Abstract
Phenolic compounds in the fruits of two diploid strawberries (Fragaria vesca f. semperflorens) inbred lines-Ruegen F7-4 (a red-fruited genotype) and YW5AF7 (a yellow-fruited genotype) were characterised using ultra-high-performance liquid chromatography coupled with tandem high-resolution mass spectrometry (UHPLC-HRMSn). The changes of anthocyanin composition during fruit development and between Ruegen F7-4 and YW5AF7 were studied. About 67 phenolic compounds, including taxifolin 3-O-arabinoside, glycosides of quercetin, kaempferol, cyanidin, pelargonidin, peonidin, ellagic acid derivatives, and other flavonols were identified in these two inbred lines. Compared to the regular octoploid strawberry, unique phenolic compounds were found in F. vesca fruits, such as taxifolin 3-O-arabinoside (both) and peonidin 3-O-malonylglucoside (Ruegen F7-4). The results provide the basis for comparative analysis of polyphenolic compounds in yellow and red diploid strawberries, as well as with the cultivated octoploid strawberries.
Keywords: Strawberry, Fragaria vesca, UHPLC, HRMS, Flavonoids, Anthocyanins
1. Introduction
Strawberries are an economically important horticultural crop and much research has been conducted on maximising fruit growth in the field and increasing postharvest fruit quality (Goulas & Manganaris, 2011; Reganold et al., 2010; Shin et al., 2008; Villa-Rojas, Lopez-Malo, & Sosa-Morales, 2011; Wojdylo, Figiel, & Oszmianski, 2009; Yang et al., 2010). In 2011, the worldwide production of strawberries was near 4.6 million tons and the value of strawberry production in the United States valued at over $2 billion (www.faostats.fao.org). Commercial strawberry Fragaria × ananassa is an octoploid (2n = 8 × = 56) hybrid of two octoploid species, Fragaria chiloensis and Fragaria virginiana, native to America (Sun & Shi, 2008). Fragaria vesca is a widely distributed diploid (2n = 2 × = 14) species whose ancestor is believed to be an ancestral genome donor to the octoploid strawberries. The small size of the F. vesca plant, its transformability with Agrobacterium, its small genome and available genome sequence, and the existing inbred lines support F. vesca as a useful reference plant for strawberry {Shulaev, 2011 #11;Slovin, 2009 #12;Kim, 2003 #16263}. Strawberry fruits contain high levels of vitamin C, folate, and phenolic compounds, and are considered to be beneficial to human health. Thus, many studies have been done on the characterisation of these secondary metabolites, their biosynthesis, and their accumulation during fruit development (Bianco et al., 2009; Zhang et al., 2011). The phenolic compounds of regular strawberry fruits, including anthocyanins, proanthocyanindins, flavonols, flavanols, and derivatives of hydroxycinnamic and ellagic acid, are well-studied (Aaby, Ekeberg, & Skrede, 2007; Aaby, Mazur, Nes, & Skrede, 2012; Aaby, Wrolstad, Ekeberg, & Skrede, 2007; Buendia et al., 2010; Hilt et al., 2003; Kelebek & Selli, 2011; Maatta-Riihinen, Kamal-Eldin, & Torronen, 2004). However, there are not much detailed polyphenol content studies on F. versa. In a recently published study on F. versa, agrimoniin was isolated in the fruit of F. vesca and identified as the one of the main ellagitannins (Vrhovsek et al., 2012). High-resolution mass spectrometry (HRMS) has gained popularity in the last few years. HRMS instruments are being used in both quantitative and confirmative food analysis (Kaufmann, 2012). The higher resolution of HRMS provides enough resolving power to calculate the molecular formula for an analyte, so a suggested structure can be confirmed or denied. For example, HRMS can reliably differentiate between a glucosyl (C6H10O5) and caffeoyl (C9H6O3), a rhamnosyl (C6H10O4) and a coumaroyl (C9H6O2), which are commonly seen substituent groups in polyphenols from strawberry while these two pairs of substituent groups exhibit the same mass weights on unit mass spectrometers.
Two inbred lines, F. vesca, YW5AF7 and Ruegen F7-4 have yellow fruit with tan achenes and red fruit with red achenes respectively, and were specifically developed for genetic and genomic studies (Kim et al., 2003). To assist the future molecular, genetic, and genomic studies of F. vesca, it is necessary to perform a detailed study on the polyphenols in these two diploid lines. Thus an ultra-high-performance liquid chromatography (UHPLC) with diode array detection (DAD) and multi-stage high-resolution mass spectrometry (HRMSn) detection method was established for this purpose, which leads to the identification of 67 phenolic compounds, including taxifolin 3-O-arabinoside, glycosides of quercetin, kaempferol, cyanidin, pelargonidin, peonidin, ellagic acid derivatives, and other flavonols.
2. Experimental
2.1. Materials
Diploid strawberry (F. vesca) inbred lines YW5AF7 and Ruegen F7-4, as well as octoploid strawberry (F. × ananassa) cv. Fort Laramie, were grown in a greenhouse with a diurnal rhythm of 16 h light and 8 h darkness following normal cultivation practices. Five fruits (octoploid) and 20 fruits (diploids) were collected from the greenhouse. The fruits at different stages were classified based upon the fruit size and colour of achenes and receptacles. Green: small fruit with green achene and receptacle; turning: fruit with white receptacles and tan (YW5AF7 and Fort Laramie) or red (F7-4) achenes; ripe: ripe fruit with yellow (YW5AF7) or red (F7-4 and Fort Laramie) receptacles (Figure S1). The fruit collection and extraction experiments were repeated at least three times. After harvest, all the fruits from different stages were washed in water, cut into quarters, immediately frozen in liquid nitrogen and kept at −80 °C for future use.
HPLC-grade methanol, acetonitrile, and formic acid were purchased from Sigma-Aldrich (St. Louis, MO). Pelargonidin 3-O-glucoside, cyanidin 3-O-glucoside, peonidin 3-O-glucoside, ellagic acid, quercetin 3-O-glucuronide, quercetin 3-O-glucoside, kaempferol 3-O-glucuronide and (+)-catechin were purchased from Chromadex Inc. (Irvine, CA).
2.2. Sample preparation
One gram of each freezed-dried sample was homogenised with 50 mL of extraction solution (methanol/water/formic acid; 60:40:1 v/v/v) using an Ultra Turrax T18 Basic Disperser (IKA Werke GmbH & Co, Staufen, Germany) for 1 min on ice followed by sonication in an ultrasound bath for 15 min (Branson 3200; Branson, Danbury, CT). The homogenates were then centrifuged at 5000g for 10 min (IEC clinical centrifuge; IEC, Needham Heights, MA). The upper layer was filtered through a 0.22-µm PTFE filter, transferred into a 2-mL HPLC vial, and 2 µL was injected for UHPLC-HRMSn analysis.
2.3. The UHPLC-HRMSn conditions
The UHPLC-HRMSn system consisted of an LTQ Orbitrap XL MS with an Accela 1250 binary pump, a PAL HTC Accela TMO autosampler, an Accela PDA detector (Thermo Fisher Scientific, San Jose, CA), and an Agilent G1316A column compartment (Agilent, Santa Clara, CA). The separation was carried out on a Hypersil Gold C18 column (200 × 2.1 mm, 1.9 µm particle size; Thermo Fisher Scientific, San Jose, CA) with a flow rate of 0.3 mL/min. Mobile phase A was H2O (0.1% formic acid) and B was acetonitrile (0.1% formic acid). The linear gradient was 4–20% B (v/v) from 0 to 40 min, to 35% B at 60 min, to 100% B at 61 min, and was held at 100% B to 65 min for column washing. The column temperature was set at 60 °C and UV/Vis spectra were recorded between 200 and 700 nm. High-accuracy mass measurements were carried out under both positive and negative ionisation modes. The MS conditions were set as follows: sheath gas at 70 (arbitrary units), auxiliary and sweep gas at 10 (arbitrary units), spray voltage at 4.5 kV for positive ionisation mode and 4 kV for negative ionisation mode, capillary temperature at 250 °C, capillary voltage at 40 V for positive ionisation mode and −50 V for negative ionisation mode, and tube lens at 150 V. For FTMS, the mass range is from m/z 100 to 1500 with a resolution of 15,000, AGC target value of 200,000 and 100,000 in full scan and FTMS/MS AGC target at 1e5, isolation width of 1 amu, and max ion injection time of 750 ms; the ion trap settings used were: AGC target value of 30,000 and 10,000 in full scan and MSn mode, respectively, maximum ion injection time of 200 ms. The most intense ion was selected for the data-dependent scan with normalisation collision energy at 35%.
