Abstract
Dysregulation of oncogenes by translocation to the IgH locus (14q32) is a seminal event in the pathogenesis of B-cell tumours1. In multiple myeloma (MM), translocations to the IgH locus have been reported at an incidence of 20–60%. For most translocations, the partner chromosome is unknown (14q+); for the others, a diverse array of chromosomal partners have been identified, with 11q13 (cyclin D1) the only chromosome that is frequently involved2–6. Recently, we developed a Southern-blot assay that detects translocation breakpoint fragments in most MM tumours, including those with no translocation detected by conventional karyotyping6. In a continuing analysis of translocations in 21 myeloma cell lines and primary tumours, we show that the novel, karyotypically silent translocation t(4;14)(p16.3;q32.3) is present in five lines and at least three of ten primary tumours. The chromosome-4 breakpoints are clustered in a 70-kb region centromeric to the fibroblast growth factor receptor 3 gene (FGFR3), the apparent dysregulated oncogene. Two lines and one primary tumour with this translocation selectively express an FGFR3 allele containing activating mutations identified previously in thanatophoric dwarfism. We propose that after the t(4;14) translocation, somatic mutation during tumour progression frequently generates an FGFR3 protein that is active in the absence of ligand.
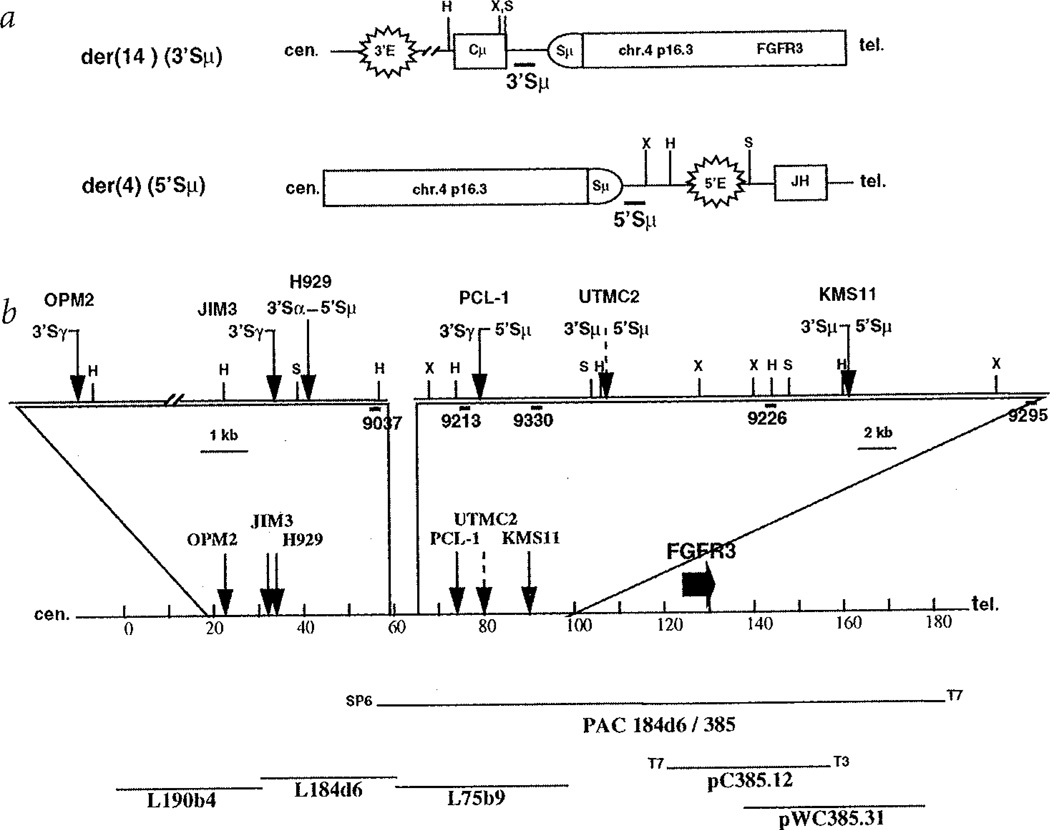
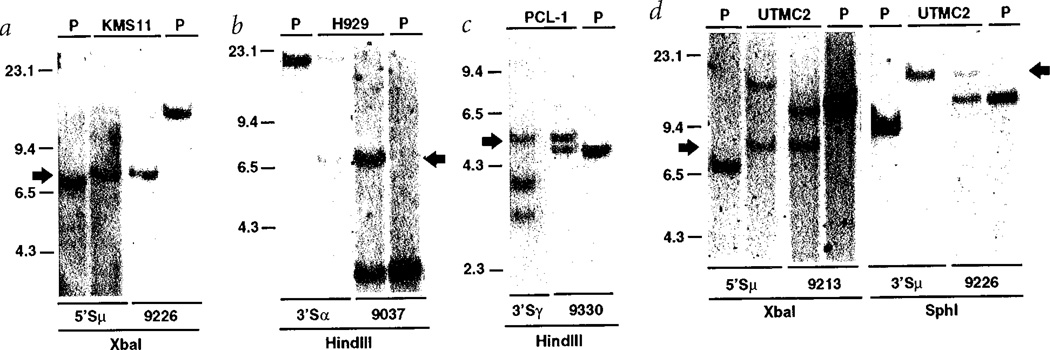
As described in detail elsewhere6, we generated paired probes immediately upstream and downstream of the repetitive sequences in each switch region—for example, 5'Sµ and 3'Sµ probes. By Southern-blot analysis, candidate translocation breakpoint fragments were identified as rearranged fragments that hybridize to only one switch probe. Fig. 1a illustrates a balanced translocation into Sµ, with the 3'Sµ probe detecting the der(14) breakpoint and the 5'Sµ probe detecting the der(4) breakpoint. Using this approach, we cloned one translocation breakpoint fragment involving chromosome 4 from each of five samples: four MM lines (KMS11, H929, OPM2 and JIM3) and one plasma cell leukaemia tumour (PCL-1). In each case, the novel non-Ig sequences at the breakpoint and at one end of the fragment were identical to sequences present in a cosmid contig that is centromeric to the fibroblast growth factor receptor 3 (FGFR3) gene at 4pl6.3 (Fig. 1)7. Having the published sequence of the entire breakpoint region, we designed specific primer pairs to PCR-amplify probes that were used in Southern-blot analyses to detect and map reciprocal breakpoint fragments for KMS11, H929 and PCL-1 (Fig. 2a–c). For example, as shown in Fig. 2a for the der(4)(5'Sµ) reciprocal breakpoint in KMS11 (Fig. 1), a 5'Sµ probe detected a rearranged 7.9-kb XbaI fragment (compared to 7.5-kb fragment in placental [P] DNA) and a 4pl6.3 probe (9226 in Fig. 1b) that detects a 13.8-kb germline fragment in the tumour and in placental DNA, co-hybridizes to the 7.9-kb XbaI fragment in KMS11. These results, together with Southern blots using other enzymes, are consistent with the der(4) (5'Sµ) translocation breakpoint occurring near the cloned der(14) (3'Sµ) translocation breakpoint for KMS11 (Fig. 1b).
