Abstract
Background
Negative energy balance (NEB), an altered metabolic state, occurs in early postpartum dairy cattle when energy demands to support lactation exceed energy intake. During NEB the liver undergoes oxidative stress and increased breakdown of fatty acids accompanied by changes in gene expression. It is now known that micro RNAs (miRNA) can have a role in mediating such alterations in gene expression through repression or degradation of target mRNAs. miRNA expression is known to be altered by metabolism and environmental factors and miRNAs are implicated in expression modulation of metabolism related genes.
Results
miRNA expression was profiled in the liver of moderate yielding dairy cattle under severe NEB (SNEB) and mild NEB (MNEB) using the Affymetrix Gene Chip miRNA_2.0 array with 679 probe sets for Bos-taurus miRNAs. Ten miRNAs were found to be differentially expressed using the ‘samr’ statistical package (delta = 0.6) at a q-value FDR of < 12%. Five miRNAs including miR-17-5p, miR-31, miR-140, miR-1281 and miR-2885 were validated using RT-qPCR, to be up-regulated under SNEB. Liver diseases associated with these miRNAs include non-alcoholic fatty liver (NAFLD) and hepatocellular carcinoma (HCC). miR-140 and miR-17-5p are known to show differential expression under oxidative stress. A total of 32 down-regulated putative target genes were also identified among 418 differentially expressed hepatic genes previously reported for the same animal model. Among these, GPR37 (G protein-coupled receptor 37), HEYL (hairy/enhancer-of-split related with YRPW motif-like), DNJA1, CD14 (Cluster of differentiation 14) and GNS (glucosamine (N-acetyl)-6-sulfatase) are known to be associated with hepatic metabolic disorders. In addition miR-140 and miR-2885 have binding sites on the most down-regulated of these genes, FADS2 (Fatty acid desaturase 2) which encodes an enzyme critical in lipid biosynthesis. Furthermore, HNF3-gamma (Hepatocyte nuclear factor 3-gamma), a hepatic transcription factor (TF) that is involved in IGF-1 expression regulation and maintenance of glucose homeostasis is a putative target of miR-31.
Conclusions
This study shows that SNEB affects liver miRNA expression and these miRNAs have putative targets in hepatic genes down-regulated under this condition. This study highlights the potential role of miRNAs in transcription regulation of hepatic gene expression during SNEB in dairy cattle.
Background
Over the past few decades improvements in milk production through genetic selection have been associated with a reduction in cow fertility and this decrease in fertility has become a major concern for dairy producers [1,2]. Reproduction is an energetically expensive process and an altered metabolic state called NEB has been established as one of the major physiological causes of decreased fertility in high yielding dairy cattle [3-5]. NEB is the result of increased energy demands to support lactation, coupled with lowered feed intake [4,5]. Immune related and hepatic functions are known to be effected by NEB [6-9] and there is an increased metabolic load on the liver to overcome the energy deficit under NEB. There is a dramatic increase in hepatic oxidation of fatty acids for energy production [9,10] and in addition there are extra demands on the liver to increase glycogenesis to meet the glucose requirements of milk production. In order to understand the complex metabolic adjustments in postpartum dairy cattle liver during NEB, a dairy cattle model based on different milking regimes was developed. Earlier in a microarray gene expression study 418 hepatic genes were reported to be differentially expressed as a result of SNEB [9]. These differentially expressed genes have roles in lipid metabolism and glycogenic processes, immune response and the somatotropic axis involved in milk production. The regulation in gene expression under SNEB, however, is yet to be fully understood.
It is now known that a class of small RNAs called miRNA, about 19 to 25 nucleotides in length, can regulate such alterations at the gene expression level. miRNAs were first discovered from the round worm C. Elegans almost 3 decades ago [11]. Since their discovery, hundreds of miRNAs have been identified across the plant and animal kingdoms. It is estimated that whereas only 1-5% of genomic transcripts in mammals code miRNAs, up to 60% of genes are regulated by miRNAs [12-14].
The biogenesis of miRNA is a multistep process that begins with miRNA gene transcription resulting in primary miRNA (pri-miRNA) in the nucleus, followed by the generation of around 70nt long stem–loop precursor miRNAs (pre-miRNAs) from pri-miRNA. Pre-miRNAs are then translocated to the cytoplasm where they are trimmed to remove the terminal loop and release ~22 nt long duplex mature miRNAs containing a guiding strand and a passenger strand. The guiding strand assembles into cytoplasmic RNA-induced silencing complex (RISC) with argonaute protein that guides the complex to their complementary mRNA targets [15,16]. The miRNA target-mRNA complementary base-pair interaction generally occurs between the target site on the 3′UTR of the mRNA and the 2nd to 8th nucleotides on the 5′UTR of the miRNA called the ‘seed region’. Generally, miRNAs repress translation through deadenylation of mRNA leading to their subsequent degradation or translation repression. miRNAs are reported to regulate a wide range of biological processes including cell cycle regulation, proliferation and differentiation as well as development, immune response, carcinogenesis and various metabolic processes [17-19]. The effects of metabolites and environmental factors including hormones, cytokines and nutrients on miRNAs is well established [20-24]. Alterations in cattle liver miRNA expression in response to external anabolic steroids was reported and miRNAs were suggested as potential biomarker for drug abuse in cattle [24]. In another study testosterone treatment was reported to alter miR-22, miR-690, miR-122, let-7a, miR-30 and let-7d expression in female rat liver [25]. Increases in expression of miR-155 and miR-132 due to oxidative stress was reported in an ALD (alchololic liver disorder) mouse model [26]. Oxidative stress of hepatocytes was also reported to alter miR-199a-5p expression [27]. Nutritional modulation of miRNA expression has been reported in various dietary intervention studies of metabolic disorders like obesity, diabetes and fatty liver [28-30]. A high fat diet was reported to alter the adipose tissue miRNA expressional profile of miR-19a, -92a, -92b, -101, -103, -106, -142–5p, and 296 in cattle [31]. miRNAs can regulate metabolism and homeostasis of high energy metabolites as well as insulin signalling and glucose homeostasis [32-34]. miR-33a and miR-33b located within the sterol regulatory element-binding proteins (SREBP), key transcription regulators of genes involved in cholesterol biosynthesis and uptake, regulate cholesterol homeostasis jointly with their host genes [35] and have roles in the regulation fatty acid metabolism and insulin signaling [36]. In addition, miR-122, a liver specific miRNA regulates hepatic fatty acid oxidation and fatty acid and cholesterol synthesis rate [32,37]. miRNAs are associated with the pathophysiology of hepatic metabolic disorders like NAFLD and NASH (non-alcoholic steatohepatitis) as well as HCC in mouse and human studies [38-41]. The potential role miRNAs have to play in NEB in the dairy cow remains to be elucidated.