3. Results and discussions
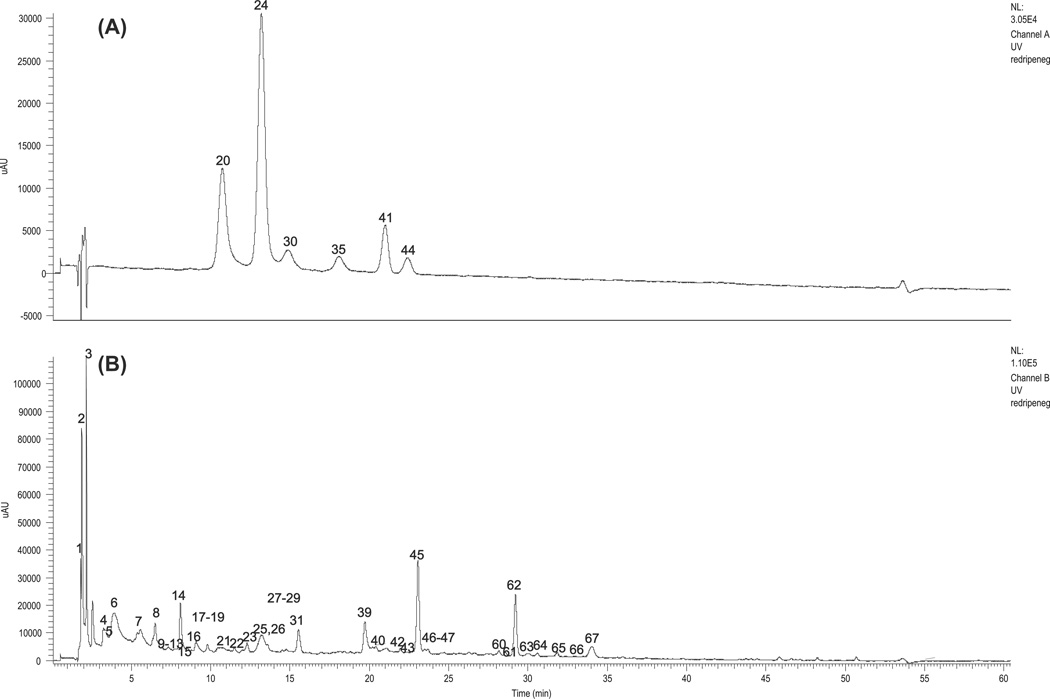
The basic structures of the above mentioned compounds are shown in Figure S2. Since different classes of phenolic compounds exhibit absorbance maxima at different wavelengths, two wavelengths were selected for real-time monitoring: 280 nm for non-anthocyanin phenolic compounds and 520 nm for anthocyanins. HRMSn detection in both the positive and the negative ionisation modes was used to obtain information on the structural features and the conjugated forms of phenolic compounds. Identification of the phenolic compounds was based on chromatographic behaviour, UV/Vis and mass spectra, accurate mass measurements, consecutive MS2–MS4 analyses, and comparison with data in the literature (Buendia et al., 2010; Kelebek & Selli, 2011; Maatta-Riihinen et al., 2004; Mikulic-Petkovsek et al., 2013; Zhang et al., 2011; Zheng, Song, Doncaster, Rowland, & Byers, 2007). Seventy-four compounds, including anthocyanins, dihydroflavonols and flavonols, flavan-3-ols, proanthocyanidins, and ellagic acid and its derivatives were identified from the two F. vesca inbred lines. The basic structures of the abovementioned compounds are shown in Fig. 1 and summarised in Table 1, where the compounds are numbered according to their retention times as shown in typical chromatograms (Fig. 1).
Fig. 1.
The HPLC chromatograms of anthocyanin (A, at 520 nm) and non-anthocyanin polyphenols (A, at 280 nm) from F × vesca var. Ruegen.
Table 1.
Compounds identified from Fragaria vesca var. Ruegen and YW5AF7.
| Peak no. |
tR(min) | m/z | Error (mmu) |
Formula | Adduct | MS2 to MS4 data | UV λmax (nm) |
Tentative identification |
|---|---|---|---|---|---|---|---|---|
| 1 | 1.82 | 341.1083 | −0.64 | C12H22O12 | [M–H]− | MS2[341]: 179(100), 161(18), 143(23), 119(13), 113(18), 101(6) MS3[341 → 179]: 161(100), 149(21), 143(97), 131(27), 125(8), 119(45), 113(18), 106(12), 101(8), 89(94), 87(11)MS4[341 → 179 → 161]: 113(100), 101(32), 71(69) | – | Hexosyl hexose |
| 2 | 1.89 | 191.0193 | −0.41 | C6H8O7 | [M–H]− | MS2[191]: 173(100), 111(64)MS3[191 → 173]: 155(27), 111(100)MS4[191 → 173 → 111]: 67(100) | – | Citric acid |
| 3 | 2.38 | 191.0194 | −0.33 | C6H8O7 | [M–H]− | MS2[191]: 173(20), 111(100)MS3[191 → 111]: 67(100) | – | Citric acid isomer |
| 4 | 3.34 | 629.0411 | −0.96 | C27H18O18 | [M–H]− | MS2[629]: 615(31), 613(9), 601(100), 599(9)MS3[629 → 601]: 573(45), 557(16), 529(6), 449(33), 439(6), 431(100), 405(15), 389(7), 387(14), 287(26), 286(13), 261(12)MS4[629 → 601 → 431]: 403(41), 387(62), 371(17), 369(12), 359(8), 343(9), 327(6), 312(6), 299(19), 287(100), 286(94), 285(16), 283(8), 273(10), 271(12), 261(98), 257(8), 245(6), 243(14), 227(9), 225(8), 217(27), 216(10), 215(13), 189(18), 187(7) | – | Gallotannin |
| 5 | 3.67 | 783.0665 | −1.3 | C34H24O22 | [M–H]− | MS2[783]: 481(31), 301(100), 275(17)MS3[783 → 301]: 301(36), 300(9), 284(39), 257(100), 229(76), 201(13), 185(27) | – | bis-HHDP-glucose |
| 6 | 3.95 | 947.0433 | 0.052 | C41H24O27 | [M–H]− | MS2[947]: 929(100), 901(73), 883(16), 875(8) | – | Unknown ellagitannin |
| 7 | 5.58 | 289.0921 | −0.761 | C12H18O8 | [M–H]− | MS2[289]: 161(100), 113(6), 101(8)MS3[289 → 161]: 143(28), 129(22), 125(12), 113(62), 101(100), 99(13), 97(14), 89(6), 85(7), 73(10), 71(29) | 275,229 | Furaneol hexoside |
| 8 | 6.34 | 957.0535 | −1.041 | C89H48O50 | [M– 2H]2− | MS2[957]: 1557(25), 1224(17), 1099(33), 1096(42), 1026(17), 986(8), 967(17), 943(33), 939(58), 937(25), 934(33), 932(17), 930(100), 928(17), 926(25), 922(17), 917(25), 916(17), 912(25), 911(25), 907(50), 901(17), 899(17), 895(50), 872(25), 842(17), 837(8), 817(8), 814(17), 778(17), 754(25), 739(8), 731(17), 717(8), 655(42), 541(8), 493(8), 469(8), 413(8), 397(17), | – | Unknown ellagitannin |
| 9 | 6.40 | 965.0516 | −0.428 | C89H48O51 | [M– 2H]2− | MS2[965]: 1859(11), 948(16), 947(16), 942(11), 929(16), 921(11), 920(11), 916(21), 903(26), 897(11), 889(11), 887(11), 885(11), 871(21), 852(37), 823(11), 797(11), 795(11), 783(100), 781(16), 764(11), 591(11), 481(11), 479(16), 421(16) | – | Unknown ellagitannin |
| 10 | 6.50 | 285.0611 | −0.144 | C12H14O8 | [M–H]− | MS2[285]: 165(7), 153(100), 152(28), 109(9)MS3[285 → 153]: 109(100) | – | 1-O-protocatechuyl-beta-xylose |
| 11 | 6.54 | 203.0822 | −0.451 | C11H12O2N2 | [M–H]− | MS2[203]: 186(10), 159(100), 142(15), 116(37)MS3[203 → 159]: 144(9), 132(30), 130(56), 129(100), 128(42), 116(58), 115(30) | 280, 230 | D or L-tryptophan |
| 12 | 6.84 | 187.0248 | −0.451 | C7H8O6 | [M–H]− | MS2[187]: 143(100)MS3[187 → 143]: 99(100), 85(9) | – | 2-methylaconitate or its isomer |
| 13 | 7.16 | 577.1346 | −0.569 | C30H26O12 | [M–H]− | MS2[577]: 559(16), 451(46), 425(100), 407(59), 299(7), 289(27), 287(11)MS3[577 → 425]: 407(100), 273(6)MS4[577 → 425 → 407]: 389(29), 363(6), 339(14), 297(45), 285(100), 284(11), 283(28), 281(88), 269(6), 256(8), 255(20), 253(17), 243(17) | – | Proanthocyanidin b1 (catechin-catechin) |
| 14 | 8.11 | 289.0713 | −0.441 | C15H14O6 | [M–H]− | MS2[289]: 247(7), 245(100), 231(7), 205(33), 179(9)MS3[289 → 245]: 227(27), 217(8), 203(100), 202(7), 188(15), 187(24), 175(9), 161(17) | 233, 280, 334 | (+)-Catechin* |
| 15 | 8.75 | 865.1973 | 1.096 | C45H38O18 | [M–H]− | MS2[865]: 848(25), 739(55), 713(39), 695(100), 587(22), 577(52), 575(33), 569(9), 557(6), 543(11), 451(19), 449(15), 425(17), 423(6), 413(8), 407(23), 405(7), 395(6), 289(6), 287(16)MS3[865 → 695]: 677(38), 586(8), 585(10), 543(100), 525(23), 451(28), 407(16), 405(33), 391(11), 387(6), 363(22), 299(11), 289(15), 243(33) | – | Proanthocyanidin C1 (catechin trimer) |
| 16 | 9.09 | 1153.2596 | 1.225 | C42H58O37 | [M–H]− | MS2[1153]: 1136(73), 1110(8), 1101(10), 1070(6), 1028(65), 1010(12), 1002(33), 991(6), 984(35), 983(20), 965(6), 917(6), 908(41), 906(8), 865(100), 863(49), 861(10), 857(12), 850(8), 847(14), 846(8), 831(20), 822(6), 814(6), 739(25), 723(6), 713(6), 701(27), 695(16), 694(10), 587(25), 577(22), 575(55), 557(24), 549(8), 533(8), 501(6), 459(6), 457(6), 455(6), 449(12), 423(22), 407(24), 405(10) | 233, 271, 360 | Proanthocyanidin tetramer |