Fig. 1.
Translocation breakpoints involving 4p16.3. a, Diagram of der(14) and der(4) breakpoints resulting from a translocation into Sµ. The centromere is to the left, and the figure is not to scale. Structural elements include enhancers (3'E and 5'E), switch region (Sµ) and coding segments (Cµ and JH). Thick horizontal lines depict 3'- and 5'Sµ probes. Vertical lines represent restriction enzyme sites, X = XbaI, H = HindIII, S = SphI. b, Distribution of 4p16.3 breakpoints within the distal 70 kb of a 2-Mb cos-mid contig spanning the Huntington's disease region, centromeric to the FGFR3 gene. The diagram is drawn to scale. Solid arrows indicate the positions of the cloned breakpoints for the OPM2, JIM3, H929 and KMS11 myeloma cell lines and for the tumour sample (PCL-1), while a dashed arrow shows where the breakpoints for UTMC2 have been mapped by Southern-blot analysis. Next to each arrow is indicated the type and orientation of the switch region associated with the breakpoint; the shaded designation indicates breakpoints determined by Southern blotting. The sites of restriction enzymes, indicated by a capital letter (H for HindIII, S for SphI, X for XbaI), as well as the position of the probes, indicated by thick lines, used for cloning the breakpoint or for Southern-blot analysis are depicted in the enlarged sections. The entire sequence (except for three short gaps in L75b9) of the most distal cosmid contig (L190b4, L184d6 and L75b9) in the Hunting-ton's disease region is known. The transcription orientation of FGFR3 gene is from the T7 end to the T3 end of pC385.12 (M.R. Alther, pers. comm.r).
Fig. 2.
Identification of chromosomal rearrangements affecting the 4p16.3 locus. Southern blots were performed on 10 µg of DNA from three MM cell lines (KMS11 in a, H929 in b, and UTMC2 in d), from a tumour sample (PCL-1 in c) and from placenta (P). The DNA was digested with the indicated restriction enzyme and hybridized sequentially with the switch and the chromosome 4p16.3 probes listed below each line. Arrows identify the rearranged bands that co-hybridize with the switch and chromosome 4p16.3 probes as a consequence of the translocations t(4;14)(p16;q32). Additional bands not seen in placental DNA or designated by the arrow represent other rearrangements unrelated to the translocation; for instance, the 15-kb band detected by the 3'Sα probe in H929 (b) identifies a productive µ to α switch recombination event. It should be noted that the absence or altered migration of germline bands in some of the samples (for example, UTMC2 probed with 5'Sµ, 2d) is due to multiple kinds of switch recombination events (for example, translocation, productive rearrangement), genetic polymorphisms, or deletion. We refer only to the bands related to the t(4;14) translocation products.