This study set out to determine a) if hepatic miRNA expression was altered as a result of SNEB in a model of moderate yielding Holstein Friesian dairy cows in the early postpartum period, and b) to integrate hepatic miRNA and mRNA expression profiles through prediction of targets of these miRNAs among a set of previously reported differential expressed hepatic genes under SNEB from the same animal model. Elucidation of the expression patterns of miRNAs and computational identification of their putative target among genes regulated under SNEB will contribute to the understanding of the roles of miRNAs in regulating gene expression during SNEB in dairy cows.
Methods
Animal model
The animal model employed in this study has been described previously [5,9]. All procedures were carried out under license in accordance with the European Community Directive, 86-609-EC. In brief, multiparous Holstein-Friesian cows were blocked according to parity, body condition score, and previous milk yield, two weeks prior to expected calving and within block were randomly allocated to two treatments; mild NEB (MNEB, n = 12) or severe NEB (SNEB, n = 12) groups based on different feeding and milking regimes. On day 2 after calving, MNEB cows were fed ad libitum grass silage with 8 kg per day of a 21% crude protein dairy concentration and milked once daily; SNEB cows were fed 25 kg of silage per day silage with 4 kg of crude protein per day and milked three times daily. Three times a day versus once a day milking was used to advance a state of SNEB in one group by increasing energy withdrawal with a concomitant lower need for differences in energy intake between groups. The chemical composition of silage and concentrate offered as previously described [5]. For sample collection, cows were selected from each group based on extremes of energy balance (MNEB, n = 6; SNEB, n = 6). Cows were slaughtered approximately 14 days postpartum (MNEB; 13.6 ± 0.75, range 11–15; SNEB 14.3 ± 0.56, range 13–16) [9]. Liver tissues were retrieved within 30 min of slaughter and snap frozen at -80°C. One MNEB cow was removed retrospectively as she was deemed to be ill during the experimental period.
RNA extraction and quality analysis
For the purpose of RNA extraction, 100 mg frozen liver tissue was directly immersed in 1 ml Trizol (Invitrogen, Stockholm, Sweden). A Precellys 24 homogeniser (Bertin Technologies, Montigny-le-Bretonneux, France) was used to homogenise the tissue (6000 rpm for 20 sec). Homogenate was left for 5 min at RT to permit the complete dissociation of nucleoprotein complexes followed by centrifugation at 12,000 g for 10 min at 4°C. Supernatant was transferred to a 2 ml tube. 2μl of Pellet paint® NF Co-Precipitant (EMD Millipore, Darmstadt, Germany) and 200 μl chloroform was added and incubated at RT for 5 min. After centrifugation for 15 min at 4°C and 14000 g the aqueous phase was transferred into a new tube and 500 μl isopropanol was added and incubated for 5 min on ice. Following centrifugation for 10 min at 4°C and 14000 g the supernatant was removed and the pellet washed with 500 μl 75% ETOH followed by centrifugation for 10 min at 4°C and 14000 g. A 100 μl aliquot of nuclease-free water was added to the pellet and incubated at RT for 10 min. After vortexing for 1 min 10 μl of 3 M sodium acetate was added. Next, 250 μl ice cold ETOH was added and incubated for 1 hr at -20°C, followed by centrifugation at 4°C for 10 min and 14000 g. The supernatant was removed and washed with 500 μl of 75% EtOH by vortexing followed by centrifugation at 4°C for 10 min and 14000 g. The supernatant was removed and the pellet was air-dried for 5 min at RT following which 30μl of RNase-free water was added and left for 5 min. RNA quality and quantity was assessed using a NanoDrop spectrophotometer (ND-1000; Wilmington, DE, USA) and Bioanalyzer 2100 (Agilent Technologies Ireland, Dublin, Ireland). Mean RIN values were > 8. Total RNA was stored at -80°C.
Gene Chip miRNAs_2.0 array hybridization
Total RNA from liver samples of 11 cows were hybridized to the Affymetrix Gene Chip miRNA_2.0 (Affymetrix, Santa Clara, CA, USA) arrays for miRNA expression profiling. This array has miRBase v15 coverage (www.mirbase.org) with 15,644 probe sets of 131 organisms including 679 probe sets for Bos-taurus miRNAs. Array hybridization was carried out at ATLAS Biolabs, Berlin, Germany. Briefly, 200 ng total RNA labelled with Flash Tag Biotin was hybridized to the miRNA Array in a Hybridization Oven 640 (Affymetrix) at 48°C for 18 h. The arrays were stained with the Fluidics Station 450 using fluidics script FS450_0003 (Affymetrix), and then scanned on a GeneChip Scanner 3000 7G (Affymetrix, Santa Clara, CA, USA).
Statistical analysis
All statistical analyses were performed in the R statistical computing environment (Version 2.14; http://www.r-project.org) with the samr package from the Bioconductor project (http://www.bioconductor.org). Data quality was assessed with the ArrayQualitymetrix package from Bioconductor [42,43]. One cel file from each group was discarded due to issues with hybridisation quality. The expresso method of the Affy package [44] was used to pre-processes the data with quantile normalization and median polish summarization. While the chip contains probes complementary to miRNA from other species only the expression intensities of 679 cattle miRNAs were used for further analysis. The Significance Analysis of Microarray (SAM) two-class unpaired method implemented in the samr Bioconductor package was used to identify differentially expressed miRNAs [45]. The samr parameter delta 0.6 was selected that classified miRNAs as differentially expressed at a fold-change > 1.25 and a q-value < 12%.
RT-qPCR validation of differentially expressed miRNAs
Seven candidate miRNAs differentially expressed on the microarray; including miR-17-5p, miR-31, miR-140, miR-1281, miR-2885, miR-296, and miR-671 were selected for RT-qPCR validation. miRNA expression was carried out with TaqMan miRNA RT-qPCR assays according to the manufacturer’s instructions (Applied Biosystems, Dublin, Ireland). miRNA-specific reverse transcription was performed on 10 ng of purified total RNA using the TaqMan MicroRNA Reverse Transcription kit according to manufacturer’s instructions. RT-qPCR reactions were performed using 1 μl of cDNA (10 ng/μl) in 9 μl of Taqman universal master mix containing TaqMan PCR primers and probes on a BioRad CFX96 real time PCR system (Bio-Rad, Hemel Hempstead, UK) using the following cycling parameters; 95°C for 10 min followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. Four biological replicates from each of the MNEB and SNEB treatment groups were used for RT-qPCR validation. Three reference miRNAs including miR-122, let-7b and RNU6B were tested with the software program geNorm version 3.5 [46] for calculating the gene expression stability measure (M value). RNU6B was found to be the most stable internal reference miRNA across treatments with an M value of 0.7. It was more stable on its own than when used in combination with the two other reference miRNA. The software package BioRad CFX manager was used for correction of the Ct values and normalization to RNU6B using the 2-ΔΔCt method [47]. Corrected Ct values were used to calculate differential expression using the PROC t-test (SAS) [48].
