91 |
| Helicase* | 72 | 95 | 84 | 97 | 90 |
| ODV-E28/PIF-4* | 73 | 96 | 85 | 98 | 89 |
| 38K* | 74 | 98 | 86 | 99 | 88 |
| LEF-5* | 75 | 99 | 87 | 100 | 87 |
| p6.9* | 76 | 100 | 88 | 101 | 86 |
| C42* | 77 | 101 | 89 | 102 | 85 |
| P12 | 78 | 102 | 90 | 103 | 84 |
| P45* | 79 | 103 | 91 | 104 | 83 |
The collinearity was shown by the ORFs orders in BusuNPV, AcMNPV, HearNPV G4, LdMNPV and CpGV. Conserved ORF of all baculovirus are marked by ‘*’.
TLP means Telokin-like protein.
Repeat Structures
Homologous repeated sequences (hrs) were supposed to be characteristic for many baculovirus genomes. The hrs are repeat regions with palindrome structure interspersed in the genome. Hrs consist of similar repeat sequence with varying length in a genome, but the hr sequence vary widely in different baculoviruses [20]. Hrs were suggested to be origins of DNA replication in baculovirus [21], [22], however, a contrasting study showed deletion individual hr had no effect on the replication of AcMNPV [23]. Other studies attributed an enhancer function to hrs. They appear to bind to ie1 in AcMNPV and promote the transactivation level of IE1 [24]–[26]. Hrs are absent from the BusuNPV genome.
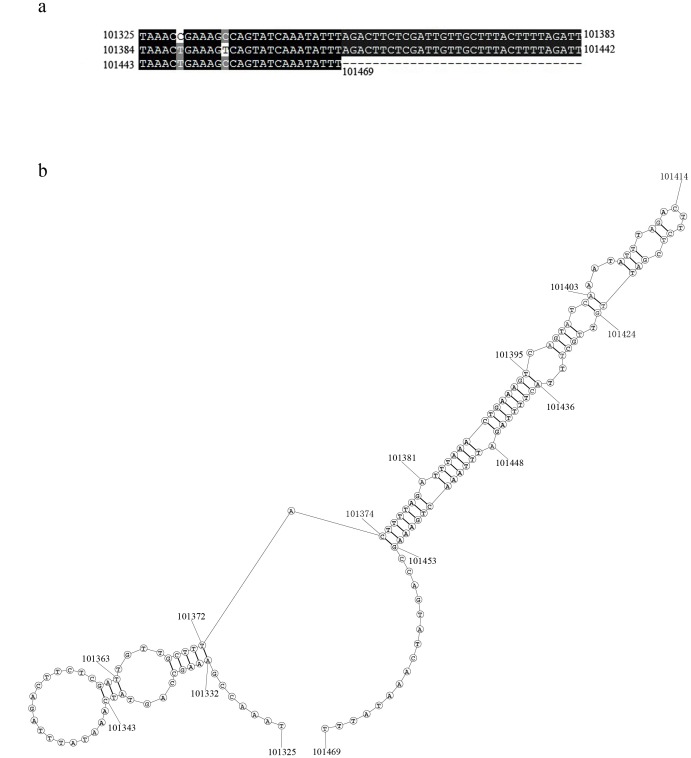
A non-hr origin was also suggested to initiate replication which contains palindromic and repetitive sequences in a complex organization [21], [27]. A repeat sequence was detected from nt 101325 to 101469 in the BusuNPV genome and contained two complete repeats and a truncated repeat. The repeat is 59 nt (Fig. 4a), high in A+T content (71.7%) and probably forms a hairpin structure (Fig. 4b). Whether this is a functional non-hr origin for BusuNPV needs further analysis.
Figure 4. Repeat structure of BusuNPV.
a. Sequence comparing of repeat regions. Blank background shows same bases between 3 compared regions, gray indicates same bases only in 2 regions. b. Predicted secondary structure of overlap repeat region. Numbers on both sides of the chains are the base position in the genome.