We used an RT-PCR assay to screen for expression of FGFR3. A prominent band (Fig. 3a) was detected in three lines (KMS11, OPM2 and H929) and one primary tumour (PCL-1) with cloned t(4;14)(pl6.3;q32.3) translocation breakpoints, and also in one additional line (UTMC2). Eight MM lines (8226, ARK, SKMM1, DELTA47, H1112, KMM1, TH and U266) generated barely detectable PCR products (data not shown). The remaining nine lines (ANBL6, FLAM76, SKMM2, JIM3, JJN3, KMS12, MM-M1, MM.1–144 and MY5) were completely negative for FGFR3 expression by this assay. In addition, a similar PCR analysis of nine intramedullary MM samples, together with normal peripheral blood and bone marrow samples, showed amplification of FGFR3 from cDNA in three intramedullary MM samples (MM.T0, MM.T1 and MM.T2; Fig. 3a and data not shown) but not in normal bone marrow or blood cells. Overexpression of FGFR3 by the MM cell lines was confirmed by northern blot and immunoprecipitation analyses. As seen in Fig. 3b, an intense 4.5-kb FGFR3 mRNA8 is present at extremely high levels in KMS11 and OPM2, at much lower levels in H929 and at barely detectable levels in UTMC2. The protein analysis (Fig. 3c) demonstrates the expected 115- and 135-kD FGFR3 protein species9 at levels concordant with the levels of mRNA. As assessed by northern and western blots, respectively, there was no detectable FGFR3 mRNA or protein in the other MM cell lines examined, including those that showed a barely detectable signal by the RT-PCR assay.
Fig. 3.
Expression of FGFR3 in MM. a, RT-PCT assay: a fragment of 598 bp has been amplified from four MM cell lines (UTMC2, KMS11, H929 and OPM2) and four tumour samples (PCL-1, JIM3-T, MM-T1 and MM-T2) by 35 cycles with a primer pair specific for the 3' half of the second immunoglobulin-like domain and the juxtapose-membrane (JM) region of the FGFR3 gene, b, Northern-blot assay: Poly(A)+ RNA from 100 µg of total RNA from six MM cell lines was fractionated on an agarose gel, blotted, and then hybridized sequentially with FGFR3 and β-actin probes. After five hours of exposure, the 4.5 kb-band corresponding to FGFR3 mRNA was detected in OPM2, H929 and KMS11, whereas a much longer exposure (63 hours) was needed for detection in UTMC2. c, Immunoblotting detection of FGFR3. Protein lysates from five MM cell lines were immunoprecipitated with a polyclonal rabbit antiserum against FGFR3. The immune complexes were separated on a 7.5% polyacrylamide SDS gel, blotted and detected with the same antiserum. Specific bands of 115 and 135 kD were detected in all four MM cell lines positive both by RT-PCT and northern-blot analysis. d, Selective expression of an FGFR3 variant allele in KMS11 by SSCP analysis. A primer pair flanking the juxta-membrane and transmembrane region of FGFR3 generate a 164-bp fragment by PCR from KMS11 genomic DNA and cDNA, and from placental genomic DNA (P). The products were then labelled with 32P-dCTP, denatured and fractionated on a native 10% polyacrylamide gel, and detected by autoradiography. The informative region of the gel is shown.
