Abstract
In conjunction with the recent American Chemical Society symposium titled “Docking and Scoring: A Review of Docking Programs” the performance of the DOCK6 program was evaluated through (1) pose reproduction and (2) database enrichment calculations on a common set of organizer-specified systems and datasets (ASTEX, DUD, WOMBAT). Representative baseline grid score results averaged over five docking runs yield a relatively high pose identification success rate of 72.5 % (symmetry corrected rmsd) and sampling rate of 91.9 % for the multi site ASTEX set (N = 147) using organizer-supplied structures. Numerous additional docking experiments showed that ligand starting conditions, symmetry, multiple binding sites, clustering, and receptor preparation protocols all affect success. Encouragingly, in some cases, use of more sophisticated scoring and sampling methods yielded results which were comparable (Amber score ligand movable protocol) or exceeded (LMOD score) analogous baseline grid-score results. The analysis highlights the potential benefit and challenges associated with including receptor flexibility and indicates that different scoring functions have system dependent strengths and weaknesses. Enrichment studies with the DUD database prepared using the SB2010 preparation protocol and native ligand pairings yielded individual area under the curve (AUC) values derived from receiver operating characteristic curve analysis ranging from 0.29 (bad enrichment) to 0.96 (good enrichment) with an average value of 0.60 (27/38 have AUC ≥ 0.5). Strong early enrichment was also observed in the critically important 1.0–2.0 % region. Somewhat surprisingly, an alternative receptor preparation protocol yielded comparable results. As expected, semi-random pairings yielded poorer enrichments, in particular, for unrelated receptors. Overall, the breadth and number of experiments performed provide a useful snapshot of current capabilities of DOCK6 as well as starting points to guide future development efforts to further improve sampling and scoring.
Keywords: Pose identification, Pose rescoring, Docking, Virtual screening, Enrichment, ROC curves, Scoring, Sampling, Rmsd, Symmetry
Introduction
Despite the many challenges [1, 2] computational docking has emerged to become a useful part of drug discovery and design [3-5]. And, with the advent of ever faster computers, publicly available databases of purchasable compounds [6], and numerous software choices [7], more and more research groups are embarking on virtual high throughput screening projects to identify potentially useful lead molecules. These facts make continued validation and refinement of docking software and associated protocols all the more important. In particular, it is important to gauge how well a given procedure can accurately generate and score known ligand binding poses [8] and rank-order known inhibitors versus property-matched decoys [9].
DOCK was the first widely used docking program [10] and its usefulness in drug discovery applications is well established [1-3, 5, 11]. Notably, the basic methodology established by Kuntz and coworkers in the early 1980s is still in use today. Briefly, orientations of the ligand relative to a receptor binding cavity are searched using a negative surface image of the target. The image is generated by filling the solvent accessible receptor surface with overlapping spheres and selecting a subset of the spheres to represent the binding site [10]. Ligand atoms and spheres are geometrically matched to sample rigid-body orientational space [12], and conformational space of the ligand is sampled in the presence of the receptor binding site via the anchor-and-grow incremental construction approach [13]. The quality of a binding geometry (termed pose) is typically determined using a physics-based energy score. The most computationally efficient such score is grid score which uses a simplified molecular mechanics force field [14].
Recent versions, starting with DOCK5, [15] have been written in C++ and designed to be modular allowing for relatively straightforward inclusion of different sampling methods and scoring functions. The extensibility of the modular platform was first demonstrated in DOCK6 [16] by the addition of several scoring functions which incorporate solvation effects using Generalized Born Surface Area (GB/SA) [16-18] or Poisson-Boltzmann Surface Area (PB/SA) [19] models. Receptor flexibility, conjugate gradient minimization, and molecular dynamics (MD) simulation capabilities using the full Amber molecular mechanics force field were also incorporated [16, 20]. DOCK6.4 included modifications to the internal energy and anchor-and-grow routines which improves ligand sampling, [8] and in DOCK6.5 a new footprint-based scoring function [21] was added which calculates intermolecular energies and hydrogen bonds on a per-residue basis. The two latter versions were extensively validated using the SB2010 database consisting of 780 protein–ligand complexes derived from the protein databank [8]. A more detailed description of the enhancements to the sampling behavior introduced starting in DOCK6.4, and a comparison with earlier DOCK versions, will be presented in a future manuscript.
In addition to its off-the-shelf utility, DOCK is designed to be a learning tool and a platform for exploring new docking methods. Ligand growth trees and growth statistics enables users to “look under the hood” of the docking engine, and online tutorials [22, 23] as well as extensive pose validation [8] and enrichment databases [9] are all geared to make the program more robust, easier to use, and ultimately more useful. It is important to note that DOCK source code has always been distributed with the program, and many users over the years have successfully modified the code for their own purposes.
The current manuscript describes docking results which were obtained in the context of preparing for a symposium titled “Docking and Scoring: A Review of Docking Programs” held at the Spring 2011 Meeting of the American Chemical Society. Organizers of the symposium invited groups to present their results using two popular validation testsets, Astex [24] and DUD [9], so that performance could be gauged across a wide variety of programs. Specifically, groups were asked to report: (1) pose identification statistics (scoring and sampling success rates) using the Astex database consisting of all viable dockable sites in each of the 85 pdb entries, and (2) enrichment statistics (ROC curves, AUC, etc.) using the standard DUD database consisting of 40 systems or a subset of 13 systems using actives from the WOMBAT [25] database. The nature and type of experiments and statistics to be reported were provided to the participants prior to the meeting. In addition, the participants were asked to use receptors that had been re-refined and provided by the organizers. The results presented here represent the combined efforts of two DOCK development groups (Rutgers University and Stony Brook University) and in some cases, both groups independently performed similar experiments, but under slightly different conditions to gauge noise and reproducibility. Continued refinement and testing of new docking methods is essential for the field to move forward as a whole; thus we applaud the organizers efforts and feel that group participation in these and related activities is important.
Theoretical methods
Pose identification
For the first primary experiment, pose identification, two criteria are examined (1) Docking Success is observed when the top scoring pose is within 2 Å heavy atom root-mean-square deviation (rmsd) of the crystal ligand, and (2) Sampling Success (or minimum rmsd) is observed when there exists in the top 32 poses a conformation within 2 Å of the crystal ligand. When native-like poses are sampled, but the top scoring pose is not within 2 Å, this is referred to as a Scoring Failure. Figure 1a–c shows representative examples for a ligand docked to a target in which a computed rmsd indicates a success, a near success, and a failure in comparison to the crystallographic reference. It is important to note that methods for examining docking accuracy depend on the rmsd algorithm. Historically, in many computational chemistry programs, such as DOCK and Amber, computation of rmsd assumes there is a single one-to-one correspondence between selections of atoms being compared (atom 1 with atom 1, atom 2 with atom 2, etc.). In this manuscript such calculations are referred to as standard rmsd. However, some ligands may transform under symmetry operations into physically indistinguishable objects. More commonly, local operations, such as rotation about an individual bond, may produce indistinguishable objects (i.e. phenyl group, t-butyl group, etc.). Thus, ideally, rmsd should be corrected to properly account for symmetry equivalent atoms. In this report, the Hungarian matching algorithm [26, 27] was used for computing symmetry corrected rmsd. Figure 1d shows a representative example in which standard rmsd indicates a failure (6.96 Å) but symmetry corrected rmsd is a success (0.48 Å). As highlighted by the lively discussion at the recent Spring 2011 ACS meeting a number of research groups are incorporating symmetry corrected rmsd in their docking calculations; however the exact procedures used are, for the most part, unfortunately not well documented. More discussion, comparison, and robust evaluation of symmetry corrected rmsd in the literature, especially with regards to docking, would be welcome. A related issue involved challenges with using organizer supplied data for rmsd calculations in which ligand references had different atom orderings and atom names compared with the molecules to be docked. To address this issue, an iterative four step mapping algorithm (see supplementary material, Figure S1) was used to find the maximum common substructure [28] between two ligands, regardless of order, so that standard and symmetry corrected rmsd could be computed.
Fig. 1.

Representative docked ligand poses (green) in comparison to a crystallographic reference (red) showing a success (a), near success (b), and a failure (c) using a 2.0 Å rmsd definition of success. d How a pose classified as failure using standard rmsd can be recovered using a symmetry corrected algorithm. Figure generated using UCSF Chimera [29]
In addition to standard rmsd and symmetry corrected rmsd, in which the deviates are differences of Cartesian coordinates of corresponding atoms, the receptor flexibility component of this study also employs the intramolecular-distance-based rmsd, termed here drmsd (Eq. 1):
| (1) |
Here, c and c′ are two conformations of a molecule with N atoms and Dij is the N × N distance matrix of all the intramolecular distances. In drmsd, the deviates are differences of corresponding interatomic distances. This measure is independent of the translational and rotational molecular degrees of freedom and derived entirely by comparing internal distances within a molecule.
Enrichment metrics
For the second primary experiment, enrichment, receiver operating characteristic (ROC) curves are used to evaluate how well methods favorably rank known active molecules compared with a large number of decoys [30]. In ROC curves, the True Positive Rate (TPR = TP/P) is plotted versus the False Positive Rate (FPR = FP/N), where TP is the number of True Positives, P is the total number of Positives (actives), FP is the number of False Positives, and N is the total number of Negatives (decoys). Figure 2 shows ROC curve examples representative of good enrichment (panel a), random enrichment (panel b), and poor enrichment (panel c). Quantitatively, the total area under the curve (AUC) of a ROC plot provides a measure of global enrichment. In a practical sense however, good early enrichment is reasonably expected to be more important for prioritizing compounds identified by virtual screening of large ligand libraries. The example in Fig. 2c illustrates ROC curve behavior with poor total enrichment but strong early enrichment. Several methods for assessing early enrichments have been reported [31]. In this report, we use four very specific definitions (%TPR, %FPR, %AUC, and FE) to assess early enrichment as outlined below.
Fig. 2.

Representative examples of ROC curves showing good (a), random (b), and poor (c) global enrichments
For early enrichment, we report %TPR, and %FPR for different percentages (0.1, 1.0, 2.0) of the ranked database. Transforming to percentages yield scaled values which are much more readable for very early enrichments (i.e. not near zero). Percentages yield %FPR and %TPR in the range [0, 100]. We also report %AUC for early enrichment which is in the range [0, 10,000]. We feel %AUC is a more meaningful metric to gauge early enrichment since the values involve the area and not a single point on the ROC curve. When reporting total AUC we report unscaled values. The panel in Fig. 3a illustrates the relationship between %FPR (Xε), %TPR (Yε), and the % of database ranked (ε = 0.1, 1.0, 2.0) for three different values. It should be noted that, the %FPR and the % of database screened are not necessarily equal. However, under most conditions, when P < < N this is in fact a reasonable assumption (Xε ≈ ε).
Fig. 3.
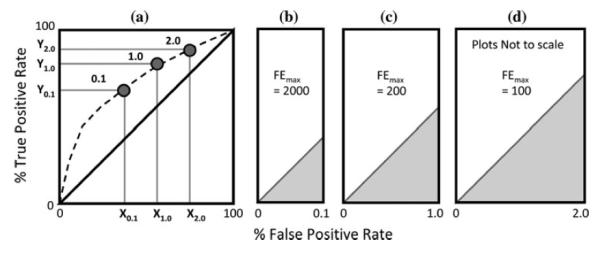
a Schematic showing possible enrichments at 0.1, 1.0, and 2.0 % of the database scanned and b–d maximum Fold Enrichment (FE) values at each of these points. FE = AUC/AUCran
The fourth early enrichment metric used here is termed fold enrichment, recently employed to assess the footprint-based scoring function [21], and defined as FE = AUC/AUCran. This quantity facilitates comparison with random behavior and is a very intuitive measure. Using the approximation Xε ≈ ε when 0.1, 1.0, and 2.0 % of the database are kept then 2,000, 200, and 100 are the best possible (FEmax) fold enrichment values (Fig. 3c, d). It is important to note that for FE it is only meaningful to compare values calculated at the same percentage of the database but not between different percentages.
Scoring functions
The main body of this work employed three physics-based scoring functions, termed grid score, Amber score, and LMOD score, to rank order docked poses. PB/SA scoring was also examined (see supplementary material). It is important to note that no additional parameterization, i.e., beyond that of the predetermined molecular mechanics parameterization, was performed, nor was there any tuning of the scoring functions for individual systems.
