Abstract
Bcl-xL plays a critical role in maintaining cell survival. However, the relationship between the potential interaction of Bcl-xL with other cytosolic proteins and the regulation of cell survival remains incompletely defined. We have identified translationally controlled tumor protein (TCTP), a multifunctional protein, as a novel antiapoptotic Bcl-xL-interacting protein. TCTP interacted in vivo and in vitro with Bcl-xL, and their sites have been mapped to an N-terminal region of TCTP and the Bcl-2 homology domain 3 of Bcl-xL. Consistent with a role in maintaining T-cell survival during activation, TCTP was significantly upregulated in murine T cells activated by T-cell antigen receptor (TCR) ligation and CD28 costimulation, which was correlated with the upregulation of Bcl-xL in activated T cells. Moreover, downregulation of TCTP expression by antisense technology in T cells results in the increase of T-cell apoptosis. Furthermore, the N-terminal region of TCTP was required for its ability to inhibit apoptosis. In conclusion, this study has demonstrated that an N-terminal region of a cytosolic protein, TCTP, is required for its binding to Bcl-xL and for its antiapoptotic activity.
Keywords: Bcl-xL, TCTP, apoptosis, protein interaction
Introduction
Bcl-x proteins play an essential role in maintaining T-cell and thymocyte survival (Boise et al., 1995; Ma et al., 1995; Motoyama et al., 1995; Yang et al., 1997b; Opferman and Korsmeyer, 2003; Marrack and Kappler, 2004) and T-cell function (Boise et al., 1995; Ye et al., 2002). The dominant antiapoptotic isoform of the Bcl-x gene, Bcl-xL, has been implicated in the prevention of cytochrome c release by maintaining the integrity of the mitochondrial membrane (Motoyama et al., 1995). The current model for Bcl-xL function requires that Bcl-xL be anchored to membranes (Ma et al., 1995; Ye et al., 2002; Marrack and Kappler, 2004). However, Bcl-xL is present in both soluble cytosolic and membrane-bound forms (Hsu et al., 1997) and may move through the cytosol before becoming attached to mitochondrial membrane (Adams, 2003; Jeong et al., 2004). Furthermore, among the five Bcl-x isoforms identified (Boise et al., 1993; Fang et al., 1994; Gonzalez-Garcia et al., 1994; Yang et al., 1997b), the antiapoptotic Bcl-x isoforms that share Bcl-2 homology domains 1–4 (BH1–4) with Bcl-xL, Bcl-xγ, Bcl-xΔTM, and Bcl-xβ have no membrane-anchoring domain and are located in the cytosolic fraction (Fang et al., 1994; Yang et al., 1997b). The functional significance of the cytosolic localization of prosurvival Bcl-x isoforms remains incompletely defined.
We hypothesized that a protein complex exists consisting of Bcl-xL and another cytosolic protein, which are temporally co-upregulated in T-cell activation and functionally collaborated for T-cell survival. To test this hypothesis, we used a yeast two-hybrid system to screen a lymphocyte cDNA library for cytosolic Bcl-xL-interacting proteins. We have identified translationally controlled tumor protein (TCTP) as a novel Bcl-xL-interacting protein. TCTP is a multifunctional protein (also see an excellent review by Bommer and Thiele, 2004). Intracellular TCTP was previously identified as a microtubule-stabilizing protein (Yarm, 2002), an antiapoptotic protein (fortilin) (Li et al., 2001), and a guanine nucleotide dissociation inhibitor regulating protein translation (Cans et al., 2003). We show that both TCTP and Bcl-xL are highly upregulated during T-cell activation induced by T-cell antigen receptor (TCR) and CD28 costimulation, suggesting a role for TCTP in unifying the cell survival function of Bcl-xL with certain pathways in T-cell activation.
Results
Bcl-xL interacts with TCTP
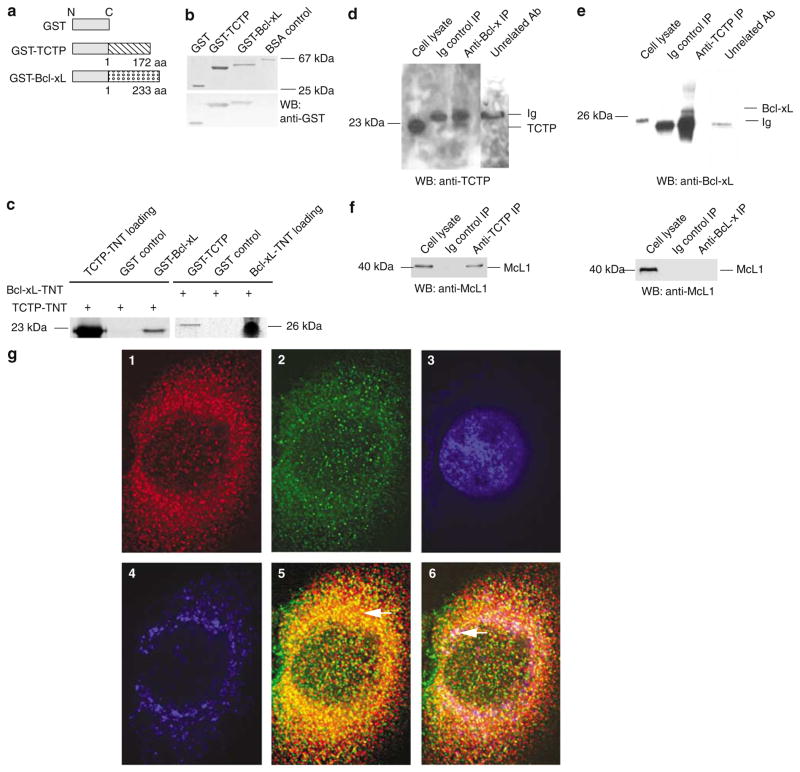
To examine whether novel cytosolic protein(s) may interact with Bcl-xL, the yeast two-hybrid system was used with full-length Bcl-xL as bait. After screening, three identical clones with a 0.8 kb cDNA insert were detected (not shown). GenBank BLAST searching identified this cDNA as TCTP (Bommer and Thiele, 2004) (GenBank accession: NM_009429). Murine TCTP cDNA had an open reading frame (ORF) of 519 bp and encoded 172 amino acids (aa). The results of Bcl-xL interaction with TCTP revealed by yeast two-hybrid screening included the positive activation of a nutritional reporter gene (HIS3) and subsequently the positive activation of a second reporter gene (β-galactosidase) (not shown), suggesting that Bcl-xL interacted with TCTP inside the cells. To test whether the interaction between Bcl-xL and TCTP is direct, GST pull-down assays were performed with 35S-labelled TCTP prepared by in vitro transcription and translation (TNT) and purified GST-Bcl-xL fusion protein. The results in Figure 1a–c show that TCTP interacts specifically with Bcl-xL but not with two negative controls including glutathione S-transferase (GST) control and glutathione Sepharose beads control (not shown). The stringency of the buffer used in the GST pull-down assay was stronger than that used in previous studies for the identification of Bcl-xL interaction proteins Rad9 (Komatsu et al., 2000) and BAR (Zhang et al., 2000), suggesting the high specificity of the interaction between TCTP and Bcl-xL. The direct interaction between TCTP and Bcl-xL was confirmed by repeating the pull-down assay with GST-TCTP and 35S-labelled TNT-prepared Bcl-xL (Figure 1c, right panel). The interaction between TCTP and Bcl-xL was further established in vivo with co-immunoprecipitation (co-IP) assays. As shown in Figure 1d, we found that anti-Bcl-xL antibodies, but neither the Ig control nor an unrelated antibody (anti-splicing factor ASF/SF2), specifically co-immunoprecipitated endogenous TCTP. Similarly, anti-TCTP antibodies, but neither the Ig control nor an unrelated antibody (anti-ASF/SF2), also specifically co-immunoprecipitated endogenous Bcl-xL (Figure 1e), suggesting that Bcl-xL interacted with TCTP in cells. Under these immunoprecipitation conditions, the anti-TCTP antibodies also co-immunoprecipitated endogenous Mcl-1, as previously reported (Figure 1f, left panel) (Zhang et al., 2002), which also served as a positive control for the co-IP experiments in identification of TCTP–Bcl-xL interaction. These results suggest that TCTP interacts not only with Mcl-1 but also with another antiapoptotic, Mcl-1 homologous, Bcl-2 family protein Bcl-xL. In addition, since our data (Figure 1f, right panel) and others’ show that Mcl-1 does not interact with Bcl-xL in cells (Bae et al., 2000), our results suggest that TCTP interacts with Bcl-xL directly, instead of mediating through Mcl-1.
