Abstract
Pathogenicity of Pseudomonas aeruginosa, a major cause of many acute and chronic human infections, is determined by tightly regulated expression of multiple virulence factors. Quorum sensing (QS) controls expression of many of these pathogenic determinants. Previous microarray studies have shown that the AmpC β-lactamase regulator AmpR, a member of the LysR family of transcription factors, also controls non-β-lactam resistance and multiple virulence mechanisms. Using RNA-Seq and complementary assays, this study further expands the AmpR regulon to include diverse processes such as oxidative stress, heat shock and iron uptake. Importantly, AmpR affects many of these phenotypes, in part, by regulating expression of non-coding RNAs such as rgP32, asRgsA, asPrrF1 and rgRsmZ. AmpR positively regulates expression of the major QS regulators LasR, RhlR and MvfR, and genes of the Pseudomonas quinolone system. Chromatin immunoprecipitation (ChIP)-Seq and ChIP–quantitative real-time polymerase chain reaction studies show that AmpR binds to the ampC promoter both in the absence and presence of β-lactams. In addition, AmpR directly binds the lasR promoter, encoding the QS master regulator. Comparison of the AmpR-binding sequences from the transcriptome and ChIP-Seq analyses identified an AT-rich consensus-binding motif. This study further attests to the role of AmpR in regulating virulence and physiological processes in P. aeruginosa.
INTRODUCTION
Pseudomonas aeruginosa is ubiquitous, and can be isolated from diverse sources including plants, animals and humans. A high degree of nutritional versatility and adaptability ensure that P. aeruginosa is able to colonize a wide range of natural and man-made habitats. In humans, P. aeruginosa is seldom part of the normal microbial flora and is found in <2–6% of individuals (1). An opportunistic, nosocomial pathogen, P. aeruginosa colonization rates in hospitalized patients, however, can be >50% and especially so in cases of mucosal or cutaneous breach, or in immunocompromised individuals (2). Pseudomonas aeruginosa is also the leading cause of morbidity and mortality in cystic fibrosis (CF) patients (3).
The pathogenic potential of P. aeruginosa is multifactorial and can be broadly classified into cell-associated and secreted virulence factors. The cell-associated virulence factors are typically structural components of the cell, such as the lipopolysaccharide, pili and flagella (4–6). The process of quorum sensing (QS) regulates expression of many of the major secreted virulence factors. QS is a mechanism of coordinating gene expression based on the population density, employed by both non-pathogenic and pathogenic bacteria (7). Quoromones (acyl-homoserine lactones) are small diffusible molecules that mediate QS communication between cells to synchronize expression of virulence genes (8). Precise signaling is ensured by the species-specific nature of quoromones, although crosstalk between related bacteria is known to occur (9,10). Pseudomonas aeruginosa employs three interdependent mechanisms of QS, namely, the Las, Rhl and Pseudomonas quinolone system (PQS). The Las system is at the top of the regulatory hierarchy, above the Rhl system, while PQS interacts with both Las and Rhl [reviewed in (11,12)]. In P. aeruginosa, the QS process controls production of secreted enzymes and toxins such as LasA, LasB and ToxA; redox-active compounds such as phenazines (12) and in the case of chronic infections, the formation of bacterial communities called biofilms (13). In addition, some efflux pumps, such as MexGHI-OpmD, which play a role in pumping out quoromones from the cytoplasm to the cell exterior, are also QS-regulated (14,15). However, a majority of the 12 putative and established RND efflux pumps in P. aeruginosa are involved in antibiotic resistance (16,17).
Antibiotic resistance is a major problem in dealing with P. aeruginosa infections. The current treatment regimen for P. aeruginosa is typically a combination therapy of β-lactams, aminoglycosides and quinolones (3,18). However, a 6-year survey by the National Nosocomial Infections Surveillance System of the Centers for Disease Control and Prevention revealed that P. aeruginosa isolates were resistant to many commonly used antibiotics in both intensive-care unit and non-intensive-care unit patients (19). The infection rates with antibiotic-resistant P. aeruginosa were as high as 36% (19).