Grid score
Grid score consists of intermolecular non-bonded van der Waals (VDW) and Coulombic energies (scaled by a distance-dependent dielectric) between the ligand and receptor [14]. The van der Waals components are generalized, and the exact attractive and repulsive exponents used are specified in the next section. Although grid generation is an expensive calculation, it is a one-time upfront cost and used to pre-compute the potential energy of the receptor to speed up the calculations while docking. Calculating the intermolecular interaction energy directly on the receptor is O(N*M) compared to O(N) on the grid, where N is the number of ligand atoms and M is the number of receptor atoms.
Amber score
Amber score is a simple MM-GB/SA approach that does not explicitly treat entropic effects [20]. A number of papers using MM-GB/SA and related MM-PB/SA methods have focused on docking [32-37]. The Amber score binding energy is calculated as EComplex − (EReceptor + ELigand), where EComplex, EReceptor, and ELigand are MM-GB/SA energies as approximated by the Amber force field. Amber score enables all or a part of the ligand-receptor complex to be flexible by defining a movable region in the DOCK input. Only atoms in the movable region are allowed to move during minimization and MD simulation. The movable region options are nothing, ligand, distance, everything and NAB [38] atom expression. So for example, for the nothing option no minimization or MD simulation occurs. For distance movable, the ligand and any receptor residues within a specified distance from the ligand are movable. If any atom is within the cutoff distance then the whole residue is selected. The ligand is represented by the active site sphere list, and thus the movable receptor residues are well defined and independent of any particular ligand molecule. Due to the size and nature of this investigation only the nothing, ligand, and distance movable regions were used.
LMOD score
LMOD score operates on the same basic principle as Amber score, but uses the LMOD [39, 40] method instead of MD. LMOD (short for LowMODe) is a way of exploring conformational space by following low-frequency vibrational modes. This can in principle be more efficient than MD since high-frequency motions (e.g. bond stretching, angle bending, individual side-chain movement) tend to complicate the overall energy landscape and frustrate searches, while low-frequency motions (i.e. motions in which the movement of many atoms are correlated) are usually of greater interest.
Briefly stated, the LMOD procedure involves (1) calculation of low-mode eigenvectors, (2) perturbation of the starting structure by uphill movement along a randomly chosen low-mode eigenvector until a barrier is crossed, (3) minimization of the perturbed structure, (4) return to step 2 and follow a different mode. Once all modes of a structure have been followed, a new structure is chosen from a pool of previously found minimum energy structures and the procedure is repeated. The LMOD score binding energy is calculated as EComplex − (EReceptor + ELigand). The calculation of EComplex with LMOD score involves an initial minimization using a conjugate gradient method followed by the LMOD procedure itself. Both the ligand and all residues within 6.0 Å of the ligand are allowed to move; all other atoms are frozen. EReceptor and ELigand are calculated using final structures obtained from LMOD with no further minimization.
One drawback of LMOD is that it is more computationally demanding than MD. For comparison, the initial iteration of LMOD is about 40 times slower than 1,000 steps of MD. It should be noted that this LMOD cost does include the most computationally demanding part of the method, which is initial calculation of the eigenvectors of the system; subsequent iterations will not be quite as expensive. Due to the high computational cost, in this study LMOD score was only used to rescore a subset of poses for certain systems.
Computational details
Structure preparation
This study employed five basic types of structural preparations (termed ASTEX PDB, ASTEX SUP, ASTEX AMB, DUD PDB, and DUD SUP) as outlined in Table 1, to create the basic structure files to setup docking, rescoring, and/or enrichment experiments. For specific details regarding each of the five definitions, including why the different preparations were performed and challenges encountered, please consult the supplementary material. Briefly, for the pose identification experiments employing systems contained in the Astex dataset, both single site and multi site sets were prepared. Single site preparations employed the first occurrence of each dockable site and were constructed from the raw pdb coordinates (ASTEX PDB) using protocols consistent with the recently described SB2010 database [8]. Multi site preparations employed all dockable sites in each entry and were constructed from re-refined coordinates supplied by the organizers (ASTEX SUP). In addition, a preparation required for Amber score calculations was generated using organizer supplied coordinates (ASTEX AMB). For enrichment studies employing DUD, as before, one set was derived from the raw pdb coordinates (DUD PDB). Another set was derived using organizer supplied structures (DUD SUP) in which hydrogen atoms were added to the receptors from the original DUD database [9].
Table 1.
Structure preparation overview for Astex and DUD datasets
| Preparation definition | Small molecules |
Proteins |
|||
|---|---|---|---|---|---|
| Charge model | Program | Charge model | Program | Comments | |
| (1) ASTEX PDB | AM1BCC | MOE, antechamber | FF99SB | LEAP | Built from PDB. Validated SB2010 protocol. MD ready (N = 84). |
| (2) ASTEX SUP | AM1BCC | Chimera | FF99SB | Chimera | Built from supplied structures. Two DOCK protocols (SB, RU). Not MD ready (N = 147) |
| (3) ASTEX AMB | AM1BCC | DOCK | FF94 | DOCK | Built from supplied structures. Employed DOCK Amberize scripts. MD ready (N = 147). |
| (4) DUD PDB | FF99SB | LEAP | Rec from PDB, SB2010 protocol. Lig from DUD MD ready (N = 38) |
||
| (5) DUD SUP | Gastig | MOE | Rec from supplied structures. Significant atom name problems. Not MD ready (N = 40) |
||
Detailed protocols
DOCK specific parameters
Regardless of which of the protocols was used to prepare the initial receptor and ligand structures for docking, a number of other steps are required to prepare the binding site(s) prior to running actual DOCK calculations. Setups typically involve a number of accessory programs including DMS, SPHGEN, sphgen_cpp, sphere_selector, SHOWBOX, and GRID which are available with the standard DOCK distributions available on the UCSF website (http://dock.compbio.ucsf.edu/). The two research groups participating in this work, denoted in this manuscript as SB (Stony Brook University) or RU (Rutgers University), used somewhat different binding site preparation protocols as noted below.
The first step involved in preparing the binding sites is the calculation of the solvent accessible surface of each receptor, without hydrogen atoms, using a probe radius of 1.4 Å with the program DMS. A negative image of the surface is then generated as a set of overlapping spheres [10] using the programs SPHGEN or sphgen_cpp. A subset of spheres near the crystallographic ligand is then selected with the aid of the program sphere_selector or an in-house perl script. For receptors with multiple binding sites each site is prepared separately. Preparation of the docking grids is then performed and follows a two step procedure. Firstly, a box around the binding site is constructed with the accessory program SHOWBOX. The box includes the selected spheres and a protocol dependent margin. Secondly, the actual grids are computed with the accessory program GRID using a 0.3 Å grid spacing, a 9,999 Å distance cutoff, and a 4r distance dependent dielectric constant. For the SB group, selected spheres were within 8 Å from ligand heavy atoms of the crystal structure and for computing the energy grids an 8 Å box margin and 6–9 VDW exponents were used. For the RU group, selected spheres were within 10 Å from the ligand and energy grids employed a 5 Å box margin and 6–12 VDW exponents. Otherwise, all preparation steps were the same for both groups. It should be noted that the optimal set of parameters to use in docking is still an active area of research. The protocols and parameter options presented here represent two reasonable starting points.
For actual docking runs, the RU group used DOCK6.5 in conjunction with docking the ASTEX SUP data set while the SB group used a soon to-be-released version termed DOCK6.6 and docked both the ASTEX SUP dataset and the ASTEX PDB data set. For rescoring with Amber Score and LMOD, the RU group converted docked ASTEX SUP results into the required ASTEX AMB formats. Figure 4 graphically outlines the overall rescoring procedure. In addition, the SB group performed enrichment studies using DUD PDB and DUD SUP.
Fig. 4.

Flowchart showing pose-rescoring protocols using three variants of Amber score and LMOD score
Enrichment datasets
DUD [9] consists of forty protein structures (39 from the PDB and one homology model, denoted here with the name MODL) divided into six families as shown in Table 2: Metalloenzyme (N = 4), Nuclear Hormone Receptor (N = 8), Kinase (N = 9), Folate Enzyme (N = 2), Serine Protease (N = 2), and finally a miscellaneous family called “Other Enzymes” (N = 14) the same breakdown as in [9]. Table 2 lists for each entry, the associated pdb code, the DUD system name, number of active ligands, number of decoys, number of WOMBAT active ligands (if applicable), and the so-called semi-random pairings for which the DUD receptors were combined with actives and decoys developed for a different system. The WOMBAT set [25] consists of active small molecules for 13 of the DUD systems. Two WOMBAT sets did not correspond to any DUD receptor and were not used. For another set, active ligands for Estrogen Receptor alpha were a mix of both agonist and antagonists. However, DUD distinguishes between agonist and antagonist receptors thus WOMBAT runs for Estrogen Receptor are not included. In summary, the DUD SUP datasets consist of 40 DUD and 10 WOMBAT sets of active compounds which were used with the 40 supplied receptors. For DUD PDB, two receptors 1L2I and 1AH3 were excluded owing to structural problems in the PDB coordinates. Therefore, the DUD PDB datasets consist of 38 DUD and 9 WOMBAT sets of active compounds, with corresponding DUD decoy ligand sets, with 38 corresponding receptors.
Table 2.
DUD systems and semi-random matching information
| Protein family | PDB name |
DUD name | No. ligands | No. decoys |
No. WOMBAT |
Semi-random pairing |
Pairing description |
|---|---|---|---|---|---|---|---|
| Metallo-enzymea | 1O86 | ace | 49 | 1,797 | ada | ||
| (N = 4) | 1NDW | ada | 39 | 927 | ace | ||
| 1H1D | comtb | 11 | 468 | pde5 | |||
| 1XP0 | pde5 | 88 | 1,978 | 101 | comt | ||
| Nuclear Hormone Receptor | 2AO6 | ar | 79 | 2,854 | 56 | rxr | |
| (N = 8) | 1L2Id | er_agonist | 67 | 2,570 | 83c | mr | |
| 3ERT | er_antagonist | 39 | 1,448 | 83c | ppar | ||
| 1M2Z | gr | 78 | 2,947 | pr | |||
| 2AA2 | mrb | 15 | 636 | er_agonist | |||
| 1FM9 | ppar | 85 | 3,127 | 43 | er_antagonist | ||
| 1SR7 | pr | 27 | 1,041 | gr | |||
| 1MVC | rxr | 20 | 750 | ar | |||
| Kinase | 1CKP | cdk2 | 72 | 2,074 | 190 | pdgfrb | |
| (N = 9) | 1M17 | egfr | 475 | 15,996 | 81 | hsp90 | |
| 1AGW | fgfr1 | 120 | 4,550 | src | |||
| 1UY6 | hsp90 | 37 | 979 | egfr | |||
| 1KV2 | p38 | 454 | 9,141 | 60 | vegfr2 | ||
| MODL | pdgfrb | 170 | 5,980 | cdk2 | |||
| 2SRC | src | 159 | 6,319 | fgfr1 | |||
| 1KIM | tkb | 22 | 891 | pnp | Unrelated | ||
| 1VR2 | vegfr2 | 88 | 2,906 | p38 | |||
| Folate Enzyme | 3DFR | dhfr | 410 | 8,367 | gart | ||
| (N = 2) | 1C2T | gart | 40 | 879 | dhfr | ||
| Serine Protease | 1F0R | fxa | 146 | 5,745 | 125 | thrombin | |
| (N = 3) | 1BA8 | thrombin | 72 | 2,456 | fxa | ||
| 1BJU | trypsin | 49 | 1,664 | hivpr | Unrelated | ||
| Other Enzymesa | 1EVE | ache | 107 | 3,892 | hmga | ||
| (N = 14) | 1AH3d | alr2 | 26 | 995 | 42 | ampc | |
| 1XGJ | ampc | 21 | 786 | alr2 | |||
| 1Q4G | cox1 | 25 | 911 | sahh | |||
| 1CX2 | cox2 | 426 | 13,289 | 88 | na | ||
| 1A8I | gpb | 52 | 2,140 | hivrt | |||
| 1HPX | hivpr | 62 | 2,038 | trypsin | Unrelated | ||
| 1RT1 | hivrt | 43 | 1,519 | 120 | gpb | ||
| 1HW8 | hmga | 35 | 1,480 | ache | |||
| 1P44 | inha | 86 | 3,266 | parp | |||
| 1A4G | na | 49 | 1,874 | cox2 | |||
| 1EFY | parp | 35 | 1,351 | inha | |||
| 1B8O | pnp | 50 | 1,036 | tk | Unrelated | ||
| 1A7A | sahh | 33 | 1,346 | cox1 |
Proteins in these groups are unrelated, and not expected to cross-enrich
The very small number of ligands makes evaluating enrichment statistics at 0.1 % problematic (<1 ligand)
ERα ligands not used for WOMBAT studies
DUD systems not used in DUD PDB prep
For each PDB code in DUD, the organizers also prepared alternative non-native pairings (termed semi-random pairings) which was described as a way to perform “null hypothesis” testing. Unrelated proteins would be expected to yield enrichments no better than random (null hypothesis). However, due to the fact that the semi-random pairings actually involve structurally related proteins (Table 2), good enrichment may not be unexpected for certain pairings. Metalloenzyme and Other Enzymes groups represent collections of unrelated proteins.