Figure 1.
TCTP interacts with Bcl-xL. (a) Schematic representation of GST, GST-Bcl-xL, and GST-TCTP fusion proteins. (b) Migration of purified GST, GST-Bcl-xL, and GST-TCTP on Coomassie blue-stained SDS–PAGE (upper panel) and their specific reactivity on Western blot (lower panel) to GST-specific antibodies with bovine serum albumin (BSA) as a negative control. (c) GST pull-down assay. On the left panel, GST-Bcl-xL, but not GST, precipitated 35S-labelled TCTP (prepared by in vitro TNT). On the right panel, GST-TCTP, but not GST, pulled down 35S-labelled Bcl-xL. (d) Co-IP of endogenous TCTP with endogenous Bcl-xL. The co-IP was performed with anti-Bcl-xL antibodies, an Ig control, and an unrelated antibody control (anti-ASF/SF2), followed by Western blot with anti-TCTP antibodies. (e) Co-IP of endogenous Bcl-xL with endogenous TCTP. The co-IP was performed with anti-TCTP antibodies, an Ig control, and an unrelated antibody control (antisplicing factor ASF/SF2), followed by Western blot with anti-Bcl-xL antibodies. (f) Co-IP of endogenous Mcl-1 with endogenous TCTP but not with Bcl-xL. As a positive control for TCTP interaction with a known partner, the co-IP was performed with anti-TCTP antibodies followed by Western blot with anti-Mcl-1 antibodies (left panel). In addition, the co-IP was performed with anti-Bcl-xL antibodies followed by Western blot with anti-Mcl-1 antibodies (right panel). (g) Colocalization of TCTP and Bcl-xL in cytosol and mitochondria demonstrated by immunofluorescence confocal microscopy. In panel g1, the subcellular location of TCTP was revealed with anti-TCTP antibodies followed by secondary antibodies conjugated to AlexaFluor594. In panel g2, the subcellular location of Bcl-xL was detected with anti-Bcl-xL antibodies followed by secondary antibodies conjugated with AlexaFluor488. In panel g3, nuclear DNA was costained with DAPI. In panel g4, mitochondria were stained with MitoTracker Deep Red 633 and transformed into blue pseudocolor, as observed by microscope. In panel g5, colocalizing TCTP-Bcl-xL (yellow areas, arrow) is more abundantly observed in the cytosol. In panel g6, TCTP-Bcl-xL colocalization is demonstrated in both cytosol (yellow areas) and mitochondria (white areas, arrow)
Next, we examined whether endogenous TCTP and Bcl-xL reside in the same cellular compartment. All images were acquired by confocal microscopy and deconvolved. We used HeLa cells to facilitate our immunofluorescence studies by taking into consideration the expression of both TCTP and Bcl-xL in HeLa cells as well as the high degree of conservation of both proteins between mice and humans. Consistent with previous reports, TCTP (red; Figure 1g1) was predominantly localized in the cytosol with a punctuated pattern (Gachet et al., 1999; Bommer and Thiele, 2004). Similar to the previous report that Bcl-xL is localized in both cytosol and mitochondria (Hsu et al., 1997), we found that Bcl-xL (green; Figure 1g2) was localized in both cytosol and mitochondria (not shown). As judged by the yellow spots localized in cytosol (Figure 1g5) after superimposing subcellular TCTP in red (Figure 1g1) with that of Bcl-xL in green color (Figure 1g2), we conclude that TCTP and Bcl-xL are partially colocalized in the cytosol. Furthermore, as judged by the white spots localized in mitochondria (Figure 1g6) after superimposing subcellular TCTP in red (Figure 1g1), that of Bcl-xL in green color (Figure 1g2) with the mitochondria in blue (Figure 1g4), we also conclude that TCTP and Bcl-xL are partially colocalized in the mitochondria.
The N-terminal region of TCTP mediates its interaction with Bcl-xL
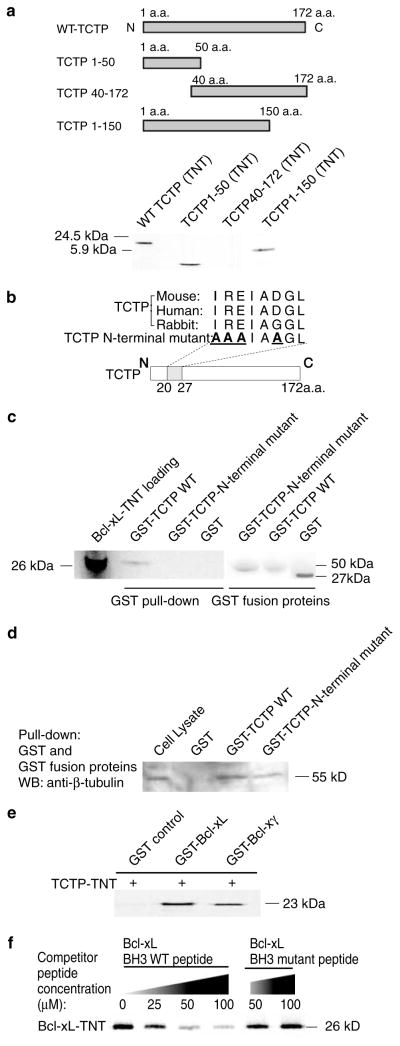
By comparing TCTP sequences from 24 eukaryotes (Thaw et al., 2001), two highly conserved TCTP signature regions were identified, including the TCTP1 signature from aa 45 to 55 and the TCTP2 signature from aa 129 to 147, both of which may mediate the interactions between TCTP and other molecules. Furthermore, several TCTP-interacting proteins have been identified with the interaction domains mapped: (1) the C-terminal self-interaction domain from aa 126 to 172 (Yoon et al., 2000); (2) the polo-like kinase interaction domain from aa 107 to 172 (Yarm, 2002); (3) the tubulin binding domain from aa 79 to 123 (Thaw et al., 2001); (4) the Ca2+ binding region from aa 81 to 112 (Kim et al., 2000); (5) the triad binding surface consisting of Glu12, Leu74, and Glu134 for binding the Rab G protein-like translation elongation factor eEF1A (Thaw et al., 2001; Cans et al., 2003); and (6) Arg21 critical for the interaction between TCTP and the Bcl-xL homolog Mcl-1 (Zhang et al., 2002). To map the Bcl-xL-interacting region in TCTP, we constructed three TCTP deletion mutants: (a) the TCTP1–50 aa mutant, (b) the TCTP1–150 aa mutant that contains both TCTP1 and TCTP2 signature regions, and (c) the TCTP40–172 aa mutant that also contains both TCTP1 and TCTP2 signature regions but lacks the N-terminus of TCTP. As shown in Figure 2a, Bcl-xL bound to the TCTP1–50 aa and the TCTP1–150 aa deletion mutants but not to the TCTP40–172 aa mutant, suggesting that the Bcl-xL interaction region was localized to the TCTP N-terminal 1–40 aa region.
Figure 2.