Replication Genes
Although the mechanism of baculovirus genome replication is not totally clear, several viral genes have been identified as important for DNA replication [28]. BusuNPV encodes genes essential for replication including DNA polymerase (Busu58), DNA helicase (Busu72), late expression factor-1 (lef-1, Busu126), lef-2 (Busu104) and lef-3 (Busu56). Other genes related to DNA replication include very late factor-1 (vlf-1, Busu61), DNA binding proteins-1,2 (dbp-1, Busu11 and dbp-2, Busu26), lef-11 (Busu28), alkaline exonuclease (alk-exo, busu116) and me-53 (Busu22) [29] have also identified in BusuNPV.
Transcription Genes
Like all other baculovirus, BusuNPV encodes all four subunits of RNA polymerase [30], lef-4 (Busu68), lef-8 (Busu50), lef-9 (Busu35) and P47 (Busu25). The lef-5 (Busu75) and vlf-1 (Busu61) are two other core genes involved in transcription. In addition, four non-core transcription related gene: 39k/pp31 (Busu29), lef-6 (Busu10), lef-11 (Busu28), Protein kinase-1 (pk-1, Busu3) are present in BusuNPV. The early transcription genes found in BusuNPV are Immediate early gene (ie-1, Busu14) and ie-0 (Busu21) [31], [32].
Structural Genes
BusuNPV contains all structural core genes identified in other baculoviruses. In the alphabaculoviruses, p6.9 (Busu76) encodes a nucleocapsid protein and participates in DNA condensation. VLF-1 (Busu61) is a structural protein in both ODV and BV required for very late genes expression and is essential for nucleocapsid production [33], [34]. Other core genes related to the viral nucleocapsid include 38K (Busu74), 49k (Busu20), odv-ec27 (Busu18), odv-e43 (Busu82), odv-e18 (Busu19), vp39 (Busu67), vp91/p95 (Busu66), vp1054 (Busu42), desmoplakin (Busu57), Ac53 (Busu45), p18 (Busu70) and gp41 (Busu63). The p33 (Busu69) encodes a type of a sulfhydryl oxidase in baculoviruses [35]. Proteins encoded by c42 (Busu77) and pp78/83 (Busu2) participate in nuclear actin polymerization [36]. Busu62 encodes a protein similar to Ha72, which be verified essential for ODV occlusion and BV production [37].
Other non-core structural proteins encompass the F protein (Busu120), which is essential for virus entry and budding and VP80 (Busu80), which is involved in nucleocapsid packaging and trafficking [38]. Busu98 is a homologue of Calyx/PEP and is the major protein of polyhedron envelope that enhances the stability of OBs [39], [40]. Busu7 encodes P10 [17] and is involved in the process of OB envelopment and nuclear lysis at the late stages of infection [41].
Oral Infectivity Factors
So far 7 conserved genes were identified to be essential for oral infectivity of baculovirus including p74 (Busu23), per os infectivity factors-1 (pif-1, Busu111), pif-2 (Busu101), pif-3 (Busu90), pif-4 (Busu73), pif-5/odv-e56 (Busu6) and pif-6(Busu55) [42], [43].
Busu34 is a homologue of the gene encoding viral enhancing factor (VEF) that dissolves the peritrophic membrane (PM) of the midgut [44]. A study in LdMNPV found it helps ODV envelopes [45].