Enrichment calculations
For a given molecule (active or decoy) that is docked to a target it is important to note that a final answer may not always be obtained [41, 42]. Such occurrences necessitate a decision as to how subsequent ROC curve analysis should be performed. For the current study, this was generally not problematic as on average a viable 3-D pose was returned for 99 % of the molecules (see Supplementary Material Tables S6–S7). From a practical standpoint, there are at least three possible ways of generating ROC curves (see Supplementary Material Figure S2 for a graphical representation) when not all molecules yield a ranked answer: (1) Generate ROC curves by ignoring molecules for which an answer was not obtained. In this case the number of actives (P, positives) and decoys (N, negatives) employed become Pdocked and Ndocked. (2) Generate ROC curves by using the initial number of actives and decoys (Pinitial and Ninitial). In this instance ROC curves may not always reach TPR = 1, FPR = 1. (3) Generate ROC curves by assuming perfect, none, or random enrichment for the molecules which do not yield a final answer. In this scenario ROC curves always reach TPR = 1, FPR = 1. In the current manuscript, ROC curves (and accompanying AUC values) were generated using method 3 (Pinitial and Ninitial) and using the reasonable assumption of random enrichment for missing molecules.
Amber score specific parameters
The calculation of each of the three energies required by Amber Score uses the same scheme: minimization with a conjugate gradient method is followed by MD simulation with a Langevin thermostat, another minimization, and a final energy evaluation that includes the surface area term. Amber score is implemented with the NAB toolkit [38] and is interfaced with the DOCK input, but it is currently implemented only for rescoring. Key default parameters [20] include 100 cycles of pre-MD-minimization, 3,000 steps of MD simulation at 300 K, 100 post-MD-minimization cycles, and a 0.01 minimization convergence criterion for the root-mean-square of the components of the gradient. The modified Onufriev-Bashford-Case GB/SA implicit solvent model [43] was used with a non-bonded cutoff of 18 Å. The surface area term is derived using the linear combinations of pairwise overlap algorithm [44].
LMOD score specific parameters
The LMOD procedure was run using the NAB [38] toolkit from AmberTools 1.5. The procedure used the same topology and final coordinates from Amber score (nothing moveable). The ligand and all residues within 6.0 Å of the ligand were fully flexible, and the GB Hawkins-Cramer-Truhlar implicit solvent model [45] was used with a cutoff of 99 Å. An initial minimization of 100 steps using a conjugate gradient method (XMIN, also implemented in NAB) was performed. A total of five low-frequency modes were then computed by LMOD for each structure. LMOD was run for three iterations, with three randomly selected low-modes used to drive LMOD moves. The final coordinates of the lowest energy structure found using this procedure were then used to obtain the separate receptor and ligand structures, and calculate the overall interaction energy.
Results and discussion
Pose reproduction using grid-based docking
Grid success rates
Table 3 displays the baseline docking and sampling success rates (≤2 Å), using grid-based scoring, for the different preparation protocols and numbers of binding sites. Both standard and symmetry corrected rmsd values are shown. For standard rmsd, there is an equal chance that any native-like poses generated, which are geometrically and energetically equivalent (some with rmsd >2 Å), could be picked as the top-scoring pose thus explaining the poorer results. With symmetry corrected rmsd this element of chance is removed and all equivalent native-like poses will have similar rmsd values. Figure 5 shows representative examples of eight molecules rescued using symmetry. Here, for system 1tz8 the effect is especially dramatic while for the other systems rescues were already relatively close to the 2 Å cutoff. As a general observation, symmetry-corrected rmsd yields improvement in success of ca. 5 % (see Table 3). However, the improvement is only ca. 2 % for sampling since symmetrical alternate poses are likely part of the sampled ensemble. For simplicity, all further discussion and analysis in this manuscript will present and employ symmetry corrected rmsd values.
Table 3.
Pose reproduction success rates for Astex single and multi sites
| Sitea | Size | Groupb | Sourcec | Docking success (%) |
Sampling success (%) |
|||
|---|---|---|---|---|---|---|---|---|
| stdd | syme | std | sym | |||||
| a | single (x) | 84 | SB | ASTEX PDB | 72.4 | 76.4 | 91.2 | 94.1 |
| b | single (m) | 84 | SB | ASTEX PDB | 66.0 | 74.2 | 91.7 | 94.1 |
| c | single (o) | 85 | SB | ASTEX SUP | 62.8 | 70.3 | 89.4 | 90.3 |
| d | single (o) | 85 | RU | ASTEX SUP | 61.4 | 65.2 | 83.5 | 85.9 |
| e | multi (o) | 147 | SB | ASTEX SUP | 66.3 | 72.5 | 91.2 | 91.9 |
| f | multi (o) | 147 | RU | ASTEX SUP | 62.4 | 66.0 | 85.3 | 87.3 |
Single = first site in pdb, multi = all viable sites, single (x) = crystallographic starting ligand coordinates, single (m) = minimized starting ligand coordinates, single (o) and multi (o) = organizer supplied ligand coordinates
SB Stony Brook Group, RU Rutgers University Group
ASTEX PDB = built from PDB, ASTEX SUP = built from supplied structures
std standard pairwise rmsd
sym symmetry corrected rmsd using Hungarian matching algorithm. All results averaged over five DOCK runs using different random seeds
Fig. 5.

Eight complexes are displayed where accounting for symmetry with the Hungarian matching scheme rescues native-like poses. Red = native pose, Green = docked pose. The PDB code and the rmsd values are displayed under each molecule. The standard rmsd is on the left and the symmetry corrected rmsd is on the right
Initial docking tests showed that when using different random seeds there could be variation of up to 5 % in success and sampling rates. Unique random seeds influence energy minimization, which in turn affects ligand growth. To partially address the issue of statistical noise, the data in Table 3 represents averages over five docking runs performed using different random seeds (1,000, 2,000, 3,000, 4,000, and 5,000). However, with the exception of Table 3, unless otherwise stated, results in the manuscript employ one DOCK run with the default random seed of 0.
For docking to the single sites (N = 84), the averaged SB/ASTEX PDB results (Table 3a) show a DOCK success rates of 76.4 % and sampling success rates of 94.1 %. The higher sampling success suggests substantial room for improvement if a more accurate scoring function were used to rank-order the poses. Docking to single sites using organizer supplied data yields a success rate of 70.3 % for the SB/ASTEX SUP runs (Table 3c) which is somewhat higher than the 65.2 % result for the RU/ASTEX SUP runs (Table 3d). Likely reasons for the lower success rates obtained using SB/ASTEX SUP or RU/ASTEX SUP versus SB/ASTEX PDB include potential ligand bias or differences in the receptor preparation (i.e., raw vs. refined).
The SB/ASTEX PDB results were obtained using crystallographic ligand poses as starting coordinates for docking which can favorably impact results given that bond lengths and angles are biased towards the binding site. Previous work by the SB group [8] showed a 2–9 % variation in success when using different energy minimization protocols and/or force fields to prepare ligands before docking. Interestingly, this is comparable to the difference obtained here of ~6 % between the SB/ASTEX PDB and SB/ASTEX SUP single site data (Table 3a vs. c). To explore the effects of removing potential ligand bias, additional SB/ASTEX PDB runs were performed in which ligands were rotated/translated roughly 30 Å from each binding site and subjected to a short energy minimization (1,000 steps, ε = 4r dielectric) and molecular dynamics (10 ps) simulation prior to docking (Table 3b). These calculations employed Amber11 with the General Amber Force Field (GAFF). As shown in Table 3b, this causes a relatively minor drop in success from 76.4 to 74.2 % (Table 3a vs. b). In any event, given the relatively small number of systems (N = 85, N = 147) used in this study relatively small differences in protocols are likely to have a greater than desired impact on overall success rates. For example, an increase or decrease in success for only 4/85 molecules changes the results by ca 5 %. For this reason, future studies should employ much larger databases (or cross-docking experiments) to assess pose reproduction success.
For the multi site set, the SB/ASTEX SUP results (Table 3e) yield 72.5 % in comparison to the RU/ASTEX SUP runs (Table 3f) which yield 66.0 %. Interestingly, data from the multi site SB runs show slight improvement relative to the analogous single site runs (72.5 vs. 70.3 %) which is likely a function of the fact that docking to different sites in the same protein can yield different docking outcomes. Differences in Table 3 between SB and RU groups are also likely a function of differences in setup protocols (see supplementary material) or DOCK-specific parameters (see Theoretical methods). The most significant difference between the SB and RU groups is the choice of van der Waals exponents (6–9 or 6–12 respectively) used in preparation of the docking grids. The softer 6–9 exponents used by the SB group, which are known to smooth out the energy landscape as a pseudo-mimic of receptor flexibility, likely explains why higher success rates in sampling are obtained for the SB group across the board (Table 3). Other possibilities include slight differences arising from different computer architectures or DOCK versions used (6.5 vs. 6.6). Additional multi site analysis is presented in Table S1.
Grid rmsd distributions and statistics
Table 4 contains more detailed rmsd statistics, using the top scoring pose, for the same protocols used in Table 3 (symmetry-corrected rmsd only). For each set, statistics here are based on only a single DOCK run (random seed 0). The SB/ASTEX PDB results are the lowest at 1.5 Å (mean) and 0.6 Å (median) reflecting the high 76.4 % success observed above in Table 3a. The min or max rmsd is not representative of the entire set, but rather highlights extreme cases (Table 4). A minimum rmsd of 0.1–0.2 Å serves as a sanity check that at least a few very low rmsd poses are being recovered to preclude systemic errors in the set. The maximum possible rmsd (12 Å [SB] or 15 Å [RU]) is bounded by the dimensions of the active sites, and these are defined by the spheres (only spheres within 8 Å [SB] or 10 Å [RU] of the native ligands are retained) and the grid box (margins and spheres). Poses are oriented to the spheres, and if a pose exceeds a grid boundary then the pose is pruned. The mean rmsd is an alternative measure of success which is not dependent on the hard 2 Å cutoff. However, a mean rmsd of less than 2 Å is only recovered for the ASTEX PDB set. The mean rmsd value as a success metric is affected by docking failures with high rmsds, where in practice a 4 Å failure may be no worse than a 15 Å failure. The standard deviations are in the range 2.1–2.9 Å. The median rmsd is likely the most representative metric in Table 4, since it is not affected detrimentally by extreme values like the mean rmsd value. The organizers noted that 25/85 systems had significant structural problems or ligands with alternate conformations, and they requested an analysis of the impact of these issues on the results. No significant impact was found; see Table S2 for further details.
Table 4.
Rmsd statistics for Astex single and multi sites
| Sitea | Size | Groupb | Sourcec | rmsd (Å) |
||||
|---|---|---|---|---|---|---|---|---|
| Min | Max | Mean | SD | Median | ||||
| Single (x) | 84 | SB | ASTEX PDB | 0.14 | 12.31 | 1.53 | 2.05 | 0.61 |
| Single (m) | 84 | SB | ASTEX PDB | 0.21 | 13.16 | 1.88 | 2.23 | 0.96 |
| Single (o) | 85 | SB | ASTEX SUP | 0.21 | 13.44 | 2.32 | 2.68 | 1.15 |
| Single (o) | 85 | RU | ASTEX SUP | 0.24 | 15.25 | 2.29 | 2.72 | 1.03 |
| Multi (o) | 147 | SB | ASTEX SUP | 0.21 | 13.44 | 2.23 | 2.64 | 1.03 |
| Multi (o) | 147 | RU | ASTEX SUP | 0.24 | 15.25 | 2.16 | 2.61 | 0.92 |
Single = first site in pdb, multi = all viable sites, single (x) = crystallographic starting ligand coordinates, single (m) = minimized starting ligand coordinates, single (o) and multi (o) = organizer supplied ligand coordinates
SB Stony Brook Group, RU Rutgers University Group
ASTEX PDB = built from PDB, ASTEX SUP = built from supplied structures
Rmsd spectrum plots
As an alternative way to characterize docking outcomes, not based solely on a hard 2.0 Å cutoff, spectrum plots for symmetry corrected rmsds are shown for SB/ASTEX PDB (single site, Fig. 6a) and SB/ASTEX SUP (multi site, Fig. 6b) results. These indicate docking outcome (docking success = blue, sampling failures = red, scoring failures = green) as a function of a variable rmsd cutoff used to define success. For each rmsd cutoff, the percentage of all three possible outcomes adds up to 100 %. This kind of analysis allows the decoupling of sampling and scoring failures in pose reproduction studies. At an rmsd cutoff of 0 Å, all poses produced are determined as sampling failures, and this rapidly decreases as the cutoff approaches more generous and realistic values. For the SB ASTEX/PDB set, at the 1 Å mark 62 % rmsd success rates are reached, compared to 76 % success rates reached at the 2 Å mark. Interestingly, for the SB ASTEX/SUP set, at the 1 Å mark 49 % rmsd success rates are reached, compared to 68 % success rates reached at the 2 Å mark. This discrepancy probably arises from the use of unbiased starting coordinates in the ASTEX SUP, where bond angles and torsions are reconstructed from scratch using 2D structures. Overall, based on the general shape of the spectrum plots, and the point at which docking success, sampling failures and scoring failures all begin to intersect, a 2 Å definition of success appears to be reasonable.