The N-terminal region in TCTP and the BH3 domain in Bcl-xL mediate the interaction of TCTP and Bcl-xL. (a) The upper panel shows schematic representation of WT TCTP and three deletion mutants of TCTP. Bcl-xL interaction was mapped to the region between aa 1 and 40 of TCTP by GST pull-down assay with 35S-labelled TCTP WT, TCTP deletion mutants, and GST-Bcl-xL. (b) Comparison of the N-terminal regions (8 aa) of TCTP from three different species. The N-terminal region mutant sequence of TCTP is also presented. The mutated amino acids (replaced with alanine, A) in TCTP N-terminal region are in bold and underlined. (c) Dependence of TCTP interaction with Bcl-xL on the N-terminal region of TCTP. 35S-labelled Bcl-xL was precipitated by GST-TCTP WT but not by an equivalent amount of GST or GST-TCTP N-terminal mutant (Coomassie blue-stained gel shown on the right). (d) Appropriate folding of GST-TCTP WT and GST-TCTP N-terminal mutant. Endogenous β-tubulin was precipitated by GST-TCTPWT and GST-TCTP N-terminal mutant but not by the GST control. (e)Mapping of TCTP binding domain in the Bcl-x N-terminal common region shared by the alternative spliced Bcl-x isoforms. The TCTP binding domain was mapped by GST pull-down assay using 35S-labelled TCTP, GST-Bcl-xL, and GST-Bcl-xγ. (f) Identification of the BH3 domain in the N-terminal Bcl-x common region of Bcl-xL as the TCTP binding domain. The binding of 35S-labelled Bcl-xL by GST-TCTP was competed away with increasing concentrations of WT Bcl-xL BH3 domain peptide, but not with Bcl-xL BH3 domain mutant peptide
We further searched the N-terminal 40 aa of TCTP for a Bcl-xL-interacting region. We found an N-terminal sequence (20IREIADGL27) in TCTP that is highly conserved among mice, rabbits, and humans (Figure 2b). This region of TCTP included the Arg21 that was critical for the TCTP–Mcl-1 interaction (Zhang et al., 2002). We mutated the amino acids Ile20, Arg21, Glu22, and Asp25 in the N-terminal region of TCTP into Ala20, Ala21, Ala22, and Ala25 (Figure 2b). The mutation from Ile20 into Ala20 was designed according to those mutations previously used in Bcl-xL BH3 mutant peptide to disrupt hydrophobic interactions between interacting partners (Shangary and Johnson, 2002). The mutations of the other three charged amino acids from Arg21, Glu22, and Asp25 into Ala21, Ala22, and Ala25, respectively, were designed using the charge-to-alanine scanning mutagenesis approach (Gibbs and Zoller, 1991) to disrupt potential protein–protein interactions while causing only minor perturbation in protein structure (Gibbs and Zoller, 1991). As shown in Figure 2c, wild-type (WT) GST-TCTP precipitated Bcl-xL, but an equivalent amount of GST-TCTP bearing the mutated N-terminal region, and GST did not precipitate Bcl-xL. Of note, the N-terminal mutant form of GST-TCTP was capable of precipitating β-tubulin (Figure 2d), suggesting that the lack of detectable interaction between Bcl-xL and the mutant TCTP was not due to potential misfolding of the latter protein. These results show that the N-terminal region of TCTP, but not the β-tubulin binding domain from aa 79 to 123 of TCTP (Gachet et al., 1999), mediates its interaction with Bcl-xL.
Bcl-xL BH3 domain is required for its interaction with TCTP
To examine whether the Bcl-xL N-terminal common region, shared by all the Bcl-x antiapoptotic isoforms, mediates the interaction of Bcl-xL with TCTP, we used both GST-Bcl-xL and GST-Bcl-xγ in GST pull-down assays. As shown in Figure 2e, both GST-Bcl-xL and GST-Bcl-xγ precipitate TCTP, suggesting that Bcl-xL interacts with TCTP via the N-terminal Bcl-x common region (aa 1–188) shared by both Bcl-x isoforms (Yang et al., 1997b) rather than the Bcl-x isoform-specific C-terminal regions. Since the BH3 domain is responsible for both hetero- and homodimerization between anti-apoptotic and proapoptotic molecules, we examined whether the BH3 domain of Bcl-xL mediates its interaction with the N-terminal region of TCTP (Diaz et al., 1997; Shangary and Johnson, 2002). We used a peptide competition assay using peptide sequences comprising the WT or mutant form of the Bcl-xL BH3 domain. The mutations in the Bcl-xL BH3 domain peptide altered some of the critical residues responsible for the Bcl-xL–TCTP interaction, decreasing the competitive capacity of the peptide (Shangary and Johnson, 2002). As shown in Figure 2f, increasing concentrations of cold peptide bearing the WT Bcl-xL BH3 domain sequence (90LREAGDEF97) attenuated the binding of 35S-labelled Bcl-xL to GST-TCTP. In contrast, the Bcl-xL BH3 domain mutant peptide 90AREAGAEF97 (mutated residues are underlined) did not competitively reduce the binding of Bcl-xL to GST-TCTP, even at high concentrations of the cold mutant peptide (Figure 2f). Taken together, these results suggest that the interaction between Bcl-xL and TCTP is specifically mediated through the Bcl-xL BH3 domain and an N-terminal region of TCTP.
TCTP is co-upregulated with Bcl-xL during T-cell activation
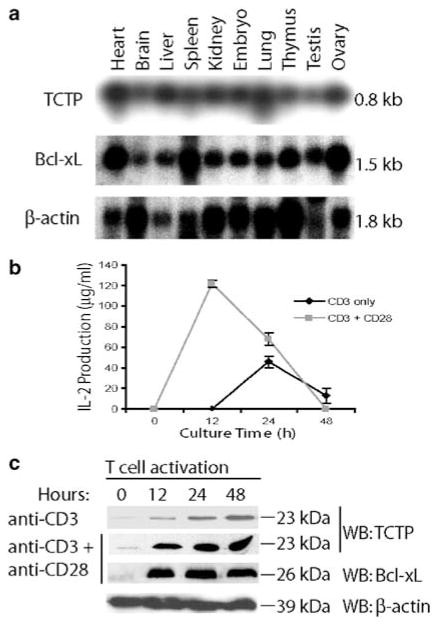
Northern blot analyses (Figure 3a) show that TCTP and Bcl-xL are both expressed in lymphoid tissues, suggesting that coexpression of antiapoptotic TCTP and Bcl-xL may play an important role in T-cell function. We reasoned that expression of TCTP may be upregulated during T-cell activation induced by TCR ligation and CD28 costimulation, similar to that of Bcl-xL (Boise et al., 1995) and IL-2 (Schwartz, 2003). Thus, we examined the expression of TCTP protein at several time points (0, 12, 24, and 48 h) following murine T-cell activation induced by TCR ligation and CD28 costimulation. IL-2 secretion was significantly upregulated during T-cell activation, suggesting that T cells were fully activated (Figure 3b). TCTP protein levels were significantly upregulated when T cells were activated with plate-bound anti-CD3 antibody alone. The upregulation of TCTP expression in T cells activated with combined anti-CD3 and anti-CD28 antibodies was higher than that in T cells activated by anti-CD3 alone (Figure 3c). The co-upregulation of Bcl-xL and TCTP suggested a novel mechanism by which activated T cells can survive.
Figure 3.