| 17 | 9.44 | 331.1033 | −0.305 | C14H20O9 | [M–H]− | MS2[331]: 313(8), 289(46), 287(7), 271(100), 235(11), 203(9), 169(24), 165(6), 127(10) | 233, 271 | Galloyl glucose |
| 18 | 9.57 | 865.197 | −1.517 | C45H38O18 | [M–H]− | MS2[865]: 848(17), 739(62), 720(8), 713(51), 695(100), 587(20), 577(64), 575(38), 557(9), 543(19), 533(6), 525(8), 451(18), 449(15), 425(25), 423(6), 413(9), 407(18), 405(12), 395(9), 363(8), 289(9), 287(27) | 233, 271 | Proanthocyanidin trimer |
| 19 | 9.81 | 561.1398 | −0.465 | C30H26O11 | [M–H]− | MS2[561]: 543(42), 435(52), 425(13), 407(17), 289(100), 271(11)MS3[561 → 289]: 247(6), 245(100), 205(33), 203(11), 179(12) | 233, 271 | Unknown |
| 20 | 10.74 | 447.0931 | 0.952 | C21H21O11+ | [M–2H]− | MS2[465]: 339(8), 285(100), 241(13)MS3[465 → 285]: 257(9), 243(21), 241(100), 217(19), 199(10), 149(9) | 233, 271, 514 | Cyanidin 3-O-glucoside* |
| 21 | 11.18 | 849.2023 | −1.353 | C45H38O17 | [M–H]− | MS2[849]: 831(24), 723(100), 697(43), 695(32), 679(19), 577(95), 571(51), 559(78), 553(10), 541(9), 517(6), 451(19), 433(19), 425(22), 407(30), 397(12), 299(7), 289(15), 287(14) | – | Propelargonidin trimer (afz-cat-cat) |
| 22 | 11.55 | 631.0565 | −0.13 | C27H20O18 | [M–H]− | MS2[631]: 613(12), 451(100)MS3[631 → 451]: 433(79), 423(9), 407(78), 405(28), 395(10), 379(37), 377(10), 367(6), 363(8), 351(100), 337(7), 335(24), 323(21), 311(28), 307(25), 295(14), 285(88), 283(7), 165(8) | 215, 233 | Castalin or its isomer |
| 23 | 12.32 | 331.1026 | 0.241 | C14H20O9 | [M–H]− | MS2[331]: 313(22), 312(6), 289(15), 288(6), 271(25), 253(21), 235(33), 211(14), 205(6), 203(32), 193(63), 181(11), 169(64), 161(10), 151(42), 127(100), 125(29), 113(11), 101(9), 97(9) | 232, 275 | Unknown |
| 24 | 13.21 | 431.0979 | 0.591 | C21H21O11 | [M–2H]− | MS2[431]: 413(10), 387(6), 269(100)MS3[431 → 269]: 241(59), 225(29), 199(12), 197(7), 147(100) | 233, 271, 500 | Pelargonidin 3-O-glucoside* |
| 25 | 13.83 | 401.1448 | −1.272 | C18H26O10 | [M-H] | MS2[401]: 383(18), 357(8), 356(6), 355(6), 293(9), 269(100), 233(9), 161(20) | – | Apigenin pentose |
| 26 | 13.90 | 635.0881 | −1.44 | C27H24O18 | [M–H]− | MS2[635]: 465(100)MS3[635 → 465]: 313(100), 295(9), 235(9), 169(19)MS4[635 → 465 → 313]: 295(21), 253(41), 241(31), 223(6), 211(6), 193(17), 169(100), 151(9), 125(15) | 233, 270 | Trigalloylglucose |
| 27 | 14.53 | 525.1964 | −0.243 | C25H34O12 | [M–H]− | MS2[525]: 363(100), 345(6), 179(6), 165(10)MS3[525 → 363]: 345(27), 315(19), 239(8), 221(7), 179(32), 165(100) | – | GA8-hexose gibberellin |
| 28 | 14.74 | 351.1292 | 0.577 | C14H24O10 | [M–H]− | MS2[351]: 333(25), 249(100), 231(10), 113(8)MS3[351 → 249]: 231(100), 189(18), 175(18), 157(7), 129(11), 115(18), 113(86), 111(21), 109(7), 99(18), 95(11), 85(29), 83(21), 75(7) | – | Unknown |
| 29 | 14.87 | 463.1227 | −0.838 | C22H23O11 | [M–H]− | MS2[463]: 301(100), 300(54)MS3[463 → 301]: 301(12), 284(14), 257(100), 229(21), 185(9) | – | Ellagic acid-hexoside |
| 30 | 14.95 | 463.0506 | −0.188 | C20H16O13 | M+ | MS2[463]: 301(100)MS3[463- > 301]: 286(100) | 233, 271, 512 | Peonidin 3-O-glucoside* |
| 31 | 15.46 | 517.1552 | 0.028 | C22H30O14 | [M–H]− | MS2[517]: 499(10), 471(8), 355(18), 337(46), 295(35), 265(49), 235(55), 193(100), 175(32), 160(14) | – | Unknown |
| 32 | 15.66 | 449.1079 | 0.227 | C21H22O11 | [M–H]− | MS2[449]: 355(100), 329(8), 287(40), 269(28), 193(13)MS3[449 → 355]: 193(100), 192(10), 165(6)MS4[449 → 355 → 193]: 165(100), 137(14) | – | Ferulic acid hexose derivative |
| 33 | 16.71 | 371.0977 | 0.417 | C16H20O10 | [M–H]− | MS2[371]: 249(100)MS3[371 → 249]: 231(87), 175(14), 113(100), 111(8), 103(7), 99(9), 95(11), 85(21) | – | Unknown |
| 34 | 17.49 | 585.2184 | −0.479 | C27H38O14 | [M–H]− | MS2[585]: 377(100), 329(13)MS3[585 → 377]: 329(100)MS4[585 → 377 → 329]: 314(100), 164(10) | – | Unknown |
| 35 | 18.07 | 535.1075 | −0.692 | C24H23O14 | M+ | MS2[535]: 287(100)MS3[535- > 287]: 287(100), 269(68), 259(28), 245(11), 241(36), 231(46), 217(6), 216(9), 213(74), 199(12), 189(16), 185(32), 175(35), 163(9), 137(17) | 233, 515 | Cyanidin 3-O-malonylglucoside |
| 36 | 18.08 | 935.0771 | −1.357 | C41H28O26 | [M–H]− | MS2[935]: 633(100), 301(41)MS3[935 → 633]: 463(8), 301(100) | – | Galloyl bis-hexahydroxydiphenoyl (HHDP)-glucose |
| 37 | 19.28 | 433.0408 | −0.479 | C19H14O12 | [M–H]− | MS2[433]: 301(100), 300(32)MS3[433 → 301]: 301(49), 284(36), 273(12), 257(100), 229(67), 213(8), 201(6), 185(32) | – | Ellagic acid pentoside |
| 38 | 19.41 | 461.2023 | −0.505 | C21H34O11 | [M–H]− | MS2[461]: 461(6), 453(9), 443(55), 430(6), 418(9), 417(15), 416(9), 415(30), 400(6), 399(16), 393(6), 376(6), 329(100), 299(10), 293(90), 233(49), 191(185), 161(13), 149(28) | – | Unknown |
| 39 | 19.72 | 300.9983 | −0.73 | C14H6O8 | [M–H]− | MS2[301]: 301(34), 300(13), 284(28), 257(100), 229(64), 201(13), 185(34)MS3[301 → 257]: 229(96), 213(23), 201(11), 185(100), 173(6) | 234, 252, 368 | Ellagic acid* |
| 40 | 20.42 | 447.0564 | −0.499 | C20H17O12 | [M–H]− | MS2[447]: 301(100), 300(19)MS3[447 → 301]: 301(15), 284(11), 257(100), 229(29), 185(10) | 233, 365 | Ellagic acid-methyl pentoside |
| 41 | 21.07 | 473.1083 | −0.142 | C23H23O11 | [M–2H]− | MS2[473]: 269(100)MS3[473 → 269]: 241(59), 225(62), 224(8), 201(7), 199(9), 147(100) | 233, 501 | Pelargonidin acetyl hexoside |
| 42 | 21.89 | 623.1247 | 0.414 | C27H28O17 | [M–H]− | MS2[623]: 608(11), 477(46), 476(8), 460(100), 314(11), 313(6)MS3[623 → 460]: 445(10), 314(35), 313(100)MS4[623 → 460 → 313]: 298(100), 285(42), 283(6) | – | Unknown |
| 43 | 22.26 | 503.1189 | 0.498 | C24H24O12 | [M–H]− | MS2[503]: 299(100)MS3[503 → 299]: 284(100), 283(22), 255(23), 240(10), 147(11) | 230, 365 | Diosmetin acetylhexoside |