Prediction of differentially expressed miRNAs targets among differentially expressed hepatic genes under SNEB
A target prediction algorithm for custom data, from the TargetScan database website [13] implemented in Perl (targetscan_61_context_scores.pl), was used to identify target sites of differentially expressed miRNAs in the 3′UTRs of 418 differentially expressed genes reported previously for the same liver tissue [9]. Using this algorithm, conserved 8mer and 7mer sites matching the seed region of each miRNA in the 3′UTRs of differentially expressed hepatic genes were identified [49]. Non-conserved sites and sites with mismatches in the seed region that were compensated by conserved 3′ pairing were also included [13]. Predictions were ranked based on the context + scores of the sites that represent the predicted efficacy of targeting [50]. GO (Gene Ontology) biological processes associations of 3 conserved miRNAs (miR-31, miR 17-5p and miR-140) were retrieved using FAME (functional assignment of miRNAs via enrichment) software [51]. No data was available for miR-1281 and miR-2885 in the FAME database. The FAME algorithm makes direct inference of a specific miRNA function using enriched subsets of the target genes sharing a common biological process or pathway. The GO biological processes associated with down-regulated putative targets were retrieved from the UniProt Gene Ontology Annotation (GOA) database (http://www.ebi.ac.uk/GOA).
Results
Ten out of 679 mature bovine miRNAs represented on the miRNA Gene Chip miRNA_2.0 array were found to be up-regulated in SNEB cows. Table 1 lists the miRNAs that showed differential expression between MNEB and SNEB groups. RT-qPCR results confirmed the differential expression of 5 out of the 7 miRNAs tested. The expression of miR17-5p, -1281, -140, 2885 and -31 were consistent between microarray and RT-qPCR whereas miR-296 and miR-671 were not significant. Table 2 lists the differentially expressed miRNAs validated by RT-qPCR. Putative targets of these five miRNAs were identified in the set of 418 (230 down- and 188 up-regulated) hepatic genes reported previously in the same animal model. Among the 418, 67 (35 up-regulated and 32 down-regulated) genes were found to have target sites in the 3′UTRs of the up-regulated miRNAs (Table 3).
Table 1.
Differentially expressed post-partum dairy cattle miRNAs under SNEB
| miRNA | miRBase accession | q-value (%) | Fold change |
|---|---|---|---|
| bta-miR-31 |
MI0004762 |
0.00 |
1.79 |
| bta-miR-1281 |
MI0010466 |
11.97 |
1.71 |
| bta-miR-2483 |
MI0011545 |
11.97 |
1.56 |
| bta-miR-2885 |
MI0013058 |
0.00 |
1.55 |
| bta-miR-296 |
MI0009786 |
11.97 |
1.52 |
| bta-miR-2316 |
MI0011336 |
11.97 |
1.40 |
| bta-miR-140 |
MI0005010 |
11.97 |
1.33 |
| bta-miR-17-5p |
MIMAT0003815 |
11.97 |
1.30 |
| bta-miR-671 |
MI0009887 |
11.97 |
1.29 |
| bta-miR-2455 | MI0011512 | 11.97 | 1.28 |
Differential expression analysis was done for microarray expression intensities of 679 bovine miRNAs with SAM (significance analysis of microarrays) method at delta parameter of 0.6 and q FDR < 12%.
Table 2.
Differentially expressed postpartum dairy cattle hepatic miRNAs under SNEB
| miRNA | Fold change | p value |
|---|---|---|
| bta-miR-31 |
4.11 |
0.033 |
| bta-miR-1281 |
3.10 |
0.042 |
| bta-miR-2885 |
2.91 |
0.006 |
| bta-miR-17-5p |
4.62 |
0.008 |
| bta-miR-140 |
3.90 |
0.030 |
| bta-miR-296 |
0.85 |
0.490 |
| bta-miR-671 | 0.92 | 0.930 |
Differential expression analysis was carried out with PROC t-test (SAS) after normalization of Ct-values to reference gene RNU6B.
Table 3.
Putative target genes of up-regulated hepatic miRNAs differentially expressed under SNEB
|
Down-regulated putative targets |
Up-regulated putative targets |
||
|---|---|---|---|
| Symbol | Gene name | Symbol | Gene name |
|
ALDH1A1 |
aldehyde dehydrogenase 1 family, member A1 |
AADAT |
aminoadipate aminotransferase |
|
BTG1 |
B-cell translocation gene 1, anti-proliferative |
ACP2 |
Acid phosphatase 2, lysosomal |
|
CCL19 |
chemokine (C-C motif) ligand 19 |
ADTRP |
Androgen-Dependent TFPI-Regulating Protein |
|
CD14 |
CD14 molecule |
ARG1 |
Arginase, liver |
|
CIB1 |
Calcium and integrin binding 1 (calmyrin) |
BIN1 |
Bridging integrator 1 |
|
DNAJA1 |
DnaJ (Hsp40) homolog, subfamily A, member 1 |
BNIP3L |
BCL2/adenovirus E1B 19 kDa interacting protein 3-like |
|
DSG1 |
desmoglein 1 |
CMPK1 |
cytidine monophosphate (UMP-CMP) kinase 1, cytosolic |
|
EPB41L5 |
Erythrocyte membrane protein band 4.1 like 5 |
CNN1 |
Calponin 1, Basic, Smooth Muscle |
|
ERRFI1 |
ERBB receptor feedback inhibitor 1 |
CPQ |
carboxypeptidase Q |
|
FADS2 |
Fatty acid desaturase 2 |
CPT1B |
carnitine palmitoyltransferase 1B (muscle) |
|
FOXA3 |
Forkhead box A3 |
DENND2D |
DENN/MADD domain containing 2D |
|
FXR1 |
Fragile X mental retardation, autosomal homolog 1 |
DPYD |
dihydropyrimidine dehydrogenase |
|
GKAP1 |
G kinase anchoring protein 1 |
HDC |
histidine decarboxylase |
|
GNS |
glucosamine (N-acetyl)-6-sulfatase |
HMGN4 |
High mobility group nucleosomal binding domain 4 |
|
GPBP1 |
GC-rich promoter binding protein 1 |
HSF2BP |
Heat shock transcription factor 2 binding protein |
|
GPR37 |
G protein-coupled receptor 37 (endothelin receptor type B-like) |
IL1A |
Interleukin 1, alpha |
|
HEYL |
hairy/enhancer-of-split related with YRPW motif-like |
LPCAT3 |
Lysophosphatidylcholine acyltransferase 3 |
|
IP6K2 |
inositol hexakisphosphate kinase 2 |
MBL2 |
Mannose-binding lectin (protein C) 2, soluble (opsonic defect) |
|
KIAA1 191 |
KIAA1 191 |
MINA |
MYC induced nuclear antigen |
|
MIEN1 |
Migration and invasion enhancer 1Bottom of Form |
NHEJ1 |
Nonhomologous end-joining factor 1 |
|
MIS 12 |
MIND kinetochore complex component, homolog (S. pombe) |
OAT |
ornithine aminotransferase (gyrate atrophy) |
|
NR2F1 |
nuclear receptor subfamily 2, group F, member 1 |
PDRG1 |
p53 and DNA-damage regulated 1 |
|
PRSS35 |
protease, serine, 35 |
POLR3GL |
polymerase (RNA) III polypeptide G (32kD)-like |
|
PTPRR |
protein tyrosine phosphatase, receptor type, R |
RCAN3 |
RCAN family member 3 |
| RNF 128 | ring finger protein 128 | REEP5 | Receptor accessory protein 5 |
Putative binding sites were identified in the 3′UTR of differentially expressed hepatic genes under SNEB with the Targetscan algorithm.