Co-upregulation of TCTP and Bcl-xL during T-cell activation. (a) The expression of TCTP in the lymphoid system (spleen and thymus) and other tissues was revealed by Northern blot with probes for TCTP, Bcl-xL, and β-actin (as control). (b) T-cell activation was induced by plate-bound anti-CD3 antibody and anti-CD28 antibody coligation. T-cell activation was measured by an IL-2-specific ELISA assay over a 2-day time course. (c) Expressions of TCTP, Bcl-xL, and β-actin were detected by Western blotting at 0, 12, 24, and 48 h after T-cell activation induced by anti-CD3 alone, or by anti-CD3 and anti-CD28
Antiapoptotic effects of TCTP require its N-terminal region
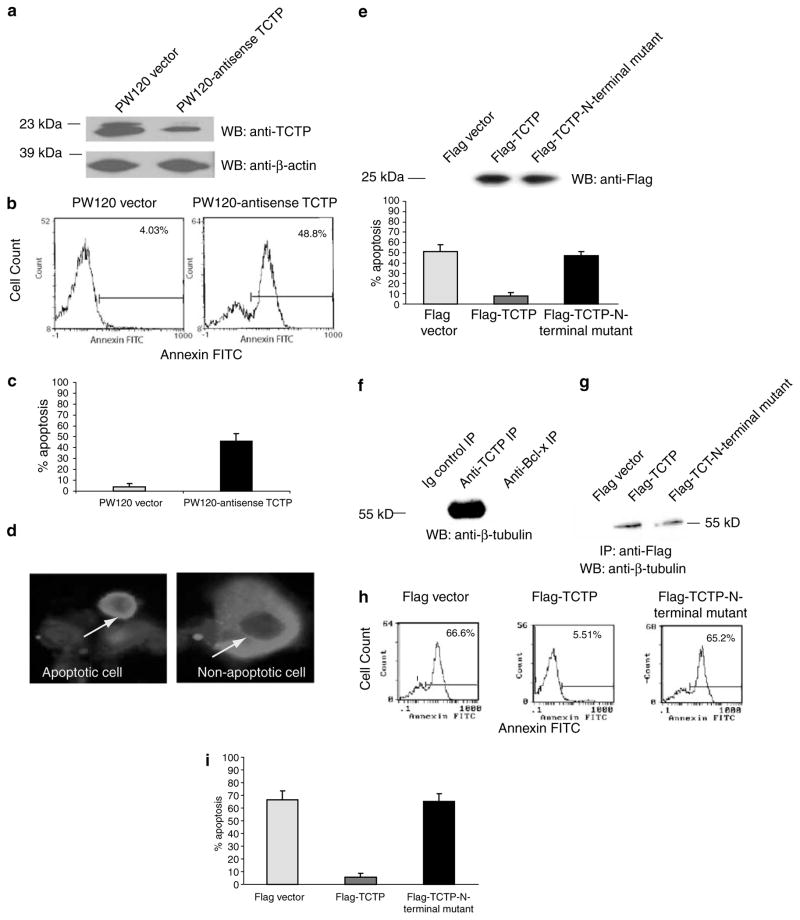
Previous reports showed that Bcl-xL (Boise et al., 1993; Ma et al., 1995; Motoyama et al., 1995) and TCTP (Li et al., 2001; Graidist et al., 2004) play critical roles in the promotion of cell survival. Since we showed that TCTP and Bcl-xL are dramatically co-upregulated in primary T cells induced by TCR ligation and CD28 costimulation (Figure 3c), we further examined whether endogenous TCTP expression levels in T cells are critical for survival of T cells. TCTP antisense cDNA was constructed in the T-cell expression vector pW120 under the control of a T-cell-specific tyrosine kinase p56lck distal promoter (Wildin et al., 1995). As shown in Figure 4a, the transfection of TCTP antisense in Jurkat T cells resulted in the partial knockdown of TCTP expression in comparison to that in the Jurkat T-cell control transfected with pW120 empty vector, as judged by Western blot with anti-TCTP antibody, similar to that described previously (Yang et al., 1997b). In contrast, β-actin expressions in the cells transfected with the vector control and transfected with pW120-TCTP antisense were comparable. More importantly, partial knockdown of endogenous TCTP expression by TCTP antisense in Jurkat T cells resulted in an increase in apoptotic cell populations by 44% in comparison to control cells transfected with the pW120 vector alone, as judged by Annexin V-FITC-stained cell populations (Figure 4b and c). These results suggest that endogenous TCTP expression is critical for maintaining T-cell survival.
Figure 4.
Dependence on the N-terminal region of TCTP for inhibition of apoptosis. (a) Knockdown of endogenous TCTP expression in Jurkat T cells by TCTP antisense. Transfection of Jurkat T cells with pW120-TCTP antisense resulted in knockdown of endogenous TCTP compared to cells transfected with the pW120 empty vector control. Bands of β-actin expression in control and TCTP antisense-transfected cells are equal. (b) Increased T-cell apoptosis by knockdown of endogenous TCTP expression in Jurkat T cells. A representative experiment is shown. Apoptosis in control pW120 vector-transfected and pW120-TCTP antisense-transfected Jurkat T cells was measured by flow cytometry using FITC-conjugated Annexin V. Apoptotic cells, as judged by Annexin V-FITC positivity, were significantly increased from 4.0% in control Jurkat T cells to 48.8% in pW120-TCTP antisense-transfected Jurkat T cells. (c) Summary of the Annexin V-FITC flow cytometry data from three independent experiments. The mean percentage of apoptotic cells and the standard deviation in each group are presented. (d) Morphological assessment of apoptosis by immunofluorescence microscopy. Cells were stained with anti-TCTP mAb followed by Texas Red-conjugated anti-mouse IgG. Apoptotic cells (arrow, left panel) were shrunken and had condensed chromatin in nucleus. Non-apoptotic cells (arrow, right panel) had unaltered cell structure and chromatin. (e) Inhibition of Taxol-induced apoptosis in HeLa cells by WT TCTP but not TCTP N-terminal region mutant. Equivalent expression levels of WT Flag-TCTP and Flag-TCTP N-terminal region mutant in transfected HeLa cells are demonstrated by anti-FLAG immunoblotting (upper panel). Apoptosis induced by Taxol was inhibited in TCTP-transfected cells, but not in the TCTP N-terminal region mutant-transfected cells. Three independent blinded observers scored for apoptotic cells by microscopy (lower panel). The mean and the standard deviation in each group were calculated and presented. (f) β-Tubulin binding to TCTP but not Bcl-xL. Endogenous β-tubulin in HeLa cells was co-immunoprecipitated by anti-TCTP antibodies, and detected by Western blot with anti-β-tubulin antibodies. In contrast, β-tubulin was not co-immunoprecipitated by anti-Bcl-xL antibodies, suggesting that Bcl-xL does not interact with β-tubulin. (g) β-Tubulin binding to WT TCTP and TCTP N-terminal region mutant. After transfection with FLAG, WT Flag-TCTP, and FLAG-TCTP N-terminal mutant cDNAs, anti-Flag antibodies were used to immunoprecipitate Flag-tagged protein. Immunoblotting with anti-β-tubulin antibodies revealed that β-tubulin co-immunoprecipitates with both WT and mutant TCTP. (h) Inhibition of etoposide-induced apoptosis in Jurkat cells by WT TCTP but not the TCTP N-terminal region mutant. A representative experiment is shown. Jurkat T cells stably expressing the Flag vector, Flag-TCTP, or Flag-TCTP N-terminal region mutant were treated with 50 μM etoposide for 24 h to induce apoptosis. Etoposide-induced apoptosis, as judged by Annexin V-FITC positivity, significantly decreased from 66.6% in Flag vector-transfected Jurkat T cells to 5.5% in Flag-TCTP (WT)-transfected Jurkat T cells, whereas the rate of the etoposide-induced apoptosis in Jurkat cells transfected with the Flag-TCTP N-terminal region mutant remained at similar levels (65.2%) as that of controls. (i) Summary of the data from three independent experiments measuring etoposide-induced apoptosis rates in vector-transfected Jurkat T cells, WT TCTP-transfected Jurkat T cells, and TCTP N-terminal region mutant-transfected Jurkat T cells. The mean and the standard deviation in each group are presented
We further examined whether TCTP inhibition of apoptosis is dependent on the intact N-terminal region of TCTP. As shown, overexpression of WT TCTP significantly inhibited apoptosis in HeLa cells induced by Taxol from 50% to less than 10%. In contrast, transfection of cells with TCTP bearing mutations within the N-terminal region failed to inhibit Taxol-mediated apoptosis in HeLa cells at the equivalent expression levels as that of WT TCTP (Figure 4d and e). Previous reports have shown that TCTP stabilizes microtubules (Yarm, 2002) and inhibits various forms of apoptosis (Li et al., 2001; Zhang et al., 2002; Bangrak et al., 2004; Graidist et al., 2004). As shown in Figure 4f, TCTP co-immunoprecipitated β-tubulin, but Bcl-xL did not. Since the β-tubulin binding domain of TCTP is located in aa 79–123 (Gachet et al., 1999), which different from the Bcl-xL binding region in the N-terminus of TCTP, as expected, we also found that TCTP N-terminal region mutant bound to β-tubulin (Figure 4g). These results suggest that the Bcl-xL BH3 domain and the N-terminal region of TCTP do not participate in binding to β-tubulin. These results also suggest that the antiapoptotic effects of TCTP result from its interaction with Bcl-xL rather than β-tubulin.