Pseudomonas aeruginosa has multiple mechanisms of antibiotic resistance (16). Resistance to the β-lactam class of antibiotics is primarily conferred by the chromosomally encoded β-lactamase AmpC (16). The MexEF-OprN efflux pump mediates quinolone resistance (20). Our recent study demonstrated that the LysR-type transcriptional regulator (LTTR) AmpR modulates expression of both ampC and mexEF-oprN (21). In addition, P. aeruginosa AmpR is a global regulator of many virulence determinants and transcriptional factors (21,22). Using DNA microarrays and complementary assays, we have demonstrated that the AmpR regulon consists of >500 genes that are involved in virulence and metabolism (21). Importantly, the analyses reveal that AmpR positively regulates many acute infection phenotypes while repressing chronic ones (21). Interestingly, the AmpR regulon included the small regulatory RNA rgRsmZ (21). Given the extensive nature of the AmpR regulon, we hypothesized that other small regulatory RNAs could have been missed, as the microarray platform is not designed to detect them. In addition, given the limited sensitivity of microarrays, other potentially AmpR-regulated genes may have escaped detection. This study uses RNA-Seq to identify other non-coding RNAs (ncRNAs) and chromatin immunoprecipitation (ChIP)-Seq to determine direct targets of AmpR. Furthermore, we assign a role for AmpR in previously unidentified critical cellular processes such as iron uptake, oxidative stress and heat shock. This study reaffirms AmpR as a critical regulator of P. aeruginosa virulence and physiological processes.
MATERIALS AND METHODS
Strains, plasmids, primers and culture conditions
The strains and plasmids used in this study are listed in Table 1. The primers used are listed in Supplementary Table S1. The wild-type P. aeruginosa PAO1 and its isogenic in-frame ampR deletion strain, PAOΔampR, are described earlier (21,23).
Table 1.
Strains and plasmids used in this study
| Strain/plasmid | Relevant characteristics | Source |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| DH5α | General purpose cloning strain; Δ(lacZ)M15 | New England Biolabs |
| DBS206 | E. coli DH5α harboring 3x-V5 tag on pCR2.1 TOPO | This study |
| DBS215 | DH5α with ampR ORF and the ampR-ampC intergenic region PCR cloned into pCR2.1 TOPO | This study |
| DBS222 | DH5α harboring ampR ORF tagged with 3x V5-tag on pCR2.1 TOPO | This study |
| DBS234 | DH5α with mini CTX2, containing 3x V5-tagged ampR | This study |
| Pseudomonas aeruginosa | ||
| PAO1 | Wild-type | (23) |
| PKM315 | PAOΔampR; in-frame deletion of ampR (PA4109) | (21) |
| DBS248 | PAOΔampR::ampR-V5; 3x-V5-tagged ampR cloned onto mini-CTX2 and moved into PKM315 | This study |
| Plasmids | ||
| pCR2.1 TOPO | TA cloning vector for PCR products; ApR, KmR; ColE1 f1 ori lacZα | Invitrogen |
| ZM747-V5 | 3x V5 tag C-term and N-term of yeast URA-3 in pBlueScript; pDBS193 | (24) |
| Mini CTX2 | Plasmid for single-copy gene integration in P. aeruginosa; TcR | (25) |
| pDBS206 | 3x V5 tag PCR-amplified from ZM747-V5 and cloned into pCR2.1 TOPO | This study |
| pDBS215 | 1112-bp ampR ORF along with the ampR-ampC intergenic region, PCR-amplified with primers DBS_ampRF1 and DBSampR; cloned into pCR2.1 TOPO | This study |