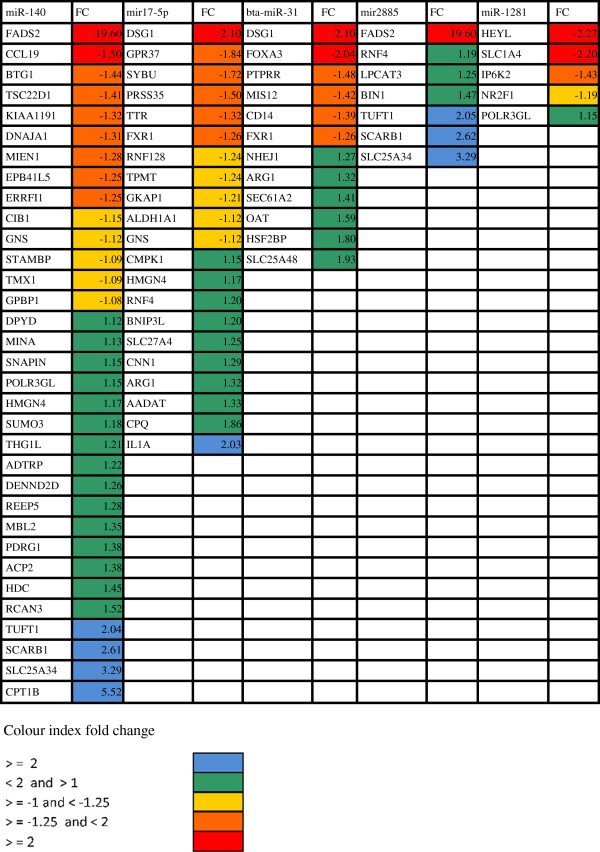
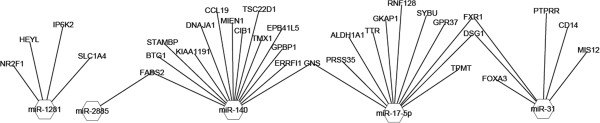
All of the five differentially expressed miRNAs validated by RT-qPCR in this study were up-regulated however there was no significant enrichment among the down-regulated putative targets genes using the Fisher test for enrichment analysis. Overall miR-140 was found to have the most putative targets among differentially expressed genes under SNEB while miR-17-5p had the second highest number of targets. As the consensus is that up-regulated miRNAs result for the most part in down-regulation of their target genes, we further selected only down-regulated genes as putative affected targets for subsequent analysis to investigate the role of differentially expressed miRNAs under SNEB. As miRNAs are thought not to switch off their targets completely but rather fine-tune their expression, all down-regulated genes were included irrespective of fold-change. Figure 1 presents the subset of the putative down-regulated targets for these up-regulated miRNAs. miR-140 had the highest number of putative down-regulated target genes followed by miR-17-5p. miR-31 was the most up-regulated miRNA under SNEB while miR-1281 had all but one of its putative target genes down-regulated under SNEB. miR-2885 has FADS2 as its only putative down-regulated target under SNEB and FADS2 is also a putative target of miR-140. DSG1 (desmoglein1) and FXR1 (fragile X mental retardation, autosomal homolog 1) are both shared targets of miR-17-5p and miR-31 while GNS (glucosamine-N-acetyl-6-sulfatase) is a shared target of mir-140 and miR-31 (Figure 2). Table 4 lists the bio-types of putative targets down-regulated under SNEB. The GO biological processes associated with down-regulated putative targets are given in Additional file 1: Table S1. The overall GO biological functions associated with three conserved miRNAs that are relevant in terms of SNEB are given in Additional file 1: Table S2. miR-31 is associated with the GO biological processes of cell growth, amino acid transport and response to stress. miR-17-5p is associated with GO biological processes include regulation of cell cycle and protein metabolism. miR-140 is associated with GO biological processes of cell proliferation, polysaccharide metabolism and signal transduction, while both miR-31 and miR-140 are associated with response to nutritional level.
Figure 1.
Predicted miRNA gene targets with fold-change under SNEB.
Figure 2.
Hexagons represent the up-regulated miRNAs with edges leading to nodes representing their putative targets down-regulated under SNEB. Nodes with two edges represent putative targets shared by two up-regulated miRNAs.
Table 4.