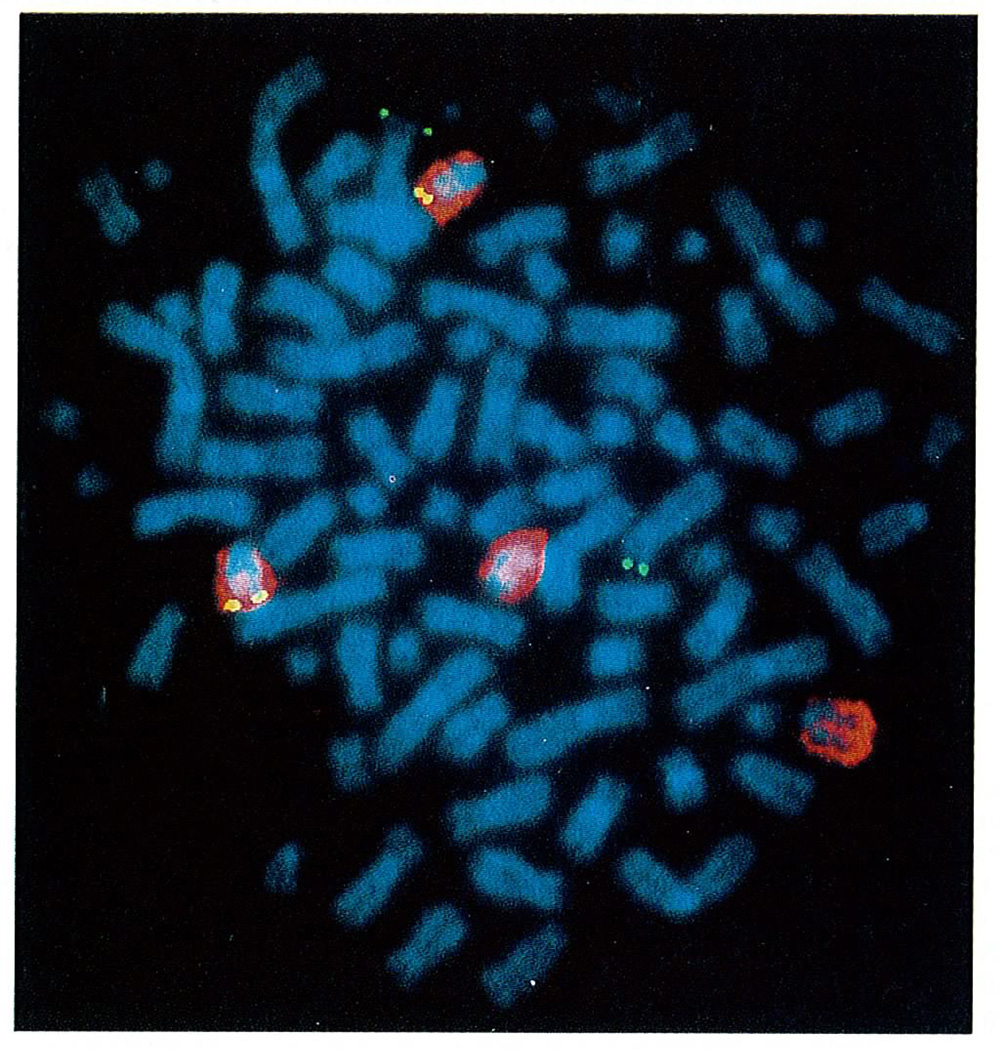
On the basis of the FGFR3 expression data, we looked for evidence of translocation t(4;14)(pl6.3;q32.3) in the UTMC2 MM cell line (Fig. 2d). We detected a rearranged 8.6-kb XbaI fragment that co-hybridizes with a 5'Sµ probe and the chromosome-4 probe 9213, and a rearranged 17.3-kb SphI fragment that co-hybridizes with a 3'Sµ probe and the chromosome-4 probe 9226—that is, the der(4) (5'Sµ) and der(14) (3'Sµ) reciprocal breakpoints (Fig. 1). This translocation was confirmed in UTMC2 by two-colour FISH analysis (Fig. 4). We also identified the t(4;14) translocation by mapping its presence in the FGFR3-expressing tumour samples MM.T1 and MM.T2 (data not shown).
Fig. 4.
Two-colour fluorescence in situ hybridization of the UTMC2 MM line with FGFR3 and chromosome-14 probes. The chromosome-14 probe generates a red signal, whereas other chromosomes are stained blue. The FGFR3 cosmid clone, located on chromosome band 4p16, generates green and yellow signals, respectively, on chromosomes that are counterstained blue and red. The FGFR3 cosmid clone hybridized to the telomeric band of two chromosomes 4 (normal map position) and to the telomeric band of two chromosomes 14 [translocation t(4;14)(p16;q32)].The other two chromosomes 14 showed only the hybridization signal of the chromosome 14–specific painting probe.
Although the translocation t(4;14)(pl6.3;q32.3) has not been described previously, the results reported here suggest that it occurs with an incidence of about 25% in MM. Conventional karyotypic analysis of the PCL-1 tumour and the five cell lines in which we have identified the translocation t(4;14), failed to detect this translocation, although JIM3 was reported to have a 14q+ translocation. These results are consistent with our recent finding that karyotypically silent translocations to the IgH locus occur frequently in MM6. In addition, this translocation is not a cell-line artifact, as we have confirmed the presence of the translocation breakpoint in six primary tumour samples: PCL-1, MM.T1, MM.T2 and the corresponding tumours of three (JIM3, H929 and UTMC2) of the five MM lines with this translocation (data not shown).