| pDBS222 | ampR ORF from pDBS215 subcloned as KpnI-SacI upstream of and inframe with 3x V5 tag in pDBS206 | This study |
| pDBS234 | 3x V5-tagged ampR ORF from pDBS222 subcloned as a KpnI-NotI fragment into mini-CTX2 | This study |
For ChIP-Seq studies, AmpR was tagged at the carboxy-terminus with a 3x-V5 epitope tag. Briefly, the 3x-V5 epitope was polymerase chain reaction (PCR)-amplified from the plasmid ZM474 (24) using primers DBS_V5F and DBS_V5R containing KpnI and NheI sites, respectively. Termination codons were included in all three reading frames with the NheI site to prevent runoff translation. The 3x-V5 amplicon was cloned into pCR2.1 TOPO (Invitrogen) to generate plasmid pDBS206 and sequenced to ensure absence of any mutations. The 1112-bp ampR ORF with the native promoter but without the stop codon was PCR-amplified using primers DBS_ampRF1 (with a KpnI site) and DBS_ampRR (with a SacI site), cloned into pCR 2.1 TOPO (pDBS215) and sequenced. The KpnI-SacI fragment was subcloned in-frame with the 3x-V5 tag in pDBS206 to generate plasmid pDBS222. The 3x V5-tagged ampR was then moved into mini-CTX2 [pDBS227; (25)] as a KpnI-NheI fragment, generating plasmid pDBS234. After confirmation by PCR and restriction digestion, the suicide plasmid pDBS234 was moved into PAOΔampR by electroporation (26). This resulted in strain DBS248 with a single chromosomal copy of tagged ampR that was then used for the ChIP-Seq studies. Functionality of the tagged AmpR in DBS248 was verified by determining the minimum inhibitory concentration (MIC) of the β-lactams, ampicillin-sulbactam and amoxicillin, and by ChIP–quantitative real-time polymerase chain reaction (qPCR).
All strains were grown in standard LB media with aeration, unless otherwise specified. Synthetic succinate medium (SSM) was used as the iron-limited media (27) and contained (g/l) K2HPO4 6.0, KH2PO4 3.0, (NH4)2SO4 1.0, MgSO4.7H2O 0.2, sodium succinate 4.0, pH 7.0. Antibiotics were used at the following concentrations: for Escherichia coli: gentamycin 15 µg/ml, tetracycline 15 µg/ml, ampicillin 100 µg/ml; for P. aeruginosa: gentamycin 75 µg/ml, tetracycline 60 µg/ml, carbenicillin 150 µg/ml.
Library preparation for RNA-Seq analysis
Total RNA was isolated from PAO1 and PAOΔampR with and without sub-MIC β-lactam stress, and harvested at the same growth phase as described previously (21) using the hot-phenol extraction protocol (28). RNA quality was analyzed on the Agilent Bioanalyzer and those with RNA Integrity Numbers of 8.0 or above were used for rRNA depletion using the MICROBExpress Kit (Ambion). After rRNA depletion, cDNA synthesis was performed using the SuperScript III First-Strand Synthesis System (Invitrogen) as per manufacturer protocols. Terminal transferase (New England Biolabs) and dATP (New England Biolabs) were used to add a 100–300-base poly-A tail to the cDNA samples following manufacturer instructions. The 3′ ends were then blocked with biotinylated ddATP (Perkin Elmer) using terminal transferase (New England Biolabs), followed by cleanup with the MinElute cleanup kit (Qiagen). Tailed and blocked samples were then processed on the HeliScope Single Molecule Sequencer at the Molecular Biology Core Facility, Dana Farber Cancer Institute, Boston, MA.