Auxiliary Genes
Ubiquitin is encoded by most baculoviruses as well as BusuNPV. Like most alphabaculoviruses and some betabaculoviruses, BusuNPV also encodes cathepsin (Busu24) and chitinase (Busu51), both are involved in liquefaction of insect and OB release [46], [47]. A fibroblast growth factor (FGF, Busu114) aids virus dissemination through the tracheal system [48], [49]. The egt gene which prevents larvae molting and pupation [50], [51] was found in BusuNPV (Busu124) [15] and the baculovirus with deficiency egt gene kill the infected larvae faster than wild type stains [52], [53]. BusuNPV also contains a sod (Busu92) and three iap genes (iap-1, Busu48; iap-2, Busu54; and iap-3, Busu93). Three Baculovirus repeated orf (bro) genes have also been found. The absence or duplication of these genes is common in baculovirus, although between stains with closer affinity [54]. A study on BmNPV showed that mutant bro-d or double mutant bro-a and bro-c could not be isolated, it suggested bro takes essential functions in BmNPV [55]. Another study indicated bro genes encode a protein with DNA binding activity, especially to single stranded DNA [56]. BusuNPV encodes poly (ADP-ribose) glycohydrolase (parg, Busu88), which is conserved in group II alphabaculoviruses with a function of poly (ADP-ribose) catabolism [29]. A study in HearNPV G4 showed it affects oral infectivity of OBs [57].
Unique Genes
Six unique ORFs (Busu5, Busu15, Busu47, Busu100, Busu 109 and Busu110) with no homology to other baculovirus ORFs were identified and potentially encode functional proteins.
The Busu100 encodes a 532 aa protein with low homology to tryptophan repeat gene family in entomopoxvirus (minimum E value = 0.012). Busu109 encodes a 155 aa protein sharing a very low homology to 5-methyltetrahydropteroyltriglutamate–homocysteine methyltransferase in some bacteria (minimum E value = 2.1).
In summary, the genome sequence revealed BusuNPV is a distinct species in group II Alphabaculovirus. Phylogenetically, it is most closely related to EcobNPV, EupsNPV, OrleNPV and ApciNPV. It does not contain typical baculovirus hrs, but contain a new repeat structure, the function of which needs to be further characterized. A 28.8 kb conserved region was identified among alpha- and betabaculoviruses.
Materials and Methods
Viral DNA Extraction
BusuNPV was propagated in B. suppressaria larvae and OBs were purified by differential centrifugation [12]. DNA was extracted as described previously [16].
Sequencing and Bioinformatic Analysis
The genome was sequenced with the Roche 454 GS FLX system by using shotgun strategy. The reads were assembled with Roche GS De Novo assembler software. Contigs assembly was assisted by previously generated restriction maps [13]. A few regions that were not assembled into the contigs were further amplified by PCR, cloned and sequenced. The genome sequence data was uploaded to GenBank (GenBank accession number: KF611977).
Hypothetical ORFs were predicted by softberry FGENESV program (http://www.softberry.com/berry.phtml) [58] to contain the standard ATG start, and a stop codon and potentially encode at least 50 amino acids. Gene-parity plot analysis [13] was performed using Microsoft Office Excel to draw scatter diagram with using BusuNPV ORFs number as the X-axis and other baculovirus ORFs as the Y-axis. Gene annotation and comparisons were done with NCBI protein-protein BLAST algorithm (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Repeat structures were detected by BLAST alignment of two sequences (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The identity among homologous genes was done with MegAlign software using clustalW with default parameters. Regulatory regions and promoter motifs were identified as described previously [29].
Restriction sites were predicted by MapDraw software. Genome map framework drawn with genomeVX [59].
Phylogenetic Analysis
The Phylogenetic analysis was based on amino acid sequences of 37 core genes form BusuNPV and the other 61 baculoviruses listed in NCBI genome database (Table S1). All the sequences were linked by a stationary order and multiple alignments using clusterW method with MEGA5 by using default settings. A phylogenetic tree was constructed by MEGA5 using Maximum Likelihood method based on the JTT matrix-based model [60], [61]. Phylogeny tested by Bootstrap method with a value of 1000 [62].
Prediction of Secondary Structure
Secondary structure was drawn by Predict a Secondary Structure online server (http://rna.urmc.rochester.edu/RNAstructureWeb/Servers/Predict1/Predict1.html) with default setting of DNA Nucleic Acid Type [63].
Supporting Information
Basic informationof all sequenced baculovirus genome in Genbank (October, 2013). NP means no published. Genomes used to build phylogeny tree marked by ‘ *’.
