Abstract
Several cellular transcription factors have been shown to be involved in IE62-mediated activation. The YY1 cellular transcription factor has activating and repressive effects on gene transcription. Analysis of the VZV genome revealed 19 postulated YY1 binding sites located within putative promoters of 16 VZV genes. Electrophoretic mobility shift assays (EMSA) confirmed the binding of YY1 to ORF10, ORF28/29 and gI promoters and the mutation of these binding sites inhibited YY1 binding and the promoter activation by IE62 alone or following VZV infection. Mutation of the ORF28/29 YY1 site in the VZV genome displayed insignificant influence on virus growth in melanoma cells; but it inhibited the virus replication significantly at day 5 and 6 post infection in HELF cells. This work suggests a novel role for the cellular factor YY1 in VZV replication through the mediation of IE62 activation of viral gene expression.
Keywords: IE62, VZV, YY1, ORF10, ORF28/29, gI, Transcription, promoters
INTRODUCTION
Varicella-zoster virus (VZV) is a neurotropic herpesvirus and member of the family alphaherpesvirinae. It causes two diseases, varicella (chickenpox) during primary infection and herpes zoster (shingles) upon reactivation from latency in sensory ganglia. The VZV genome consists of a 125 kb linear double-stranded DNA molecule that encodes at least 71 genes (Cohen et al., 2007). All of the VZV genes are believed to be expressed during lytic infection in three kinetic classes, immediate early (IE), early and late. Like other herpesviruses, VZV uses the host cell RNA polymerase II (RNA Pol II) and the general transcription apparatus of the cell for viral gene transcription. A few VZV proteins, including IE62, IE4, ORF61, IE63 and ORF10 are responsible for efficient viral gene expression (Cohen et al., 2007). IE62, the primary viral transactivator, regulates the expression of genes from all three putative kinetic classes (Kinchington et al., 2000).
Several cellular transcription factors have been shown to be involved in the regulation of VZV gene expression mediated by IE62. Sp1 family members target the GC rich sequence within VZV promoters and interact with IE62 (Peng et al., 2003; and Ruyechan et al., 2003). The presence of a GC rich sequence that binds Sp1 has been found in gE, gI and ORF28/29 promoters (Beraraducci et al., 2008; Peng et al., 2003; and Yang et al., 2004). Sp1 and Sp3 bind to the downstream region of VZV oriS and to the ORF3 promoter; mutation of their binding sites inhibited ORF62, ORF63 and ORF3 expression in reporter gene assays (Khalil et al., 2012; and Khalil et al., 2013).
The upstream sequence factor, USF, is another cellular factor involved in IE62-mediated gene expression (Rahaus et al., 2003). The USF consensus binding sequence (5′-CACGTG-3′) is present in some VZV promoters and IE62 interacts with USF (Rahaus et al., 2003). The ORF10 and ORF28/29 promoter sequences have USF sites and mutation of these sites inhibited gene expression in reporter gene assays (Che et al., 2007; Yang et al., 2004; and Yang et al., 2006). Yang et al., (2008) showed that the human mediator complex is also an essential component for efficient VZV gene expression. The physical interaction between the N-terminal acidic activation domain (TAD) of IE62 and the factors involved in the formation of the human mediator complex has been demonstrated (Yang et al., 2008 and Yamamoto et al., 2009).
Other classes of mammalian transcription factors have been identified but their role in transcriptional regulation is not well understood. Yin Yang1 or YY1 is a cellular transcription factor discovered about 22 years ago (Shi et al., 1991). YY1 belongs to the GLI-Kruppel class of zinc finger proteins that are ubiquitously expressed and play an important role in biological processes such as embryogenesis, differentiation, replication and cellular proliferation (Yang et al., 1995; and Gordon et al., 2006). The YY1 protein contains four C2H2 type zinc finger domains near the C-terminal half of the protein responsible for the DNA binding activity and two acidic activation domains near the N-terminal half that are capable of transcriptional activation (Hyde-DeRuyscher et al., 1995; and Yant et al., 1995). YY1 protein is responsible for activating or repressing a diverse group of promoters depending on the promoter architecture, which is the origin of the name Yin Yang (Shi et al., 1991). YY1 has also been shown to interact with other cellular factors including Sp1, Histone deacetylase2, ATF6 and Notch1 (Seto et al., 1993; Yao et al., 2001; Li et al., 2000; and Yeh et al., 2003).
Two YY1 protein consensus binding sites were identified recently in the VZV genome; the first one is in the downstream region of VZV oriS (5′CAAATGGCG-3′) and the other is within the ORF3 promoter (5′CCCATATAT-3′) (Khalil et al., 2008; Khalil et al., 2012; and Khalil et al., 2013). The mutation of the YY1 site downstream of VZV oriS had no significant effect on VZV DNA replication or ORF62 and ORF63 transcription efficiency in DpnI replication and reporter gene assays respectively although it increased the VZV growth in skin xenografts slightly at day 21 post-infection (Khalil et al., 2012). In contrast, mutation of the YY1 site in the ORF3 promoter inhibited expression in reporter gene assays (Khalil et al., 2013).
In the work presented here, we investigated the role of YY1 binding sites within VZV promoters in activation of IE62-dependent gene expression in order to better understand mechanisms that IE62 uses to exploit the host cell environment for efficient viral gene expression. The binding of YY1 to sites within the putative VZV promoters of ORF10, ORF28/29 and gI was established using EMSA and supershift assays. The mutation of these YY1 sites ablated formation of the YY1 specific complex and inhibited activation of these promoters. A VZV recombinant virus with a mutation of the YY1 site in the ORF28/29 promoter was constructed and effects on virus growth were studied. This mutation was found to have no significant effect on VZV replication in melanoma cells and on the ORF29 expression. On the other hand, this mutation inhibited VZV replication in HELF significantly at day 5 and 6 post infection. This work suggests that the cellular transcription factor YY1 acts as a cellular factor involved in the IE62-mediated activation of VZV genes.
MATERIALS AND METHODS
Cells and viruses
MeWo cells, a human melanoma cell line, and HELF cells, a human embryonic lung fibroblast cell line, were grown in Eagle’s minimal essential medium supplemented with 10% fetal bovine serum (Spengler et al., 2000). VZV strains MSP and pOka were propagated in MeWo cell and HELF cell monolayers as described by Lynch et al. (2002) and Peng et al. (2003).
Nuclear and whole cell lysate preparation and immunoblot analysis
Nuclear extracts of VZV infected MeWo cells were prepared as previously described (Lynch et al., 2002). MeWo cells were incubated in buffer A (10 mM HEPES, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol) at 4 °C for 15 min to lyse the cells and release the cytoplasmic fraction. After centrifugation, the crude nuclear pellet was incubated on ice in buffer C (20 mM HEPES, pH 7.9, 25% (v/v) glycerol, 0.42 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride, 0.5 mM dithiothreitol). After centrifugation, the nuclear extract was dialyzed against buffer D (20mM HEPES, pH 7.9, 20% (v/v) glycerol, 0.1 M KCl, 0.2 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride, 0.5 mM dithiothreitol).
Whole cell lysates of VZV infected MeWo cells were prepared in lysis buffer (50 mM Tris-HCl, pH 7.5, 0.15 M NaCl, 1 mM EDTA, 0.1% Triton X-100 and protease inhibitor cocktail (Roche, Mannheim, GE) added per the manufacturer’s instructions) and analyzed for ORF29 by immunoblot. Rabbit polyclonal antisera against the C-terminus half of the ORF29 protein by immunoblot (10% SDS-PAGE) using rabbit polyclonal antibody against the C-terminus of ORF29 protein (Peng et al., 2003; and Kinchington et al., 1988). Mouse monoclonal antibody against α-tubulin was obtained from Sigma-Aldrich (St. Louis, MO). Quantification of the relative amounts of ORF29 and α-tubulin was done using a BioRad GS700 Imaging Densitometer (BioRad Hercules, CA). Statistical significance was determined by one-way ANOVA analysis of variance followed by Tukey’s post hoc test.