Intracellular location and bio-types of down-regulated putative gene targets of up-regulated miRNAs
| Symbol | Entrez gene name | Location | Type |
|---|---|---|---|
| KIAA1191 |
KIAA1191 |
Cytoplasm |
Other |
| MIEN 1 |
Migration and invasion enhancer 1 |
Cytoplasm |
Other |
| MIS 12 |
MIS 12, MIND kinetochore complex component, homolog (S. pombe) |
Nucleus |
Other |
| NR2F1 |
Nuclear receptor subfamily 2, group F, member 1 |
Nucleus |
Ligand-dependent nuclear receptor |
| PRSS35 |
Protease, serine, 35 |
Extracellular space |
peptidase |
| PTPRR |
Protein tyrosine phosphatase, receptor type, R |
Plasma membrane |
phosphatase |
| RNF 128 |
Ring finger protein 128, E3 ubiquitin protein ligase |
Cytoplasm |
Enzyme |
| SLC1A4 |
Solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
Plasma membrane |
Transporter |
| STAMBP |
STAM binding protein |
Nucleus |
Enzyme |
| SYBU |
Syntabulin (syntaxin-interacting) |
Unknown |
Other |
| TMX1 |
Thioredoxin-related transmembrane protein 1 |
Cytoplasm |
Enzyme |
| TPMT |
Thiopurine S-methyltransferase |
Cytoplasm |
Enzyme |
| TTR |
Transthyretin1 |
Cytoplasm |
Transporter |
| ALDH1A1 |
Aldehyde dehydrogenase 1 family, member A1 |
Cytoplasm |
Enzyme |
| BTG1 |
B-cell Translocation gene 1, anti-proliferative |
Nucleus |
Transcription Regulator |
| CCL19 |
chemokine (C-C motif) ligand 19 |
Extracellular space |
Cytokine |
| CD14 |
CD14 molecule |
Plasma Membrane |
Transmembrane receptor |
| CIB1 |
Calcium and integrin binding 1 (calmyrin) |
Nucleus |
Other |
| DNAJA1 |
DnaJ (Hsp40) homolog, subfamily A, member 1 |
Nucleus |
Other |
| DSG1 |
Desmoglein 1 |
Plasma membrane |
Membrane protein |
| EPB4 1 L5 |
Erythrocyte membrane protein band 4.1 like 5 |
Plasma membrane |
Membrane protein |
| ERRFI1 |
ERBB receptor feedback inhibitor 1 |
Cytoplasm |
Other |
| FADS2 |
Fatty acid desaturase 2 |
Plasma membrane |
Enzyme |
| FOXA3 |
Forkhead box A3 |
Nucleus |
Transcription regulator |
| FXR1 |
Fragile X mental retardation, autosomal homolog 1 |
Cytoplasm |
Other |
| GKAP1 |
G kinase anchoring protein 1 |
Cytoplasm |
Other |
| GNS |
Glucosamine (N-acetyl)-6-sulfatase |
Cytoplasm |
Enzyme |
| GPBP1 |
GC-rich promoter binding protein 1 |
Nucleus |
Transcription regulator |
| GPR37 |
G protein-coupled receptor 37 (endothelin receptor type B-like) |
Plasma membrane |
G-protein coupled receptor |
| HEYL |
Hairy/enhancer-of-split related with YRPW motif-like |
Nucleus |
Transcription regulator |
| IP6K2 | Inositol hexakisphosphate kinase 2 | Cytoplasm | Kinase |
Discussion
Dairy cows enter a state of NEB postpartum when the energy supply is prioritised towards the mammary tissues for milk production while the energy intake is reduced. The principal physiological response to the energy deficit is mobilization of non-esterified fatty acids (NEFA) from adipose tissue. NEFAs are broken down in the liver to release energy and any incomplete breakdown results in production of BHB (β-hydroxybutyrate) or TAGs (triacylglycerides). Accumulation of TAGs, BHB and NEFAs results in the onset of oxidative stress in the liver that can lead to fatty liver or lipidosis. In addition components of the somatotrophic axis like IGF (insulin like growth factor) and GH (growth hormones) that have roles in the control of milk production are also affected by SNEB [8-10]. Reduced IGF1 expression is also negatively correlated with fertility in dairy cows [52]. Previous nutritional supplementation of animals in NEB with lipogenic and glycogenic nutrients partly succeeded in compensating for energy partitioning between mammary tissues and the rest of the body [4,53]. However, the regulation of such alterations is yet to be explored. In this study, five miRNAs were altered in response to SNEB. Two of these miRNAs have been reported to be altered under oxidative stress in humans [41,54] and miR-17-5p has been associated with oxidative stress in HCC [41]. In addition alterations in miR-140 expression under chemical and oxidative stress has also been reported [54]. Three of the miRNAs, miR- 17, miR-31 and miR-140, found to be up-regulated under SNEB have been previously associated with hepatic disorders [55-58] and miR-140 has been associated with NAFLD [40,57]. A regulatory role for miR-17-5p has been reported in HCC [55,56] while miR-31 has also been associated with both NAFLD and HCC [58-60].
A number of the putative down-regulated targets of the differentially expressed miRNAs in this study are known to have roles in hepatic disorders. These include the miR-17-5p target GPR37 (G protein-coupled receptor 37) a member of the G-protein coupled receptor-1 family associated with NAFLD [61]. The miR-1281 target HEYL (hairy/enhancer-of-split related with YRPW motif-like) is associated with HCC [62,63]. The miR-140 target DNJA1 that has a role in lipid intake is associated with the oxidative stress response of high fat induced NAFLD [28,30]. The miR-31 target, CD14 (Cluster of differentiation 14) involved in transmission and release of polysaccharides, is also implicated in ALD, hepatic cholestasis and hepatic fibrosis [64,65]. The miR-17-5p target GNS (glucosamine (N-acetyl)-6-sulfatase) that metabolises glucosamine sulphate has been implicated in hepatocellular dysplasia, cirrhosis of the liver and hepatic fibrosis [66]. Moreover, the miR-17-5p target TTR, a carrier protein for transport of lipid soluble vitamins from the liver to the circulation is also reduced under SNEB. Reduced TTR expression has been reported in a study of periparturient period Holstein and Jersey dairy cows in NEB [67]. In addition we were able to postulate a role for miR-31 in SNEB through its putative target hepatic transcription factor FOXA3 also known as HNF3-gamma. Bovine IGF-I has binding sites for HNF3-gamma in its promoter region [68]. While another recent human study reported target sites of miR-31 on the 3′UTR of IGF-1[69]. HNF-gamma is also known to be involved in glucose homeostasis in hepatocytes through regulation of GLUT2 (glucose transporter 2) [70,71]. However, the most interesting putative target of all was the gene encoding the lipogenesis enzyme FADS2 which is critical for long-chain polyunsaturated fatty acids biosynthesis [72]. In this study we found binding sites for two up-regulated miRNAs miR-140 and miR-2885 in the 3′UTR of FADS2, whereas miR-140 has been previously implicated in the regulation of FADS2 homolog FADS1[73].
Conclusion
This study suggests a role for hepatic miRNAs in lipid and energy metabolism through the identification of a subset of their putative targets associated with important metabolic processes. Moreover hepatic miRNAs associated with genes involved in the somatotropic axis can be a possible link between reduced reproductive performance and SNEB in the early postpartum dairy cow. Further direct functional studies of selected miRNA-mRNA putative pairs in the liver of such models will help to improve our understanding of the molecular mechanisms and pathways involved in SNEB in postpartum dairy cattle.
Availability of supporting data
The data sets supporting the results of this article are available in Gene Expression Omnibus (GEO) repository (GSE51658).
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
DM, CS and SW conceived and designed the experiments. AF and PO performed the experiments. AF, CS and DM analyzed the data. AF, DM and CS drafted the manuscript. All authors read and approved the final manuscript.
Authors’ information
Cathal Seoighe and Dermot G Morris are joint senior authors.
Supplementary Material
GO biological processes associated with putative target genes down-regulated under SNEB. Table S2. FAME GO biological processes associated with up-regulated miRNAs under SNEB.
Contributor Information
Attia Fatima, Email: attia.fatima@gmail.com.
Sinead Waters, Email: sinead.waters@teagasc.ie.