Fig. 6.

Rmsd spectrum plots for a SB/ASTEX PDB and b SB/ASTEX SUP runs showing Blue = Pose Reproduction Success, Red = Sampling Failure, Green = Scoring Failure
The effect of clustering
For reporting sampling success, the organizers requested that the ensemble size be at most 32 docked poses. An upper limit of 5,000 was selected for ensemble size, with clustering via either a 1 Å cutoff (RU) or a 2 Å cutoff (SB) and the 32 best-scored poses were retained for reporting sampling success. As shown in Table 5 for the RU/ASTEX SUP runs, sampling success for the ensemble of 32 extracted from the clustered pool of poses is substantially better (83.5 %) than that without clustering (78.8 %) and begins to approach the maximum ensemble size (89.4 %). Over all systems, the actual maximum ensemble sizes were in the range of 40 to 2,361, with a mean of 430 and a median of 251 for the ASTEX SUP/RU set. It is important to note that the clustering cutoff and the number of poses retained affect the pose diversity of the ensemble available for rescoring and that clustering in DOCK is performed with standard rmsd calculations.
Table 5.
Pose reproduction statistics for different clustering protocols using grid scoring for the single site set (N = 85)
| Ensemble sizea | Clustering cutoff | Rmsd (Å) |
Sampling success (%) | ||||
|---|---|---|---|---|---|---|---|
| Min | Max | Mean | SD | Median | |||
| 32 | None | 0.16 | 8.54 | 1.36 | 1.59 | 0.68 | 78.8 |
| 32 | 1 Å | 0.21 | 8.50 | 1.23 | 1.41 | 0.72 | 83.5 |
| All | None | 0.16 | 6.38 | 0.97 | 0.98 | 0.61 | 89.4 |
| All | 1 Å | 0.21 | 6.38 | 1.03 | 0.96 | 0.68 | 88.2 |
Data from RU/ASTEX SUP docking runs. Single site set
Rescoring with Amber score
Poses sampled using grid score were subsequently rescored with three variants of Amber score (see Fig. 4) to determine if a potentially more accurate function could be used to identify native-like poses (Table 6). Here, the three Amber score types are distinguished by their allowable movable regions, (nothing, ligand, and distance) sampled during the energy minimizations and molecular dynamics. Thus, the sequence of movable regions in Table 6 from nothing → ligand → distance is one of increasing flexibility, and a simple paradigm for Amber rescoring is that movability enables relaxation with respect to the Amber force field and ultimately accommodation of the induced fit binding model. For each site an ensemble of ligand poses (Table 5, ensemble size all and clustering cutoff none) were obtained via the RU/ASTEX SUP protocol from the DOCK run with the default random seed of 0. As a baseline, grid score yields a docking success of 68.0 % and a sampling success of 89.4 % for this run. Here, sampling success provides a soft upper limit, since additional sampling is being performed. Docking successes are included in Table 6 for rmsd (sym) but not drmsd (see Theoretical methods) because a 2 Å cutoff is not appropriate for this metric.
Table 6.
Astex pose reproduction statistics of the rmsds of the top scored poses
| Score | Sourcea | Rmsd (Å) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Type | Min | Max | Mean | SD | Median | Success (%) | |||
| a | Grid | ASTEX SUP | sym | 0.24 | 15.25 | 2.16 | 2.61 | 0.92 | 68.0 |
| b | Nothing | ASTEX AMB | sym | 0.21 | 18.46 | 3.26 | 4.41 | 1.00 | 67.3 |
| c | Ligand | ASTEX AMB | sym | 0.18 | 17.21 | 2.88 | 4.06 | 1.07 | 69.4 |
| d | Distance | ASTEX AMB | sym | 0.15 | 39.18 | 4.26 | 5.49 | 1.46 | 56.5 |
| e | Grid | ASTEX SUP | drmsd | 0.04 | 2.30 | 0.41 | 0.36 | 0.32 | |
| f | Nothing | ASTEX AMB | drmsd | 0.04 | 2.97 | 0.45 | 0.44 | 0.35 | |
| g | Ligand | ASTEX AMB | drmsd | 0.02 | 3.11 | 0.48 | 0.50 | 0.30 | |
| h | Distance | ASTEX AMB | drmsd | 0.04 | 2.45 | 0.49 | 0.46 | 0.31 | |
Data from RU/ASTEX SUP docking runs and RU/ASTEX AMB rescoring runs. Multi site set (N = 147)
Nothing movable protocol
Rescoring with Amber score nothing movable yields a docking success of 67.3 % (Table 6b), which is close to grid score’s 68.0 % (Table 6a). This is a strict single point rescoring: no ligand flexibility and no minimization or additional sampling is performed. Here, 16 binding sites are rescued by rescoring and 17 sites that are successes for grid score are failures for rescoring. This indicates that the two scoring functions (grid vs. Amber score nothing movable) have system dependent strengths and weaknesses which should be explored. The 1.00 Å median observed here is almost equal to the 0.92 Å in grid score, but the 3.26 Å mean is substantially different from the 2.16 Å in grid score, indicating that some docking failures for the rescoring are more severe (see Supplementary Material for a comparison with PB/SA rescoring, Tables S3–S4).
Ligand movable protocol
Rescoring with Amber score ligand movable (Table 6c) yields a docking success of 69.4 % which is slightly better than grid score’s 68.0 % (Table 6a). However, grid score has a somewhat narrower distribution of top scored rmsds; for example, the mean and median are 2.16 and 0.92 Å for grid score but 2.88 and 1.07 Å for Amber score ligand movable. This is a flexible ligand rescoring, but the sampling successes are equal because 1yvf and 2br1 are successes for ligand movable but failures for grid score and 1w1p sites 1 and 2 are failures for ligand movable but successes for grid score. With regards to docking success, 19 binding sites are rescued by rescoring, and 17 different sites that are successes for grid score are failures for rescoring. Again, the two functions (grid vs. ligand movable) correctly rank different systems from among the 147 total. In comparison with nothing movable (Table 6b), there are 8 successes that are failures with ligand movable, and 11 nothing movable failures that are successes with ligand movable. The mean rmsd for ligand movable (2.88 Å) is somewhat smaller than that for the nothing movable protocol (3.26 Å) although there is a slight increase in median rmsd (1.07 Å from 1.00 Å respectively). Thus, ligand flexibility reduces the severity of some of the nothing movable docking failures, but the distribution of top scored poses shifts to higher rmsds.
Distance movable protocol
For the distance movable region option, the receptor-ligand distance threshold was initially set to 2 Å. However, this was insufficient to guarantee at least one movable receptor residue for every binding site: 26 sites required a 2.1 Å threshold, five required 2.2 Å, and one required 2.3 Å. Table 7 contains various statistics for the numbers of receptor residues showing that for the final thresholds used at least one residue (Min column) and at most seven residues (Max column) were movable. The large maximum value of 2,778 is from 1of6 which is composed of eight chains and has eight binding sites. The numbers of movable residues are small compared to the total numbers of residues. An alternative subset, termed computationally inexpensive subset (defined below), with an extended distance movable cutoff (3 Å) was also examined.
Table 7.
Astex pose reproduction statistics of the numbers of receptor residues for the multi site set and a computationally inexpensive subset of nine binding sites
| Residue cutoff | Count | Min | Max | Mean | SD | Median | Mode |
|---|---|---|---|---|---|---|---|
| ~2 Å movablea | 147 | 1 | 7 | 2.6 | 1.6 | 2 | 1 |
| All residuesa | 147 | 131 | 2,778 | 822 | 615 | 622 | 2,778 |
| ~2 Å movableb | 9 | 1 | 6 | 1.8 | 1.6 | 1 | 1 |
| 3 Å movableb | 9 | 28 | 54 | 41.9 | 7.7 | 42 | None |
| All residuesb | 9 | 131 | 515 | 350 | 117 | 344 | None |
All sites in the set
Computationally inexpensive subset (see text for definition)
Amber score distance movable, with a docking success of 56.5 % (Table 6d), performs poorly compared to the other Amber scores as well as to grid score. Because the intent of the small distance threshold (2–3 Å) is to enable a relaxation of the few residues close to the binding site, the following analysis focuses on comparing the distance movable results to the ligand movable results. Interestingly the maximum rmsd for this protocol goes from 15.25 to 39.18 Å (Table 6a vs. d) which is an indication that some poses within the distance movable ensemble have drastically changed from their original starting coordinates. With regards to sampling success, there is little difference: 1mzc and 1t9b are failures for distance but successes for ligand. On the other hand, for docking success 5 sites are successes for distance movable but failures for ligand movable and 24 sites are failures for distance but successes for ligand. The distribution of top scored poses shifts to much higher rmsds: the mean and median for distance movable are 4.26 and 1.46 Å respectively compared to 2.88 and 1.07 Å for ligand movable (Table 6). These poorer docking successes (Table 6d) and rmsd statistics are the opposite of what would be expected for a more rigorous methodology for estimating scoring and may indicate that the current relaxation models are overly simplistic.
One possible explanation is that there is significant movement of the movable receptor residues, which necessarily affects the binding site. In that case the coordinate-based rmsds of the ligands might be unfair measures of binding pose reproduction because a given ligand might need to translate or rotate to follow a deformable active site. To remove rigid body effects and examine how different Amber score protocols might influence just the internal ligand geometries, drmsd values (see Theoretical methods) were also computed as shown in Table 6e–h. Interestingly, despite being the protocol potentially allowing the largest movement overall (including greater ligand variation), Amber score distance movable yields the second smallest drmsd range (0.04–2.45 Å). Further, the mean drmsd of 0.49 Å for distance movable, although the largest, is similar to those of the other Amber score methods. Thus, the distance movable protocol does not show a substantially larger variation in ligand internal geometry, and consequently, rigid body motion is the most likely cause of the lower success rates (56.5 %). On the other hand, the fact that grid score has the smallest drmsd range (0.04–2.30 Å) and the smallest mean (0.41 Å) indicates that the different allowable movable region protocols have more severe docking failures.
Previous work on RNA-ligand complexes [16] studied the effect of systematically increasing the distance threshold; however, no overall improvement in docking success was found. To examine how increasing site flexibility would affect the present results, calculations for a subset of binding sites (N = 9) using an extended distance threshold of 3 Å were performed (Table 8). Termed the computationally inexpensive subset, this refers to the nine systems (1n2v, 1oq5, 1pmn, 1tow, 1tt1, 1uou, 1x8x, 1y6b, 1yqy) with the fastest runtimes from the 2 Å experiments. Table 7 indicates that a 3 Å threshold includes at least 28 residues and at most 54 residues in the movable region. However, despite the increased size of the movable region the success rates become worse (Table 8). This is a challenging subset for docking success; only four of nine are successful for ligand movable, three for 2 Å distance movable, and only two for 3 Å distance movable. Note that the sampling success is the same for all three scores. Structure 1oq5 looks like it might be rescued by a further increase in the movable region because its rmsd drops from 14.24 to 2.67 Å when the threshold increases from 2 to 3 Å, but an additional run using a threshold of 4 Å yielded a worse top scored rmsd of 3.89 Å. This behavior is typical of that seen in the previous work [16].
Table 8.