| 44 | 22.43 | 549.1228 | −1.022 | C25H25O14 | M+ | MS2[549]: 301(100)MS3[549- > 301]: 286(100) | 233, 515 | Peonidin 3-O-malonylglucoside |
| 45 | 23.05 | 435.0931 | −0.145 | C20H20O11 | [M–H]− | MS2[435]: 303(100), 285(34)MS3[435 → 303]: 285(100), 177(11), 125(7)MS4[435 → 303 → 285]: 257(11), 243(17), 241(100), 217(13), 199(23), 175(56) | 217, 234, 289 | Taxifolin 3-O-arabinofuranoside |
| 46 | 23.50 | 463.0876 | −0.609 | C21H20O12 | [M–H]− | MS2[463]: 301(100), 300(23)MS3[463 → 301]: 283(6), 273(16), 257(15), 229(8), 179(100), 151(63) | 233, 364 | Quercetin 3-O-glucoside* |
| 47 | 23.61 | 477.067 | −0.444 | C21H18O13 | [M–H]− | MS2[477]: 315(100)MS3[477 → 315]: 300(100)MS4[477 → 315 → 300]: 300(100), 283(6), 272(24), 271(21), 244(53), 243(12), 228(13), 216(17), 200(22) | 217, 233, 271 | Methylellagic acid hexose |
| 48 | 23.73 | 521.2017 | −1.115 | C26H34O11 | [M–H]− | MS2[521]: 503(9), 359(100)MS3[521 → 359]: 344(100)MS4[521 → 359 → 344]: 329(34), 328(8), 313(100), 255(16), 203(14), 191(10), 189(52), 173(11), 159(41) | 215, 233, 271 | Tetramethylellagic acid hexose |
| 49 | 24.55 | 491.0831 | −0.014 | C22H20O13 | [M–H]− | MS2[491]: 476(20), 328(100), 313(9)MS3[491 → 328]: 313(100)MS4[491 → 328 → 313]: 298(100), 285(54) | 215, 233, 280 | Dimethylellagic acid hexose |
| 50 | 24.73 | 521.2016 | −1.235 | C26H34O11 | [M–H]− | MS2[521]: 503(9), 359(100)MS3[521 → 359]: 344(100)MS4[521 → 359 → 344]: 329(32), 328(10), 313(100), 255(16), 203(16), 191(10), 189(45), 173(11), 159(45) | 216, 233, 271 | Tetramethylellagic acid hexose |
| 51 | 25.32 | 709.1252 | −0.526 | C30H30O20 | [M–H]− | MS2[709]: 709(7), 691(10), 665(84), 663(9), 625(8), 563(100), 545(13), 519(83), 477(18), 461(8), 447(7), 357(21), 315(46), 301(27), 300(13) | 217, 233, 356 | Unknown |
| 52 | 25.60 | 607.1298 | −0.638 | C27H28O16 | [M–H]− | MS2[607]: 461(100)MS3[607 → 461]: 314(100), 299(6)MS4[607 → 461 → 314]: 313(31), 299(97), 286(66), 285(100), 284(21), 283(25) | 215, 233 | Unknown |
| 53 | 26.20 | 567.2076 | −0.674 | C27H36O13 | [M–H]− | MS2[567]: 567(10), 558(11), 550(7), 549(14), 545(7), 523(10), 521(40), 499(7), 359(77), 341(100), 329(87) | 215, 233 | Unknown |
| 54 | 26.28 | 505.0983 | −0.434 | C23H22O13 | [M–H]− | MS2[505]: 463(22), 301(100), 300(46)MS3[505 → 301]: 283(17), 273(29), 257(10), 229(7), 193(7), 179(100), 151(63) | – | Quercetin acetyl hexoside |
| 55 | 26.51 | 327.1234 | −0.447 | C19H20O5 | [M–H]− | MS3[327 → 312]: 295(22), 284(47), 283(30), 281(48), 267(100), 256(12), 253(26), 240(11), 145(30) | – | Unknown |
| 56 | 26.67 | 447.0563 | −0.599 | C20H16O12 | [M–H]− | MS2[447]: 315(100)MS3[447 → 315]: 300(100)MS4[447 → 315 → 300]: 300(100), 283(12), 272(24), 271(12), 244(69), 243(29), 228(33), 216(47), 200(35), 188(6), 172(10) | – | Methylellagic acid pentose |
| 57 | 27.34 | 461.0724 | −0.189 | C21H18O12 | [M–H]− | MS2[461]: 328(6), 315(100)MS3[461 → 315]: 300(100)MS4[461 → 315 → 300]: 300(19), 283(59), 272(100), 271(18), 244(39), 228(40), 200(20), 172(17) | – | Methylellagic acid methyl pentose |
| 58 | 27.68 | 519.0779 | −0.148 | C23H20O14 | [M–H]− | MS2[519]: 315(100)MS3[519 → 315]: 300(100)MS4[519 → 315 → 300]: 300(100), 272(24), 271(18), 244(67), 243(12), 228(12), 216(21), 200(12), 172(6), 151(6) | – | Methylellagic acid acetyl hexose |
| 59 | 27.88 | 939.1095 | −1.404 | C41H32O26 | [M–H]− | MS2[939]: 787(8), 769(100), 617(9)MS3[939 → 769]: 725(16), 617(100), 601(37), 599(33), 511(7), 465(6), 447(21), 431(11), 429(14), 403(6)MS4[939 → 769 → 617]: 465(100), 447(37), 423(18), 313(8), 295(6), 211(6) | – | Pentagalloyl hexose |
| 60 | 28.16 | 447.0932 | −0.115 | C21H20O11 | [M–H]− | MS2[447]: 327(18), 285(92), 284(100), 255(14)MS3[447 → 284]: 255(100), 227(13)MS4[447 → 284 → 255]: 255(12), 227(100), 211(57), 183(8), 167(6) | – | Kaempferol 3-O-hexoside |
| 61 | 28.55 | 519.0779 | −0.148 | C23H20O14 | [M–H]− | MS2[519]: 315(100), 300(10)MS3[519 → 315]: 300(100)MS4[519 → 315 → 300]: 300(100), 283(11), 272(27), 271(29), 244(80), 243(17), 228(23), 216(19), 200(34), 172(10) | 217, 233, 366 | Methylellagic acid acetyl hexoside |
| 62 | 29.22 | 461.0718 | −0.738 | C21H18O12 | [M–H]− | MS2[461]: 315(100), 314(6)MS3[461 → 315]: 300(100)MS4[461 → 315 → 300]: 300(100), 283(17), 272(36), 271(24), 244(74), 243(21), 228(23), 216(24), 200(39), 172(10) | 243, 375 | Methylellagic acid rhamnoside |
| 63 | 30.00 | 477.1035 | −0.399 | C22H22O12 | [M–H]− | MS2[477]: 459(6), 357(23), 315(31), 314(100), 299(6), 285(8), 271(7)MS3[477 → 314]: 299(18), 286(36), 285(100), 271(72), 257(11), 243(24) | – | Methylellagic acid hexose |
| 64 | 30.56 | 461.0725 | −0.069 | C21H18O12 | [M–H]− | MS2[461]: 315(100)MS3[461 → 315]: 300(100)MS4[461 → 315 → 300]: 300(100), 283(10), 272(17), 271(21), 244(65), 243(18), 228(18), 216(18), 200(24), 172(7) | 217, 233, 365 | Methylellagic acid hexose |
| 65 | 31.81 | 489.1038 | −0.029 | C23H22O12 | [M–H]− | MS2[489]: 285(100), 284(7)MS3[489 → 285]: 267(43), 257(100), 256(9), 243(6), 241(29), 240(10), 239(13), 229(49), 223(10), 213(15), 211(9), 199(10), 197(20), 195(9), 163(17) | – | Kaempferol acetylhexoside |
| 66 | 32.45 | 341.1393 | −0.107 | C20H22O5 | [M–H]− | MS2[341]: 326(100)MS3[341 → 326]: 311(100)MS4[341 → 326 → 311]: 293(6), 283(100), 267(12), 266(14), 252(7) | – | Unknown |
| 67 | 34.05 | 519.1142 | −0.214 | C24H24O13 | [M–H]− | MS2[519]: 315(100)MS3[519 → 315]: 300(100), 287(6), 272(6)MS4[519 → 315 → 300]: 272(43), 271(100), 255(66) | 215, 233, 329 | Methylellagic acid acetyl hexoside |
Confirmed with reference standards, - weak absorbance.
3.1. Identification of anthocyanins in F. vesca var. Ruegen F7-4
Fig. 1(A) shows the HPLC-UV (520 nm) chromatogram of Ruegen F7-4 ripe fruits (stage 3). Previous reports of anthocyanins in strawberries were used to assist in the identification of the anthocyanins in Table 1 in addition to UV and HRMSn data (Aaby et al., 2007; Buendia et al., 2010; Hilt et al., 2003; Kelebek & Selli, 2011; Zhang et al., 2011).