We asked next whether the N-terminal region of TCTP was critical for resistance to apoptosis induced by other proapoptotic agents in T cells. The topoisomerase II inhibitor etoposide induces DNA damage and is widely used as a pro-apoptotic agent (Li and Liu, 2001). To determine whether TCTP exerts antiapoptotic effects in Jurkat T cells, Jurkat T cells stably expressing the Flag vector, Flag-TCTP, or Flag-TCTP N-terminal region mutant were established and treated with 50 μM etoposide for 24 h to induce apoptosis. As shown in Figure 4g and h, WT TCTP inhibited etoposide-induced Jurkat cell apoptosis, but the TCTP N-terminal region mutant, expressed at the equivalent levels as WT TCTP (not shown), could not. Of note, in addition to Annexin V-FITC staining, the results were similar with the propidium iodide (PI) staining (not shown) described previously (Yang et al., 1997b). These results suggest that TCTP inhibits apoptosis induced by various proapoptotic agents in different cell types, including T cells. In light of our identification of the N-terminal region of TCTP as a Bcl-xL-interacting domain, our results demonstrate that the N-terminal region of TCTP is required for its antiapoptotic activity.
Discussion
TCTP is a novel Bcl-xL-interacting antiapoptotic protein
Having identified a novel interaction between Bcl-xL and TCTP by yeast two-hybrid screening, we confirmed the interaction with a variety of biochemical approaches, including GST pull-down assay complemented with site-directed mutagenesis, co-IP assay, subcellular colocalization assay, and peptide competition assay. We have further demonstrated that the TCTP N-terminal region and the Bcl-xL BH3 domain mediate the interaction. Our results also show that the interaction of TCTP with Bcl-xL via the TCTP N-terminal region is critical for its antiapoptotic properties. Of note, Taxol was reported using a range of concentrations from 1 nM to 100 μM and was found to affect cell growth and survival ranging from (a) a slight inhibition of cell growth at the doses lower than 5 nM, (b) mitotic arrest and cell death at the low or clinical relevant doses of 5–200 nM, to (c) the immediate effects including lipopolysaccharide (LPS)-like activity at high doses of 3–100 μM (Blagosklonny and Fojo, 1999). Our results suggest that TCTP plays a critical role in inhibiting Taxol-induced apoptosis in Jurkat T cells. The concentration of Taxol used in our studies (100 nM) has been shown to induce apoptosis through the phosphorylation and inactivation of Bcl-xL (Biswas et al., 2001; Basu and Haldar, 2003). Therefore, our results raise the interesting possibility that TCTP may inhibit T-cell apoptosis by preventing the phosphorylation/inactivation of Bcl-xL. This mechanism is currently under investigation. In agreement with the cytoprotective effects observed by TCTP overexpression in HeLa cells and Jurkat T cells examined in this study, the antisense knockdown of endogenous TCTP from Jurkat T cells was shown to promote apoptosis. Our findings corroborate the antiapoptotic effects of TCTP described in HeLa cells and U2OS cells (Li et al., 2001; Graidist et al., 2004), glioma cells (Baudet et al., 1998), and shrimps cells (Bangrak et al., 2004). Taken together, these studies demonstrate that TCTP plays an important role in maintaining survival of a variety of cell types, including T cells.
Not all of the Bcl-2/Bcl-xL family members interact with the same set of proteins (Sedlak et al., 1995). For instance, a recent report has shown that Tankyrase 1 interacts with Mcl-1 but not with other Bcl-2/Bcl-xL family proteins (Bae et al., 2003). Therefore, the previous identification of an interaction between TCTP and the Bcl-2/Bcl-xL family member Mcl-1 (Zhang et al., 2002) does not necessarily predict the possibility of TCTP interacting with Bcl-xL. In fact, the previous studies did not identify an interaction between TCTP and Bcl-xL (Zhang et al., 2002), presumably due to the differences in the methods and antibodies used. For instance, the previous studies used Mcl-1 as bait in the yeast two-hybrid screening (Zhang et al., 2002), whereas we used Bcl-xL as bait in the yeast two-hybrid screening. In this report, we identify for the first time a novel interaction between TCTP and Bcl-xL. Since Mcl-1 was not found to interact with Bcl-xL in cells (Bae et al., 2000), our results suggest that TCTP interacts with Bcl-xL directly, instead of mediating through Mcl-1. Furthermore, we describe for the first time an N-terminal region of TCTP that mediates its binding to Bcl-xL and its inhibition of apoptosis. This finding corresponds to a previous report that has shown that Arg21 in the N-terminal region of TCTP was critical for TCTP binding to Mcl-1 (Zhang et al., 2002).
TCTP is a unique multifunctional Bcl-xL-interacting protein
The multiple functions attributed to TCTP suggest the intriguing possibility that the Bcl-xL–TCTP complex may function in multiple signalling pathways. For instance, TCTP is a pluripotent cytokine (Vonakis et al., 2003) known to promote the growth of B cells (Kang et al., 2001), to induce a calcium response, and the secretion of IL-4, IL-8, and IL-13 from basophils (Vonakis et al., 2003; Bommer and Thiele, 2004). TCTP does not bear a classical signal peptide, suggesting that it may be released via a nonclassical ER–Golgi-independent secretory pathway facilitated by TSAP6 (Amzallag et al., 2004), analogous to the secretion of IL-1β (Nickel, 2003). The potential mechanism of how Bcl-xL affects TCTP secretion is being examined in our laboratory. Of note, we found that TCTP is colocalized with Bcl-xL in cytosol and mitochondria. In contrast to Bcl-xL, which has a C-terminal transmembrane domain and can be anchored on mitochondrial membrane (Borner, 2003), our bioinformatic analyses have not revealed any mitochondrial targeting sequence of TCTP (Mihara, 2000). The issue is currently under investigation that whether TCTP is localized to mitochondria via its interaction with Bcl-xL. TCTP has also been implicated in cell growth and differentiation (Bommer and Thiele, 2004), as knockdown of TCTP by RNA interference (RNAi) in Caenorhabditis elegans results in a slow-growth phenotype (Kamath et al., 2003). TCTP has also been described as a protein with multiple functions, including cytosolic Ca2+ binding (Kim et al., 2000), microtubule stabilization (Yarm, 2002), and Na/K-ATPase inhibition (Jung et al., 2004). It is not clear how these intracellular pathways affect the regulatory function of TCTP in cell growth and survival. Recently, the biochemical mechanism of TCTP function in cell growth began to be revealed. TCTP interacts with translation elongation factor eEF1A, and is functional as a guanine nucleotide dissociation inhibitor, and promotes the efficiency of protein synthesis (Cans et al., 2003), which is due to the structural similarity of TCTP to the guanine nucleotide-free chaperone (GFC) structural superfamily with the Mss4/Dss4 family of proteins (Thaw et al., 2001). As an addition to its multiple roles, our study suggests a novel mechanism by which the antiapoptotic pathway mediated by TCTP– Bcl-xL interaction may interplay with the TCTP function in the promotion of protein synthesis and other signalling pathways, and enhance cell proliferation (Tuynder et al., 2004) in a synergistic manner.
Co-upregulation of TCTP and Bcl-xL in T-cell activation induced by TCR and CD28 costimulation suggests their functional collaboration in maintaining activated T-cell survival
Bcl-xL plays an important role in the survival of T cells activated by TCR and CD28 (Boise et al., 1995; Yang et al., 2002; Ye et al., 2002; Marrack and Kappler, 2004). Among all the Bcl-xL-interacting proteins identified so far (Opferman and Korsmeyer, 2003), TCTP is one of the few antiapoptotic proteins that are dramatically upregulated by TCR ligation and CD28 costimulation (Boise et al., 1995; Yang et al., 1997b), implying an important role for TCTP in T-cell survival. Our in silico analysis further confirms a role for TCTP in T-cell activation. The TCTP promoter contains a TATA box at −30 and potential binding sites for transcription factors that are activated in response to T-cell activation such as Sp1, NF1, AP1, c-Ets1, CP2, MZF1, and others. We also note that murine TCTP mRNA has an AU-rich element (ARE) consisting of AUUUA repeats (not shown), which destabilize mRNAs, and is a common feature in the 3′ untranslated region of short-lived T-cell cytokines, such as IL-2 (Shim and Karin, 2002). With our identification of TCTP as a physical and functional interacting partner of Bcl-xL, we have shown that the Bcl-xL–TCTP-containing complex may play an important role in maintaining T-cell survival during activation.