Plasmids
The luciferase reporter plasmids containing the ORF10, ORF28/29 and gI promoters were constructed as described (Che et al., 2007; Yang et al., 2004 and White et al., 2010). The wild type ORF10-Luc was constructed by inserting a 242 bp intergenic region between ORF9 and ORF10 into the pGL3 basic vector flanked by firefly luciferase. The wild type R28/29F was constructed by inserting a 221 bp intergenic region between ORF28 and ORF29 into the basic pGL2 luciferase vector containing the Renilla and firefly luciferase reporters. The wild type gI-Luc was constructed by inserting the gI (VZV ORF67) promoter sequence into the pGL2 basic vector flanked by firefly luciferase.
The plasmids containing the YY1 site specific mutations within the ORF10, ORF28/29 and gI promoters were generated from the wild type plasmids containing the wild type promoter sequences using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, LaJolla, CA). The primer sets for these mutations were: ORF10 promoter YY1 site: 5′-TCAGTTGCTACCAAACAAACCAAATTAGACGGCGGGTTTTGATAA-3′ and 5′-TTATCAAAACCCGCCGTCTAATTTGGTTTGTTTGGTAGCAACTGA-3′; ORF28/29 promoter YY1 site: 5′-TTGACCCTGCCAACAACCCCAAATTATTACGAGT ACTTCACCAAA-3′ and 5′-TTTGGTGAAGTACTCGTAATAATTTGGGGTTGTTGGC AGGGTCAA-3′; and gI promoter YY1 site: 5′-AACTTAATACAGAGTCACGCCCCGTT ACAACAAGGATAAAACACG-3′ and 5′-CGTGTTTTATCCTTGTTGTAACGGGGCGT GACTCTGTATTAAGTT-3′. The mutated nucleotides are indicated in bold. All primers were synthesized by IDT (Coralville, IA). The mutations were verified by sequencing at the Roswell Park Cancer Institute sequencing facility, Buffalo NY.
The pCMV62 plasmid expressing ORF62 under the control of the cytomegalovirus immediate-early (IE) promoter has been described previously (Perera et al., 1992 and 1993).
Reporter gene assays
Luciferase reporter gene assay experiments were performed in MeWo cells as previously described (Yang et al., 2004). Transfections were performed using 12-well plates. 2 × 105 MeWo cells were seeded in each well 24 h before transfection. Cells were transfected with one microgram of each reporter vector using Lipofectamine reagent (Invitrogen, Carlsbad, CA), along with 5 ng of pEF1α-RL plasmid (Promega, Madison, WI) for ORF10 and gI promoter experiments or 0.4 μg of β-galactosidase (β-Gal)-expressing plasmid (Invitrogen, Carlsbad, CA) for ORF28/29 promoter experiments as a control of transfection efficiency. The cells were super infected with VZV MSP 24 hr post transfection 0.4 infected cells per 1 uninfected cell.
In the experiments done in the presence of IE62, the reporter plasmids were co-transfected with 5 ng of pCMV-ORF62 expressing plasmids. Dual luciferase activities were normalized to the Renilla luciferase activities or β-Gal. pcDNA was transfected along with the pCMV62 plasmid to equalize the amounts of both total DNA and the promoter construct in each set of transfections.
The cells were lysed 48 h post transfection or super-infection in 250 μl of lysis buffer (50 mM HEPES, pH 7.4, 250 mM NaCl, 1% NP-40, 1 mM EDTA). Control experiments without transfection of pCMV-ORF62 or VZV infection were done for each plasmid to determine basal expression levels. Dual-luciferase assays were performed according to the manufacturer’s instruction. Transfection experiments were repeated at least three times.
EMSA and supershift analyses
40 bp oligonucleotide probes (IDT, Coralville, IA) containing wild type and mutant YY1 binding sites were used in electrophoretic mobility shift assays (EMSAs). Probes were end labeled with ATP [α-32P] using T4 kinase (Invitrogen, Carlsbad, CA). One hundred femtomoles of the 40 bp labeled probes containing either the wild type or the mutant sequences (~1 × 105 dpm) were incubated with 15 μg of VZV infected MeWo cell nuclear extract in a 10 μl reaction mixture in binding buffer: 40 mM HEPES, pH 7.9, 100 mM NaCl, 10 mM MgCl2, 200 μg/ml bovine serum albumin (BSA), 12% glycerol, 0.05% NP-40, 1 mM dithiothreitol, and 3 μg poly (dI.dC). The samples were analyzed by electrophoresis on a 5% polyacrylamide (37.5:1 acrylamide/bisacrylamide) gel followed by autoradiography. In competition assays, the ratio of cold probe to labeled probe was 100:1. In the supershift assays, rabbit polyclonal YY1 antiserum, generously supplied by Dr. Te-Chung Lee (University at Buffalo) was added in 2 μl (YY1) aliquots to reaction mixtures before the addition of the labeled probe to the nuclear extract-antibody mixture. Quantification of the relative amounts of YY1 complexes was done using a BioRad GS700 Imaging Densitometer (BioRad Hercules, CA).
Generation and growth kinetics of a pOka recombinant virus with mutation in the YY1 site of the ORF28/29 promoter
Recombinant virus was produced using the self-excisable pOka-BAC (pOka – parental Oka) as described previously (Oliver et al., 2009; and Tischer et al., 2007). The mutagenic primers for the YY1 site were: ORF28/29 mut-F 5′-ACAGACTGGGTTTTGGGTGGTCATTTGACCCTGCCAACAA CCCCAAATTATTACGAGTACTTCACAGGATGACGACGATAAGTAGGG-3′ and ORF28/29 mut-R 5′-AGTCTTCTGAGTATTTTCCATTTTGGTGAAGTACTCGTAAT AATTTGGGGTTGTTGGCAGGGTCACAACCAATTAACCAATTCTGATTAG-3′. The changed nucleotides are indicated in bold italics. The primers were used to amplify the Kanr gene from the pKan-EP-S vector using Accuprime Pfx polymerase (Invitrogen, Carlsbad, CA). The PCR products were cloned into pCR4-TOPO after the addition of adenosine overhangs using recombinant Taq polymerase (Invitrogen, Carlsbad, CA). Clones were sequenced to determine that the VZV specific sequences contained the desired changes and did not have any unexpected deletions or substitutions. The Kanr cassette flanked with the VZV sequences were amplified from the pCR4 vectors using short primers (ORF28/29 promoter-F PCR 5′-ACAGACTGGGTTTTGGGTGGTCATTTGACC-3′ and ORF28/29 promoter-R PCR. 5′-AGTCTTCTGAGTATTTTCCATTTTGGTG-3′ to generate high yields of the PCR product. PCR products were gel purified (QIAGEN, Inc., Valencia, CA) and then used for recombination. The recombination steps were screened by PCR for the addition and subsequent removal of the Kanr cassette. Positive clones were purified using a Large-construct kit (QIAGEN, Inc., Valencia, CA). All purified BACs were digested with HindIII to ensure the expected DNA fragments were present and sequenced to verify the changed nucleotides.
Transfections were done as previously described (Niizuma et al., 2003). DNA was extracted from infected cells using DNazol (Invitrogen) and used as a template for PCR using the following primers PCR ORF28-29 F 5′-TAAGTGTACCGACGTGA ACC-3′ and PCR ORF28-29 R 5′ ACCCGTAGTGCGTGCTCCAG-3′ and the mutation in the ORF28/29 promoter was verified by sequencing using the above PCR primers (Quintarabio. Berkeley, CA). Excision of the Mini F vector from the Bac-derived virus was done as previously described (Oliver et al., 2009).