Dominant, activating mutations in different domains of FGFR3 are implicated in dwarfism, including (in order of increasing severity) hypochondroplasia, achondroplasia and thanatophoric dysplasia type I and II (TDI-II)10–13. We used RT-PCT SSCP analysis to screen for FGFR3 mutations in all twelve MM cell lines (KMS11, OPM2, H929, UTMC2, H1112, TH, KMM1, 8226, ARK, SKMM1, Delta 47 and U266) and five tumour samples (JIM3-T, PCL-1, MM.TO, MM.T1 and MM.T2) that express FGFR3 mRNA. For example, as shown in Fig. 3d, KMS11 contains wild-type and variant alleles but expresses only the variant allele, which was found to contain a Y373C [A1157G] mutation. Missense mutations of FGFR3 were also identified in the OPM2 cell line (K650E [A1987G]) and in the MM.T1 primary tumour (K650M [A1988T]), both of which have t(4;14) translocations. Each of these missense mutations occurs in thanatophoric dysplasia14–16; for one of them (K650E), the constitutive activation of FGFR3 in the absence of ligand has been proved by transfection experiments11,12.
Although we cannot exclude the possibility that other genes are dysregulated by the translocation t(4;14) in MM, we have identified the FGFR3 gene as an apparent oncogene that is dysregulated by this translocation into an IgH switch region. First, the FGFR3 gene is localized no more than about 100 kb from the most centromeric breakpoint at 4pl6.3, and is located on the der(14) chromosome that contains the 3' IgH enhancer(s) but not the intronic enhancer (Fig. 1a). This is similar to the situation for cyclin Dl, which is localized 100–400 kb from the breakpoint in the translocation t(l1;14)(ql3; q32.3) that occurs in mantle-cell lymphoma and MM tumours5,17–19. Second, FGFR3 mRNA is overexpressed in all but one cell line (JIM3) with the translocation t(4;14), but primary tumour cells (JIM3-T) from the excepted line do express FGFR3 mRNA (Fig. 3a). It is possible that the translocation in JIM3 was initially associated with dysregulated expression of FGFR3, but that silencing of this gene occurred during tumour evolution after another genetic event that eliminated the need for FGFR3 expression. Conversely, all MM lines that overexpress FGFR3 have a translocation t(4;14). It was somewhat surprising to us that the level of expression of FGFR3 mRNA or protein in the UTMC2 line is approximately 30 times lower than found for the KMS11 line, even though Southern blotting indicates no amplification of the FGFR3 gene in any of the lines with a translocation (data not shown). However, consistent with this extreme variability in expression (Fig. 3c), the expression of c-myc in Burkitt lymphoma lines that have an t(8; 14) translocation can vary over an approximately 100-fold range (M. Zajac-Kaye, personal communication). Third, Iwo cell lines (KMS11, OPM2) and two tumours (PCL.1, MM.T1) contain two genetically distinguishable FGFR3 alleles, but each selectively expresses one allele, as predicted if the translocation is responsible for dysregulation of FGFR3 (Fig. 3 and data not shown). Fourth, the variant, missense alleles in KMS11, OPM2 and MM.T1 must result from a somatic mutation, since these mutations in the germline cause thanatophoric dysplasia14–16, the most severe form of dwarfism, which results in fetal or neonatal death. The activating mutations of FGFR3 seem to occur frequently in MM—that is, two of five lines and one of three primary tumours in which we have identified a t(4;14) translocation by cloning or mapping. We suggest that the translocation t(4;14) occurred at an early stage of tumorigenesis, and the subsequent mutation in the dysregulated FGFR3 gene during tumour progression contributed to tigand independent growth. Finally, FGFs have been shown to have different effects on cell growth, survival and differentiation, depending on the cell type20. More specifically, FGFR3 can transduce signals that either inhibit or stimulate cell proliferation. For example, FGFR3 inhibits proliferation of chondrocytes in normal individuals or excessively inhibits growth of chondrocytes in individuals that have FGFR3 alleles with activating mutations21–23. Alternatively, it has been shown that an IL3-dependent lymphoid cell line expressing a transfected FGFR3 gene can be mitogenically stimulated by FGFs plus heparin in the absence of IL3 (ref. 24). It has been postulated by others that growth and/or survival of MM cells is mediated by interaction with bone marrow stromal cells25. We propose that dysregulated expression of FGFR3 provides a continuous oncogenic signal for MM cells juxtaposed to stromal cells that are known to express FGFs26.
Methods
Cell culture
Human MM cell lines, as described in detail previously, were grown in Petri dishes in RPMI 1640 medium supplemented with 10% fetal calf serum6. For three cell lines, we obtained corresponding primary tumour cells—that is, pleural effusion cells for the JIM3 (JIM3-T), H929 and UTMC2 cell lines. Primary tumour samples reported previously include PCL.1 and MM.T0, corresponding to patient #1 and patient #2, respectively6. Additional intramedullary MM tumor specimens are designated MM.T1 to MM.T8.
Cosmid clones
L190b4, L184d6, L75b9, pWC385.31 and pC385.12 cosmids were kindly provided to us by M.R. Altherr, Los Alamos National Laboratory.