Peak 20 with M+ at m/z 449.1071 (C21H21O11, −1.53 ppm) and a product ion at m/z 287 (−162 amu, hexose moiety) was identified as cyanidin 3-O-glucoside. Peak 24, the major peak in Ruegen F7-4 with M+ at m/z 433.1122 (C21H21O10, 1.58 ppm) and a major MS2 product ion at m/z 271(−162 amu, hexose moiety), was identified as pelargonidin 3-O-glucoside. Peak 30 with M+ at m/z 463.1228 (C22H23O11, 1.49 ppm) and a MS2 product ion at m/z 301(−162 amu, hexose moiety) was identified as peonidin 3-O-glucoside. Peak 35 with [M]+ at m/z 535.1072 (C24H23O14, −2.0 ppm) and two major product ions at m/z 449 and 287 (−86 amu, and then −162 amu, corresponding to malonyl and glucose moiety, respectively) was identified as cyanidin 3-O-malonylglucoside. Peak 41 showed the M+ ion at m/z 519.1126 (C24H23O13, −1.38 ppm) and a neutral loss of 248 amu (malonyl-hexosyl residue) for its product ion and was identified as pelargonidin 3-O-malonylglucoside. Peak 44 showed the M+ at m/z 549.1226 (C25H25O14, −1.38 ppm) and a neutral loss of 248 amu (malonyl-hexosyl residue) for its product ion and was identified as peonidin 3-O-malonylglucoside. Cyanidin and pelargonidin glycosides are commonly found in cultivated strawberry (F. × ananassa var. Fort Laramie), but peonidin 3-O-glucoside and peonidin 3-O-malonylglucoside were identified in F. vesca var. Ruegen F7-4 for the first time. However, YW5AF7 ripe fruits (stage 3) only contain pelargonidin 3-O-glucoside.
3.2. Identification of non-anthocyanin phenolic compounds in Ruegen F7-4 and YW5AF7
Fig. 1 (B) shows the HPLC-UV profiles at 280 nm of Ruegen F7-4 ripe fruits (stage 3). More than sixty non-anthocyanin phenolic compounds were identified. Unlike anthocyanins, the Ruegen F7-4 and YW5AF7 exhibited very similar profiles and all non-anthocyanin phenolic compounds were found in both genotypes (details discussed below). However, the phenolic profiles differed from those of cultivated strawberries previously reported in the literature (Bianco et al., 2009; Buendia et al., 2010; Kelebek & Selli, 2011; Zhang et al., 2011).
3.2.1. Dihydroflavonol and flavonols
Peak 45 exhibited UV/Vis absorption maxima at about 234 and 290 nm. The HRMS gave a deprotonated [M–H]− ion at m/z 435.0321, suggesting the formula of C20H19O11 (0.33 ppm). The MS2 major product ion was m/z 303 (−132 amu, pentose), the MS3 and the MS4 spectra of peak 45 were consistent with the MS2 and the MS3 spectra of taxifolin. Hence this compound was identified as taxifolin 3-O-arabinoside, a compound previously reported in cultivated strawberry roots but not in fruits (Ishimaru, Omoto, Asai, Ezaki, & Shimomura, 1995). Peak 29 with m/z at 447.0927 (C21H19O11, −1.22 ppm) and a major product ion at m/z 285 (−162 amu: hexose) was identified as kaempferol 3-O-glucoside. Peak 46 (C21H20O12) with [M–H]− ion at m/z 463, a major MS2 product ion at m/z 301, and corresponding MS3 product ions at m/z 151 and 179, was identified as quercetin 3-O-glucoside. Similarly, peak 54 (C23H22O13) was identified as quercetin-acetyl-hexoside; and peaks 65 and 68 were identified as kaempferol 3-O-acetylhexosides.
3.2.2. Flavan-3-ols and proanthocyanidins
(+) Catechin, B type proanthocyanidin dimers, B type proanthocyanidin trimers, and B type proanthocyanidin tetramers were found in these two genetically improved strawberries using HRMSn data, UV spectral data, and literature reports. Peak 14, with a deprotonated molecule ion [M–H]− at m/z 289 (C15H13O6) and characteristic MS2 ions at m/z 245, 205, 231, and 179, was identified as (+)-catechin. Peak 13 ([M–H]− at m/z 577.1346, C30H25O12, 0.95 ppm, primary MS2 ion at m/z 577, −152 amu via a characteristic fragmentation pathway by retro Diels–Alder reaction) was identified as a proanthocyanidin dimer of the B type catechin–catechin (Aaby et al., 2007). Peak 15 was identified as a B type proanthocyanidin trimer. Its [M–H]− ion was at m/z 865.1970 (C45H37O18, −1.82 ppm) and MS2 ions were at m/z 695, 739, 713, 577, 425, and 287. The MS3 ions of m/z 695 gave fragment ions at m/z 543, 451, 289, and 243. Using the fragment pattern, the sequence of this trimer was epicatechin–epicatechin–epicatechin. Peak 16 had an [M–H]− ion at m/z 1153.2596 (C60H4s9O24, −1.98 ppm) and was tentatively identified as an isomer of a B type proanthocyanidin tetramer according to its characteristic MS2 ions at m/z 865, 1135, 1027, 983, 695, 575 and 407. It was composed of four epicatechin units. Peak 19 had [M–H]– at m/z 561.1393 (C30H25O11) and MS2 ions at m/z 289, 543, and 435. The compound was identified as epiafzelechin–epicatechin. The fragmentation pathway of peak 19 was different from that of the B-catechin dimer in strawberries described in a previous study (Aaby et al., 2007). Peak 21 had [M–H]− at m/z 849.2023 (C45H37O17), characteristic MS2 ions at m/z 801, 697, 577 (base peak obtained after a loss of 272 amu), 559, 425, 407, and 287. The MS3 spectra of the MS2 base peak (m/z 577) gave ions at 425, 407, and 289 (−288 amu, epi catechin) and corresponded to B type proanthocyanidin trimer of the type epiafzele-chin−epicatechin−epicatechin (Aaby et al., 2007; Hilt et al., 2003).
3.2.3. Ellagic acid and its Derivatives
Peak 39 had [M–H]− at m/z 300.9983 (C14H5O8, −0.97 ppm) and MS2 fragmentation ions at m/z 257, 229,185, and 157. It was identified as ellagic acid. The identity was confirmed with an ellagic acid reference standard. The UV/Vis spectra of peaks 30 and 37 suggested glycosylated forms of ellagic acid (33). Peak 30 with [M–H]– at m/z 463 (C20H15O13) and the main MS2 ion at m/z 301 (−162, hexose) was tentatively identified as an ellagic acid hexoside. Peak 37 had the [M–H]– at m/z 447 (C19H13O12). Its main MS2 product ion was at m/z 301 (MS3 ions at m/z 257, 229, and 185) and corresponded to ellagic acid. Peak 37 was identified as ellagic acid methyl pentoside. A similar compound has been previously reported in strawberries (Aaby et al., 2007).
Peak 62 (m/z 461.0725, C21H17O12, −0.107 ppm) is the major compound observed in the HPLC-DAD profiles for both Ruegen and YW5AF7. The maximum UV absorptions were at 243 nm and 375 nm. The characteristic MS2 product ion at m/z 315 (−146 amu, methyl pentose) and its MS3 product ions at m/z 300 and its MS4 product ions at m/z 272, 271, 244 confirmed the identity of methylellagic acid. Hence, Peak 62 was identified as methylella-gic acid methyl pentoside. Peaks 57 and 64 had the same m/z (461.0725) and exhibited similar fragmentation behaviour to that of peak 62, except that the relative abundance of product ion at m/z 315 was different. These two compounds were tentatively identified as methylellagic acid methyl pentoside isomers. Peaks 61 and 67 had [M–H]− ions at m/z 519 (C23H19O14) with the MS2 product ions corresponding to kaempferol glucoside (m/z 315) after the loss of the acetyl and hexosyl moieties (162 + 42 amu). They were identified as methylellagic acid 3-O-acetyl-hexosides. Similarly, Peak 56 (m/z 447.0563) was identified as methylellagic acid 3-O-pentoside. Similar compounds (methylellagic acid-pen-tose conjugates) have been previously reported (Aaby et al., 2007).
Peaks 5, 6, 8, 9,17, 26, and 36 were identified as ellagitannins. Ellagitannins are hydrolysable tannins, since they are esters of hexahydroxydiphenic acid (HHDP: 6,6’-dicarbonyl-2,2′,3,3′,4,4′-hexahydroxybiphenyl moiety) and a polyol, usually glucose, and in some cases gallic acid, which are commonly found in strawberries (Aaby et al., 2007; Kelebek & Selli, 2011; Vrhovsek et al., 2012). Typical losses during fragmentation of ellagitannins are galloyl (152 amu), HHDP (302 amu), galloyl-glucose (332 amu), HHDP-glucose (482 amu), and galloyl-HHDP-glucose (634 amu). Recent research found that agrimoniin is one of the most abundant ellagitannins and sanguiin H-6 and lambertianin C are minor compounds in both F. vesca and F. ananassa D. (Vrhovsek et al., 2012). However, we did not find any of these three compounds in our two strawberry lines. Peak 8 had a [M–2H]2− ion at m/z 957.0535 (its isotopic distribution suggests it to be a double-charged ion), implying the formula of C82H52O55. Fragmentation of the double-charged ions gave single-charged MS2 product ions at m/z 1557, 1224, 1099, 1096, 943, 930, 451, and 301. Thus Peak 8 has two more oxygen atom substituted in the structures than that of sanguiin H-6/agrimoniin. Similarly, Peak 9 (C82H52O56) had three more oxygen atoms substituted in the structure in comparison to literature reports for sanguiin H-6/agrimoniin (Aaby et al., 2007; Vrhovsek et al., 2012).
Peak 5, with [M–H]− at m/z 783 (C34H23O22) and MS2 fragmentations at m/z 481 (−302 amu, loss of HHDP) and 301 (−482 amu, loss of HHDP-glucose) was identified as bis-HHDP-glucose, previously reported in strawberries (Aaby et al., 2007). Peak 36 was identified as galloyl-bis-HHDP-glucose with [M–H]− at m/z 935 and MS2 product ions at m/z 633 −02 amu, loss of HHDP) and 301 (332 amu, loss of galloyl glucose).