Astex pose reproduction rmsds of the top scored poses for a computationally inexpensive subset of nine systems
| System | rmsd (Å) |
||
|---|---|---|---|
| Ligand | Distance 2 Å | Distance 3 Å | |
| 1n2v | 1.44 | 1.49 | 10.92 |
| 1oq5 | 13.27 | 14.24 | 2.67 |
| 1pmn | 14.64 | 7.34 | 9.39 |
| 1tow | 1.10 | 0.62 | 1.28 |
| 1tt1 | 0.89 | 0.89 | 1.07 |
| 1uou | 14.72 | 14.16 | 16.46 |
| 1x8x | 0.29 | 10.46 | 12.70 |
| 1y6b | 2.79 | 2.98 | 6.07 |
| 1yqy | 12.98 | 11.13 | 11.46 |
Although the ligand movable results (69.4 %) are on par with the baseline grid score results (68.0), surprisingly, none of the more sophisticated Amber rescoring methods yield significantly enhanced success rates (Table 6). No single explanation is apparent but there is a long list of potential contributors including: (1) initial coordinates that have not been relaxed and equilibrated with the Amber force field, (2) the very short minimization/MD/minimization protocol applied in this study, (3) the strengths and weaknesses of the particular GB solvation model used, and (4) more fundamental issues, such as, the use of an implicit solvent approach, the neglect of entropic terms, and even the use of an empirically derived Hamiltonian. The good news is that this benchmarking study opens the door to detailed investigations of these sources of error.
Rescoring with LMOD score
At this time, the high computational expense associated with the LMOD procedure precluded application to all of the 85 systems (and their associate docked ensembles) studied in this manuscript. To keep costs tractable, LMOD was used to rescore only a subset of systems (N = 19, ~32 poses each) from the Amber score nothing moveable protocol (see Fig. 4). Ideally, all 85 systems with their complete ensemble of poses should be evaluated. Although LMOD score does not depend on Amber score and could be used to rescore results from grid score directly, the structures from Amber score nothing movable were used for two reasons: (1) like Amber score, LMOD score requires Amber-type topology and coordinates, so using those already generated for Amber score cut down on preparation time, and (2) since Amber score nothing movable does not allow movement of the receptor or ligand, the starting structures are the same as those used in grid score, so that LMOD score is rescoring of grid score, not Amber score. The systems chosen for LMOD rescoring were pdb codes 1gm8, 1ke5, 1l2s, 1nv2, 1oq5, 1pmn, 1sj0, 1tow, 1tt1, 1uml, 1uou, 1v0p, 1v48, 1x8x, 1xoz, 1y6b, 1yqy, 1ywr, and 1z95 (systems picked at random, see supplemental material). For each system the top ten scored poses, the top ten lowest ligand rmsd poses, and additional poses with higher rmsds were included to ensure the results would not be too biased towards low-rmsd structures. The number of initial poses ranged from 23 to 40 (average 32) as shown in Table S5. It should be noted that for some systems, some poses in the ensemble are identical given that clustering was not used during the docking. Table 9 gives an overview of scoring and sampling successes of LMOD score in comparison to Amber score and grid score. For comparison purposes, the same initial set of starting poses was used in all experiments.
Table 9.
RMSD of the top scored ligand pose and number of poses with rmsd less than 2.0 Å for grid, Amber, and LMOD score for select poses from a subset of 19 systems, along with overall scoring and sampling success percentages
| System | N poses | Top scored pose rmsd (Å)a |
No. of poses with rmsd < 2 Åb |
||||
|---|---|---|---|---|---|---|---|
| Grid | Amber | LMOD | Grid | Amber | LMOD | ||
| 1gm8 | 26 | 3.02 | 3.16 | 2.96 | 4 | 4 | 1 |
| 1ke5 | 34 | 0.41 | 0.34 | 0.49 | 15 | 15 | 16 |
| 1l2s | 33 | 0.74 | 0.74 | 1.33 | 13 | 13 | 15 |
| 1n2v | 26 | 2.19 | 1.91 | 2.75 | 15 | 15 | 7 |
| 1oq5 | 40 | 3.93 | 14.60 | 3.87 | 10 | 5 | 5 |
| 1pmn | 30 | 7.77 | 15.25 | 4.45 | 0 | 0 | 0 |
| 1sj0 | 23 | 11.67 | 11.67 | 0.75 | 0 | 0 | 2 |
| 1tow | 25 | 0.65 | 0.65 | 6.35 | 5 | 5 | 5 |
| 1tt1 | 34 | 1.03 | 0.51 | 0.75 | 15 | 15 | 15 |
| 1uml | 30 | 2.04 | 12.48 | 1.41 | 4 | 4 | 4 |
| 1uou | 40 | 2.56 | 15.09 | 0.79 | 10 | 6 | 18 |
| 1v0p | 23 | 0.74 | 1.55 | 1.57 | 19 | 19 | 19 |
| 1v48 | 40 | 0.92 | 0.95 | 0.47 | 20 | 20 | 22 |
| 1x8x | 34 | 0.64 | 0.64 | 3.72 | 18 | 18 | 22 |
| 1xoz | 30 | 6.99 | 6.60 | 0.24 | 4 | 4 | 4 |
| 1y6b | 30 | 2.75 | 3.07 | 2.76 | 0 | 0 | 0 |
| 1yqy | 31 | 0.47 | 13.40 | 3.84 | 10 | 10 | 10 |
| 1ywr | 36 | 2.09 | 0.51 | 0.69 | 18 | 18 | 22 |
| 1z95 | 40 | 0.40 | 0.48 | 0.27 | 20 | 20 | 23 |
| No. of success | 9 | 10 | 11 | 16 | 16 | 17 | |
| % success | 47.37 | 52.63 | 57.89 | 84.21 | 84.21 | 89.47 | |
Rmsd < 2.0 Å indicates docking success
Number of poses in the top 32 with rmsd < 2.0 Å (>0 indicates sampling success). All results from RU/ASTEX SUP docking (grid) runs and RU/ASTEX AMB rescoring (Amber, LMOD) runs. Amber score used nothing movable protocol
Encouragingly, for this dataset, LMOD (scoring = 57.89 %, sampling = 89.47 %) outperforms both Amber score nothing movable (scoring = 52.63 %, sampling = 84.21 %) and grid score (scoring = 47.37 %, sampling = 84.21 %) in terms of both scoring and sampling success (Table 9). And, the average rmsd of the top scored pose from LMOD score (2.08 Å) is lower than either grid score (2.68 Å) or Amber score (5.45 Å). In particular, for 1sj0 use of LMOD leads to both sampling and docking success (Table 9) where Amber score and grid score do not (see discussion below). In addition, LMOD achieves a docking success for 1xoz, 1uml and 1uou. There are only four cases in which Amber score and/or grid score achieved docking success where LMOD did not (1n2v, 1tow, 1x8x, and 1yqy). For 1n2v and 1tow Amber score was successful, for 1yqy grid score was successful, and for 1x8x both Amber and grid score were successful. An additional important observation is the fact that LMOD is often able to find a lower energy conformation beyond the initial energy minimization (100 steps of conjugate gradient). Across all poses from all structures (607 total), LMOD sampling was able to find a lower energy conformation than the starting pose 72 % of the time. The median individual LMOD success rate (# of poses with LMOD success for system/# poses evaluated for system) among all systems was 77 %.
As noted above, LMOD outperforms Amber score and grid score for 1sj0 in both sampling and scoring. Neither Amber score nor grid score were able to locate a pose with an rmsd of less than 2.0 Å which illustrates the potential superior sampling power of the LMOD method. Figure 7 makes the point graphically, showing ligand rmsd (Å) plotted versus score (kcal/mol) for each pose evaluated using all the functions. Despite none of the starting poses having an rmsd < 2.0 Å, LMOD not only finds two poses with low rmsd (0.75 and 0.81 Å) but also ranks them correctly as shown in Fig. 7 (black to red circles connected by purple lines). In particular, LMOD resulted in a range of low energy conformational space being sampled (ca –55 kcal/mol and 0.75 to 2.3 Å rmsd) although the poses originated from a group of three identical poses (ca +45 kcal/mol and 3 Å rmsd). In general, after rescoring with LMOD score the lower rmsd poses obtain better scores, while the high rmsd poses are either scored very poorly or do not have their score change that much.
Fig. 7.

Score (kcal/mol) versus ligand heavy-atom rmsd (Å) for the subset of poses of system 1sj0 for which LMOD score was run; Amber score (nothing), LMOD score, and grid score values are shown. Green lines indicate transitions from Amber score poses (black circles) to LMOD poses (red circles). Purple lines indicate 3 transitions for which 2 yielded results <2 Å
In some cases, a scoring failure can occur due to an outlier. For example, LMOD score has a docking failure for 1oq5 because the rmsd of the top pose is 3.87 Å (Table 9); however, the next top 4 scored poses all have rmsds less than 2.0 Å. In order to examine the effects of potential outliers on the results for each scoring method, the average ligand heavy-atom rmsds for the top five scored poses were calculated (Table 10). Here, docking failures using LMOD for systems 1yqy and 1x8x are likely caused by outliers, as the average of the top five scored rmsds are less than 2.0 Å for both. Overall, LMOD score generates lower average rmsds for top-ranked poses for 10 out of the 19 structures, in some cases significantly so (1oq5, 1sj0, 1uou, and 1xoz). Amber score generates lower average rmsds for 5 structures, and grid score generates lower average rmsds for 4 structures; these numbers are consistent with the docking success percentages reported in Table 9.
Table 10.
Average ligand rmsd (Å) and standard deviation for the top 5 scored poses according to grid, Amber, and LMOD scores
| Structure | Avg rmsd (Å) |
||
|---|---|---|---|
| Grid | Ambera | LMOD | |
| 1gm8 | 3.13 ± 0.09 | 3.11 ± 0.08 | 2.96 ± 0.00 |
| 1ke5 | 0.38 ± 0.04 | 0.34 ± 0.04 | 0.47 ± 0.04 |
| 1l2s | 1.05 ± 0.17 | 1.12 ± 0.20 | 1.32 ± 0.02 |
| 1n2v | 1.97 ± 0.15 | 1.61 ± 0.22 | 2.64 ± 0.22 |
| 1oq5 | 3.90 ± 0.05 | 15.19 ± 0.36 | 1.75 ± 1.06 |
| 1pmn | 7.43 ± 0.95 | 11.79 ± 2.49 | 7.09 ± 1.59 |
| 1sj0 | 11.80 ± 0.26 | 11.80 ± 0.26 | 1.79 ± 0.84 |
| 1tow | 2.56 ± 2.39 | 2.87 ± 2.72 | 5.94 ± 0.49 |
| 1tt1 | 0.86 ± 0.23 | 0.86 ± 0.23 | 0.70 ± 0.25 |
| 1uml | 1.88 ± 0.15 | 10.63 ± 2.13 | 2.91 ± 1.19 |
| 1uou | 2.57 ± 0.03 | 14.29 ± 0.41 | 0.88 ± 0.10 |
| 1v0p | 1.27 ± 0.44 | 1.38 ± 0.32 | 1.47 ± 0.32 |
| 1v48 | 0.75 ± 0.15 | 0.99 ± 0.06 | 0.71 ± 0.20 |
| 1x8x | 0.58 ± 0.11 | 0.52 ± 0.12 | 1.26 ± 1.23 |
| 1xoz | 7.09 ± 0.26 | 6.87 ± 0.62 | 1.36 ± 0.64 |
| 1y6b | 2.89 ± 0.14 | 2.97 ± 0.18 | 2.90 ± 0.13 |
| 1yqy | 0.50 ± 0.07 | 11.82 ± 1.41 | 1.65 ± 1.10 |
| 1ywr | 1.18 ± 0.72 | 0.52 ± 0.02 | 0.52 ± 0.13 |
| 1z95 | 0.40 ± 0.01 | 0.45 ± 0.03 | 0.30 ± 0.03 |
| Overall avg | 2.75 | 5.22 | 2.03 |
Amber score with nothing movable protocol
In general LMOD score yields top 5 average rmsds that are lower than or comparable to Amber and grid score; only in the case of 1tow does LMOD perform significantly worse than Amber score or grid score. The overall average improvement using LMOD score (2.03 Å) versus Amber score (5.22 Å) is likely due to more efficient sampling of the conformational space of the ligand/protein complex given that the underlying energy function is the same. The fact that LMOD tends to find low energy conformations beyond initial minimization combined with the fact that the ligand rmsd tends to show improvement shows that rescoring poses with LMOD can improve both scoring and sampling success. Although the high computational cost of LMOD currently prevents its use as a sole scoring function, based on the data in Tables 9, 10 and Fig. 7, the method shows great promise when used as a tool for rescoring.