Translocation breakpoint fragments
The cloned chromosome 4 translocation breakpoint fragments included a 7.6-kb HindIII fragment detected with a 3' Sµ probe (KMS11); 2.0-kb (OPM2) and 3.8-kb (JIM3) HindIII fragments detected with a 3' Sγ probe; and 4.0-kb SphI (H929) and 2.6-kb HindIII (PCL-1) fragments detected with a 5' Sµ probe.
Isolation and characterization of PAC184d6/385
A PAC human genomic library (Genome Systems) was screened with a telomeric L75b9 probe (#9295, 237 bp) PCR amplified using the following primers: 5'–TTGCT-GTCCCGTAAGTTGCTAAG–3' and 5' –TCTGCTATTGTGATACCGCAT-GAC–3' (56 °C). A single clone was obtained, and its two ends were sequenced. The SP6 end overlaps L184d6 by 5 kb, whereas the telomeric T7 end has no homology to known sequences and does not overlap any of the cosmids in this region. Sequence from the T3 and T7 ends of pC385.12 (which contains the FGFR3 gene) enabled us to make primer pairs for each end of the insert (5'–GATATTGCGGAAGATACTAAGGC–3' and 5'–GTAAGTGGGTCTCACTATGTTGC–3'; 5'-CAGACAATACTAAACC-GAGCTTTC–3' and 5'–CGCTGCCTCAATCTATGCTCTCAG–3'). These primers were used in a PCR assay to demonstrate that both ends of the pC385.12 insert are contained in the PAC clone, while only the T3 end overlaps the pC385.31 cosmid clone. The FGFR5 gene appears to be located approximately 100 kb telomeric to the most centromeric OPM2 translocation breakpoint.
Primers and probes
Switch region probes upstream (for example, 5'Sµ) and downstream (for example, 3'Sµ) of the repetitive sequences in each switch region were generated. The sequence of PCR primers used to amplify these probes is reported elswhere6, with the exception of the 5'Sµ probe (260 bp), which was amplified with the following primers: 5'–-GGGACCT-GCTCATTTTTATC–3' and 5'–TCAGCTAAAGCCATCTCATTGCC–3'(56 °C). The 9037 probe was a 211-bp fragment PCR amplified from L184d6 cosmid with the following primers: 5'–CTTGCTTGA-CAACGTCAGGCTAC–3' and 5'–ACCTGGCTGAGGAGACAGTAA-GTG–3' (56 °C). The 9213 (151 bp), 9226 (328 bp) and 9330 (264 bp) probes were PCR amplified from L75b9 cosmid by 5'–TGCAGTAAGTC-CTCACTCTACCAAC–3' and 5'–CACAACTGATGGTGGTTCAAATC–3' (61 °C), 5'–CTGCTGCATGTACCTGGCTAAAG–3' and 5'–TGCTGGGAA-GTATGATGAAGGAAG–3' (61°C), 5'–CTTAGTGTCTGCTCGAGCTTT–3' and 5'–ACAACTCAACCCATACAGGGG–3' (59 °C), respectively: the FGFR3 probe used in the northern-blot assay was a PCR amplified 262-bp fragment, extending from the middle of exon IIIC to the middle of the transmembrane-spanning domain27. The β-actin probe was a 2.0-kb BamHI rabbit cDNA fragment. Information regarding the oligonucleotides used to generate PCR fragments from FGFR3 cDNA or genomic DNA for SSCP analysis are available on request.
RT-PCT assays
The efficiency of cDNA synthesis was monitored by measuring incorporation of labelled dCTP, and the quality of the cDNA was also verified by PCR amplification of an 804-bp c-myc fragment (except for cell line U266, which does not express c-myc mRNA). An RT-PCT assay, utilizing the primer pairs described by Johnston et al.28 and 10 ng of cDNA, was used to assess the expression of the exon IIIB and IIIC alternatively spliced forms of FGFR3 mRNA, both of which showed similar patterns of expression. Product was detected by ethidium bromide staining of a 2% agarose gel after 30 to 40 cycles, and also by hybridization with an internal oligonucleotide probe (5'–GAGTCCAACGCGTCCATGAGC–3') to detect product blotted from a gel after 23 cycles.
Immunoprecipitation and immunoblotting
Ten million cells were harvested and lysed in RIPA buffer (PBS, 1% NP-40, 0.5% sodium deoxy-cholate, 0.1% SDS, 10 µg/ml aprotinin, 1 mM sodium orthovanadate), and 2.5 mg of the total proteins obtained was immunoprecipitated with antiserum specific for the Cinus terminus of FGFR3 (Santa Cruz Biotechnology). The immune complexes were collected on protein A/G PLUS-Agarose beads (Santa Cruz Biotechnology), washed extensively, electrophoresed through a 7.5% SDS-PAGE gel and transferred to nitrocellulose. The filter was incubated with anti-FGFR3 antiserum and horse-radish peroxidase-conjugated goat anti-rabbit IgG (Amersham) and developed with the ECL system (Amersham).