Peak 26 had an [M–H]− at m/z 635 (C27H23O18), and a major MS2 product ion at m/z 465 (−170 amu, gallic acid). The MS3 ion of m/z 465 was at m/z 313 (−152 amu, loss of galloyl unit). The ion at m/z 313 could be further fragmented into an MS4 product ion at m/z 169. Peak 26 was identified as tri-galloyl-glucose.
Peak 36 had a [M–H]− ion at 935.0771, and ions at m/z 633,m/z 463 and m/z 301 in the MS2 to MS3 specta, indicating the structure of galloyl-bis-HHDP-glucose.
3.2.4. Other compounds
Peak 12 (C7H8O6) was identified as benzoic acid with the main MS2 ion at m/z 143 (loss of CO2). It was reported previously (Russell, Scobbie, Labat, & Duthie, 2009). Peak 32 was identified as a ferulic acid hexose derivative with [M–H]− at m/z 449 and MS2 ions at m/z 355, 329, 287, 269, and 193 (base peak 355) (Aaby et al., 2007). The major MS3 ion of m/z 355 was at m/z 193 (loss of a hexose unit). The fragmentation patterns were in agreement with previously published data (Aaby et al., 2007). Possible composition of Peak 32 could be ferulic acid, hexose, and a C6H6O group. Peak 1 (C12H22O12) was identified as hexosyl-hexose and it is the sugar form that widely exists in fruits and vegetables (Kallio, Hakala, Pel-kkikangas, & Lapvetelainen, 2000). Peaks 2 and 3 (C6H8O7) were tentatively identified as citric acid and its isomer (Kelebek & Selli, 2011). Peak 7 (C12H18O8) was tentatively identified as furaneol glucoside, a compound previously reported in detached ripening strawberry fruits (Roscher, Bringmann, Schreier, & Schwab, 1998). Peak 11 (C11H12O2N2) was identified as tryptophan, which was reported in Chilean strawberry F. × chiloensis ssp. chiloensis (Cheel et al., 2005).
3.3. The chemical differences of F. vesca Ruegen F7-4 and YW5AF7 and F. × ananassa cv. Fort Laramie
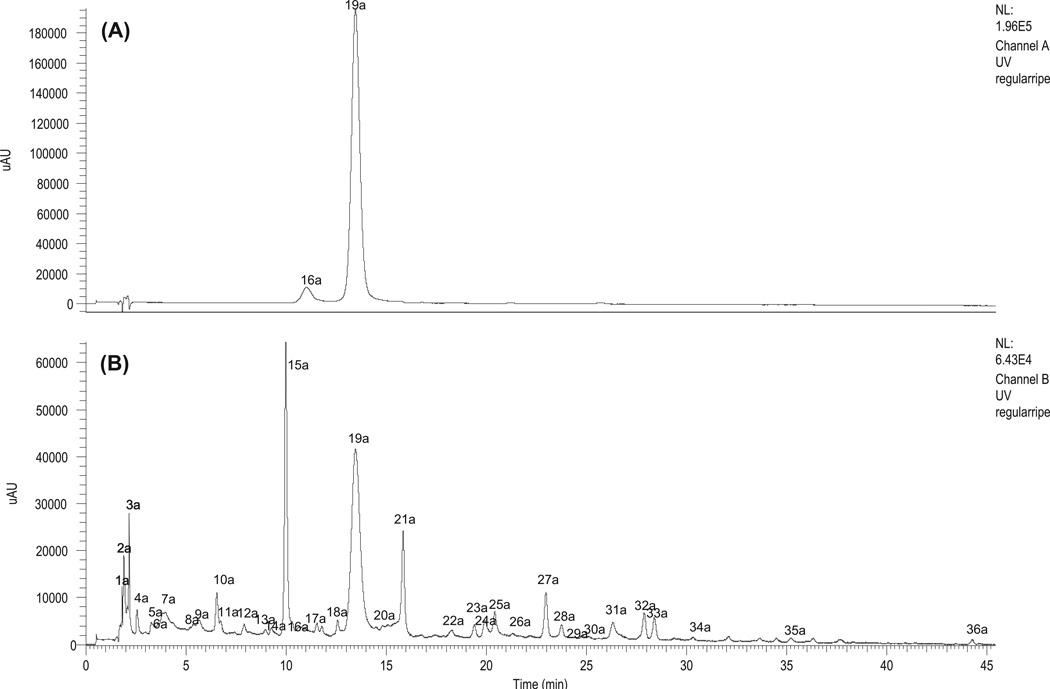
The metabolite profiles of strawberry fruits are strongly affected by developmental, genetic, and environmental factors (Carbone et al., 2009). In previous publications, the difference between wild and cultivated strawberry species in the production of specific volatile terpenoid flavour components was studied using a combination of molecular and biochemical tools. The results suggest that domestication of strawberry has involved selection for specific alleles in the cultivated species which contribute to a strongly modified flavour profile (Aharoni et al., 2004). In this investigation, we also found a significant difference between wild and cultivated strawberry species as shown by their phenolic profiles. The constituents found in octoploid strawberry F. × ananassa cv. Fort Laramie are shown in Fig. 2 and Table 2. The two major anthocyanins of Fort Laramie, cyanidin 3-O-glucoside and pelargonidin 3-O-glucoside, accounted for almost 100% of the total peak area in its HPLC chromatogram at 520 nm; on the other hand, there are many other anthocyanins in diploid strawberry Ruegen F7-4, such as peonidin 3-O-glucoside, peonidin 3-O-malonylglucoside cyanidin 3-O-malonylglucoside, and pelargonidin 3-O-malonylglucoside.
Fig. 2.
The HPLC chromatograms of anthocyanins (A, at 520 nm) and non-anthocyanin polyphenols (B, at 280 nm) from F × ananassa cv. LAR.
Table 2.
Compounds identified from Fragaria × ananassa cv. Fort Laramie.
| Peak no. |
tR (min) |
m/z | Error (mmu) |
Formula | Adduct | MS2 to MS4 data | UV λmax (nm) |
Possible identification |
|---|---|---|---|---|---|---|---|---|
| 1a | 1.76 | 341.1083 | −0.635 | C12H22O11 | [M–H]− | MS2[341]: 179(100), 161(20), 143(21), 131(7), 119(15), 113(19), 101(6) | – | Hexosyl hexose |
| 2a | 1.89 | 191.0193 | −0.406 | C6H8O7 | [M–H]− | MS2[191]: 173(25), 111(100) | – | Citric acid or its isomer |
| 3a | 2.11 | 191.0194 | −0.326 | C6H8O7 | [M–H]− | MS2[191]: 173(25), 111(100) | – | Citric acid isomer |
| 4a | 2.59 | 191.0194 | −0.286 | C6H8O7 | [M–H]− | MS2[191]: 173(25), 111(100) | – | Citric acid isomer |
| 5a | 3.28 | 865.1965 | −2.007 | C45H38O18 | [M–H]− | MS2[865]: 848(11), 739(15), 713(29), 695(100), 587(8), 577(32), 575(28), 543(8), 451(10), 449(10), 425(11), 413(8), 407(12), 405(6), 363(6), 287(13)MS3[865 → 695]: 677(30), 651(11), 586(15), 585(9), 543(100), 525(15), 451(37), 407(15), 405(51), 391(12), 387(7), 363(48), 299(10), 289(27), 243(46) | 234 | Proanthocyanidin trimer |
| 6a | 3.33 | 865.196 | −2.497 | C45H38O18 | [M–H]− | – | – | Proanthocyanidin trimer |
| 7a | 3.98 | 783.0670 | −1.685 | C34H24O22 | [M–H]− | MS2[783]: 481(28), 301(100), 275(8) | – | bis-HHDP hexose |
| 8a | 5.39 | 783.0682 | −0.465 | C34H24O22 | [M–H]− | MS2[783]: 481(28), 301(100), 275(81) | – | bis-HHDP Hexose |
| 9a | 5.62 | 289.0924 | −0.511 | C12H18O8 | [M–H]− | MS2[289]: 245(6), 161(100), 101(8)MS3[289 → 161]: 143(9), 125(9), 113(61), 101(100), 99(13), 97(11), 89(9), 85(11), 73(7), 71(30) | 232, 271 | Furaneol hexoside |
| 10a | 6.54 | 285.0614 | −0.211 | C12H14O8 | [M–H]− | MS2[285]: 165(6), 153(100), 152(31), 109(7), 108(6)MS3[285 → 153]: 109(100) | 231, 283 | Dihydroxybenzoic acid xylose |
| 11a | 6.73 | 951.0743 | −0.178 | C41H28O27 | [M–H]− | MS2[951]: 907(100), 783(25) | Geraniin | |
| 12a | 7.91 | 865.1976 | −0.967 | C45H38O18 | [M–H]− | MS2[865]: 847(21), 739(64), 713(31), 695(100), 677(6), 587(31), 577(57), 575(33), 543(18), 533(7), 451(21), 449(21), 425(21), 423(8), 413(11), 407(26), 405(11), 289(10), 287(17)MS3[865 → 695]: 677(29), 585(6), 543(100), 525(27), 451(49), 407(11), 405(28), 391(7), 387(8), 363(13), 299(9), 289(14), 243(29) | 234, 278 | Proanthocyanidin trimer |
| 13a | 8.98 | 865.1976 | −0.967 | C45H38O18 | [M–H]− | MS2[865]: 847(27), 739(59), 713(49), 695(100), 587(19), 577(63), 575(33), 559(7), 557(6), 543(17), 533(7), 451(22), 449(22), 425(20), 423(8), 413(11), 407(21), 405(10), 395(6), 289(10), 287(18)MS3[865 → 695]: 678(34), 585(6), 543(100), 542(15), 525(28), 451(36), 407(8), 405(26), 391(11), 363(26), 299(9), 289(23), 243(49) | 234, 278 | Proanthocyanidin trimer |
| 14a | 9.25 | 1153.261 | −1.425 | C42H58O37 | [M–H]− | MS2[1153]: 1135(50), 1028(90), 1009(7), 1002(38), 1001(17), 983(85), 982(6), 965(8), 907(24), 865(100), 863(53), 857(7), 847(25), 846(11), 821(8), 739(50), 738(21), 701(32), 695(25), 694(7), 683(9), 588(6), 577(40), 575(55), 569(6), 560(11), 451(11), 449(18), 423(9), 407(26), 405(11), 395(7) | 234, 278 | Proanthocyanidin tetramer |