The replication kinetics of recombinant viruses was assessed by an infectious focus assay with immunostaining to detect plaques as previously described (Chaudhuri et al., 2008; and Moffat et al., 1995). Briefly, 6-well assay plates and 24-well titer plates were seeded with MeWo and HELF cells. Assay plates were incubated for variable times, and several dilutions of the samples were taken for infectious focus assay. Titer plates were incubated for 4 days then fixed with 4% paraformaldehyde for immunohistochemical staining using anti-VZV monoclonal antibody (Meridian). Statistical differences in growth kinetics were determined by Student’s t test.
RESULTS
Predicted YY1 binding sites within putative VZV promoters
As the first step in studying the role of YY1 in regulation of viral gene expression, the number of predicted YY1 binding sites in putative VZV promoters was determined. Since there is no degenerate consensus binding site for the YY1 protein, we used the YY1 binding sites previously identified in the VZV genome as the template in the bioinformatics search. These were 5′-CAAATGG-3′ in the downstream region of VZV oriS and 5′-CCCATATAT-3′ in the ORF3 promoter (Khalil et al., 2008; Khalil et al., 2013). The search was done with NCBI Blastn program (http://blast.ncbi.nlm.nih.gov/Blast.cgi). In order to estimate the number of predicted YY1 sites, we used a conservative promoter size of 200 to 300 bp upstream from the translational start sites of the VZV genes. Nineteen YY1 sites were identified within putative promoter regions (Table 1). Three of the 19 promoters that had been studied previously, including those of ORF10, ORF28/29 and gI genes were selected to study the binding of YY1 and the influence of these predicted YY1 binding sites on regulation of the expression of these genes.
Table 1.
List of the VZV promoters containing predicted YY1 binding site.
| VZV gene | No. of YY1 site | YY1 site Sequence |
|---|---|---|
| ORF3 | 1 | CCCATATA |
| ORF4 | 1 | AATGGG |
| ORF7 | 1 | CCCATTTTT |
| ORF10 | 1 | CCCCATT |
| ORF11 | 1 | CCCATATA |
| ORF15 | 1 | CCCCCCCATTTT |
| ORF28/29 | 1 | CCCCCCATT |
| ORF30 | 1 | CCCATAT |
| ORF35 | 1 | AAAAATGGG |
| ORF42 | 1 | CCCATTT |
| ORF49 | 1 | CCCATT |
| ORF61 | 1 | CCCATTT |
| ORF63, 70 | 3 | CCCCCATT, CCATTTT, AAATGG |
| ORF67 | 1 | CCCCATT |
YY1 binds to the predicted YY1 binding sites within ORF10, ORF28/29 and gI promoters
In the first set of experiments, we used EMSA and supershift assays to establish the binding of YY1 to its predicted binding sites within VZV ORF10, ORF28/29 and gI promoters. We performed our experiments using 40 bp duplex oligonucleotides containing wild type and mutant YY1 sites within these promoters and nuclear extracts from VZV infected MeWo cells. 30 bp duplex oligonucleotides containing the origin binding protein (OBP) box A sequence in the ORF62/ORF63 intergenic region was used as the nonspecific competitor (Khalil et al., 2008). The sequence of the duplex oligonucleotides used in the EMSA and supershift assays are listed in Table 2.
Table 2.
List and sequences of the duplex oligonucleotides used in the EMSA and supershift assays.
| Oligonucleotide | Sequence |
|---|---|
|
| |
| Wild Type ORF10 | AGTTGCTACCAAACAAACCCCATTAGACGGCGGGTTTTGA |
| YY1 Mutant ORF10 | AGTTGCTACCAAACAAACCAAATTAGACGGCGGGTTTTGA |
| Wild Type ORF28/29 | GACCCTGCCAACAACCCCCCATTATTACGAGTACTTCACC |
| YY1 Mutant ORF28/29 | GACCCTGCCAACAACCCCAAATTATTACGAGTACTTCACC |
| Wild Type gI probe | CTTAATACAGAGTCACGCCCCATTACAACAAGGATAAAAC |
| YY1 Mutant gI probe | CTTAATACAGAGTCACGCCCCGTTACAACAAGGATAAAAC |
| Box A probe | GTCCAACCACCGTTCGCACTTTCTTTCTAT |
YY1 site indicated in bold. Mutated nucleotides indicated in bold italics.
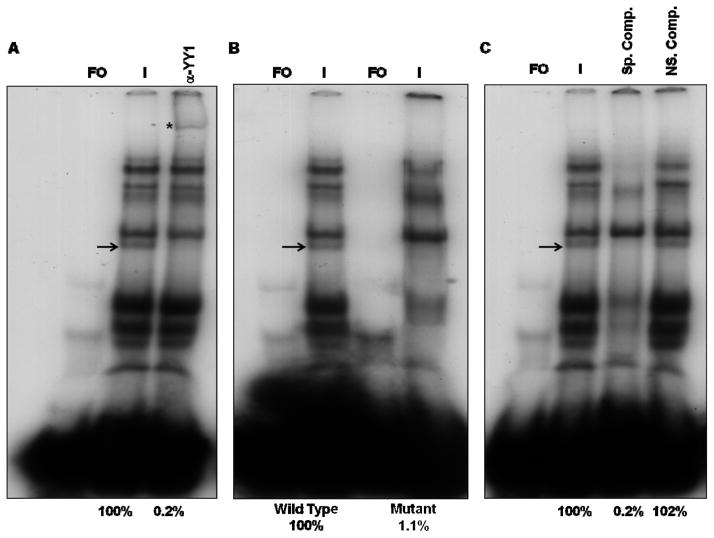
EMSA and supershift assays using the oligonucleotides containing the predicted YY1 binding site of the ORF10 promoter revealed multiple complexes including two major faster migrating complexes and several minor slowly migrating complexes (Fig. 1A). Antibody supershift assays were then performed to assess the binding of the YY1 cellular transcription factors to this site. The anti-YY1 antibody supershifted one of the minor complexes formed (Fig. 1A).
FIG. 1.
EMSA and supershift assays using 40 bp oligonucleotides containing the wild-type YY1 site of the ORF10 promoter. A) Supershift assays were done in the presence of VZV-infected cell nuclear extracts using polyclonal antibodies against YY1. B) EMSA assays using wild type and YY1 mutant oligonucleotides. C) EMSA and competition assays using Box A containing oligonucleotides as non specific competitor. Lanes: FO, free oligonucleotide; I, VZV-infected nuclear extract. The positions of the YY1 containing complexes are indicated by arrows and the positions of the supershifted bands are indicated by asterisks. The numbers under the figures represent the relative amounts of YY1 complexes, assessed by densitometry.
In the next series of experiments, EMSAs were performed using oligonucleotides containing the mutation in the YY1 binding site of the ORF10 promoter that AliBaba2.1 and Patch programs (http://www.gene-regulation.com/pub/programs.html; Biobase) predicted to inhibit the binding of YY1. The mutant probe has a substitution of the CC residues with AA in the YY1 binding site. As shown in Fig. 1B, this mutation inhibited the formation of the identified YY1 complex and also affected the formation of some of the major complexes suggesting that these complexes may need the presence of the YY1 binding site.
To test the specificity of the formation of the YY1 containing complex with ORF10 containing nucleotides, competition EMSA experiments were done using unlabeled oligonucleotides containing the YY1 binding site of the ORF10 promoter as the specific competitor and a 30-bp oligonucleotides containing the origin binding protein (OBP) box A sequence described above. As shown in Fig. 1C, the cold specific competitor efficiently competed away the formation of the YY1 containing complex and some major complexes, while the presence of the nonspecific competitor had no effect on the shift pattern. These experiments indicated that cellular transcription factor YY1 binds specifically to the ORF10 promoter element.