(DOCX)
The ORF positions in the genomeof BusuNPV. E or L means early or late promoter motif and ORF directionrepresented by+ or –.* stands for stain HearNPV G4. a, position of granulin in CpGV genome. b, BJDP stands for DnaJ domain protein. c, PKIP stands forProtein kinase interacting.
(DOCX)
Acknowledgments
The authors would like to thank Qian Chen, Lei Wen, Leiping Zeng, Na Li and Yanbo Ye for their valuable suggestions.
Funding Statement
The research was supported in part by the National Science Foundation of China (31130058,31321001), Chinese Academy of Sciences visiting professorship for senior international scientists (2012T1S0019 to B.A.), and Wuhan Institute of Virology for Virus Resources and Bioinformatics Center. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hayakawa T, Ko R, Okano K, Seong SI, Goto C, et al. (1999) Sequence analysis of the Xestia c-nigrum granulovirus genome. Virology 262: 277–297. [DOI] [PubMed] [Google Scholar]
- 2. Lauzon HA, Lucarotti CJ, Krell PJ, Feng Q, Retnakaran A, et al. (2004) Sequence and organization of the Neodiprion lecontei nucleopolyhedrovirus genome. J Virol 78: 7023–7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Keddie BA, Aponte GW, Volkman LE (1989) The pathway of infection of Autographa californica nuclear polyhedrosis virus in an insect host. Science 243: 1728–1730. [DOI] [PubMed] [Google Scholar]
- 4. Garavaglia MJ, Miele SA, Iserte JA, Belaich MN, Ghiringhelli PD (2012) The ac53, ac78, ac101, and ac103 genes are newly discovered core genes in the family Baculoviridae. J Virol 86: 12069–12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Miele SA, Garavaglia MJ, Belaich MN, Ghiringhelli PD (2011) Baculovirus: molecular insights on their diversity and conservation. Int J Evol Biol 2011: 379424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carstens EB, Ball LA (2009) Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2008). Arch Virol 154: 1181–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jehle JA, Blissard GW, Bonning BC, Cory JS, Herniou EA, et al. (2006) On the classification and nomenclature of baculoviruses: a proposal for revision. Arch Virol 151: 1257–1266. [DOI] [PubMed] [Google Scholar]
- 8. Hefferon KL, Oomens AG, Monsma SA, Finnerty CM, Blissard GW (1999) Host cell receptor binding by baculovirus GP64 and kinetics of virion entry. Virology 258: 455–468. [DOI] [PubMed] [Google Scholar]
- 9. WF IJ, Westenberg M, Goldbach RW, Blissard GW, Vlak JM, et al. (2000) A novel baculovirus envelope fusion protein with a proprotein convertase cleavage site. Virology 275: 30–41. [DOI] [PubMed] [Google Scholar]
- 10. Monsma SA, Oomens AG, Blissard GW (1996) The GP64 envelope fusion protein is an essential baculovirus protein required for cell-to-cell transmission of infection. J Virol 70: 4607–4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pearson MN, Groten C, Rohrmann GF (2000) Identification of the lymantria dispar nucleopolyhedrovirus envelope fusion protein provides evidence for a phylogenetic division of the Baculoviridae. J Virol 74: 6126–6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hu ZH, Liu MF, Jin F, Wang ZX, Liu XY, et al. (1993) Nucleotide sequence of the Buzura suppressaria single nucleocapsid nuclear polyhedrosis virus polyhedrin gene. J Gen Virol 74: 1617–1620. [DOI] [PubMed] [Google Scholar]