| 15a | 10.01 | 325.0929 | −1.267 | C15H18O8 | [M–H]− | MS2[325]: 265(19), 235(7), 205(7), 187(50), 163(90), 145(100), 119(9)MS3[325 → 145]: 117(100) | 219, 234, 314 | Coumaroyl glucose |
| 16a | 11.05 | 449.1074 | −0.148 | C21H21O11 | M+ | MS2[449]: 287(100) | 276, 428, 500 | Cyanidin 3-O-glucoside* |
| 17a | 11.57 | 849.2031 | −0.563 | C45H38O17 | [M–H]− | MS2[849]: 831(22), 724(90), 723(13), 714(7), 697(34), 695(30), 679(12), 577(100), 571(45), 559(74), 553(15), 543(10), 451(19), 433(14), 425(38), 407(25), 397(11), 289(15), 287(20) | 234, 278 | Epiafzelechin-(4β → 8)-epicatechin-(4β → 8)-epicatechin |
| 18a | 12.61 | 577.1343 | −0.859 | C30H26O12 | [M–H]− | MS2[577]: 559(15), 451(54), 425(100), 407(52), 299(10), 289(26), 287(13) | 233, 278 | Proanthocyanidin dimer |
| 19a | 13.32 | 433.1125 | −0.463 | C21H21O10 | M+ | MS2[433]: 271(100) | 234, 276, 428, 499 | Pelargonidin 3-O-glucoside* |
| 20a | 14.87 | 865.1970 | −1.577 | C45H38O18 | [M–H]− | MS2[865]: 847(15), 739(63), 714(34), 713(17), 695(100), 677(6), 587(28), 577(60), 575(29), 569(10), 561(11), 559(8), 543(19), 451(20), 449(12), 425(29), 413(9), 407(33), 405(7), 289(7), 287(18)MS3[865 → 695]: 677(31), 652(7), 569(23), 543(86), 525(49), 451(15), 449(11), 407(100), 405(19), 363(9), 289(12), 243(24) | 233, 278 | Proanthocyanidin trimer |
| 21a | 15.84 | 449.1089 | 0.015 | C21H22O11 | [M–H]− | MS2[449]: 355(100), 329(12), 287(36), 269(29), 193(13) MS3[449 → 355]: 193(100), 192(12) | 234, 330 | Ferulic acid hexose |
| 22a | 18.18 | 935.0782 | 1.414 | C41H28O26 | [M–H]− | MS2[935]: 633(100), 301(43)MS3[935 → 633]: 463(7), 301(100) | 234, 278 | Galloyl bis-HHDP Hexose |
| 23a | 19.40 | 433.0406 | −0.659 | C19H14O12 | [M–H]− | MS2[433]: 415(6), 301(100), 300(30)MS3[433 → 301]: 301(42), 284(47), 257(100), 229(70), 185(27) | 216, 234, 278 | Ellagic acid pentoside |
| 24a | 19.95 | 300.9985 | −0.45 | C14H6O8 | [M–H]− | MS2[301]: 301(43), 300(6), 284(29), 283(20), 273(15), 257(100), 245(9), 229(69), 213(8), 201(9), 185(39) | 233, 366 | Ellagic acid* |
| 25a | 20.44 | 447.0563 | −0.559 | C20H16O12 | [M–H]− | MS2[447]: 301(100), 300(30)MS3[447 → 301]: 301(13), 284(11), 257(100), 229(27), 185(12) | 234, 370 | Ellagic acid deoxyhexoside |
| 26a | 21.31 | 447.0566 | −0.289 | C20H16O12 | [M–H]− | MS2[447]: 301(100), 300(30)MS3[447 → 301]: 301(25), 300(12), 284(14), 283(40), 271(6), 257(100), 255(6), 245(7), 229(50), 185(23) | – | Ellagic acid deoxyhexoside |
| 27a | 22.99 | 477.0673 | −0.204 | C21H18O13 | [M–H]− | MS2[477]: 301(100)MS3[477 → 301]: 273(20), 257(15), 179(100), 151(69)MS4[477 → 301 → 179]: 151(100) | 216, 233, 350 | Quercetin 3-O-glucuronide* |
| 28a | 23.75 | 463.088 | −0.219 | C21H20O12 | [M–H]− | MS2[463]: 301(100), 300(21)MS3[463 → 301]: 283(8), 273(28), 257(14), 239(6), 193(8), 179(100), 151(75), 107(6) | – | Quercetin 3-glucoside* |
| 29a | 24.41 | 935.0782 | −1.354 | C41H28O26 | [M–H]− | MS2[935]: 918(6), 633(100), 463(7), 301(61) | – | Galloyl-bis-HHDP-glucose |
| 30a | 24.81 | 355.1031 | −0.395 | C16 H19 O9 | [M–H]− | MS2[355]: 337(23), 311(10), 309(100), 207(35), 147(50) | 216, 233, 282 | Feruloyl hexoside |
| 31a | 26.28 | 934.0712 | −0.609 | C82H52O54 | [M– 2H]2− | MS2[934]: 1568(100), 1567(9), 1265(25), 1085(26), 916(31), 915(93), 897(84), 783(32), 633(41), 301(86) | 233 | Agrimoniin |
| 32a | 27.89 | 461.0725 | −0.039 | C21H18O12 | [M–H]− | MS2[461]: 285(100)MS3[461 → 285]: 267(42), 257(100), 243(8), 241(23), 240(17), 239(21), 229(41), 223(11), 213(24), 211(10), 199(16), 197(16), 195(7), 185(7), 163(14), 151(8 | 217, 233, 265, 350 | Kaempferol 3-O-glucuronide* |
| 33a | 28.40 | 447.0934 | 0.065 | C21H20O11 | [M–H]− | MS2[447]: 327(23), 301(7), 285(90), 284(100), 255(15)MS3[447 → 284]: 255(100), 227(13) | 218, 233, 350 | Kaempferol 3-O-glucoside |
| 34a | 30.33 | 505.0983 | –0.494 | C23H22O13 | [M–H]− | MS2[505]: 487(10), 463(25), 343(6), 301(100), 300(53) | – | Quercetin 3-acetylglucoside |
| 35a | 32.04 | 489.1039 | 0.031 | C23H22O12 | [M–H]− | MS2[489]: 285(100)MS3[489 → 285]: 267(59), 257(100), 256(16), 241(25), 240(12), 239(25), 229(57), 223(12), 213(26), 199(26), 197(21), 195(10), 189(6), 165(6), 163(23) | – | Kaempferol 3-acetylglucoside |
| 36a | 44.26 | 593.1299 | −0.124 | C30H25O13 | [M–H]− | MS2[593]: 447(10), 307(6), 285(100) | – | Kaempferol 3-coumaroylglucoside |
Confirmed with reference standards.
The non-anthocyanin polyphenols in Fort Laramie are mainly coumaroyl-glucose, ferulic acid hexose, quercetin, and kaempferol derivatives while in Ruegen F7-4 and YW5AF7, taxifolin 3-O-arabi-noside, methylellagic acid rhamnoside, and ellagic acid are the major constituents. Understanding the chemical differences between cultivated strawberry Fort Laramie and wild strawberry Ruegen F7-4/YW5AF7 may help target modification of strawberries for quality improvement.
4. Conclusion
This study was the first chemical investigation to describe the phenolic composition of two diploid inbred lines, Ruegen F7-4 and YW5AF7, which will facilitate genetic and biochemical studies of the enzymes catalysing the biosynthesis of these important compounds. The UHPLC-DAD-HRMS analysis identified the presence of a variety of anthocyanins, dihydroflavonols, flavonols, fla-van-3-ols, proanthocyanidins, free and conjugated forms of ellagic acid, and ellagitannins. A total of 78 phenolic compounds were identified. The results demonstrate the differences in anthocyanin composition in Ruegen F7-4 and cultivated strawberry Fort Laramie are mainly due to peonidin 3-O-glucoside and peonidin 3-O-malonylglucoside. The identification of phenolic compounds revealed some interesting compounds newly found in strawberry. Taxifolin 3-O-arabinoside and methylellagic acid glycosides in Ruegen F7-4 and YW5AF7 were reported in strawberry fruits for the first time. The high diversity of the phenolic com pounds found in wild diploid strawberry samples may have potential beneficial effects for human health. Further studies should be carried out for quantification of major compounds and biochemical and genetic characterisation of biosynthetic pathways.
Supplementary Material
Acknowledgments
This research is supported by the Agricultural Research Service of the U.S. Department of Agriculture and an Interagency Agreement with the Office of Dietary Supplements (ODS) of the National Institutes of Health (NIH).