Padraig O’Boyle, Email: padraic.oboyle@teagasc.ie.
Cathal Seoighe, Email: cathal.seoighe@nuigalway.ie.
Dermot G Morris, Email: dermot.morris@teagasc.ie.
Acknowledgements
The authors would like to acknowledge a Teagasc Walsh Fellowship and funding for AF.
References
- Diskin MG, Morris DG. Embryonic and early foetal losses in cattle and other ruminants. Reprod Domest Anim. 2008;15(s2):260–267. doi: 10.1111/j.1439-0531.2008.01171.x. [DOI] [PubMed] [Google Scholar]
- Lucy MC. Fertility in high-producing dairy cows: reasons for decline and corrective strategies for sustainable improvement. Reprod Domest Ruminants. 2007;15(1):237–254. doi: 10.5661/RDR-VI-237. [DOI] [PubMed] [Google Scholar]
- Butler WR. Energy balance relationships with follicular development, ovulation and fertility in postpartum dairy cows. Livest Prod Sci. 2003;15(2):211–218. [Google Scholar]
- Van Knegsel ATM, Van den Brand H, Dijkstra J, Van Straalen WM, Heetkamp MJW, Tamminga S, Kemp B. Dietary energy source in dairy cows in early lactation: energy partitioning and milk composition. J Dairy Sci. 2007;15(3):1467–1476. doi: 10.3168/jds.S0022-0302(07)71632-6. [DOI] [PubMed] [Google Scholar]
- Patton J, Kenny DA, Mee JF, O’Mara FP, Wathes DC, Cook M, Murphy JJ. Effect of milking frequency and diet on milk production, energy balance, and reproduction in dairy cows. J Dairy Sci. 2006;15(5):1478–1487. doi: 10.3168/jds.S0022-0302(06)72215-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wathes DC, Cheng Z, Chowdhury W, Fenwick MA, Fitzpatrick R, Morris DG, Patton J, Murphy JJ. Negative energy balance alters global gene expression and immune responses in the uterus of postpartum dairy cows. Physiol Genomics. 2009;15(1):1–13. doi: 10.1152/physiolgenomics.00064.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris DG, Waters SM, McCarthy SD, Patton J, Earley B, Fitzpatrick R, Murphy JJ, Diskin MG, Kenny DA, Brass A, Wathes DC. Pleiotropic effects of negative energy balance in the postpartum dairy cow on splenic gene expression: repercussions for innate and adaptive immunity. Physiol Genomics. 2010;15(1):28–37. doi: 10.1152/physiolgenomics.90394.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenwick MA, Fitzpatrick R, Kenny DA, Diskin MG, Patton J, Murphy JJ, Wathes DC. Interrelationships between negative energy balance (NEB) and IGF regulation in liver of lactating dairy cows. Domest Anim Endocrinol. 2008;15(1):31–44. doi: 10.1016/j.domaniend.2006.10.002. [DOI] [PubMed] [Google Scholar]
- McCarthy SD, Waters SM, Kenny DA, Diskin MG, Fitzpatrick R, Patton J, Wathes DC, Morris DG. Negative energy balance and hepatic gene expression patterns in high-yielding dairy cows during the early postpartum period: a global approach. Physiol Genomics. 2010;15(3):188–199. doi: 10.1152/physiolgenomics.00118.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe M, Waters S, Morris D, Kenny D, Lynn D, Creevey C. RNA-seq analysis of differential gene expression in liver from lactating dairy cows divergent in negative energy balance. BMC Genomics. 2012;15(1):193. doi: 10.1186/1471-2164-13-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;15(5):843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- Friedman JR, Kaestner KH. The Foxa family of transcription factors in development and metabolism. Cell Mol Life Sci CMLS. 2006;15(19–20):2317–2328. doi: 10.1007/s00018-006-6095-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;15(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Julie Li Y-S, Huang H-D, Shyy JYJ, Chien S. microRNA: a master regulator of cellular processes for bioengineering systems. Annu Rev Biomed Eng. 2010;15:1–27. doi: 10.1146/annurev-bioeng-070909-105314. [DOI] [PubMed] [Google Scholar]
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;15(5):376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;15(9):597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer. 2006;15(4):259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Hoefig KP, Heissmeyer V. MicroRNAs grow up in the immune system. Curr Opin Immunol. 2008;15(3):281–287. doi: 10.1016/j.coi.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Carleton M, Cleary MA, Linsley PS. MicroRNAs and cell cycle regulation. Cell Cycle. 2007;15(17):2127–2132. doi: 10.4161/cc.6.17.4641. [DOI] [PubMed] [Google Scholar]
- Rottiers V, Naarr AM. MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol. 2012;15(4):239–250. doi: 10.1038/nrm3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Yu X, Guo X, Tian Z, Su M, Long Y, Huang C, Zhou F, Liu M, Wu X. miR-143 is downregulated in cervical cancer and promotes apoptosis and inhibits tumor formation by targeting Bcl-2. Mol Med Rep. 2012;15(3):753–760. doi: 10.3892/mmr.2011.696. [DOI] [PubMed] [Google Scholar]
- Liu J, Liu W, Ying H, Zhao W, Zhang H. Analysis of microRNA expression profile induced by AICAR in mouse hepatocytes. Gene. 2012;15:364–372. doi: 10.1016/j.gene.2012.09.118. [DOI] [PubMed] [Google Scholar]
- Li S, Chen X, Zhang H, Liang X, Xiang Y, Yu C, Zen K, Li Y, Zhang C-Y. Differential expression of microRNAs in mouse liver under aberrant energy metabolic status. J Lipid Res. 2009;15(9):1756–1765. doi: 10.1194/jlr.M800509-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker C, Riedmaier I, Reiter M, Tichopad A, Pfaffl MW, Meyer HHD. Changes in the miRNA profile under the influence of anabolic steroids in bovine liver. Analyst. 2011;15(6):1204–1209. doi: 10.1039/c0an00703j. [DOI] [PubMed] [Google Scholar]
- Deli D, Grosser C, Dkhil M, Al-Quraishy S, Wunderlich F. Testosterone-induced upregulation of miRNAs in the female mouse liver. Steroids. 2010;15(12):998–1004. doi: 10.1016/j.steroids.2010.06.010. [DOI] [PubMed] [Google Scholar]