Fluorescence in situ hybridization
Metaphase chromosome preparation, chromosome-14 painting probe generation, hybridization and detection protocols are described elsewhere29. The pC385.12 cosmid, containing the entire FGFR3 gene, was labelled by nick-translation and is localized to 4pl6.3.
Other procedures
Construction of genomic libraries, screening, isolation and sequencing of recombinant clones, Southern and northern blot analysis5, RT-PCT and SSCP analysis30 are described elsewhere31.
GenBank accession numbers for translocation breakpoint sequences
U73660 for JIM3, U73661 for PCL-1, U73662 for H929, U73663 for KMS11 and AF006657 for OPM2.
Acknowledgements
We are indebted to Dr. S. Kirby for providing the PCL.1 primary tumour sample, and to Drs. I.A. Kirsch, I. MacLennan and A. Solomon for providing primary tumour material corresponding to the H929, JIM3 and UTMC2 MM cell lines, respectively. Finally, we would like to thank M. R. Altherr (Los Alamos National Laboratory) for providing materials and for helpful discussion about FGFR3 mapping, and the many investigators who supplied multiple myeloma cell lines. This work was supported in part by generous contributions from the Dorothy Rodbell Cohen Foundation and the Melvin Savin Memorial Fund.
References
- 1.Korsmeyer SJ. Chromosomal translocations in lymphoid malignancies reveal novel proto-oncogenes. Annu. Rev. Immunol. 1992;10:785–807. doi: 10.1146/annurev.iy.10.040192.004033. [DOI] [PubMed] [Google Scholar]
- 2.Taniwaki M, et al. Nonrandom chromosomal rearrangements of 14q32.3 and 19p13.3 and preferential deletion of 1p in 21 patients with multiple myeloma and plasma cell leukemia. Blood. 1994;84:2283–2290. [PubMed] [Google Scholar]
- 3.Sawyer JR, Waldron JA, Jagannath S, Barlogie B. Cytogenetic findings in 200 patients with multiple myeloma. Cancer Genet. Cytogenet. 1995;82:41–49. doi: 10.1016/0165-4608(94)00284-i. [DOI] [PubMed] [Google Scholar]
- 4.Lai JL, et al. Improved cytogenetics in multiple-myeloma—a study of 151 patients including 117 patients at diagnosis. Blood. 1995;85:2490–2497. [PubMed] [Google Scholar]
- 5.Chesi M, et al. Dysregulation of Cyclin D1 by translocation into an IgH gamma switch region in two multiple myeloma cell lines. Blood. 1996;88:674–681. [PubMed] [Google Scholar]
- 6.Bergsagel PL, et al. Promiscuous translocations into IgH switch regions in multiple myeloma. Proc. Natl. Sci. Acad. USA. 1996;93:13931–13936. doi: 10.1073/pnas.93.24.13931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baxendale S, et al. A cosmid contig and high resolution restriction map of the 2 megabase region containing the Huntington's disease gene. Nature Genet. 1993;4:181–186. doi: 10.1038/ng0693-181. [DOI] [PubMed] [Google Scholar]
- 8.Keegan K, Johnson D, William LT, Hayman MJ. Isolation of an additional member of the fibroblast growth factor receptor family, FGFR-3. Proc. Natl. Acad. Sci. USA. 1991;88:1095–1099. doi: 10.1073/pnas.88.4.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keegan K, Meyer S, Hayman MJ. Structural and biosynthetic characterization of the fibroblast growth factor receptor 3 (FGFR-3) protein. Oncogene. 1991;6:2229–2236. [PubMed] [Google Scholar]
- 10.Muenke M, Schell U. Fibroblast-growth-factor-receptor mutations in human sketetal disorders. Trends Genet. 1995;11:308–313. doi: 10.1016/s0168-9525(00)89088-5. [DOI] [PubMed] [Google Scholar]
- 11.Naski MC, Wang Q, Xu J, Ornitz DM. Graded activation of fibroblast growth factor receptor 3 by mutations causing achondroplasia and thanatophoric dysplasia. Nature Genet. 1996;13:233–237. doi: 10.1038/ng0696-233. [DOI] [PubMed] [Google Scholar]