Materials and methods
Yeast two-hybrid system
The 0.7 kb cDNA encoding full-length Bcl-xL was subcloned in-frame to the C-terminus of the GAL4 DNA binding domain in the pAS2-1 vector (BD Clontech, Palo Alto, CA, USA). A mouse lymphocyte MATCHMAKER cDNA library (BD Clontech), constructed as fusion proteins to the activation domain in the pACT2 vector, was screened for approximately 1.5×106 independent clones. The primary His+ transformants were further tested for β-galactosidase activity (the second reporter gene) using a colony lift filter assay to eliminate false positives.
DNA sequencing and bioinformatic analyses
DNA sequencing was performed by SeqWright Company (Houston, TX, USA). Sequence analyses were performed using GenBank databases with nucleotide–nucleotide BLAST and protein–protein BLAST searches (http://www.ncbi.nlm.nih.gov/BLAST/), NCBI Conserved Domain Database (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), the PROSITE database of protein families and domains, (http://us.expasy.org/prosite/) as well as ExPASy Proteomics Tools.
Northern blotting
Northern blot with multiple mouse tissues was prepared with purified polyadenylated RNA (Ambion, Austin, TX, USA). Hybridizations were conducted as previously described (Yang et al., 1997b).
In vitro transcription and translation
PCDNA3.1/V5-His TOPO TA expression plasmid vectors (Invitrogen, Carlsbad, CA, USA) containing the cDNAs encoding TCTP, Bcl-xL, and Bcl-xγ were used in in vitro TNT experiments using a TNT T7-coupled system (Promega, Madison, WI, USA) (Yang et al., 1997b).
Reverse transcription–PCR and PCR cloning
RNA was prepared using RNAzole (Tel-Test, Friendswood, TX, USA). Reverse transcription (RT) was performed by using the Reverse Transcription System (Promega). The amplified full-length TCTP PCR product was subcloned into PCDNA3.1/V5-His TOPO TA expression plasmid (Invitrogen), and confirmed by DNA sequencing. An mRNA sense primer specific for the 5′ end of the TCTP ORF (5′ ATG ATCATCTACCGGGACCTCATC3′) and a cDNA sense primer specific for the 3′ end of the TCTP ORF (5′ACA TTTCTCCATCTCTAAGCCATCCTT3′) were used in PCR cloning.
Construction of three TCTP deletion mutants
The insert of the first TCTP C-terminal deletion mutant TCTP1–50 aa was prepared by high-fidelity PCR (BD Clontech) using the TCTP mRNA sense primer GST-TCTP5 specific for the 5′ end of the TCTP ORF (5′GGCCGCGA ATTCGATGATCATCTACCGGGACCTCATC3′) to pair with the antisense primer (5′TTATCCACCGATGAGCGAG TCATCGAT3′) specific for the region around aa 50. Similarly, the insert of the second TCTP C-terminal deletion mutant TCTP1–150aa was prepared by high-fidelity PCR also using the TCTP mRNA sense primer GST-TCTP5 to pair with the antisense primer (5′TTAGTCCAGGAGAGCAACCATACC ATC3′) specific for the region around aa 150. The TCTP N-terminal deletion mutant was prepared by PCR with a sense primer TCTP (5′ACAATGGGTGCCATCGATGACTCGC TCATC3′) specific for the region beginning at aa 40 and the antisense primer GST-TCTP3 specific for the 3′ end of the TCTP ORF. The amplified PCR products were subcloned into TOPO TA plasmid (Invitrogen) and confirmed by DNA sequencing.
Preparation of GST and GST fusion proteins
Full-length cDNAs of Bcl-xL, Bcl-xγ, and TCTP were previously fused in-frame to the C-terminus of GST by subcloning into the GST vector pGEX-3X (Amersham Biosciences, Piscataway, NJ, USA) (Yang et al., 1997b). Full-length TCTP was prepared by using a high-fidelity PCR (BD Clontech) with the TCTP cDNA as the template and two TCTP primers, including the sense primer GST-TCTP5 specific for the 5′ end of the TCTP ORF and the antisense primer GST-TCTP3 specific for the 3′ end of the TCTP ORF (5′GGCCGGGAATTCTTAACATTTCTCCATCTCTAAG CCATC3′). The underlined sequences were designed for the purpose of subcloning into EcoRI site. A GST-TCTP N-terminal domain mutant was constructed from the WT pGEX-3X-TCTP vector using a QuickChange Site-directed Mutagenesis kit (Stratagene, La Jolla, CA, USA). The sense mutagenic primer Tpt-BH5 (5′TCCGACATCTACAAGGCCGCGGCG ATCGCGGCCGGATCCTGCCTGGAGGTGGAG3′) and the antisense mutagenic primer Tpt-BH3 (5′CTCCACCTCCA GGCAGGATCCGGCCGCGATCGCCGCGGCCTTGTAG ATGTCGGA3′) were synthesized. The underlined sequences were designed for mutation of the N-terminal region of TCTP. Both mutagenic primers were complementary to each other and specific for the N-terminal region of TCTP between aa 20 and 27. All the recombinant vectors were confirmed by DNA sequencing. GST and GST fusion proteins were purified as previously described (Yang et al., 2001).
GST Pull-down
GST pull-down experiments were performed as described previously (Einarson and Orlinick, 2002). Briefly, 10 μl of 35S-labelled proteins was mixed with either 10 μg GST protein or GST fusion proteins GST-Bcl-xL, GST-Bcl-xγ, or GST-TCTP, together with glutathione Sepharose 4B bead slurry and EBC-immunoprecipitation buffer with 0.5% NP-40 (Adams et al., 2002) (Roche Applied Science, Indianapolis, IN, USA) (0.5% NP-40, 50mM Tris-HCl, pH 8.0, 120mM NaCl complemented with the complete proteinase inhibitor cocktail (Roche)). After incubation and washes, fusion proteins and any interacting proteins were eluted, separated by SDS—PAGE, and detected by autoradiography (Eastman Kodak, Rochester, NY, USA).
Competitive inhibition of the interaction between Bcl-xL and TCTP with Bcl-xL BH3 peptide and Bcl-xL BH3 domain mutant peptide
Competitive inhibition assays were performed as previously reported (Shangary and Johnson, 2002). Bcl-xL BH3 domain peptide (N-MAAVKQALREAGDEFELRYRR-C) and Bcl-xL BH3 domain mutant peptide (N-MAAVKQAAREAGAEFELRYRR-C) were purchased from ABGENT Company (San Diego, CA, USA). The underlined amino-acid sequences in Bcl-xL BH3 mutant peptide were mutated.
Co-immunoprecipitation and Western blot
Polyclonal anti-Bcl-xL antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Polyclonal anti- TCTP antibodies were generously provided by Dr Susan M MacDonald at Johns Hopkins or purchased from Medical and Biological Laboratories (Naka-Ku Nagoya, Japan). Monoclonal anti-TCTP antibody F4M101 was generously provided by Drs E Guillaume and JC Sanchez at Hopital Cantonal Universitaire de Geneve in Switzerland. Co-IP reactions were performed as previous published (Adams et al., 2002). A 2 μg portion of the respective antibodies was used for each immunoprecipitation. Monoclonal antibodies to Bcl-xL and TCTP were used in the Western blots.
T-cell activation and IL-2 assay
Mouse splenic T cells were prepared as previously reported (Ye et al., 2002) and activated by either plate-bound anti-CD3 (145.2C11 clone, 1 μg/ml, BD PharMingen, San Diego, CA, USA) alone, or plate-bound anti-CD3 (1 μg/ml) and anti- CD28 (37.51 clone, 1 μg/ml, BD PharMingen) (Ye et al., 2002). Splenic T cells were cultured and collected at 0, 12, 24, and 48 h (Yang et al., 1997b). The IL-2 levels in the supernatant of the activated splenic T cells were determined by using a mouse IL-2 BD Opt EIA ELISA kit (BD PharMingen).