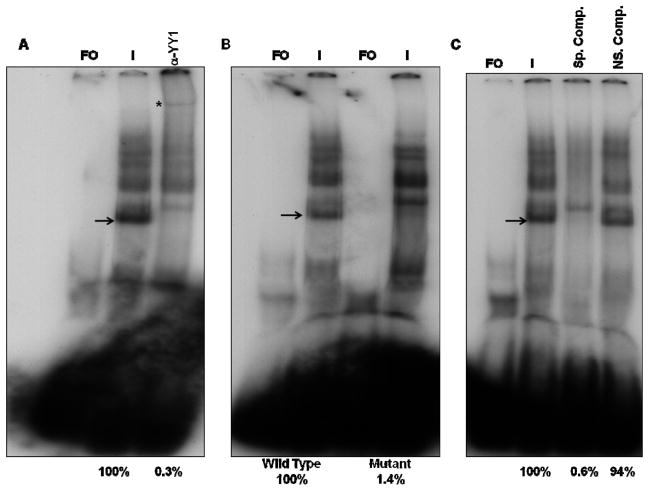
Next, a similar series of experiments was carried out to detect the binding of YY1 to the ORF28/29 promoter and to test the specificity of this binding. Numerous complexes formed using the YY1 site-containing oligonucleotides from the ORF28/29 promoter, of which the major complex is the fastest migrating complex (Fig. 2A). Antibody supershift assays were then performed to determine the binding of the YY1 cellular transcription factors to these sequences. The anti-YY1 antibody supershifted the major complex (Fig. 2A).
FIG. 2.
EMSA and supershift assays using 40 bp oligonucleotides containing the wild-type YY1 site of the ORF28/29 promoter. A) Supershift assays were done in the presence of VZV-infected cell nuclear extracts using polyclonal antibodies against YY1. B) EMSA assays using wild type and YY1 mutant oligonucleotides. C) EMSA and competition assays using Box A containing oligonucleotides as non specific competitor. Lanes: FO, free oligonucleotide; I, VZV-infected nuclear extract. The positions of the YY1 containing complexes are indicated by arrows and the positions of the supershifted bands are indicated by asterisks. The numbers under the figures represent the relative amounts of YY1 complexes, assessed by densitometry.
In the next series of experiments, EMSAs were performed using oligonucleotides containing a mutation in the YY1 binding site predicted to inhibit the binding of YY1. The mutant probe contained a substitution of CC residues to AA in the YY1 binding site similar to that in the ORF10 promoter. As shown in Fig. 2B, this mutation not only inhibited the formation of the identified YY1 complex as predicted but also enhanced the formation of minor complexes suggesting that these factors are competing with YY1 for binding to this probe.
Next, competition EMSA experiments were done using unlabeled 40-bp oligonucleotides containing the YY1 binding site from the ORF28/29 promoter as the specific competitor, and the Box A containing oligonucleotides as non-specific competitor. As shown in Fig. 2C, the cold specific competitor not only competed away the formation of the YY1 containing complex but also some minor complexes, while the presence of the nonspecific competitor did not influence the formation of any of the complexes formed with the wild type oligonucleotides. These experiments indicated that cellular transcription factor YY1 also binds specifically to the ORF28/29 promoter.
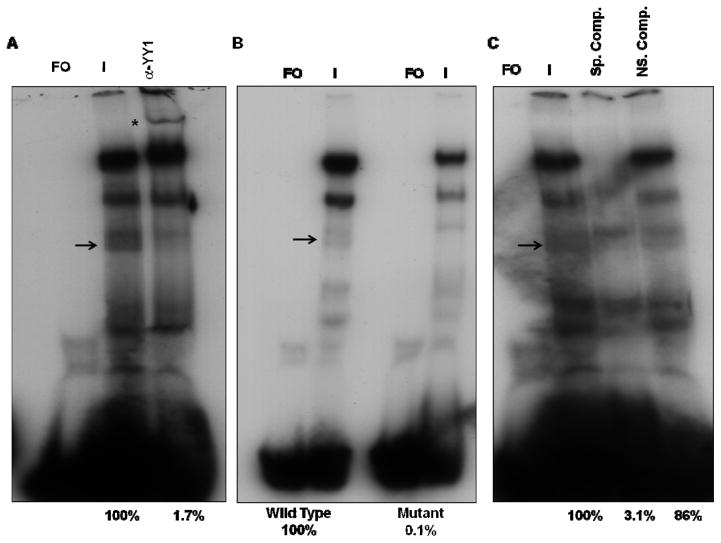
Lastly, the binding of YY1 to the gI promoter was determined using the same strategy. Several complexes formed using the oligonucleotide containing the YY1 site of the gI promoter, including two major slowly migrating complexes and two faster migrating minor complexes (Fig. 3A). The presence of anti-YY1 antibody supershifted one of the minor complexes formed (Fig. 3A).
FIG. 3.
EMSA and supershift assays using 40 bp oligonucleotides containing the wild-type YY1 site of the gI promoter. A) Supershift assays were done in the presence of VZV-infected cell nuclear extracts using polyclonal antibodies against YY1. B) EMSA assays using wild type and YY1 mutant oligonucleotides. C) EMSA and competition assays using Box A containing oligonucleotides as non specific competitor. Lanes: FO, free oligonucleotide; I, VZV-infected nuclear extract. The positions of the YY1 containing complexes are indicated by arrows and the positions of the supershifted bands are indicated by asterisks. The numbers under the figures represent the relative amounts of YY1 complexes, assessed by densitometry.
A mutant probe containing the mutation that is predicted to inhibit the binding of YY1 to the gI promoter sequence was next used in EMSA assays. The probe has a substitution of the first C in the GC rich sequence representing the Sp1 site of the gI promoter to G in the YY1 binding site. As shown in Fig. 3B, this mutation disrupted the formation of the identified YY1 complex with no significant effect on the formation of the other complexes.
EMSA experiments were also done using unlabeled specific competitor 40-bp oligonucleotides containing the predicted YY1 binding site of the gI promoter and the non-specific competitor 30-bp oligonucleotides containing the origin binding protein (OBP) box A sequence. As shown in Fig. 3C, the cold specific competitor efficiently competed away the formation of the YY1 containing complex and also the two major complexes formed, while the presence of the nonspecific competitor had no influence on the formation of any protein-DNA complex. These experiments indicated the specificity of the YY1 binding to the gI promoter.
YY1 site mutations inhibited the promoter activities of ORF10, ORF28, ORF29 and gI genes
In this set of experiments, reporter gene assays were done in the context of VZV super-infection and with pCMV-62 transfection. We used wild type and YY1 mutant reporters to determine the influence of the YY1 binding site mutations within the ORF10, ORF28/29 and gI promoters on the expression levels of these genes. The mutations in the reporters used in these experiments are the same as those that inhibited the binding of YY1 to the YY1 binding sites in the EMSA assays. Luciferase activities obtained from each reporter plasmid in the absence of VZV super-infection and ORF62 transfection represent the basal levels from this plasmid and were normalized to 1. Reporter gene activities in the presence of VZV super-infection and ORF62 transfection were reported as induction (n-fold) of luciferase activities in reference to the basal activity.
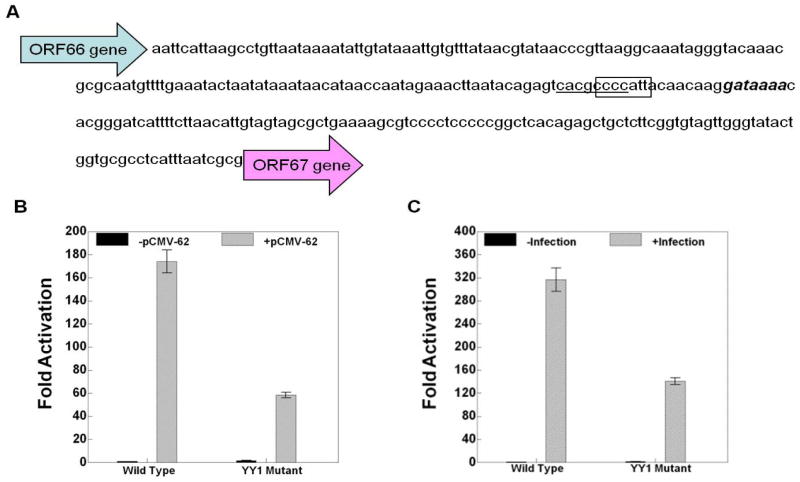
The luciferase reporter of the ORF10 promoter contains the 242 bp intergenic region between ORF9 and ORF10 (Fig. 4A). The ORF28/29 promoter reporter includes the 221 bp intergenic region between ORF28 and ORF29 flanked by Renilla luciferase in place of the ORF28 gene and firefly luciferase in place of the ORF29 gene (Fig. 5A). The gI promoter reporter includes 120 nucleotides before the translation start site of the ORF67 gene (Fig. 7A).