- Bala S, Marcos M, Szabo G. Emerging role of microRNAs in liver diseases. World J Gastroenterol. 2009;15(45):5633–5640. doi: 10.3748/wjg.15.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai BH, Geng L, Wang Y, Sui CJ, Xie F, Shen RX, Shen WF, Yang JM. microRNA-199a-5p protects hepatocytes from bile acid-induced sustained endoplasmic reticulum stress. Cell Death Dis. 2013;15(4):e604. doi: 10.1038/cddis.2013.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallanat B, Anderson S, Brown-Borg H, Ren H, Kersten S, Jonnalagadda S, Srinivasan R, Corton JC. Analysis of the heat shock response in mouse liver reveals transcriptional dependence on the nuclear receptor peroxisome proliferator-activated receptor (PPAR) BMC Genomics. 2010;15(1):16. doi: 10.1186/1471-2164-11-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomofuji T, Ekuni D, Sanbe T, Irie K, Azuma T, Maruyama T, Tamaki N, Murakami J, Kokeguchi S, Yamamoto T. Effects of vitamin C intake on gingival oxidative stress in rat periodontitis. Free Radical Bio Med. 2009;15(2):163–168. doi: 10.1016/j.freeradbiomed.2008.09.040. [DOI] [PubMed] [Google Scholar]
- Kirpich IA, Gobejishvili LN, Homme MB, Waigel S, Cave M, Arteel G, Barve SS, McClain CJ, Deaciuc IV. Integrated hepatic transcriptome and proteome analysis of mice with high-fat diet-induced nonalcoholic fatty liver disease. J Nutr Biochem. 2011;15(1):38–45. doi: 10.1016/j.jnutbio.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romao JM, Jin W, He M, McAllister T. Altered MicroRNA expression in bovine subcutaneous and visceral adipose tissues from cattle under different diet. Plos One. 2012;15(7):e40605. doi: 10.1371/journal.pone.0040605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;15(2):87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Esau C, Kang X, Peralta E, Hanson E, Marcusson EG, Ravichandran LV, Sun Y, Koo S, Perera RJ, Jain R. MicroRNA-143 regulates adipocyte differentiation. J Biol Chem. 2004;15(50):52361–52365. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- Jordan SD, Krager M, Willmes DM, Redemann N, Wunderlich FT, Branneke HS, Merkwirth C, Kashkar H, Olkkonen VM, Battger T. Obesity-induced overexpression of miRNA-143 inhibits insulin-stimulated AKT activation and impairs glucose metabolism. Nat Cell Biol. 2011;15(4):434–446. doi: 10.1038/ncb2211. [DOI] [PubMed] [Google Scholar]
- Rayner KJ, Suárez Y, Dávalos A, Parathath S, Fitzgerald ML, Tamehiro N, Fisher EA, Moore KJ, Fernández-Hernando C. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;15(5985):1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davalos A, Goedeke L, Smibert P, Ramarez CM, Warrier NP, Andreo U, Cirera-Salinas D, Rayner K, Suresh U, Pastor-Pareja JC. et al. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc Natl Acad Sci. 2011;15(22):9232–9237. doi: 10.1073/pnas.1102281108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X, Ye YF, Chen SH, Yu CH, Liu J, Li YM. MicroRNA expression pattern in different stages of nonalcoholic fatty liver disease. Dig Liver Dis. 2009;15(4):289–297. doi: 10.1016/j.dld.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Ahn J, Lee H, Chung CH, Ha T. High fat diet induced downregulation of microRNA-467b increased lipoprotein lipase in hepatic steatosis. Biochem Biophys Res Commun. 2011;15(4):664–669. doi: 10.1016/j.bbrc.2011.09.120. [DOI] [PubMed] [Google Scholar]
- Ahn J, Lee H, Jung CH, Ha T. Lycopene inhibits hepatic steatosis via microRNA-21induced downregulation of fatty acid binding protein 7 in mice fed a high fat diet. Mol Nutr Food Res. 2012;15(11):1665–1674. doi: 10.1002/mnfr.201200182. [DOI] [PubMed] [Google Scholar]
- Alisi A, Da Sacco L, Bruscalupi G, Piemonte F, Panera N, De Vito R, Leoni S, Bottazzo GF, Masotti A, Nobili V. Mirnome analysis reveals novel molecular determinants in the pathogenesis of diet-induced nonalcoholic fatty liver disease. Lab Invest. 2011;15(2):283–293. doi: 10.1038/labinvest.2010.166. [DOI] [PubMed] [Google Scholar]
- Yang F, Yin Y, Wang F, Wang Y, Zhang L, Tang Y, Sun S. miR17-5p Promotes migration of human hepatocellular carcinoma cells through the p38 mitogen-activated protein kinase heat shock protein 27 pathway. Hepatology. 2010;15(5):1614–1623. doi: 10.1002/hep.23566. [DOI] [PubMed] [Google Scholar]
- Dee S, Getts RC. Next-Generation MicroRNA Expression Profiling Technology. Springer New York Dordrecht Heidelberg London: Springer; 2012. MicroRNA expression analysis using the Affymetrix Platform; pp. 117–129. ISSN 1064-3745 e-ISSN 1940-6029, ISBN 978-1-61779-426-1 e-ISBN 978-1-61779-427-8. doi:10.1007/978-1-61779-427-8. [DOI] [PubMed] [Google Scholar]
- Kauffmann A, Gentleman R, Huber W. arrayQualityMetrics - a bioconductor package for quality assessment of microarray data. Bioinformatics. 2009;15(3):415–416. doi: 10.1093/bioinformatics/btn647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA. affy analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;15(3):307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci. 2001;15(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkens T, Van Poucke M, Vandesompele J, Goossens K, Van Zeveren A, Peelman L. Development of a new set of reference genes for normalization of real-time RT-PCR data of porcine backfat and longissimus dorsi muscle, and evaluation with PPARGC1A. BMC Biotechnol. 2006;15(1):41. doi: 10.1186/1472-6750-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;15(6):1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Yuan JS, Reed A, Chen F, Stewart CN. Statistical analysis of real-time PCR data. BMC Bioinformatics. 2006;15(1):85. doi: 10.1186/1471-2105-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;15(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Grimson A, Farh KK-H, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;15(1):91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Laurent LC, Shamir R. Towards computational prediction of microRNA function and activity. Nucleic Acids Res. 2010;15(15):e160. doi: 10.1093/nar/gkq570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor VJ, Cheng Z, Pushpakumara PGA, Wathes DC, Beever DE. Relationships between the plasma concentrations of insulin-like growth factor-I in dairy cows and their fertility and milk yield. Veterinary Rec. 2004;15(19):583–588. doi: 10.1136/vr.155.19.583. [DOI] [PubMed] [Google Scholar]