Immunocytochemistry
Immunocytochemical experiments were performed as previously published (Aumais et al., 2003). Briefly, HeLa cells grown on coverslips were fixed with 3.7% formaldehyde in PBS for 45 min at room temperature (Gachet et al., 1999). Fixed cells were subsequently stained with mouse anti-Bcl-xL monoclonal antibodies (BD PharMingen) or rabbit polyclonal TCTP antibodies for 1 h. After 3×10 min washes with PBS– 0.1% Tween 20, the following secondary antibodies were applied. AlexaFluor488-conjugated goat anti-mouse and AlexaFluor594-conjugated goat anti-rabbit (Molecular Probes, Eugene, OR, USA) antibodies were used to detect Bcl-xL and TCTP, respectively. Mitochondria were stained with the MitoTracker Deep Red 633 (Molecular Probes). Cells were mounted using the SlowFade Light Antifade kit with 4′,6- diamidino-2-phenylindole (DAPI) (Molecular Probes). The mounted coverslips were analysed by a laser scanning microscope LSM510 (Carl Zeiss Ltd, Herts, UK) at Baylor College of Medicine Microscopy Core.
Inhibition of paclitaxel (Taxol)-induced apoptosis and etoposide-induced apoptosis by TCTP
Full-length cDNAs of TCTP and the TCTP N-terminal region mutant were fused in-frame to the C-terminus of the 3x FLAG expression tag sequence in the p3xFLAG-CMV-10 expression vector (Sigma, St Louis, MO, USA). HeLa cells were transfected, respectively, with the p3xFLAG-CMV-10 vector (Flag vector, empty control), p3xFLAG-CMV-10-TCTP (Flag-TCTP), or p3xFLAG-CMV-10-TCTP N-terminal region mutant (Flag-TCTP N-terminal region mutant) and treated with Taxol (100 nM) for 24 h, as described previously (Biswas et al., 2001). The cells were fixed, permeabilized, and stained with anti-FLAG M2 antibodies (Sigma) and Texas red-conjugated anti-mouse IgG. The nuclei were counter-stained with DAPI and analysed by immunofluorescence microscopy. Three independent observers blindly scored FLAG-positive cells for apoptotic morphology. Data are expressed as the percentage of apoptotic cells in total counted cells.
Jurkat T cells stably expressing WT TCTP, TCTP N-terminal region mutant, or vector control were established by transfection followed by selection for neomycin resistance (800 μg/ml) (Invitrogen) for 2 weeks. Transfected Jurkat T cells were challenged with 50 μM etoposide (Yang et al., 1997a) (Sigma) for 24 h and stained with Annexin V-FITC (BD PharMingen). Cells were analysed for apoptotic fractions by flow cytometry on the Beckman-Coulter EPICS XL-MCL flow cytometer (Miami, FL, USA) at the Baylor Flow Cytometry Core.
Promotion of T-cell apoptosis by transfection of TCTP antisense cDNA
An antisense-oriented TCTP cDNA was subcloned into the BamHI site of the expression vector pW120, in which the expression of TCTP antisense was driven by the T-cell-specific tyrosine kinase p56lck distal promoter (Wildin et al., 1995). The pW120 empty vector and pW120-TCTP antisense were transfected into Jurkat T cells with the Nucleofection Kit (Amaxa Biosystems, MD, USA). At 24 h after transfection, the apoptosis rates of transfected Jurkat T cells were measured by flow cytometry using the Annexin V-FITC Apoptosis Detection Kit II (BD PharMingen).
Acknowledgments
We are grateful to Drs DP Huston, L-Y Yu-Lee, TH Tan, S Zhang, B Ng, H Lu, and JP Aumais at Baylor, H Cantor and J Ritz at Harvard, Dr SM MacDonald at the Johns Hopkins, and Drs E Guillaume and JC Sanchez at Hopital Cantonal Universitaire de Geneve for suggestion or reagents, Drs MA Mancini and F Kheradmand at Baylor for the use of confocal and fluorescence microscopes, and Ms K Franks and her associates in our section for assistance. This work was partially supported by funds from NIH Grants AI054514, P30 DK56238, and P20 CA103698, development support at Baylor, the Kostas Foundation, the Law Foundation, the Myositis Association of America, the American Heart Association-Texas Affiliate, and the Leukemia & Lymphoma Society and Myeloproliferative Disorders Foundation. X-F Yang is a Chao Family Scholar of Medicine.
Abbreviations
- TCTP
translationally controlled tumor protein
- BH3 domain
Bcl-2 homology domain 3
- TNT
in vitro transcription and translation
- GST
glutathione S-transferase
References
- Adams JM. Genes Dev. 2003;17:2481–2495. doi: 10.1101/gad.1126903. [DOI] [PubMed] [Google Scholar]
- Adams P, Seeholzer S, Ohh M. In: A Molecular Cloning Manual: Protein–Protein Interactions. Golemis E, editor. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2002. pp. 59–74. [Google Scholar]
- Amzallag N, Passer BJ, Allanic D, Segura E, Thery C, Goud B, Amson R, Telerman A. J Biol Chem. 2004;279:46104–46112. doi: 10.1074/jbc.M404850200. [DOI] [PubMed] [Google Scholar]
- Aumais JP, Williams SN, Luo W, Nishino M, Caldwell KA, Caldwell GA, Lin SH, Yu-Lee LY. J Cell Sci. 2003;116:1991–2003. doi: 10.1242/jcs.00412. [DOI] [PubMed] [Google Scholar]
- Bae J, Donigian JR, Hsueh AJ. J Biol Chem. 2003;278:5195–5204. doi: 10.1074/jbc.M201988200. [DOI] [PubMed] [Google Scholar]
- Bae J, Leo CP, Hsu SY, Hsueh AJ. J Biol Chem. 2000;275:25255–25261. doi: 10.1074/jbc.M909826199. [DOI] [PubMed] [Google Scholar]
- Bangrak P, Graidist P, Chotigeat W, Phongdara A. J Biotechnol. 2004;108:219–226. doi: 10.1016/j.jbiotec.2003.12.007. [DOI] [PubMed] [Google Scholar]
- Basu A, Haldar S. FEBS Lett. 2003;538:41–47. doi: 10.1016/s0014-5793(03)00131-5. [DOI] [PubMed] [Google Scholar]
- Baudet C, Perret E, Delpech B, Kaghad M, Brachet P, Wion D, Caput D. Cell Death Differ. 1998;5:116–125. doi: 10.1038/sj.cdd.4400327. [DOI] [PubMed] [Google Scholar]
- Biswas RS, Cha HJ, Hardwick JM, Srivastava RK. Mol Cell Biochem. 2001;225:7–20. doi: 10.1023/a:1012203110027. [DOI] [PubMed] [Google Scholar]
- Blagosklonny MV, Fojo T. Int J Cancer. 1999;83:151–156. doi: 10.1002/(sici)1097-0215(19991008)83:2<151::aid-ijc1>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- Boise LH, Gonzalez-Garcia M, Postema CE, Ding L, Lindsten T, Turka LA, Mao X, Nunez G, Thompson CB. Cell. 1993;74:597–608. doi: 10.1016/0092-8674(93)90508-n. [DOI] [PubMed] [Google Scholar]
- Boise LH, Minn AJ, Noel PJ, June CH, Accavitti MA, Lindsten T, Thompson CB. Immunity. 1995;3:87–98. doi: 10.1016/1074-7613(95)90161-2. [DOI] [PubMed] [Google Scholar]
- Bommer UA, Thiele BJ. Int J Biochem Cell Biol. 2004;36:379–385. doi: 10.1016/s1357-2725(03)00213-9. [DOI] [PubMed] [Google Scholar]