ChIP-Seq sample preparation
For ChIP-Seq studies, the 3x-V5-tagged AmpR containing strain (DBS248) was grown and exposed to sub-MIC β-lactam stress as described previously (21). Protein–DNA interactions were then cross-linked in vivo with formaldehyde (final concentration of 1%) at room temperature for 20 min and quenched for 15 min with 0.25 M glycine. After three washes with 1× PBS, the cells were resuspended in 1 ml lysis buffer (10 mM Tris pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% deoxycholic acid and 0.5% N-laurylsarcosine) containing a protease inhibitor cocktail (Roche). After chilling on ice, the cells were sonicated to shear the DNA to a size range of 0.5–1 kb. Cellular debris were removed by centrifugation and a 3 -µl aliquot of the supernatant was checked on an agarose gel. A 50 -µl aliquot of the supernatant was stored as the input DNA and the rest was immunoprecipitated using DynaBeads Protein G (Life Technologies), which was previously equilibrated and bound with anti-V5 monoclonal antibody (Sigma) as per manufacturer instructions. After immunoprecipitation overnight, the beads were washed five times with RIPA buffer (50 mM HEPES pH 7.5, 500 mM LiCl, 1 mM EDTA, 1% NP40, 0.7% deoxycholic acid, 50 mM NaCl in 1× TE). The beads were resuspended in 100 µl elution buffer (50 mM Tris-HCl, 10 mM EDTA, 1% SDS) at 65°C for 30 min, centrifuged to remove residual beads and incubated at 65°C overnight to reverse the cross-link. TE buffer was then added (100 µl) and the samples were treated with RNase (37°C for 2 h). The immunoprecipitated proteins were removed with proteinase K (55°C for 2 h), and the DNA was cleaned using the Qiagen Mini Reaction Cleanup kit. RNase and proteinase K treatment was also performed for the input DNA. DNA concentrations were determined using Quant-iT PicoGreen dsDNA Kit (Life Technologies). Before proceeding further, AmpR occupancy of the ampC promoter was determined using qPCR as described previously (21). The DNA samples were then poly-A-tailed, blocked with biotinylated ddATP, purified and processed on the Helicos sequencer as described in the RNA-Seq section.
Data analysis for RNA-Seq and ChIP-Seq
The raw data files for both RNA-Seq and ChIP-Seq experiments from the Helicos sequencing runs were first filtered based on read length, and converted to the FASTA format using Helisphere Open Source project. CLC Genomics Workbench, version 5 (CLC Bio), was used to map the sequence reads to the P. aeruginosa PAO1 genome (NCBI reference sequence NC_002516.2) and for all further data analyses.
For RNA-Seq analysis, the total number of reads per gene between samples was normalized using RPKM ([reads/kb gene]/[million reads aligning to the genome]) (29). Pair-wise comparisons were performed using the RPKM values to obtain differential gene expression data. Significance of the differentially expressed genes was determined using the chi-square test on the RPKM values. Bonferroni correction was then performed and false discovery rate P-values were calculated. Only genes with a proportions fold-change of ≥2.0 and with Bonferroni-corrected P-value of ≤0.05 were considered for further analyses.
For ChIP-Seq samples, after mapping the reads to the reference genome, the peaks were detected using a window size of 500, and were shifted sequentially based on read length. The peaks that were identified were filtered on a maximum probability of 1.0E–04 for identical locations of forward and reverse reads.
Comparison of microarray and RNA-Seq
To get a comprehensive picture of the AmpR regulon in P. aeruginosa, transcriptomics studies using DNA microarrays (21) and RNA-Seq (this study) were performed. Both studies used the same two strains (PAO1 and PAOΔampR), under identical conditions (without and with sub-MIC β-lactam exposure). Both EdgeR (30) and CLC genomics were used to analyze the data. Compared with our CLC analysis, the number of genes that were identified in each of the four conditions was much lower with EdgeR (data not shown). In addition, genes of some of the phenotypes that we have confirmed previously did not show up with the EdgeR analysis. Even though EdgeR has been shown to be a better package (30), RPKM analysis worked better for our dataset. It remains to be understood why such a discrepancy exists. However, the use of RPKM for the analysis of both RNA-Seq and ChIP-Seq data is well established (31–34). Further, the CLC Genomics Workbench has RPKM in-built, the widely used software for high-throughput expression data analysis. The normalized data from the microarrays and RNA-Seq were also compared to compute the extent of correlation. The Pearson correlation coefficients were determined for the four different conditions.