Fig. 4.
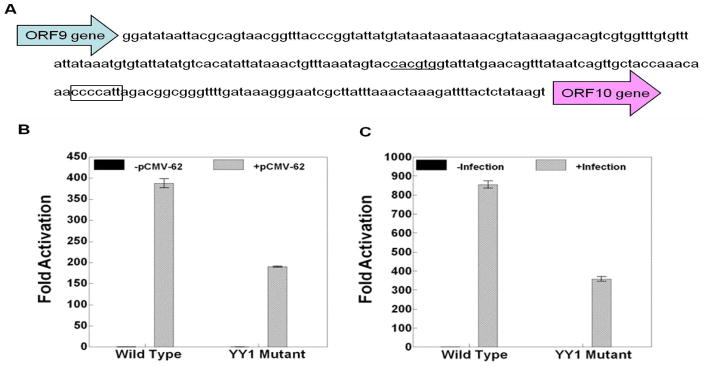
The effect of YY1 site mutation on ORF10 gene expression. A) The nucleotide sequence of the ORF9-ORF10 intergenic region. The YY1 site is boxed. The USF site reported to be important for ORF10 expression is underlined. B and C) The results of triplicate assays assessing the effects of the presence of the YY1 site mutation in the ORF10 promoter on the expression levels of the firefly luciferase reporter in the context of B) pCMV-ORF62 transfection and C) VZV super-infection. The promoter activities resulting from the presence of transfected ORF62 or VZV super-infection are reported as induction (n-fold) of luciferase activity over basal (without ORF62 transfection or VZV super-infection respectively). Statistical significance was determined by a one-way ANOVA analysis of variance followed by Tukey’s post hoc test. Error bars indicate standard error.
Fig. 5.
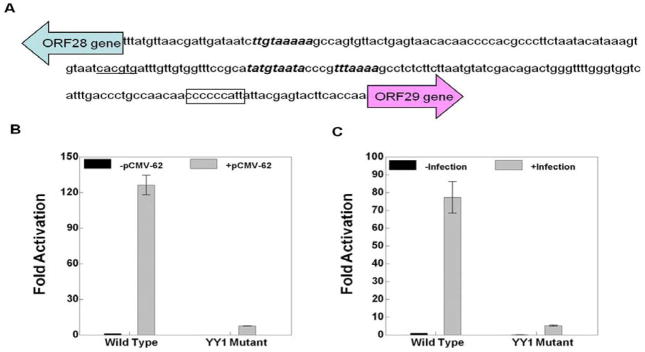
The effect of YY1 site mutation in the ORF28/29 promoter on ORF28 gene expression. A) The nucleotide sequence of the ORF28-ORF29 intergenic region. The YY1 site is boxed. The USF site reported to be important for ORF28 and ORF29 expression is underlined. The TATA boxes appear in bold italics. B and C) The results of triplicate assays assessing the effects of the presence of the YY1 site mutation in the ORF28 promoter on the expression levels of the Renilla luciferase reporter in the context of B) pCMV-ORF62 transfection and C) VZV super-infection. The promoter activities resulting from the presence of transfected ORF62 or VZV super-infection are reported as induction (n-fold) of luciferase activity over basal (without ORF62 transfection or VZV super-infection respectively). Statistical significance was determined by a one-way ANOVA analysis of variance followed by Tukey’s post hoc test. Error bars indicate standard error.
Fig. 7.
The effect of YY1 site mutation in the gI promoter on gI gene expression. A) The nucleotide sequence of the gI promoter. The YY1 site is boxed. The USF and the Sp1 sites reported to be important for gI expression is underlined. The TATA box appears in bold italics. B and C) The results of triplicate assays assessing the effects of the presence of the YY1 site mutation in the gI promoter on the expression levels of the firefly luciferase reporter in the context of B) pCMV-ORF62 transfection and C) VZV super-infection. The promoter activities resulting from the presence of transfected ORF62 or VZV super-infection are reported as induction (n-fold) of luciferase activity over basal (without ORF62 transfection or VZV super-infection respectively). Statistical significance was determined by a one-way ANOVA analysis of variance followed by Tukey’s post hoc test. Error bars indicate standard error.
The YY1 site mutation inhibited ORF10 promoter activation by about 50–60% in the experiments done using pCMV-ORF62 transfection and VZV super-infection (Fig. 4B and 4C). More strikingly, the YY1 site mutations inhibited both ORF28 and ORF29 promoter activation by about 90% in experiments done using either pCMV-ORF62 transfection or VZV super-infection (Figs. 5 and 6). Lastly, The YY1 site mutations inhibited gI expression in the reporter gene assays by about 60–70% in experiments done using pCMV-ORF62 transfection and VZV super-infection (Fig. 7B and BC).
Fig. 6.
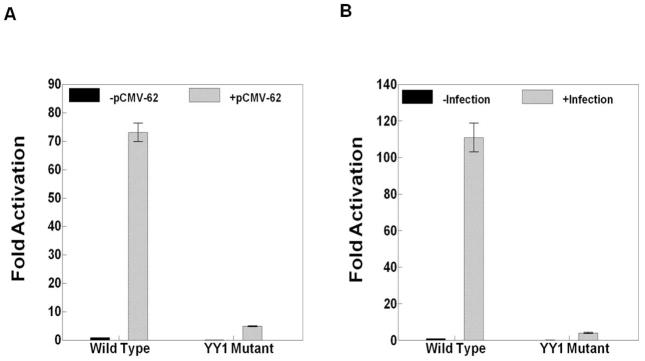
The effect of YY1 site mutation in the ORF28/29 promoter on ORF29 gene expression. A and B) The results of triplicate assays assessing the effects of the presence of the YY1 site mutation in the ORF29 promoter on the expression levels of the firefly luciferase reporter in the context of A) pCMV-ORF62 transfection and B) VZV super-infection. The promoter activities resulting from the presence of transfected ORF62 or VZV super-infection are reported as induction (n-fold) of luciferase activity over basal (without ORF62 transfection or VZV super-infection respectively). Statistical significance was determined by a one-way ANOVA analysis of variance followed by Tukey’s post hoc test. Error bars indicate standard error.
These results suggested the involvement of this YY1 site in the activation of the ORF10, ORF28, ORF29 and gI promoters mediated by IE62 or by VZV super-infection.
Influence of YY1 mutation on ORF28/29 promoter on VZV replication in vitro
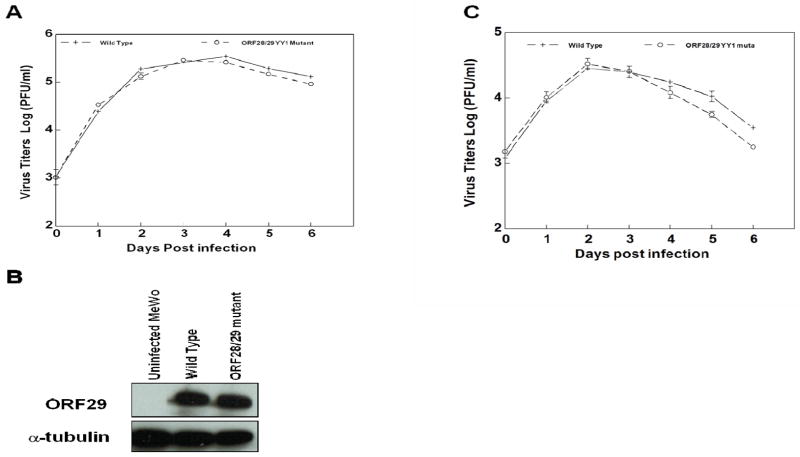
A VZV recombinant virus with the YY1 site mutation was generated as described in the materials and methods section. When the growth kinetics of pOka and the ORF28/29 promoter mutant were evaluated in MeWo cells, titers were equivalent over the first three days post infection; then there was a slight insignificant decrease in growth from day 4 to day 6 for the mutant virus (Fig. 8A). We then looked at expression of the ORF29 protein in infected MeWo cells. This YY1 site mutation in the promoter had no significant effect on the expression of ORF29 protein at 36 hrs post infection compared to ORF29 expression in the wild type pOka at the same time point (Fig. 8B).