- 13. Hu ZH, Arif BM, Jin F, Martens JW, Chen XW, et al. (1998) Distinct gene arrangement in the Buzura suppressaria single-nucleocapsid nucleopolyhedrovirus genome. J Gen Virol 79: 2841–2851. [DOI] [PubMed] [Google Scholar]
- 14. Hu Z, Luijckx T, van Dinten LC, van Oers MM, Hajos JP, et al. (1999) Specificity of polyhedrin in the generation of baculovirus occlusion bodies. J Gen Virol 80: 1045–1053. [DOI] [PubMed] [Google Scholar]
- 15. Hu ZH, Broer R, Westerlaken J, Martens JW, Jin F, et al. (1997) Characterization of the ecdysteroid UDP-glucosyltransferase gene of a single nucleocapsid nucleopolyhedrovirus of Buzura suppressaria. Virus Res 47: 91–97. [DOI] [PubMed] [Google Scholar]
- 16. Hu ZH, Arif BM, Sun JS, Chen XW, Zuidema D, et al. (1998) Genetic organization of the HindIII-I region of the single-nucleocapsid nucleopolyhedrovirus of Buzura suppressaria. Virus Res 55: 71–82. [DOI] [PubMed] [Google Scholar]
- 17. van Oers MM, Hu Z, Arif BM, van Strien EA, van Lent JW, et al. (1998) The single-nucleocapsid nucleopolyhedrovirus of Buzura suppressaria encodes a P10 protein. J Gen Virol 79 (Pt 6): 1553–1562. [DOI] [PubMed] [Google Scholar]
- 18.Yang XY, Wu J (1981) A check list of the forest insects of china. Beijing: China forestry publishing house. 188 p. [Google Scholar]
- 19. Thumbi DK, Eveleigh RJ, Lucarotti CJ, Lapointe R, Graham RI, et al. (2011) Complete sequence, analysis and organization of the Orgyia leucostigma nucleopolyhedrovirus genome. Viruses 3: 2301–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferrelli ML, Berretta MF, Belaich MN, Ghiringhelli PD, Sciocco-Cap A, et al.. (2012) The Baculoviral Genome. 10p.
- 21. Kool M, Voeten JT, Goldbach RW, Tramper J, Vlak JM (1993) Identification of seven putative origins of Autographa californica multiple nucleocapsid nuclear polyhedrosis virus DNA replication. J Gen Virol 74 (Pt 12): 2661–2668. [DOI] [PubMed] [Google Scholar]
- 22. Hilton S, Winstanley D (2008) The origins of replication of granuloviruses. Arch Virol 153: 1527–1535. [DOI] [PubMed] [Google Scholar]
- 23. Carstens EB, Wu Y (2007) No single homologous repeat region is essential for DNA replication of the baculovirus Autographa californica multiple nucleopolyhedrovirus. J Gen Virol 88: 114–122. [DOI] [PubMed] [Google Scholar]
- 24. Guarino LA, Gonzalez MA, Summers MD (1986) Complete Sequence and Enhancer Function of the Homologous DNA Regions of Autographa californica Nuclear Polyhedrosis Virus. J Virol 60: 224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Choi J, Guarino LA (1995) The baculovirus transactivator IE1 binds to viral enhancer elements in the absence of insect cell factors. J Virol 69: 4548–4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rodems SM, Friesen PD (1995) Transcriptional enhancer activity of hr5 requires dual-palindrome half sites that mediate binding of a dimeric form of the baculovirus transregulator IE1. J Virol 69: 5368–5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu Y, Carstens EB (1996) Initiation of baculovirus DNA replication: early promoter regions can function as infection-dependent replicating sequences in a plasmid-based replication assay. J Virol 70: 6967–6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mikhailov V (2003) Replication of the baculovirus genome. Molecular Biology 37: 250–259. [PubMed] [Google Scholar]
- 29.Rohrmann GF (2011) Baculovirus molecular biology. Bethesda (MD): NationalLibrary of Medicine (US), National Center for Biotechnology Information; 2011.