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.foodchem.2013.08.089.
References
- Aaby K, Ekeberg D, Skrede G. Characterization of phenolic compounds in strawberry (Fragaria x ananassa) fruits by different HPLC detectors and contribution of individual compounds to total antioxidant capacity. Journal of Agriculture and Food Chemistry. 2007a;55(11):4395–4406. doi: 10.1021/jf0702592. [DOI] [PubMed] [Google Scholar]
- Aaby K, Mazur S, Nes A, Skrede G. Phenolic compounds in strawberry (Fragaria x ananassa Duch.) fruits: Composition in 27 cultivars and changes during ripening. Food Chemistry. 2012;132(1):86–97. doi: 10.1016/j.foodchem.2011.10.037. [DOI] [PubMed] [Google Scholar]
- Aaby K, Wrolstad RE, Ekeberg D, Skrede G. Polyphenol composition and antioxidant activity in strawberry purees; impact of achene level and storage. Journal of Agriculture and Food Chemistry. 2007b;55(13):5156–5166. doi: 10.1021/jf070467u. [DOI] [PubMed] [Google Scholar]
- Aharoni A, Giri AP, Verstappen FW, Bertea CM, Sevenier R, Sun Z, et al. Gain and loss of fruit flavor compounds produced by wild and cultivated strawberry species. Plant Cell. 2004;16(11):3110–3131. doi: 10.1105/tpc.104.023895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco L, Lopez L, Scalone AG, Di Carli M, Desiderio A, Benvenuto E, et al. Strawberry proteome characterization and its regulation during fruit ripening and in different genotypes. Journal of Proteomics. 2009;72(4):586–607. doi: 10.1016/j.jprot.2008.11.019. [DOI] [PubMed] [Google Scholar]
- Buendia B, Gil MI, Tudela JA, Gady AL, Medina JJ, Soria C, et al. HPLC-MS analysis of proanthocyanidin oligomers and other phenolics in 15 strawberry cultivars. Journal of Agriculture and Food Chemistry. 2010;58(7):3916–3926. doi: 10.1021/jf9030597. [DOI] [PubMed] [Google Scholar]
- Carbone F, Preuss A, De Vos RC, D’Amico E, Perrotta G, Bovy AG, et al. Developmental, genetic and environmental factors affect the expression of flavonoid genes, enzymes and metabolites in strawberry fruits. Plant Cell Environment. 2009;32(8):1117–1131. doi: 10.1111/j.1365-3040.2009.01994.x. [DOI] [PubMed] [Google Scholar]
- Cheel J, Theoduloz C, Rodriguez J, Saud G, Caligari PDS, Schmeda-Hirschmann G. E-cinnamic acid derivatives and phenolics from Chilean strawberry fruits, Fragaria chiloensis ssp chiloensis. Journal of Agriculture and Food Chemistry. 2005;53(22):8512–8518. doi: 10.1021/jf051294g. [DOI] [PubMed] [Google Scholar]
- Goulas V, Manganaris GA. The effect of postharvest ripening on strawberry bioactive composition and antioxidant potential. Journal of the Science of Food and Agriculture. 2011;91(10):1907–1914. doi: 10.1002/jsfa.4406. [DOI] [PubMed] [Google Scholar]
- Hilt P, Schieber A, Yildirim C, Arnold G, Klaiber I, Conrad J, et al. Detection of phloridzin in strawberries (Fragaria x ananassa Duch.) by HPLC-PDA-MS/MS and NMR spectroscopy. Journal of Agriculture and Food Chemistry. 2003;51(10):2896–2899. doi: 10.1021/jf021115k. [DOI] [PubMed] [Google Scholar]
- Ishimaru K, Omoto T, Asai I, Ezaki K, Shimomura K. Taxifolin 3-Arabinoside from Fragaria-X-Ananassa. Phytochemistry. 1995;40(1):345–347. doi: 10.1016/0031-9422(95)00200-q. [DOI] [PubMed] [Google Scholar]
- Kallio H, Hakala M, Pelkkikangas AM, Lapvetelainen A. Sugars and acids of strawberry varieties. European Food Technology. 2000;212(1):81–85. [Google Scholar]
- Kaufmann A. The current role of high-resolution mass spectrometry in food analysis. Analytical and Bioanalytical Chemistry. 2012;403(5):1233–1249. doi: 10.1007/s00216-011-5629-4. [DOI] [PubMed] [Google Scholar]
- Kelebek H, Selli S. Characterization of Phenolic Compounds in Strawberry Fruits by Rp-Hplc-Dad and Investigation of Their Antioxidant Capacity. Journal of Liquid Chromatography and Related Technologies. 2011;34(20):2495–2504. [Google Scholar]
- Kim SH, Shin DH, Choi IG, Schulze-Gahmen U, Chen S, Kim R. Structure-based functional inference in structural genomics. Journal of Structural and Functional Genomics. 2003;4(2–3):129–135. doi: 10.1023/a:1026200610644. [DOI] [PubMed] [Google Scholar]
- Maatta-Riihinen KR, Kamal-Eldin A, Torronen AR. Identification and quantification of phenolic compounds in berries of Fragaria and Rubus species (family Rosaceae) Journal of Agriculture and Food Chemistry. 2004;52(20):6178–6187. doi: 10.1021/jf049450r. [DOI] [PubMed] [Google Scholar]
- Mikulic-Petkovsek M, Schmitzer V, Slatnar A, Weber N, Veberic R, Stampar F, et al. Alteration of the Content of Primary and Secondary Metabolites in Strawberry Fruit by Colletotrichum nymphaeae Infection. Journal of Agriculture and Food Chemistry. 2013;2:2–19. doi: 10.1021/jf402105g. [DOI] [PubMed] [Google Scholar]
- Reganold JP, Andrews PK, Reeve JR, Carpenter-Boggs L, Schadt CW, Alldredge JR, et al. Fruit and soil quality of organic and conventional strawberry agroecosystems. PLoS ONE. 2010;5(9) doi: 10.1371/journal.pone.0012346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roscher R, Bringmann G, Schreier P, Schwab W. Radiotracer studies on the formation of 2,5-dimethyl-4-hydroxy-3(2H)-furanone in detached ripening strawberry fruits. Journal of Agriculture and Food Chemistry. 1998;46(4):1488–1493. [Google Scholar]
- Russell WR, Scobbie L, Labat A, Duthie GG. Selective bio-availability of phenolic acids from Scottish strawberries. Molecular Nutrition & Food Research. 2009;53(Suppl 1):S85–91. doi: 10.1002/mnfr.200800302. [DOI] [PubMed] [Google Scholar]
- Shin Y, Ryu JA, Liu RH, Nock JF, Polar-Cabrera K, Watkins CB. Fruit quality, antioxidant contents and activity, and antiproliferative activity of strawberry fruit stored in elevated CO2 atmospheres. Journal of Food Science. 2008;73(6):S339–S344. doi: 10.1111/j.1750-3841.2008.00857.x. [DOI] [PubMed] [Google Scholar]
- Sun G, Shi C. The overall quality control of Radix scutellariae by capillary electrophoresis fingerprint. Journal of Chromatographic Science. 2008;46(5):454–460. doi: 10.1093/chromsci/46.5.454. [DOI] [PubMed] [Google Scholar]
- Villa-Rojas R, Lopez-Malo A, Sosa-Morales ME. Hot water bath treatments assisted by microwave energy to delay postharvest ripening and decay in strawberries (Fragaria x ananassa) Journal of the Science of Food and Agriculture. 2011;91(12):2265–2270. doi: 10.1002/jsfa.4449. [DOI] [PubMed] [Google Scholar]
- Vrhovsek U, Guella G, Gasperotti M, Pojer E, Zancato M, Mattivi F. Clarifying the identity of the main ellagitannin in the fruit of the strawberry, Fragaria vesca and Fragaria ananassa Duch. Journal of Agriculture and Food Chemistry. 2012;60(10):2507–2516. doi: 10.1021/jf2052256. [DOI] [PubMed] [Google Scholar]
- Wojdylo A, Figiel A, Oszmianski J. Effect of drying methods with the application of vacuum microwaves on the bioactive compounds, color, and antioxidant activity of strawberry fruits. Journal of Agriculture and Food Chemistry. 2009;57(4):1337–1343. doi: 10.1021/jf802507j. [DOI] [PubMed] [Google Scholar]
- Yang FM, Li HM, Li F, Xin ZH, Zhao LY, Zheng YH, et al. Effect of nano-packing on preservation quality of fresh strawberry (Fragaria ananassa Duch. cv Fengxiang) during storage at 4 degrees C. Journal of Food Science. 2010;75(3):C236–240. doi: 10.1111/j.1750-3841.2010.01520.x. [DOI] [PubMed] [Google Scholar]
- Zhang J, Wang X, Yu O, Tang J, Gu X, Wan X, et al. Metabolic profiling of strawberry (Fragaria x ananassa Duch.) during fruit development and maturation. Journal of Experimental Botany. 2011;62(3):1103–1118. doi: 10.1093/jxb/erq343. [DOI] [PubMed] [Google Scholar]
- Zheng Q, Song J, Doncaster K, Rowland E, Byers DM. Qualitative and quantitative evaluation of protein extraction protocols for apple and strawberry fruit suitable for two-dimensional electrophoresis and mass spectrometry analysis. Journal of Agriculture and Food Chemistry. 2007;55(5):1663–1673. doi: 10.1021/jf062850p. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.