Enrichment studies
Global enrichment statistics (total AUC)
Table 11 shows the overall global enrichment results (AUC values) for both the native and semi-random pairing using receptor structures derived from the protein databank (PDB) or supplied by the organizers (SUP) using actives and decoys from DUD or actives from WOMBAT. AUC values for individual systems are shown as bar plots in Fig. 8. The results for the native pairings yield, in the case of the DUD PDB runs (Fig. 8a; Table 11), individual AUC values which range from 0.29 (bad enrichment) to 0.96 (good enrichment). For the DUD SUP results the max AUC value at 0.90 is not as high (Fig. 8b; Table 11). Interestingly, both receptor preps using DUD actives and decoys yield nearly identical average AUC values of 0.60 and 0.59. This is a somewhat surprising result as the different preps use different partial charge assignments (FF99SB [46] vs. Gasteiger [47]), hydrogen orientation, and protonation states of the receptor (as discussed further below). For the WOMBAT results, average enrichment for the native pairings is worse than random (DUD PDB = 0.45, DUD SUP = 0.42). And, min AUC values are significantly lower (0.13 and 0.16) than the corresponding DUD values (0.29 and 0.21). This is likely a function of the fact that two of the WOMBAT ROC curves (pde5 and ppar) perform significantly worse than random, a phenomenon not generally seen in the other ROC curves, as is discussed in more detail in the subsequent subsection (see ROC Curve Analysis). In addition, the much smaller WOMBAT dataset size (~10) vs. DUD (~40) exacerbates this difference.
Table 11.
Global enrichment (total AUC) for native and semi-random pairings
| AUCa | Native pairing |
Semi-random pairing |
||||||
|---|---|---|---|---|---|---|---|---|
| DUD lig |
WOMBAT lig |
DUD lig |
WOMBAT lig |
|||||
| PDB | SUP | PDB | SUP | PDB | SUP | PDB | SUP | |
| Avg | 0.60 | 0.59 | 0.42 | 0.45 | 0.48 | 0.46 | 0.39 | 0.40 |
| SD | 0.17 | 0.17 | 0.17 | 0.16 | 0.18 | 0.17 | 0.15 | 0.15 |
| Median | 0.56 | 0.56 | 0.41 | 0.50 | 0.48 | 0.46 | 0.41 | 0.44 |
| Max | 0.96 | 0.90 | 0.60 | 0.61 | 0.84 | 0.85 | 0.61 | 0.62 |
| Min | 0.29 | 0.21 | 0.13 | 0.16 | 0.15 | 0.10 | 0.08 | 0.09 |
Total AUC calculated using FPR and TPR [0, 1]
Fig. 8.
Bar plots showing AUC values using (a) DUD PDB native, (b) DUD SUP native, (c) DUD PDB semi-random, and (d) DUD SUP semirandom protocols. Results sorted from high to low using ordering from panel (a)
As expected, results from semi-random experiments (Table 11; Fig. 8c, d) show average AUC values which are consistently worse than random (avg AUC < 0.50). However, as discussed further below, for many semi-random pairings good cross-enrichment is observed, especially for receptors within the same protein family. This is reflected in the relatively high max AUC values which are in the range 0.84–0.85 (Table 11) although they are lower than the native pairing (0.90–0.96). For DUD actives, average AUC drops from the higher 0.59–0.60 for the native pairing to 0.46–0.48 for the semi-random pairing. However, for WOMBAT actives, average AUCs are only marginally higher for the native pairing (0.42–0.45), compared to the semi-random pairing (0.39–0.40). This could be due to the fact that 7/10 of WOMBAT semi-random pairings are between proteins in the same family. As with the native pairings, there is striking agreement for the semi-random set, between DUD PDB and DUD SUP results, despite differences in the two different structure preparations.
Individual AUC values and group-based analysis (heatmaps)
Figure 8 shows bar plots of individual AUC values, comprising the total values in Table 11, for all pairings for both structure preparations. Here, the plots are sorted by descending total AUC using the DUD PDB native pairing results. Figure 9 shows a matrix representation of the data (heatmap) for the DUD PDB results which facilitates, for example, determining if a related group (or family) yields good or poor enrichment. Similar results are seen for the DUD SUP runs (Figure S3). In Fig. 9, receptors are labeled according to the pdb code (and group) on the y-axis and ligands are labeled according to the DUD system name on the x-axis. The entries in Fig. 9 indicate native pairings (diagonal elements) and semi-random pairings (off diagonal elements). Red = worse than random (AUC < 0.5), green = greater than or equal to random (AUC ≥ 0.5), and white = non-paired systems. See Supplementary Material (Table S8) for a detailed breakdown of systems which fall into the different categories (better, equal, or worse than random). Figure 9 groupings highlight the fact that many of the so called semi-random pairings are in fact not random but are pairings between related protein structures. Thus, for many semi-random pairings, good enrichment would not be unexpected. Exceptions would likely include the “other enzyme” group, and to a lesser extent the metalloenzyme group. Ideally, experiments using all receptors paired with all active-decoys sets should be performed (entire matrix) to derive better statistics.
Fig. 9.

Heatmaps showing enrichment study which employ SB/DUD PDB results and DUD actives and decoys. Receptors are labeled by the pdb code and family on the y axis. Ligands are labeled by the DUD system name on the x axis. Red = worse than random (AUC <0.5), green = greater than or equal to random (AUC ≥ 0.5), and white = non-paired systems
As evident by the median value of 0.56 (27/38 have AUC ≥ 0.5) for the DUD PDB results in Table 11, most systems yield better than random enrichment (green squares) for the native pairings occurring on the diagonal: metalloenzymes = 3/4, nuclear hormone receptor = 4/7, kinase = 4/9, folate enzyme = 2/2, serine protease = 3/3, other enzymes = 11/13. For the analogous off-diagonal experiments the median AUC value 0.48 (15/38 have AUC ≥ 0.5) indicates a roughly even split between good and bad enrichment: metalloenzymes = 3/4, nuclear hormone receptor = 3/7, kinase = 2/8, folate enzyme = 1/2, serine protease = 2/2, other enzymes = 1/11, misc pairings = 3/4. Importantly, off-diagonal elements for the “other enzymes” group, consisting of mostly unrelated proteins, yields poor enrichment which is to be expected. This group is probably the most useful overall as an actual null hypothesis test set for evaluating virtual screening.
Of all the groups evaluated, the serine protease group yields the best overall enrichment (5/5) for the five pairings evaluated (3 diagonal, 2 off-diagonal). Cross-enrichment was also observed for serine proteases in the original DUD manuscript (Table 2 in Ref [9]). This is not surprising given that trypsin, factor Xa and thrombin are very similar proteins. In fact, trypsin has been successfully used as a template for developing thrombin and factor Xa inhibitors [48, 49]. Interestingly, while the trypsin receptor (1bju) with HIV protease actives (hivpr) shows no enrichment (1bju-hivpr pairing, Figs. 8c, d, 9) the use of the HIV protease receptor (1hpx) with trypsin actives (trypsin) does show enrichment (1hpx-trypsin pairing, Figs. 8c, d, 9).
In contrast to serine protease, the kinase group shows poor native and semi-random enrichment. An interesting observation is that epidermal growth factor receptor (egfr) ligands show enrichment with the heat shock protein (1uy6) receptor while the converse pairing (1m17-hsp90) is worse than random enrichment for both preparations (Figs. 8a–d, 9, S4). However, since the 1uy6-hsp90 native pairing also had sub-random enrichment (Figs. 8a, b, 9) this result may not be unexpected if the set of actives and decoys dominate enrichment behavior as discussed below. Surprisingly, when thymidine kinase is paired with purine nucleoside phosphorylase actives, and vice versa, (1kim-pnp and 1b8o-tk pairings, Figs. 8c, d, 9), these two unrelated systems enrich one another. The authors of the original DUD paper also noted that these two enzymes yielded cross-enrichment (Table 2 in ref [9]) in addition to thymidine kinase being a promiscuous target.
System-specific analysis: DUD PDB versus DUD SUP preparations
Although the overall average AUC (0.59, 0.60) for native pairings is essentially the same, examination of the individual results in Fig. 8a, b reveals that there can be differences in enrichment depending on which structure preparation (DUD PDB vs. DUD SUP) is used. The DUD PDB prep starts with raw pdb files from scratch (see Theoretical methods) while the DUD SUP structures, with the exception of hydrogens added by the organizers, were from the original DUD database [9]. Examples include the good enrichment seen for system 1a8i but only when using the DUD SUP receptor prep (Fig. 8a vs. b, 0.43 vs. 0.75). Examination of the glycogen phosphorylase receptor in this system show that the DUD SUP prep contains active site waters in the binding site (Fig. 10b) which are absent in the DUD PDB prep. The known actives may use the water-mediated interactions to their advantage although a more detailed study should be performed to determine the actual importance. For certain systems, binding site water is known to play critically important roles. Another, more subtle example is shown in Fig. 10a for 1hw8. Here, the native pairings appear to favor the DUD PDB receptor prep (Fig. 8a vs. b, 0.42 vs. 0.21) although the average AUC values are both below random. The hmg coa reductase receptor in this system has four chains, labeled A-D in the original pdb structure, with four occupied binding sites. For the DUD PDB prep, the binding site used was at the interface of chains A-B in contrast to the DUD SUP prep which was at the interface of chains C-D. Differences between the two sites (see Fig. 10a) involve a conformational change of methionine and alternate orientations of two cysteine thiol hydrogens which appear to favor the DUD PDB prep. These structural differences are likely a result of a nearby co-factor originally present in the C-D site.
Fig. 10.

Differences between DUD PDB (red) and DUD SUP (green) structure preps for (a) 1a8i and (b) 1hw8. Native ligands are shown in cyan. Binding site waters in the DUD SUP prep for 1a8i indicated by blue circles
Differences are also observed for systems 1f0r and 1a4g. For factor Xa (1f0r), protonation state differences of a histidine near the binding site could influence the computed enrichments (DUD SUP = 0.78, DUD PDB = 0.62). For neuraminidase (1a4g), electrostatics are known to be especially important [50], thus the use of dramatically different charge models (F99SB [46] vs. Gasteiger [47]) would likely influence the results (DUD PDB = 0.85, DUD SUP = 0.59). Finally, although differences in a binding site environment would normally be expected to affect enrichment, for some systems this is not always observed. For example, for catechol o-methyl transferase in 1h1d, high enrichment is observed using both preps (AUC 0.85, 0.87) despite the fact the DUD SUP prep is missing part of the co-factor (S-adenyl methionine, SAM) included in the DUD PDB prep. Overall, the results highlight how use of different receptor sites or structure preparations, and by analogy alternative crystallographic coordinates of the same receptor can influence enrichment. Similar results were observed in the multiple binding site breakdown presented in Table S2.
ROC curve analysis
Figures 11a shows standard ROC curves for the native DUD pairings (see Figure S4 for semi-random pairings). The curves are sorted from high to low according to total AUC using SB/DUD PDB data with SB/DUD PDB shown in black and SB/DUD SUP shown in gray. These comparisons allow, at a glance, which systems yield overall good, reasonable, poor, or early enrichments. Most of the curves have strikingly similar shapes despite the fact that two different structure preps were evaluated. In contrast, 1hw8 and 1a8i show dramatically different shaped ROC plots which ultimately lead to the larger differences discussed above. Systems with poor overall enrichment may still have good early enrichment which is characterized by relatively steep upward sloping curves starting on the lower left hand corner of the ROC plots which then decrease as more and more of the database is covered. Although the left part of the curve will be above the random line the right part of the curve may approach the random (1m17, 1kv2) or even dip below (1hpx, 1o86). This phenomena, interestingly, seems especially true for kinases 2src, 1vr2, 1xp0, and 1agw. Indirectly, the inherent plasticity of kinase binding sites could adversely affect enrichment due to known induced fit effects which can be ligand dependent. For example, the few actives which might favor a particular kinase conformation could be scored favorably but in absence of receptor flexibility the remaining pool of actives might not yield favorable scores thus accounting for the observed early, but not global, enrichment. Other systems with short lasting early enrichment include MODL (based on a homology model) and 1sr7. For DUD vs. WOMBAT native pairings two systems in particular, 1xp0 and 1fm9, stand out as having significantly different ROC curves (Fig. 11b). The ROC curve for 1fm9 shows good enrichment behavior when using DUD but a sub-random curve with WOMBAT. For 1xp0, although the overall DUD results are not much better than random, the WOMBAT results are always sub-random. Additional analysis would be required to determine the cause of these enrichment differences.