- 30. Guarino LA, Xu B, Jin J, Dong W (1998) A virus-encoded RNA polymerase purified from baculovirus-infected cells. J Virol 72: 7985–7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Luria N, Lu L, Chejanovsky N (2012) Conserved structural motifs at the C-terminus of baculovirus protein IE0 are important for its functions in transactivation and supporting hr5-mediated DNA replication. Viruses 4: 761–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chisholm GE, Henner DJ (1988) Multiple early transcripts and splicing of the Autographa californica nuclear polyhedrosis virus IE-1 gene. J Virol 62: 3193–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yang S, Miller LK (1998) Expression and mutational analysis of the baculovirus very late factor 1 (vlf-1) gene. Virology 245: 99–109. [DOI] [PubMed] [Google Scholar]
- 34. Vanarsdall AL, Okano K, Rohrmann GF (2006) Characterization of the role of very late expression factor 1 in baculovirus capsid structure and DNA processing. J Virol 80: 1724–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wu W, Passarelli AL (2010) Autographa californica multiple nucleopolyhedrovirus Ac92 (ORF92, P33) is required for budded virus production and multiply enveloped occlusion-derived virus formation. J Virol 84: 12351–12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li K, Wang Y, Bai H, Wang Q, Song J, et al. (2010) The putative pocket protein binding site of Autographa californica nucleopolyhedrovirus BV/ODV-C42 is required for virus-induced nuclear actin polymerization. J Virol 84: 7857–7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang H, Wang M, Deng F, Hou D, Arif BM, et al.. (2013) Baculovirus core gene ha72 is essential for BV production and ODV occlusion and amino acid K22 plays an important role in its function. Journal of virology: JVI. 02281–02213. [DOI] [PMC free article] [PubMed]
- 38. Marek M, Merten OW, Galibert L, Vlak JM, van Oers MM (2011) Baculovirus VP80 protein and the F-actin cytoskeleton interact and connect the viral replication factory with the nuclear periphery. J Virol 85: 5350–5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gombart AF, Pearson MN, Rohrmann GF, Beaudreau GS (1989) A baculovirus polyhedral envelope-associated protein: genetic location, nucleotide sequence, and immunocytochemical characterization. Virology 169: 182–193. [DOI] [PubMed] [Google Scholar]
- 40. Whitt MA, Manning JS (1988) A phosphorylated 34-kDa protein and a subpopulation of polyhedrin are thiol linked to the carbohydrate layer surrounding a baculovirus occlusion body. Virology 163: 33–42. [DOI] [PubMed] [Google Scholar]
- 41. Williams GV, Rohel DZ, Kuzio J, Faulkner P (1989) A cytopathological investigation of Autographa californica nuclear polyhedrosis virus p10 gene function using insertion/deletion mutants. J Gen Virol 70 (Pt 1): 187–202. [DOI] [PubMed] [Google Scholar]
- 42. Nie Y, Fang M, Erlandson MA, Theilmann DA (2012) Analysis of the Autographa californica multiple nucleopolyhedrovirus overlapping gene pair lef3 and ac68 reveals that AC68 is a per os infectivity factor and that LEF3 is critical, but not essential, for virus replication. J Virol 86: 3985–3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Huang H, Wang M, Deng F, Wang H, Hu Z (2012) ORF85 of HearNPV encodes the per os infectivity factor 4 (PIF4) and is essential for the formation of the PIF complex. Virology 427: 217–223. [DOI] [PubMed] [Google Scholar]
- 44. Wang P, Granados RR (1997) An intestinal mucin is the target substrate for a baculovirus enhancin. Proceedings of the National Academy of Sciences 94: 6977–6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Slavicek JM, Popham HJ (2005) The Lymantria dispar nucleopolyhedrovirus enhancins are components of occlusion-derived virus. J Virol 79: 10578–10588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chukhry MG (1989) The process of baculovirus deproteinization. Acta Virol 33: 559–564. [PubMed] [Google Scholar]