- Van Knegsel ATM, Van den Brand H, Dijkstra J, Kemp B. Effects of dietary energy source on energy balance, metabolites and reproduction variables in dairy cows in early lactation. Theriogenology. 2007;15:S274–S280. doi: 10.1016/j.theriogenology.2007.04.043. [DOI] [PubMed] [Google Scholar]
- Read DE, Gupta A, Cawley K, Gupta S. Endoplasmic Reticulum Stress in Health and Disease. Springer Dordrecht Heidelberg New York London: Springer; 2012. Regulation of ER Stress Responses by microRNAs; pp. 143–161. ISBN 978-94-007-4350-2 ISBN 978-94-007-4351-9 (eBook). DOI 10.1007/978-94-007-4351-9. [Google Scholar]
- Shan SW, Lee DY, Deng Z, Shatseva T, Jeyapalan Z, Du WW, Zhang Y, Xuan JW, Yee S-P, Siragam V. MicroRNA MiR-17 retards tissue growth and represses fibronectin expression. Nat Cell Biol. 2009;15(8):1031–1038. doi: 10.1038/ncb1917. [DOI] [PubMed] [Google Scholar]
- Shan SW, Fang L, Shatseva T, Rutnam ZJ, Yang X, Lu W-Y, Xuan JW, Deng Z, Yang BB. Mature MiR-17-5p and passenger miR-17-3p induce hepatocellular carcinoma by targeting PTEN, GalNT7, and vimentin in different signal pathways. J Cell Sci. 2013;15(Pt6):1517–1530. doi: 10.1242/jcs.122895. [DOI] [PubMed] [Google Scholar]
- Michelotti GA, Machado MV, Diehl AM. NAFLD, NASH and liver cancer. Nat Rev Gastroenterol Hepatol. 2013;15:656–665. doi: 10.1038/nrgastro.2013.183. [DOI] [PubMed] [Google Scholar]
- Hill-Baskin AE, Markiewski MM, Buchner DA, Shao H, DeSantis D, Hsiao G, Subramaniam S, Berger NA, Croniger C, Lambris JD. Diet-induced hepatocellular carcinoma in genetically predisposed mice. Hum Mol Genet. 2009;15(16):2975–2988. doi: 10.1093/hmg/ddp236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Lv G, Sheng J, Yang Y. Effect of miRNA10b in regulating cellular steatosis level by targeting PPARα expression, a novel mechanism for the pathogenesis of NAFLD. J Gastroenterol Hepatol. 2010;15(1):156–163. doi: 10.1111/j.1440-1746.2009.05949.x. [DOI] [PubMed] [Google Scholar]
- Karakatsanis A, Papaconstantinou I, Gazouli M, Lyberopoulou A, Polymeneas G, Voros D. Expression of microRNAs, miR-21, miR-31, miR-122, miR-145, miR-146a, miR-200c, miR-221, miR-222, and miR-223 in patients with hepatocellular carcinoma or intrahepatic cholangiocarcinoma and its prognostic significance. Mol Carcinog. 2013;15(4):297–303. doi: 10.1002/mc.21864. [DOI] [PubMed] [Google Scholar]
- Sharma MR, Polavarapu R, Roseman D, Patel V, Eaton E, Kishor PB, Nanji AA. Transcriptional networks in a rat model for nonalcoholic fatty liver disease: A microarray analysis. Exp Mol Pathol. 2006;15(3):202–210. doi: 10.1016/j.yexmp.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Huang E-J, Wu C-C, Huang H-P, Liu J-Y, Lin C-S, Chang Y-Z, Lin JA, Lin J-G, Chen L-M, Lee S-D. Apoptotic and anti-proliferative effects of 17 beta -estradiol and 17 beta -estradiol-like compounds in the Hep3B cell line. Mol Cell Biochem. 2006;15(1–2):1–7. doi: 10.1007/s11010-005-9000-y. [DOI] [PubMed] [Google Scholar]
- Xie G, Karaca G, Swiderska-Syn M, Michelotti GA, Kruger L, Chen Y, Premont RT, Choi SS, Diehl AM. Cross talk between notch and hedgehog regulates hepatic stellate cell fate in mice. Hepatology. 2013;15(5):1801–1813. doi: 10.1002/hep.26511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki E, Brenner DA. Toll like receptors and adaptor molecules in liver disease: Update. Hepatology. 2008;15(1):322–335. doi: 10.1002/hep.22306. [DOI] [PubMed] [Google Scholar]
- Kolios G, Valatas V, Kouroumalis E. Role of Kupffer cells in the pathogenesis of liver disease. World J Gastroenterol. 2006;15(46):7413. doi: 10.3748/wjg.v12.i46.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Finegold MJ. Biopsy diagnosis of inherited liver disease. Surg Pathol Clin. 2010;15(3):743–768. doi: 10.1016/j.path.2010.06.006. [DOI] [PubMed] [Google Scholar]
- Rezamand P, Hoagland TA, Moyes KM, Silbart LK, Andrew SM. Energy status, lipid-soluble vitamins, and acute phase proteins in periparturient Holstein and Jersey dairy cows with or without subclinical mastitis. J Dairy Sci. 2007;15(11):5097–5107. doi: 10.3168/jds.2007-0035. [DOI] [PubMed] [Google Scholar]
- Eleswarapu S, Ge X, Wang Y, Yu J, Jiang H. Growth hormone-activated STAT5 may indirectly stimulate IGF-I gene transcription through HNF-3 gamma. Mol Endocrinol. 2009;15(12):2026–2037. doi: 10.1210/me.2009-0178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosakhani N, Guled M, Lahti L, Borze I, Forsman M, Paakkonen V, Ryhanen J, Knuutila S. Unique microRNA profile in Dupuytren’s contracture supports deregulation of beta-catenin pathway. Mod Pathol. 2010;15(11):1544–1552. doi: 10.1038/modpathol.2010.146. [DOI] [PubMed] [Google Scholar]
- Costa RH, Kalinichenko VV, Holterman AXL, Wang X. Transcription factors in liver development, differentiation, and regeneration. Hepatology. 2003;15(6):1331–1347. doi: 10.1016/j.hep.2003.09.034. [DOI] [PubMed] [Google Scholar]
- Shen W, Scearce LM, Brestelli JE, Sund NJ, Kaestner KH. Foxa3 (hepatocyte nuclear factor 3) is required for the regulation of hepatic GLUT2 expression and the maintenance of glucose homeostasis during a prolonged fast. J Biol Chem. 2001;15(46):42812–42817. doi: 10.1074/jbc.M106344200. [DOI] [PubMed] [Google Scholar]
- Lattka E, Eggers S, Moeller G, Heim K, Weber M, Mehta D, Prokisch H, Illig T, Adamski J. A common FADS2 promoter polymorphism increases promoter activity and facilitates binding of transcription factor ELK1. J Lipid Res. 2010;15(1):182–191. doi: 10.1194/jlr.M900289-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park WJ, Kothapalli KSD, Lawrence P, Brenna JT. FADS2 function loss at the cancer hotspot 11q13 locus diverts lipid signaling precursor synthesis to unusual Eicosanoid fatty acids. Plos One. 2011;15(11):e28186. doi: 10.1371/journal.pone.0028186. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
GO biological processes associated with putative target genes down-regulated under SNEB. Table S2. FAME GO biological processes associated with up-regulated miRNAs under SNEB.