Fig. 11.
a ROC curves for the 40 DUD families. b ROC curves for the 10 Wombat families (Wombat ligands + DUD decoys). ROC curves sorted from high to low according to total AUC using SB/DUD PDB data with SB/DUD PDB in black and SB/DUD SUP in gray
Comparing native versus semi-random
An interesting observation is that a given ligand set (actives + decoys) can yield similarly shaped ROC curves in two receptors suggesting that shape, in some instances, can be driven by properties inherent in the ligand set. Two examples of this phenomenon are the kinase pair egfr and hsp90 (Fig. 12a) and the serine proteases pair fxa and thrombin (Fig. 12b). The egfr ligand set shows strong early enrichment with both receptors. Conversely, the hsp90 ligand set yields curves that are slightly below random. As previously mentioned, fxa and thrombin have strong enrichment with the DUD SUP performing better than DUD PDB for all four combinations. On the other hand, this observation does not appear to hold when the receptors are unrelated. As an example, hivpr (1hpx) more strongly enriches the trypsin ligands than its cognate ligands (Fig. 12c). Conversely, the trypsin receptor (1bju) performs worse than random for the hivpr ligands as noted above in the heatmap discussion. More studies are needed to explore these issues in greater detail. For completeness, Figure S4 contains ROC curves for all semi-random pairings and may be used to aid in making additional comparisons.
Fig. 12.
ROC curves for 3 pairs of DUD families a egfr and hsp90, b fxa and thrombin, c trypsin and hivpr. Top panels show the native pairing. Bottom panels show the semi-random pairing
Early enrichment
Table 12 lists average early enrichment statistics, in terms of 0.1, 1.0, and 2.0 % of the database examined, using several metrics including %TPR, %FPR, %AUC, and FE (see Theoretical methods for definitions). Which is the best metric for quantifying early enrichment is still an active area of research. Here, to help gauge enrichment, values for the expected random and best possible cases are also reported. Results from the 0.1 % bracket are reported at the request of the organizers but in some cases may be too small to be meaningful considering the small number of molecules contained in some DUD datasets. Analysis presented below is focused on results obtained using DUD at 1.0 and 2.0 % of the database. WOMBAT data is provided for completeness.
Table 12.
Averaged early enrichment at 0.1, 1.0, and 2.0 % of the databases examined
| Database (%) | Metrica | Bestb | Ranc | Native pairing |
Semi-random pairing |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| DUD |
WOMBAT |
DUD |
WOMBAT |
||||||||
| PDB | SUP | PDB | SUP | PDB | SUP | PDB | SUP | ||||
| 0.1 | %TPR | 100 | 0.1 | 2.57 | 2.34 | 2.59 | 2.29 | 1.90 | 1.40 | 2.90 | 2.36 |
| 0.1 | %FPR | 0 | 0.1 | 0.03 | 0.03 | 0.06 | 0.06 | 0.04 | 0.06 | 0.05 | 0.06 |
| 0.1 | %AUC | 10 | 0.005 | 0.01 | 0.01 | 0.10 | 0.07 | 0.01 | 0.00 | 0.07 | 0.06 |
| 0.1 | FE | 2,000 | 1.0 | 2.00 | 2.88 | 19.51 | 13.50 | 1.73 | 0.83 | 13.00 | 11.79 |
| 1.0 | %TPR | 100 | 1.0 | 15.14 | 13.02 | 9.69 | 9.29 | 8.58 | 7.71 | 8.43 | 7.65 |
| 1.0 | %FPR | 0 | 1.0 | 0.56 | 0.62 | 0.78 | 0.75 | 0.77 | 0.79 | 0.76 | 0.82 |
| 1.0 | %AUC | 100 | 0.5 | 4.99 | 4.17 | 5.14 | 4.49 | 3.53 | 3.17 | 4.10 | 3.91 |
| 1.0 | FE | 200 | 1.0 | 9.99 | 8.34 | 10.29 | 8.97 | 7.06 | 6.35 | 8.20 | 7.83 |
| 2.0 | %TPR | 100 | 2.0 | 20.40 | 17.33 | 11.86 | 12.38 | 11.47 | 10.68 | 10.65 | 9.98 |
| 2.0 | %FPR | 0 | 2.0 | 1.43 | 1.52 | 1.67 | 1.66 | 1.71 | 1.71 | 1.69 | 1.77 |
| 2.0 | %AUC | 200 | 2.0 | 20.19 | 17.45 | 14.26 | 13.74 | 12.49 | 11.63 | 13.12 | 12.46 |
| 2.0 | FE | 100 | 1.0 | 10.09 | 8.72 | 7.13 | 6.87 | 6.24 | 5.82 | 6.56 | 6.23 |
Metrics for %TPR and %FPR indicate percentages on [0,100] while %AUC is on [0, 10,000] calculated using %FPR and %TPR, FE is unitless
Best best possible enrichment
Ran random enrichment. Best and Ran values are estimations used for comparison purposes
Importantly, average early enrichment is always significantly better than the hypothetical random results. For example, comparison of FE values using DUD PDB native pairings (Table 12) yields: 9.99 (1.0 %), and 10.09 (2.0 %). This is an order of magnitude improvement over random FE (1.0) regardless of the theoretical maximum which will decrease as larger percentages of the database are examined (Fig. 3; Table 12). The much better than random FE results in these early regions mirror the visual trends seen in the ROC curves shown above (Fig. 11) and should favorably benefit virtual screening efforts. The same trends are observed for the other three metrics (%TPR, %FPR, %AUC) with the computed enrichments being consistently better than random.
A comparison of results using the two different structure preps similarly shows consistently improved early enrichments for the DUD PDB prep. For example, %TPR values for DUD PDB versus DUD SUP yield: 15.14 versus 13.02 (1.0 %) and 20.40 versus 17.33 (2.0 %). Similar trends are observed for %AUC: 4.99 versus 4.17 (1.0 %) and 20.19 versus 17.45 (2.0 %) as well as FE: 9.99 versus 8.34 (1.0 %) and 10.09 versus 8.72 (2.0 %). In addition, %FPR values are lower, which is desirable: 0.56 versus 0.62 (1.0 %), and 1.43 versus 1.52 (2.0 %). Overall, use of the DUD PDB prep appears to yield better early enrichment results despite the overall average AUCs being the same at 0.60 and 0.59 (Table 11). Finally, the native pairings yield higher early enrichment values compared to the semi-random pairings which is to be expected. For example, native versus semi-random DUD PDB %AUC results yield 4.99 versus 3.53 (1.0 %) and 20.19 versus 12.49 (2.0 %). The %TPR results for these runs yield the same trend: 15.14 versus 8.58 (1.0 %) and 20.40 versus 11.47 (2.0 %).
Conclusions
The primary goals of this study were to evaluate the performance of the DOCK6 program for (1) pose identification and (2) database enrichment. The reported results represent the combined effort of two DOCK development groups and were generated as a result of participating in the recent symposium titled “Docking and Scoring: A Review of Docking Programs” held at the Spring 2011 Meeting of the American Chemical Society.
Pose identification success was examined using the standard DOCK grid scoring function (Tables 3, 4, 5), three variants of the Amber scoring function (Tables 6, 7, 8), and the new LMOD scoring procedure (Tables 9, 10). Systems for docking and scoring were derived from the single site (N = 85) or multi site Astex database (N = 147) with structure preparation protocols employing organizer supplied coordinates (ASTEX SUP) or those downloaded from the protein data bank (ASTEX PDB). For select experiments, two different docking protocols (RU, SB) were employed. Not surprisingly, use of different docking protocols, preparation procedures, and in particular, different scoring functions, yield different results.
As a baseline, the SB/ASTEX SUP grid score results (five run average, multi site set, organizer supplied structures, Table 3e) yields a symmetry corrected rmsd success rate of 72.5 % which should be comparable across different docking programs and platforms employed in the symposium. The analogous sampling success rate for the same docking runs is much higher at 91.9 % which indicates substantial room for improvement should a more accurate scoring function be used. Additional pose identification analysis includes evaluation of ligand starting conditions (Table 3), the effects of ligand symmetry (Table 3; Fig. 5), multiple binding site analysis (Table S2), rmsd statistics (Table 4), rmsd spectrum plots (Fig. 6), and the effects of clustering (Table 5). A potential problem regarding pose identification statistics is the relatively small number of systems used (N = 85 pdb codes). To minimize noise the current results are based on averages obtained over five independent docking runs using different random seeds. A recommendation is that future work should make use of much larger publically available datasets such as SB2010 (N = 780 systems).
Rescoring grid-based results with potentially more accurate Amber scoring protocols yielded generally lower success rates (Table 6). However, use of the ligand only movable protocol yielded a slightly improved success rate (69.4 %) as the comparable grid results for the same ensemble (68.0 %). Interestingly, although the numerical value was similar, the group of sites ranked successfully was different suggesting that the grid and Amber score functions have system dependent strengths and weaknesses which should be explored. Immediate future work planned will address relaxation with the Amber force field by starting with the larger MD-ready SB2010 dataset, and the protocol and the general treatment of systems will be examined by employing the NAB atom expression feature for specifying movable receptor residues to a few specific targets. Alternatively, receptor flexibility could be introduced through use of multiple-docking grids which is currently under investigation.
Testing of a new scoring procedure based on low mode sampling (LMOD score) was also examined (Table 9, 10; Fig. 7). Encouraging results were obtained with LMOD score (57.89 %) when compared to grid score (47.37 %) or Amber score (52.63 %) when using the same starting ensemble of structures for a subset of 19 Astex systems although given the relatively small dataset size employed more comprehensive studies should be performed. An important feature of LMOD is the ability to sample large regions of conformational space relatively quickly which can lead to improvements in both scoring and sampling. Representative results for system 1sj0 shows the potential utility of the method for generating multiple low-energy poses, with native-like rmsds, starting from a single high-energy structure with a non-native-like rmsd (Fig. 7). Additional LMOD studies are planned.
To evaluate the ability of DOCK6 to enrich actives versus decoy molecules, the standard DOCK grid scoring function was used to screen 38-40 systems (Table 11; Fig. 8, 9) contained in the DUD database using either organizer supplied receptor coordinates (DUD SUP) or the protein data bank (DUD PDB). In addition to the standard DUD sets a subset of 10 systems employing WOMBAT actives was studied. DUD PDB results (Fig. 8a; Table 11) using native pairings yield AUC values obtained from ROC curve analysis (Fig. 11) ranging from 0.29 (bad enrichment) to 0.96 (good enrichment) with an average AUC of 0.60 (27/38 have AUC ≥ 0.5). ROC curve analysis visually indicates good early enrichment for most systems which was quantified using several metrics including, %TPR, %FPR, %AUC, and FE, at 0.1, 1.0, and 2.0 % of the database examined (Table 12). In particular, at 1.0, and 2.0 % an order of magnitude improvement over random FE (1.0) is observed for the DUD PDB native pairing results. Overall, the early enrichment trends are encouraging and consistent with previous studies [1-3, 5, 11] in which DOCK was used to successfully identify active lead molecules through virtual screening.
As expected, analogous DUD PDB results for semi-random pairings show a lower average AUC (0.48). However, appreciable enrichment is observed among groups of related receptors (Fig. 9). In contrast, the miscellaneous other enzyme group, comprised of unrelated receptors, shows only 1/11 systems with AUC > 0.5. Some system specific analysis revealed possible contributors to differential enrichments according to which of the two receptor preparation protocols was used (DUD PDB vs. DUD SUP protocols). Observed differences include alternate side chain conformations, numbers of binding site waters, partial atomic charges, and protonation states.
In summary, the breadth and number of validation experiments presented here is notable and the results and analysis provide a useful starting point for future work to improve both sampling and scoring in DOCK. It is worth repeating that the baseline grid results were determined using only a simple two-term scoring function, and the same DOCK protocols were used for pose identification and enrichment. No fine tuning was performed to tweak parameters or scoring for any particular system or class of systems. Nevertheless, although such a simple function yields remarkably good results, there is clearly room for improvement. Community efforts are especially welcome in this regard since they attempt to provide a level field for comparing various production versions of docking software using a common set of validation tests. Participation provides an opportunity for development groups to share knowledge, thus facilitating the incorporation of new features, for example symmetry corrected rmsd which was previously missing from DOCK. While the best metrics for quantifying success and enrichment may continue to be debated, continued participation in activities such as these is important to help drive the field forward.