Fig. 8.
The effect of VZV pOka-YY1 site mutation in the ORF28/29 promoter on VZV replication. A) Growth kinetics of pOka and pOka-YY1 mutant virus in MeWo cells. Cells were inoculated at 103 PFU/ml with wild type and mutant viruses and infectious virus yields were determined for 6 days after inoculation. B) Effect of YY1 site mutation on ORF29 expression levels in MeWo cells during VZV infection. Western blot analyses show the expression levels of ORF29 and α-tubulin at 36 hrs post-infection. α-tubulin was used as a loading control in the experiments. The blots were scanned by densitometry to obtain quantitative data (in triplicate). Statistical significance was determined by a one-way ANOVA analysis of variance followed by Tukey’s post hoc test. C) Growth kinetics of pOka and pOka-YY1 mutant virus in HELF cells. Cells were inoculated at 103 PFU/ml with wild type and mutant viruses and infectious virus yields were determined for 6 days after inoculation.
On the other hand, the growth kinetics of this mutant virus in HELF cells showed significant inhibition in virus replication at day 5 and 6 post infection (Fig. 8C).
DISCUSSION
Like other herpesviruses, the VZV genome does not encode any RNA polymerase function or gene alternative to the TATA-binding protein (TBP) in binding to the TATA sites within the VZV promoters (Ruyechan et al., 2003; Ruyechan, 2010). This suggests that the basal cellular transcription machinery is required for viral gene expression. Most VZV promoters require TATA-elements for their expression (Ruyechan et al., 2003) with the exception of the ORF3 and ORF10 promoters which have been shown to be expressed in the absence of any functional TATA box sites (Khalil et al., 2013 and Che et al., 2007). All of the VZV promoters studied to date also contain predicted binding sites for cellular transcription factors including Sp1, USF and ATF. Several reports have verified that these factors contribute to expression of VZV genes both in vitro and in vivo (Berarducci et al., 2007; Che et al., 2007; Ito et al., 2003; Meier et al., 1994; Narayanan et al., 2005; Peng et al., 2003; Rahaus and Wolff, 2003; Rahaus et al., 2003; Wang et al., 2009; Yang et al., 2004). IE62 is the major VZV trans-activator, the product of ORF62 and its complement ORF71. The deletion of both copies of ORF62 and ORF71 blocked VZV replication (Sato et al., 2003).
IE62 has been shown to interact physically with cellular factors Sp1 and USF, as well as TBP (Peng et al., 2003; Rahaus et al., 2002; and Perera et al., 2000). IE62 has also been shown to interact with the human mediator complex through its N-terminal acidic activation domain (Yang et al., 2008 and Yamamoto et al., 2009). These physical interactions have been shown to be important for the expression of some VZV promoters in reporter gene assays. Mutations of the binding sites for these cellular factors in ORF61 and ORF10 promoters have been shown to inhibit VZV replication in skin xenografts (Wang et al., 2009 and Che et al., 2007).
YY1 is a zinc-finger protein that has the ability to repress and activate gene transcription in several mammalian viruses including adeno-associated virus, human papillomavirus, parvovirus B19, HSV-1 and Moloney murine leukemia virus (Lee et al., 1992; Lee et al., 1998; Pajunk et al., 1997; and Shi et al., 1998). We had previously identified two binding sites for YY1 in the VZV genome. Mutation of one these YY1 sites in the downstream region of VZV oriS had no significant effect on either origin-dependent DNA replication or expression of the ORF62 and ORF63 flanking genes. However, mutation of this YY1 site in a VZV recombinant virus was associated with an increase in VZV genome copies in MeWo cells and viral replication in skin xenografts (Khalil et al., 2008; and Khalil et al., 2012). Mutation of the YY1 site in the ORF3 promoter inhibited ORF3 expression in reporter gene assays (Khalil et al., 2013).
In this study, we identified additional putative YY1 sites and expanded our analysis to the role played by YY1 in IE62 mediated activation of VZV gene expression. We investigated binding of YY1 to the promoters of VZV ORF10, ORF28/29 and gI genes. These three promoters are analyzed previously and selected for the presence of the postulated YY1 binding sites within the sequences that had been shown to be required for the activation of the promoter (Che et a., 2007; Yang et al., 2004; and Peng et al., 2003). The mutation of these YY1 binding sites not only inhibited the formation of the YY1 complexes but also inhibited expression in reporter gene assays in the context of IE62 transfection and VZV superinfection. These results suggested a possible role for YY1 in lytic gene expression and possibly also in VZV replication.
The mutation of the YY1 binding site inhibited the expression from ORF28/29 promoter by about 90%, showing that this YY1 site is required for the expression of these genes. Previous analysis of this intergenic region showed the presence of a USF binding site which appeared to be required for activity from these promoters (Yang et al., 2004). The presence of this USF site; however, failed to compensate for the absence of the YY1 binding site.
On the other hand, the mutation of the YY1 binding site in ORF10 and gI promoters inhibited the expression of these genes by only about 60–70%. Analysis of the ORF10 promoter showed the presence of an important USF site and previous analysis of the gI promoter had shown the presence of an Sp1 site adjacent to the YY1 binding site (Che et al., 2007; and Peng et al., 2003). It is quite possible that either or both of these factors could partially compensate for the absence of the YY1 binding sites in these promoters.
The physical interaction between IE62 and YY1 was tested using GST-pulldown assays (data not shown). This interaction requires the same domain of IE62 (aa 1–299) that was shown to be required for the interaction of IE62 with Sp1 and USF (Peng et al., 2003 and Rahaus et al., 2003). Both IE62 and YY1 have been shown to physically interact with Sp1 (Peng et al., 2003; Lee et al., 1993; Seto et al., 1993; and Shi et al., 1997), raising the possibility that binding could occur between YY1 and IE62 at their respective sites in the VZV promoters, either directly or bridged by the presence of Sp1. This physical interaction may be important for the recruitment of the basal cellular transcription machinery at the VZV promoters or perhaps for stabilizing its involvement.
A recombinant virus carrying the YY1 mutation in the ORF28/29 promoter grew as well as the wild type virus in melanoma cells, with similar expression level of the ORF29 protein. On the other hand, this mutation significantly inhibited virus replication in HELF cells at later times of infection (day 5 and 6 postinfection).
This moderate inhibition with this mutation in HELF cells might well be attributed to the involvement of the ORF29 protein in the origin-dependent DNA replication process (Khalil et al., 2011). We have shown in our previous work that ORF29 protein binds to a sequence downstream of VZV oriS in VZV infected cells. The binding of ORF29 protein to this sequence downregulated oriS dependent DNA replication and enhanced ORF62 and ORF63 expression. The mutation of the YY1 site in the ORF28/29 promoter inhibited ORF29 expression significantly as shown in reporter gene assays. This decrease in ORF29 expression should thus lead to some upregulation of oriS dependent DNA replication, leading to a possible increase in the number of virus genome copies in cells infected with the mutant recombinant viruses. This effect might moderate or mask the effect of ORF29 transcription downregulation due to the YY1 mutation.