- Borner C. Mol Immunol. 2003;39:615–647. doi: 10.1016/s0161-5890(02)00252-3. [DOI] [PubMed] [Google Scholar]
- Cans C, Passer BJ, Shalak V, Nancy-Portebois V, Crible V, Amzallag N, Allanic D, Tufino R, Argentini M, Moras D, Fiucci G, Goud B, Mirande M, Amson R, Telerman A. Proc Natl Acad Sci USA. 2003;100:13892–13897. doi: 10.1073/pnas.2335950100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz JL, Oltersdorf T, Horne W, McConnell M, Wilson G, Weeks S, Garcia T, Fritz LC. J Biol Chem. 1997;272:11350–11355. doi: 10.1074/jbc.272.17.11350. [DOI] [PubMed] [Google Scholar]
- Einarson M, Orlinick JR. Identification of Protein–Protein Interactions with Glutathione-S-Transferase Fusion Proteins. Cold Spring Habor Laboratory Press; Cold Spring Harbor, NY: 2002. [DOI] [PubMed] [Google Scholar]
- Fang W, Rivard JJ, Mueller DL, Behrens TW. J Immunol. 1994;153:4388–4398. [PubMed] [Google Scholar]
- Gachet Y, Tournier S, Lee M, Lazaris-Karatzas A, Poulton T, Bommer UA. J Cell Sci. 1999;112 (Part 8):1257–1271. doi: 10.1242/jcs.112.8.1257. [DOI] [PubMed] [Google Scholar]
- Gibbs CS, Zoller MJ. J Biol Chem. 1991;266:8923–8931. [PubMed] [Google Scholar]
- Gonzalez-Garcia M, Perez-Ballestero R, Ding L, Duan L, Boise LH, Thompson CB, Nunez G. Development. 1994;120:3033–3042. doi: 10.1242/dev.120.10.3033. [DOI] [PubMed] [Google Scholar]
- Graidist P, Phongdara A, Fujise K. J Biol Chem. 2004;279:40868–40875. doi: 10.1074/jbc.M401454200. [DOI] [PubMed] [Google Scholar]
- Hsu YT, Wolter KG, Youle RJ. Proc Natl Acad Sci USA. 1997;94:3668–3672. doi: 10.1073/pnas.94.8.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong SY, Gaume B, Lee YJ, Hsu YT, Ryu SW, Yoon SH, Youle RJ. EMBO J. 2004;23:2146–2155. doi: 10.1038/sj.emboj.7600225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J, Kim M, Kim MJ, Kim J, Moon J, Lim JS, Lee K. J Biol Chem. 2004;279:49868–49875. doi: 10.1074/jbc.M400895200. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, Zipperlen P, Ahringer J. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kang HS, Lee MJ, Song H, Han SH, Kim YM, Im JY, Choi I. J Immunol. 2001;166:6545–6554. doi: 10.4049/jimmunol.166.11.6545. [DOI] [PubMed] [Google Scholar]
- Kim M, Jung Y, Lee K, Kim C. Arch Pharm Res. 2000;23:633–636. doi: 10.1007/BF02975253. [DOI] [PubMed] [Google Scholar]
- Komatsu K, Miyashita T, Hang H, Hopkins KM, Zheng W, Cuddeback S, Yamada M, Lieberman HB, Wang HG. Nat Cell Biol. 2000;2:1–6. doi: 10.1038/71316. [DOI] [PubMed] [Google Scholar]
- Li F, Zhang D, Fujise K. J Biol Chem. 2001;276:47542–47549. doi: 10.1074/jbc.M108954200. [DOI] [PubMed] [Google Scholar]
- Li TK, Liu LF. Annu Rev Pharmacol Toxicol. 2001;41:53–77. doi: 10.1146/annurev.pharmtox.41.1.53. [DOI] [PubMed] [Google Scholar]
- Ma A, Pena JC, Chang B, Margosian E, Davidson L, Alt FW, Thompson CB. Proc Natl Acad Sci USA. 1995;92:4763–4767. doi: 10.1073/pnas.92.11.4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrack P, Kappler J. Annu Rev Immunol. 2004;22:765–787. doi: 10.1146/annurev.immunol.22.012703.104554. [DOI] [PubMed] [Google Scholar]
- Mihara K. BioEssays. 2000;22:364–371. doi: 10.1002/(SICI)1521-1878(200004)22:4<364::AID-BIES6>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Motoyama N, Wang F, Roth KA, Sawa H, Nakayama K, Negishi I, Senju S, Zhang Q, Fujii S, Loh DY. Science. 1995;267:1506–1510. doi: 10.1126/science.7878471. [DOI] [PubMed] [Google Scholar]
- Nickel W. Eur J Biochem. 2003;270:2109–2119. doi: 10.1046/j.1432-1033.2003.03577.x. [DOI] [PubMed] [Google Scholar]
- Opferman JT, Korsmeyer SJ. Nat Immunol. 2003;4:410–415. doi: 10.1038/ni0503-410. [DOI] [PubMed] [Google Scholar]
- Schwartz RH. Annu Rev Immunol. 2003;21:305–334. doi: 10.1146/annurev.immunol.21.120601.141110. [DOI] [PubMed] [Google Scholar]
- Sedlak TW, Oltvai ZN, Yang E, Wang K, Boise LH, Thompson CB, Korsmeyer SJ. Proc Natl Acad Sci USA. 1995;92:7834–7838. doi: 10.1073/pnas.92.17.7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shangary S, Johnson DE. Biochemistry. 2002;41:9485–9495. doi: 10.1021/bi025605h. [DOI] [PubMed] [Google Scholar]
- Shim J, Karin M. Mol Cells. 2002;14:323–331. [PubMed] [Google Scholar]
- Thaw P, Baxter NJ, Hounslow AM, Price C, Waltho JP, Craven CJ. Nat Struct Biol. 2001;8:701–704. doi: 10.1038/90415. [DOI] [PubMed] [Google Scholar]
- Tuynder M, Fiucci G, Prieur S, Lespagnol A, Geant A, Beaucourt S, Duflaut D, Besse S, Susini L, Cavarelli J, Moras D, Amson R, Telerman A. Proc Natl Acad Sci USA. 2004;101:15364–15369. doi: 10.1073/pnas.0406776101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonakis BM, Sora R, Langdon JM, Casolaro V, MacDonald SM. J Immunol. 2003;171:3742–3750. doi: 10.4049/jimmunol.171.7.3742. [DOI] [PubMed] [Google Scholar]
- Wildin RS, Wang HU, Forbush KA, Perlmutter RM. J Immunol. 1995;155:1286–1295. [PubMed] [Google Scholar]
- Yang J, Liu X, Bhalla K, Kim CN, Ibrado AM, Cai J, Peng TI, Jones DP, Wang X. Science. 1997a;275:1129–1132. doi: 10.1126/science.275.5303.1129. [DOI] [PubMed] [Google Scholar]
- Yang XF, Weber GF, Cantor H. Immunity. 1997b;7:629–639. doi: 10.1016/s1074-7613(00)80384-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XF, Wu CJ, McLaughlin S, Chillemi A, Wang KS, Canning C, Alyea EP, Kantoff P, Soiffer RJ, Dranoff G, Ritz J. Proc Natl Acad Sci USA. 2001;98:7492–7497. doi: 10.1073/pnas.131590998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X-F, Ye Q, Press B, Han RZ, Bassing CH, Sleckman BP, Alt FW, Cantor H. Mol Immunol. 2002;39:45–55. doi: 10.1016/s0161-5890(02)00049-4. [DOI] [PubMed] [Google Scholar]
- Yarm FR. Mol Cell Biol. 2002;22:6209–6221. doi: 10.1128/MCB.22.17.6209-6221.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q, Press B, Kissler S, Yang XF, Lu L, Bassing CH, Sleckman BP, Jansson M, Panoutsakopoulou V, Trimble LA, Alt FW, Cantor H. J Exp Med. 2002;196:87–95. doi: 10.1084/jem.20012084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon T, Jung J, Kim M, Lee KM, Choi EC, Lee K. Arch Biochem Biophys. 2000;384:379–382. doi: 10.1006/abbi.2000.2108. [DOI] [PubMed] [Google Scholar]
- Zhang D, Li F, Weidner D, Mnjoyan ZH, Fujise K. J Biol Chem. 2002;277:37430–37438. doi: 10.1074/jbc.M207413200. [DOI] [PubMed] [Google Scholar]
- Zhang H, Xu Q, Krajewski S, Krajewska M, Xie Z, Fuess S, Kitada S, Godzik A, Reed JC. Proc Natl Acad Sci USA. 2000;97:2597–2602. doi: 10.1073/pnas.97.6.2597. [DOI] [PMC free article] [PubMed] [Google Scholar]