Supplementary Material
Acknowledgments
Greg Warren, Neysa Nevins, and Georgia McGauhey are thanked for organizing the special Docking and Scoring symposium. William J. Allen and Jiangyang Liu are thanked for code development and Steve Skiena is thanked for helpful discussions regarding implementation of symmetry corrected rmsd using the Hungarian matching algorithm. This work was supported in part by NIH grants GM57513 (D.A.C.), R01GM083669 (R.C.R.), and F31CA134201 (T.E.B.), as well as the Stony Brook University Office of the Vice President for Research and the New York State Office of Science Technology and Academic Research (NYSTAR). S.R.B. gratefully acknowledges the use of computational facilities at the Ohio Supercomputer Center and thanks OpenEye Scientific Software for an academic license. This work also used resources at the New York Center for Computational Sciences at Stony Brook University/Brookhaven National Laboratory supported by the US Department of Energy under Contract No. DE-AC02-98CH10886 and by the State of New York. Molecular graphics and analyses were performed with the UCSF Chimera package. Chimera is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from the National Institutes of Health (National Center for Research Resources grant 2P41RR0 01081, National Institute of General Medical Sciences grant 9P41GM103311).
Footnotes
Scott R. Brozell and Sudipto Mukherjee contributed equally to this work.
Electronic supplementary material The online version of this article (doi:10.1007/s10822-012-9565-y) contains supplementary material, which is available to authorized users.
References
- 1.Klebe G. Virtual ligand screening: strategies, perspectives and limitations. Drug Discov Today. 2006;11(13–14):580–594. doi: 10.1016/j.drudis.2006.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kitchen DB, Decornez H, Furr JR, Bajorath J. Docking and scoring in virtual screening for drug discovery: methods and applications. Nat Rev Drug Discov. 2004;3(11):935–949. doi: 10.1038/nrd1549. [DOI] [PubMed] [Google Scholar]
- 3.Kuntz ID. Structure-based strategies for drug design and discovery. Science. 1992;257(5073):1078–1082. doi: 10.1126/science.257.5073.1078. [DOI] [PubMed] [Google Scholar]
- 4.Jorgensen WL. The many roles of computation in drug discovery. Science. 2004;303(5665):1813–1818. doi: 10.1126/science.1096361. [DOI] [PubMed] [Google Scholar]
- 5.Shoichet BK. Virtual screening of chemical libraries. Nature. 2004;432(7019):862–865. doi: 10.1038/nature03197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Irwin JJ, Shoichet BK. ZINC—a free database of commercially available compounds for virtual screening. J Chem Inf Model. 2004;45(1):177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cross JB, Thompson DC, Rai BK, Baber JC, Fan KY, Hu Y, Humblet C. Comparison of several molecular docking programs: pose prediction and virtual screening accuracy. J Chem Inf Model. 2009;49(6):1455–1474. doi: 10.1021/ci900056c. [DOI] [PubMed] [Google Scholar]
- 8.Mukherjee S, Balius TE, Rizzo RC. Docking validation resources: protein family and ligand flexibility experiments. J Chem Inf Model. 2010;50(11):1986–2000. doi: 10.1021/ci1001982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang N, Shoichet BK, Irwin JJ. Benchmarking sets for molecular docking. J Med Chem. 2006;49(23):6789–6801. doi: 10.1021/jm0608356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuntz ID, Blaney JM, Oatley SJ, Langridge R, Ferrin TE. A geometric approach to macromolecule-ligand interactions. J Mol Biol. 1982;161(2):269–288. doi: 10.1016/0022-2836(82)90153-x. [DOI] [PubMed] [Google Scholar]
- 11.DesJarlais RL, Seibel GL, Kuntz ID, Furth PS, Alvarez JC, Ortiz de Montellano PR, DeCamp DL, Babé LM, Craik CS. Structure-based design of nonpeptide inhibitors specific for the human immunodeficiency virus 1 protease. Proc Nat Acad Sci. 1990;87(17):6644–6648. doi: 10.1073/pnas.87.17.6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ewing TJA, Kuntz ID. Critical evaluation of search algorithms for automated molecular docking and database screening. J Comput Chem. 1997;18(9):1175–1189. [Google Scholar]
- 13.Ewing TJA, Makino S, Skillman AG, Kuntz ID. DOCK 4.0: search strategies for automated molecular docking of flexible molecule databases. J Comput Aided Mol Des. 2001;15(5):411–428. doi: 10.1023/a:1011115820450. [DOI] [PubMed] [Google Scholar]
- 14.Meng EC, Shoichet BK, Kuntz ID. Automated docking with grid-based energy evaluation. J Comput Chem. 1992;13(4):505–524. [Google Scholar]
- 15.Moustakas DT, Lang PT, Pegg S, Pettersen E, Kuntz ID, Brooijmans N, Rizzo RC. Development and validation of a modular, extensible docking program: DOCK 5. J Comput Aided Mol Des. 2006;20(10):601–619. doi: 10.1007/s10822-006-9060-4. [DOI] [PubMed] [Google Scholar]
- 16.Lang PT, Brozell SR, Mukherjee S, Pettersen EF, Meng EC, Thomas V, Rizzo RC, Case DA, James TL, Kuntz ID. DOCK 6: combining techniques to model RNA-small molecule complexes. RNA. 2009;15(6):1219–1230. doi: 10.1261/rna.1563609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zou X, Yaxiong S, Kuntz ID. Inclusion of solvation in ligand binding free energy calculations using the generalized-born model. J Am Chem Soc. 1999;121(35):8033–8043. [Google Scholar]
- 18.Liu H-Y, Kuntz ID, Zou X. Pairwise GB/SA scoring function for structure-based drug design. J Phys Chem B. 2004;108(17):5453–5462. [Google Scholar]
- 19.Grant JA, Pickup BT, Nicholls A. A smooth permittivity function for Poisson–Boltzmann solvation methods. J Comput Chem. 2001;22(6):608–640. [Google Scholar]
- 20.Graves AP, Shivakumar DM, Boyce SE, Jacobson MP, Case DA, Shoichet BK. Rescoring docking hit lists for model cavity sites: predictions and experimental testing. J Mol Biol. 2008;377(3):914–934. doi: 10.1016/j.jmb.2008.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balius TE, Mukherjee S, Rizzo RC. Implementation and evaluation of a docking-rescoring method using molecular footprint comparisons. J Comput Chem. 2011;32(10):2273–2289. doi: 10.1002/jcc.21814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.SBU DOCK Tutorials 2012 Mar 01; http://ringo.ams.sunysb.edu/index.php/DOCK_Tutorials. Last accessed.
- 23.UCSF DOCK Tutorials 2012 Mar 01; http://dock.compbio.ucsf.edu/DOCK_6/tutorials/index.htm. Last accessed.
- 24.Hartshorn MJ, Verdonk ML, Chessari G, Brewerton SC, Mooij WTM, Mortenson PN, Murray CW. Diverse, high-quality test set for the validation of protein-ligand docking performance. J Med Chem. 2007;50(4):726–741. doi: 10.1021/jm061277y. [DOI] [PubMed] [Google Scholar]
- 25.Good A, Oprea T. Optimization of CAMD techniques 3. Virtual screening enrichment studies: a help or hindrance in tool selection? J Comput Aided Mol Des. 2008;22(3):169–178. doi: 10.1007/s10822-007-9167-2. [DOI] [PubMed] [Google Scholar]
- 26.Kuhn HW. The Hungarian method for the assignment problem. Nav Res Logist Quart. 1955;2:83–97. [Google Scholar]
- 27.Munkres J. Algorithms for the assignment and transportation problems. J Soc Ind Appl Math. 1957;5(1):32–38. [Google Scholar]
- 28.Raymond JW, Willett P. Maximum common subgraph isomorphism algorithms for the matching of chemical structures. J Comput Aided Mol Des. 2002;16(7):521–533. doi: 10.1023/a:1021271615909. [DOI] [PubMed] [Google Scholar]
- 29.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 30.Triballeau N, Acher F, Brabet I, Pin JP, Bertrand HO. Virtual screening workflow development guided by the ‘‘receiver operating characteristic’’ curve approac. Application to highthroughput docking on metabotropic glutamate receptor subtype 4. J Med Chem. 2005;48(7):2534–2547. doi: 10.1021/jm049092j. [DOI] [PubMed] [Google Scholar]
- 31.Jain A, Nicholls A. Recommendations for evaluation of computational methods. J Comput Aided Mol Des. 2008;22(3):133–139. doi: 10.1007/s10822-008-9196-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Srinivasan J, Cheatham TE, Cieplak P, Kollman PA, Case DA. Continuum solvent studies of the stability of DNA, RNA, and phosphoramidate—DNA helices. J Am Chem Soc. 1998;120(37):9401–9409. [Google Scholar]
- 33.Kollman PA, Massova I, Reyes C, Kuhn B, Huo S, Chong L, Lee M, Lee T, Duan Y, Wang W, Donini O, Cieplak P, Srinivasan J, Case DA, Cheatham TE. Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc Chem Res. 2000;33(12):889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- 34.Rastelli G, Rio AD, Degliesposti G, Sgobba M. Fast and accurate predictions of binding free energies using MM-PBSA and MM-GBSA. J Comput Chem. 2010;31(4):797–810. doi: 10.1002/jcc.21372. [DOI] [PubMed] [Google Scholar]
- 35.Kuhn B, Gerber P, Schulz-Gasch T, Stahl M. Validation and use of the MM-PBSA approach for drug discovery. J Med Chem. 2005;48(12):4040–4048. doi: 10.1021/jm049081q. [DOI] [PubMed] [Google Scholar]
- 36.Thompson DC, Humblet C, Joseph-McCarthy D. Investigation of MM-PBSA rescoring of docking poses. J Chem Inf Model. 2008;48(5):1081–1091. doi: 10.1021/ci700470c. [DOI] [PubMed] [Google Scholar]
- 37.Hou T, Wang J, Li Y, Wang W. Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J Chem Inf Model. 2011;51(1):69–82. doi: 10.1021/ci100275a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Macke TJ, Case DA. Modeling unusual nucleic acid structures. ACS Sym Ser. 1997;682:379–393. [Google Scholar]
- 39.Kolossvary I, Guida WC. Low mode search. An efficient, automated computational method for conformational analysis: application to cyclic and acyclic alkanes and cyclic peptides. J Am Chem Soc. 1996;118(21):5011–5019. [Google Scholar]
- 40.Kolossvary I, Keseru GM. Hessian-free low-mode conformational search for large-scale protein loop optimization: application to c-jun N-terminal kinase JNK3. J Comput Chem. 2001;22(1):21–30. [Google Scholar]
- 41.Sheridan RP, McGaughey GB, Cornell WD. Multiple protein structures and multiple ligands: effects on the apparent goodness of virtual screening results. J Comput Aided Mol Des. 2008;22(3–4):257–265. doi: 10.1007/s10822-008-9168-9. [DOI] [PubMed] [Google Scholar]
- 42.Truchon JF, Bayly CI. Evaluating virtual screening methods: good and bad metrics for the ‘‘early recognition’’ problem. J Chem Inf Model. 2007;47(2):488–508. doi: 10.1021/ci600426e. [DOI] [PubMed] [Google Scholar]
- 43.Onufriev A, Bashford D, Case DA. Exploring protein native states and large-scale conformational changes with a modified generalized born model. Proteins. 2004;55(2):383–394. doi: 10.1002/prot.20033. [DOI] [PubMed] [Google Scholar]
- 44.Weiser J, Shenkin PS, Still WC. Approximate atomic surfaces from linear combinations of pairwise overlaps (LCPO) J Comput Chem. 1999;20(2):217–230. [Google Scholar]
- 45.Hawkins GD, Cramer CJ, Truhlar DG. Parametrized models of aqueous free energies of solvation based on pairwise descreening of solute atomic charges from a dielectric medium. J Phys Chem. 1996;100(51):19824–19839. [Google Scholar]
- 46.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65(3):712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gasteiger J, Marsili M. Iterative partial equalization of orbital electronegativity—a rapid access to atomic charges. Tetrahedron. 1980;36(22):3219–3228. [Google Scholar]
- 48.Maignan S, Guilloteau JP, Pouzieux S, Choi-Sledeski YM, Becker MR, Klein SI, Ewing WR, Pauls HW, Spada AP, Mikol V. Crystal structures of human factor Xa complexed with potent inhibitors. J Med Chem. 2000;43(17):3226–3232. doi: 10.1021/jm000940u. [DOI] [PubMed] [Google Scholar]
- 49.Nar H, Bauer M, Schmid A, Stassen JM, Wienen W, Priepke HW, Kauffmann IK, Ries UJ, Hauel NH. Structural basis for inhibition promiscuity of dual specific thrombin and factor Xa blood coagulation inhibitors. Structure. 2001;9(1):29–37. doi: 10.1016/s0969-2126(00)00551-7. [DOI] [PubMed] [Google Scholar]
- 50.Chachra R, Rizzo RC. Origins of resistance conferred by the R292K neuraminidase mutation via molecular dynamics and free energy calculations. J Chem Theory Comput. 2008;4(9):1526–1540. doi: 10.1021/ct800068v. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.