The study of this mutant virus influence over VZV replication in other cells and tissues will be a point of interest in the future, since we have clearly shown differences in response in two separate human cell lines (MeWo and HELF). Also, generation of viral mutants to test the influence of mutations in YY1 sites in other promoters (e.g. ORF10 and gI) will be important future point for more extensive evaluation of the role of YY1 in the VZV replication. The study of cellular factors influencing VZV replication is important for developing antiviral drug strategies, through generation of inhibitors to interfere with the functions of these factors leading to inhibition of virus replication. Such studies are also essential to a better understanding how the lytic and latent phases of virus replication are controlled. In this report, we have established the importance of the cellular factor YY1, in mediating VZV IE62 transcriptional activation, and in controlling viral gene expression.
Research Highlights.
19 postulated YY1 binding sites identified within putative VZV promoters.
YY1 is binding to ORF10, ORF28/29 and gI promoters using EMSA assays.
The mutation of these YY1 binding sites inhibited YY1 binding and the transcription from ORF10, ORF28/29 and gI promoters.
YY1 is influencing the IE62 mediated transcriptional activation.
Acknowledgments
This work was supported by grants AI018449, AI053846 and AI020459 from the National Institutes of Health, and grants from the John R. Oishei Foundation and the National Shingles Foundation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bauknecht T, Shi Y. Overexpression of C/EBPbeta represses human papillomavirus type 18 upstream regulatory region activity in HeLa cells by interfering with the binding of TATA-binding protein. J Virol. 1998;72:2113–2124. doi: 10.1128/jvi.72.3.2113-2124.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berarducci B, Sommer M, Zerboni L, Rajamani J, Arvin A. Cellular and viral factors regulate the varicella-zoster virus gE promoter during viral replication. J Virol. 2007;81:10258–10267. doi: 10.1128/JVI.00553-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chaudhuri V, Sommer M, Rajamani J, Zerboni L, Arvin AM. Functions of varicella-zoster virus ORF23 capsid protein in viral replication and the pathogenesis of skin infection. J Virol. 2008;82:10231–10246. doi: 10.1128/JVI.01890-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Che X, Berarducci B, Sommer M, Ruyechan WT, Arvin AM. The ubiquitous cellular transcriptional factor USF targets the varicella-zoster virus open reading frame 10 promoter and determines virulence in human skin xenografts in SCIDhu mice in vivo. J Virol. 2007;81:3229–3239. doi: 10.1128/JVI.02537-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen JI, Straus SE, Arvin AM. Varicella-zoster virus. In: Fields BN, Knipe DM, Howley PM, editors. Fields of Virology. Vol. 2. Wolter Kluwer Health/Lippincott Williams & Wilkins Publishers; Philadelphia, PA: 2007. pp. 2773–2818. [Google Scholar]
- 6.Gordon S, Akopyan G, Garban H, Bonavida B. Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene. 2006;25:1125–1142. doi: 10.1038/sj.onc.1209080. [DOI] [PubMed] [Google Scholar]
- 7.Hyde-DeRuyscher RP, Jennings E, Shenk T. DNA binding sites for the transcriptional activator/repressor YY1. Nucleic Acids Res. 1995;23:4457–4465. doi: 10.1093/nar/23.21.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ito H, Sommer MH, Zerboni L, He H, Boucaud D, Hay J, Ruyechan W, Arvin AM. Promoter sequences of varicella-zoster virus glycoprotein I targeted by cellular transactivating factors Sp1 and USF determine virulence in skin and T cells in SCIDhu mice in vivo. J Virol. 2003;77:489–498. doi: 10.1128/JVI.77.1.489-498.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khalil MI, Hay J, Ruyechan WT. The cellular transcription factors Sp1 and Sp3 suppress varicella zoster virus origin-dependent DNA replication. J Virol. 2008;82:11723–11733. doi: 10.1128/JVI.01322-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khalil MI, Arvin A, Jones J, Ruyechan WT. A sequence within the varicella-zoster virus (VZV) oriS is a negative regulator of DNA replication and is bound by a protein complex containing the VZV ORF29 protein. J Virol. 2011;85:12188–12200. doi: 10.1128/JVI.05501-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khalil MI, Robinson M, Sommer M, Arvin A, Hay J, Ruyechan WT. An Sp1/Sp3 site in the downstream region of varicella zoster virus oriS influences origin-dependent DNA replication and flanking gene transcription and is important for VZV replication in vitro and in human skin. J Virol. 2012:86. doi: 10.1128/JVI.01538-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khalil MI, Sommer M, Arvin A, Hay J, Ruyechan WT. Regulation of the Varicella-zoster virus ORF3 promoter by cellular and viral factors. Virology. 2013;440:171–181. doi: 10.1016/j.virol.2013.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kinchington PR, Inchauspe G, Subak-Sharpe JH, Robey F, Hay J, Ruyechan WT. Identification and characterization of a varicella-zoster virus DNA-binding protein by using antisera directed against a predicted synthetic oligopeptide. J Virol. 1988;62:802–809. doi: 10.1128/jvi.62.3.802-809.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kinchington P, Fite K, Turse SE. Nuclear accumulation of IE62, the varicella-zoster virus (VZV) major transcriptional regulatory protein, is inhibited by phosphorylation mediated by the VZV open reading frame 66 protein kinase. J Virol. 2000;74:2265–2277. doi: 10.1128/jvi.74.5.2265-2277.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee JS, Galvin KM, Shi Y. Evidence for physical interaction between the zinc-finger transcription factors YY1 and Sp1. Proc Natl Acad Sci. 1993;90:6145–6149. doi: 10.1073/pnas.90.13.6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee KY, Broker TR, Chow LT. Transcription factor YY1 represses cell-free replication from human papillomavirus origins. J Virol. 1998;72:4911–4917. doi: 10.1128/jvi.72.6.4911-4917.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee TC, Shi Y, Schwartz RJ. Displacement of BrdUrd-induced YY1 by serum response factor activates skeletal alpha-actin transcription in embryonic myoblasts. Proc Natl Acad Sci. 1992;89:9814–9818. doi: 10.1073/pnas.89.20.9814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li M, Baumeister P, Raoy B, Phan T, Foti D, Luo S, Lee AS. ATF6 as a transcription activator of the endoplasmic reticulum stress element: thapsigargin stress-induced changes and synergistic interactions with NF-Y and YY1. Mol Cell Biol. 2000;20:5096–5106. doi: 10.1128/mcb.20.14.5096-5106.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lynch JM, Kenyon TK, Grose C, Hay J, Ruyechan WT. Physical and functional interaction between the varicella zoster virus IE63 and IE62 proteins. Virology. 2002;302:71–82. doi: 10.1006/viro.2002.1555. [DOI] [PubMed] [Google Scholar]
- 20.Meier JL, Luo X, Sawadogo M, Straus SE. The cellular transcription factor USF cooperates with varicella-zoster virus immediate-early protein 62 to symmetrically activate a bidirectional viral promoter. Mol Cell Biol. 1994;14:6896–6906. doi: 10.1128/mcb.14.10.6896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moffat JF, Stein MD, Kaneshima H, Arvin AM. Tropism of varicella-zoster virus for human CD4+ and CD8+ T lymphocytes and epidermal cells in SCID-hu mice. J Virol. 1995;69:5236–5242. doi: 10.1128/jvi.69.9.5236-5242.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Narayanan A, Nogueira ML, Ruyechan WT, Kristie TM. Combinatorial transcription of herpes simplex virus and varicella zoster virus immediate early genes is strictly determined by the cellular coactivator HCF-1. J Biol Chem. 2005;280:1369–1375. doi: 10.1074/jbc.M410178200. [DOI] [PubMed] [Google Scholar]