- 12.Webster MK, D'Avis PY, Robertson SC, Donoghue DJ. Profound ligand-independent kinase activation of fibroblast growth factor receptor 3 by the activation loop mutation responsible for a lethal skeletal dysplasia, thanatophoric dysplasia type II. Mol. Cell. Biol. 1996;16:4081–4087. doi: 10.1128/mcb.16.8.4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Webster MK, Donoghue DJ. Constitutive activation of fibroblast growth factor receptor 3 by the transmembrane domain point mutation found in achondroplasia. EMBO J. 1996;15:520–527. [PMC free article] [PubMed] [Google Scholar]
- 14.Rousseau F, et al. Missense FGFR3 mutations create cysteine residues in thanatophoric dwarfism type I (TD1) Hum. Mol. Genet. 1996;5:509–512. doi: 10.1093/hmg/5.4.509. [DOI] [PubMed] [Google Scholar]
- 15.Francomano CA, et al. A new skeletal dysplasia with severe tibial bowing, profound developmental delay and acanthosis nigricans is caused by a Lys 650 Met mutation in fibroblast growth factor receptor 3 (FGFR3) Am. J. Hum. Genet. 1996;59:A25. doi: 10.1086/302275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tavormina PL, et al. Thanatophoric dysplasia (types I and II) caused by distinct mutations in fibroblast growth factor receptor 3. Nature Genet. 1995;9:321–328. doi: 10.1038/ng0395-321. [DOI] [PubMed] [Google Scholar]
- 17.Williams ME, Swerdlow SH, Meeker TC. Chromosome t(11;14)(q13;q32) breakpoints in centrocytic lymphoma are highly localized at the bcl-1 major translocation cluster. Leukemia. 1993;7:1437–1440. [PubMed] [Google Scholar]
- 18.Tsujimoto Y, et al. Clustering of breakpoints on chromosome 11 in human B-cell neoplasms with the t(11;14) chromosome translocation. Nature. 1985;315:340–343. doi: 10.1038/315340a0. [DOI] [PubMed] [Google Scholar]
- 19.de Boer CJ, et al. Multiple breakpoints within the BCL-1 locus in B-cell lymphoma: rearrangements of the cyclin D1 gene. Cancer Res. 1993;53:4148–4152. [PubMed] [Google Scholar]
- 20.Mason IJ. The ins and outs of fibroblast growth factors. Cell. 1994;78:547–552. doi: 10.1016/0092-8674(94)90520-7. [DOI] [PubMed] [Google Scholar]
- 21.Su WS, et al. Activation of Stat1 by mutant fibroblast growth-factor receptor in thanatophoric dysplasia type II dwarfism. Nature. 1997;386:288–292. doi: 10.1038/386288a0. [DOI] [PubMed] [Google Scholar]
- 22.Colvin JS, Bohne BA, Harding GW, McEwen DG, Ornitz DM. Skeletal overgrowth and deafness in mice lacking fibroblast growth factor receptor 3. Nature Genet. 1996;12:390–397. doi: 10.1038/ng0496-390. [DOI] [PubMed] [Google Scholar]
- 23.Deng C, Wynshaw-Boris A, Zhou F, Kuo A, Leder P. Fibroblast growth factor receptor 3 is a negative regulator of bone growth. Cell. 1996;84:911–921. doi: 10.1016/s0092-8674(00)81069-7. [DOI] [PubMed] [Google Scholar]
- 24.Ornitz DM, Leder P. Ligand specificity and heparin dependence of fibroblast growth factor receptors 1 and 3. J. Biol. Chem. 1992;267:16305–16311. [PubMed] [Google Scholar]
- 25.Caligaris-Cappio F, et al. Role of bone marrow stromal cells in the growth of human multiple myeloma. Blood. 1991;77:2688–2693. [PubMed] [Google Scholar]
- 26.Allouche M. Basic fibroblast growth factor and hematopoiesis. Leukemia. 1995;9:937–942. [PubMed] [Google Scholar]
- 27.Shiang R, et al. Mutations in the transmembrane domain of FGFR3 cause the most common genetic form of dwarfism, achondroplasia. Cell. 1994;78:335–342. doi: 10.1016/0092-8674(94)90302-6. [DOI] [PubMed] [Google Scholar]
- 28.Johnston CL, Cox HC, Gomm JJ, Coombes RC. Fibroblast growth factor receptors (FGFRs) localize in different cellular compartments. J. Biol. Chem. 1995;270:30643–30650. doi: 10.1074/jbc.270.51.30643. [DOI] [PubMed] [Google Scholar]
- 29.Schröck E, et al. Multicolor spectral karyotyping of human chromosomes. Science. 1996;273:494–497. doi: 10.1126/science.273.5274.494. [DOI] [PubMed] [Google Scholar]
- 30.Kuehl WM, Brents LA, Chesi M, Bergsagel PL. Selective expression of one c-mycallele in two human myeloma cell lines. Cancer Res. 1996;56:4370–4373. [PubMed] [Google Scholar]
- 31.Davis LG, Kuehl WM, Battey JF, et al. Basic Methods in Molecular Biology. Norwalk, Connecticut: Appleton & Lange; 1994. [Google Scholar]