- 47. Slack JM, Kuzio J, Faulkner P (1995) Characterization of v-cath, a cathepsin L-like proteinase expressed by the baculovirus Autographa californica multiple nuclear polyhedrosis virus. J Gen Virol 76 (Pt 5): 1091–1098. [DOI] [PubMed] [Google Scholar]
- 48. Means JC, Passarelli AL (2010) Viral fibroblast growth factor, matrix metalloproteases, and caspases are associated with enhancing systemic infection by baculoviruses. Proc Natl Acad Sci U S A 107: 9825–9830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Detvisitsakun C, Cain EL, Passarelli AL (2007) The Autographa californica M nucleopolyhedrovirus fibroblast growth factor accelerates host mortality. Virology 365: 70–78. [DOI] [PubMed] [Google Scholar]
- 50. O'Reilly DR, Miller LK (1990) Regulation of expression of a baculovirus ecdysteroid UDPglucosyltransferase gene. Journal of virology 64: 1321–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. O'Reilly DR, Miller LK (1989) A baculovirus blocks insect molting by producing ecdysteroid UDP-glucosyl transferase. Science 245: 1110–1112. [DOI] [PubMed] [Google Scholar]
- 52. Georgievska L, Hoover K, Werf Wvd, Muñoz D, Caballero P, et al. (2010) Dose dependency of time to death in single and mixed infections with a wildtype and egt deletion strain of Helicoverpa armigera nucleopolyhedrovirus. Journal of invertebrate pathology 104: 44–50. [DOI] [PubMed] [Google Scholar]
- 53. Simon O, Williams T, Lopez-Ferber M, Caballero P (2012) Deletion of egt is responsible for the fast-killing phenotype of natural deletion genotypes in a Spodoptera frugiperda multiple nucleopolyhedrovirus population. J Invertebr Pathol 111: 260–263. [DOI] [PubMed] [Google Scholar]
- 54. Zhou JB, Li XQ, De-Eknamkul W, Suraporn S, Xu JP (2012) Identification of a new Bombyx mori nucleopolyhedrovirus and analysis of its bro gene family. Virus Genes 44: 539–547. [DOI] [PubMed] [Google Scholar]
- 55. Kang W, Suzuki M, Zemskov E, Okano K, Maeda S (1999) Characterization of baculovirus repeated open reading frames (bro) in Bombyx mori nucleopolyhedrovirus. J Virol 73: 10339–10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zemskov EA, Kang W, Maeda S (2000) Evidence for nucleic acid binding ability and nucleosome association of Bombyx mori nucleopolyhedrovirus BRO proteins. J Virol 74: 6784–6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Luo S, Zhang Y, Xu X, Westenberg M, Vlak JM, et al. (2011) Helicoverpa armigera nucleopolyhedrovirus occlusion-derived virus-associated protein, HA100, affects oral infectivity in vivo but not virus replication in vitro. J Gen Virol 92: 1324–1331. [DOI] [PubMed] [Google Scholar]
- 58. Solovyev VV, Salamov AA (1999) INFOGENE: a database of known gene structures and predicted genes and proteins in sequences of genome sequencing projects. Nucleic Acids Res 27: 248–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Conant GC, Wolfe KH (2008) GenomeVx: simple web-based creation of editable circular chromosome maps. Bioinformatics 24: 861–862. [DOI] [PubMed] [Google Scholar]
- 60. Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8: 275–282. [DOI] [PubMed] [Google Scholar]
- 61. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sanderson MJ, Wojciechowski MF (2000) Improved bootstrap confidence limits in large-scale phylogenies, with an example from Neo-Astragalus (Leguminosae). Syst Biol 49: 671–685. [DOI] [PubMed] [Google Scholar]
- 63. Reuter JS, Mathews DH (2010) RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics 11: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Basic informationof all sequenced baculovirus genome in Genbank (October, 2013). NP means no published. Genomes used to build phylogeny tree marked by ‘ *’.
(DOCX)
The ORF positions in the genomeof BusuNPV. E or L means early or late promoter motif and ORF directionrepresented by+ or –.* stands for stain HearNPV G4. a, position of granulin in CpGV genome. b, BJDP stands for DnaJ domain protein. c, PKIP stands forProtein kinase interacting.
(DOCX)