- 23.Niizuma T, Zerboni L, Sommer MH, Ito H, KHinchliffe S, Arvin AM. Construction of varicella-zoster virus recombinants from parent Oka cosmids and demonstration that ORF65 protein is dispensable for infection of human skin and T cells in the SCID-hu mouse model. J Virol. 2003;77:6062–6065. doi: 10.1128/JVI.77.10.6062-6065.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oliver SL, Sommer M, Zerboni L, Rajamani J, Grose C, Arvin AM. Mutagenesis of varicella-zoster virus glycoprotein B: putative fusion loop residues are essential for viral replication, and the furin cleavage motif contributes to pathogenesis in skin tissue in vivo. J Virol. 2009;83:7495–7506. doi: 10.1128/JVI.00400-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pajunk HS, May C, Pfister H, Fuchs PG. Regulatory interactions of transcription factor YY1 with control sequences of the E6 promoter of human papillomavirus type 8. J Gen Virol. 1997;78:3287–3295. doi: 10.1099/0022-1317-78-12-3287. [DOI] [PubMed] [Google Scholar]
- 26.Peng H, He H, Hay J, Ruyechan WT. Interaction between the varicella zoster virus IE62 major transactivator and cellular transcription factor Sp1. J Biol Chem. 2003;278:38068–38075. doi: 10.1074/jbc.M302259200. [DOI] [PubMed] [Google Scholar]
- 27.Perera LP, Mosca JD, Ruyechan WT, Hay J. Regulation of varicella-zoster virus gene expression in human T lymphocytes. J Virol. 1992;66:5298–5304. doi: 10.1128/jvi.66.9.5298-5304.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perera LP, Mosca JD, Sadeghi-Zadeh M, Ruyechan WT, Hay J. The varicella-zoster virus immediate early protein, IE62, can positively regulate its cognate promoter. Virology. 1992;191:346–354. doi: 10.1016/0042-6822(92)90197-w. [DOI] [PubMed] [Google Scholar]
- 29.Perera LP, Mosca JD, Ruyechan WT, Hayward GS, Straus SE, Hay J. A major transactivator of varicella-zoster virus, the immediate-early protein IE62, contains a potent N-terminal activation domain. J Virol. 1993;67:4474–4483. doi: 10.1128/jvi.67.8.4474-4483.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perera LP. The TATA motif specifies the differential activation of minimal promoters by varicella zoster virus immediate-early regulatory protein IE62. J Biol Chem. 2000;275:487–496. doi: 10.1074/jbc.275.1.487. [DOI] [PubMed] [Google Scholar]
- 31.Rahaus M, Desloges N, Yang M, Ruyechan WT, Wolff MH. Transcription factor USF, expressed during the entire phase of varicella-zoster virus infection, interacts physically with the major viral transactivator IE62 and plays a significant role in virus replication. J Gen Virol. 2003;84:2957–2967. doi: 10.1099/vir.0.19335-0. [DOI] [PubMed] [Google Scholar]
- 32.Rahaus M, Wolff MH. Reciprocal effects of varicella-zoster virus (VZV) and AP1: activation of jun, fos and ATF-2 after VZV infection and their importance for the regulation of viral genes. Virus Res. 2003;92:9–21. doi: 10.1016/s0168-1702(02)00310-6. [DOI] [PubMed] [Google Scholar]
- 33.Ruyechan WT, Peng H, Yang M, Hay J. Cellular factors and IE62 activation of VZV promoters. J Med Virol. 2003;70:S90–S94. doi: 10.1002/jmv.10328. [DOI] [PubMed] [Google Scholar]
- 34.Ruyechan WT. Roles of cellular transcription factors in VZV replication. Curr Top Microbiol Immunol. 2010;342:43–65. doi: 10.1007/82_2010_42. [DOI] [PubMed] [Google Scholar]
- 35.Sato B, Ito H, Hinchliffe S, Sommer MH, Zerboni L, Arvin AM. Mutational analysis of open reading frames 62 and 71, encoding the varicella-zoster virus immediate-early transactivating protein, IE62, and effects on replication in vitro and in skin xenografts in the SCID-hu mouse in vivo. J Virol. 2003;77:5607–5620. doi: 10.1128/JVI.77.10.5607-5620.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seto E, Lewis B, Shenk T. Interaction between transcription factors Sp1 and YY1. Nature. 1993;365:462–464. doi: 10.1038/365462a0. [DOI] [PubMed] [Google Scholar]
- 37.Shi Y, Seto E, Chang LS, Shenk T. Transcriptional repression by YY1, a human GLI-Krüppel-related protein, and relief of repression by adenovirus E1A protein. Cell. 1991;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- 38.Shi Y, Lee JS, Galvin KM. Everything you have ever wanted to know about Yin Yang 1. Biochim Biophys Acta. 1997;1332:F49–F66. doi: 10.1016/s0304-419x(96)00044-3. [DOI] [PubMed] [Google Scholar]
- 39.Spengler ML, Ruyechan WT, Hay J. Physical interaction between two varicella zoster virus gene regulatory proteins, IE4 and IE62. Virology. 2000;272:375–381. doi: 10.1006/viro.2000.0389. [DOI] [PubMed] [Google Scholar]
- 40.Tischer BK, Kaufer BB, Sommer M, Wussow S, Arvin AM, Osterrieder N. A self-excisable infectious bacterial artificial chromosome clone of varicella-zoster virus allows analysis of the essential tegument protein encoded by ORF9. J Virol. 2007;81:13200–13208. doi: 10.1128/JVI.01148-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang L, Sommer M, Rajamani J, Arvin AM. Regulation of the ORF61 promoter and ORF61 functions in varicella-zoster virus replication and pathogenesis. J Virol. 2009;83:7560–7572. doi: 10.1128/JVI.00118-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.White K, Peng H, Hay J, Ruyechan WT. Role of the IE62 consensus binding site in transactivation by the varicella-zoster virus IE62 protein. J Virol. 2010;84:3767–3779. doi: 10.1128/JVI.02522-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamamoto S, Eletsky A, Szyperski T, Hay J, Ruyechan WT. Analysis of the varicella-zoster virus IE62 N-terminal acidic transactivating domain and its interaction with the human mediator complex. J Virol. 2009;83:6300–6305. doi: 10.1128/JVI.00054-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang M, Hay J, Ruyechan WT. The DNA element controlling expression of the varicella-zoster virus open reading frame 28 and 29 genes consists of two divergent unidirectional promoters which have a common USF site. J Virol. 2004;78:10939–10952. doi: 10.1128/JVI.78.20.10939-10952.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang M, Peng H, Hay J, Ruyechan WT. Promoter activation by the varicella-zoster virus major transactivator IE62 and the cellular transcription factor USF. J Virol. 2006;80:7339–7353. doi: 10.1128/JVI.00309-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang M, Hay J, Ruyechan WT. Varicella-zoster virus IE62 protein utilizes the human mediator complex in promoter activation. J Virol. 2008;82:12154–12163. doi: 10.1128/JVI.01693-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang WM, Inouye CJ, Seto E. Cyclophilin A and FKBP12 interact with YY1 and alter its transcriptional activity. J Biol Chem. 1995;270:15187–15193. doi: 10.1074/jbc.270.25.15187. [DOI] [PubMed] [Google Scholar]
- 48.Yant SR, Zhu W, Millinoff D, Slightom JL, Goodman M, Gumucio DL. High affinity YY1 binding motifs: identification of two core types (ACAT and CCAT) and distribution of potential binding sites within the human beta globin cluster. Nucleic Acids Res. 1995;23:4353–4362. doi: 10.1093/nar/23.21.4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yao YL, Yang WM, Seto E. Regulation of transcription factor YY1 by acetylation and deacetylation. Mol Cell Biol. 2001;21:5979–5991. doi: 10.1128/MCB.21.17.5979-5991.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yeh TS, Lin YM, Hsieh RH, Tseng MJ. Association of transcription factor YY1 with the high molecular weight Notch complex suppresses the transactivation activity of Notch. J Biol Chem. 2003;278:41963–41969. doi: 10.1074/jbc.M304353200. [DOI] [PubMed] [Google Scholar]