Enrichment of functional categories
The gene sets that were positively and negatively regulated by AmpR without and with β-lactam stress were functionally categorized based on the Pseudomonas Genome Database (35). Gene distribution under individual categories in PAO1 was considered as 100% and the relative distributions in each of the four gene sets were plotted. Enrichment of specific functional categories in the individual datasets was determined using GOEAST (36). The log odds-ratio was calculated for the GOEAST enrichment. The larger the value of the ratio, the higher the relative abundance of the GO term as compared with a random condition.
Quantitative real-time PCR assays
Genes that were differentially regulated in the RNA-Seq analysis and those that had not been tested in our previous transcriptome studies (21) were selected for qPCR confirmation. RNA isolation and cDNA synthesis were performed as described previously (21). Ten nanograms of cDNA were used per reaction well in the qPCR assays. Expression of the test genes was normalized to clpX (PA1802).
Hydrogen peroxide sensitivity
To determine differences in hydrogen peroxide (H2O2) sensitivities of PAO1 and PAOΔampR, gradient plate assay was used (37). Briefly, 37 ml of LB agar, held at 50°C, was supplemented with 4–8 µM H2O2 and poured onto tilted 90-mm petri dishes. After solidification, 37 ml of LB agar without H2O2 was poured onto the plates on a flat surface to generate a gradient. OD600-normalized overnight LB cultures of the strains were then used to form a 75-mm streak across the gradient using a cotton swab. H2O2 sensitivity was scored as the extent of growth into the gradient, compared with the control (on LB plates without H2O2). The assay was performed in triplicate and representative results are shown.
Growth in iron-limited media
The differential growth abilities of the ampR mutant vis-à-vis the wild-type strain were determined on SSM (27). The cultures used for the growth curve assays were grown overnight in SSM from fresh LB plates. For the assay, the overnight cultures were normalized to an OD600 of 0.02 in SSM and the ODs of 200 -µl aliquots were monitored for 17 h at 37°C in 96-well flat-bottom tissue culture plates (Nunc). Conditions were made iron-replete by supplementing SSM with 100 µM FeCl3 (27).
Temperature sensitivity assays
The ability of PAO1 and PAOΔampR to tolerate elevated temperatures was examined. Briefly, cells grown in LB broth to either the log (OD600 of 0.6–0.8) or stationary (OD600 of >2.0) phase at 30°C were exposed to 50°C for 1–3 h. Control aliquots were maintained at 30°C. Cell counts were determined before and after exposure by plating for colony forming units (CFUs).
Statistical analyses
All the data from qPCR and phenotypic assays were examined for statistical significance using the unpaired two-tailed t-test on GraphPad analysis software (www.graphpad.com). RNA-Seq and ChIP-Seq data were analyzed for significance on CLC Genomics Workbench as described earlier.
RESULTS
RNA-Seq analysis expands the AmpR regulon
To study the function of AmpR in P. aeruginosa, a clean in-frame deletion strain (PAOΔampR) was constructed in PAO1 (21). PAO1 and PAOΔampR were used for gene expression profiling using RNA-Seq. To be able to compare results from the two studies, the experimental conditions for RNA-Seq were identical to the microarray experiments that were performed earlier (21). PAO1 and PAOΔampR were subjected to sub-MIC β-lactam stress before RNA isolation and cDNA synthesis as described in the Methods section.
Using RNA-Seq, the transcription profiles of PAO1 and PAOΔampR were compared in the presence (induced) and absence (uninduced) of sub-MIC β-lactam exposure. After data normalization between the replicates under each condition, the expression values across the entire genome for PAOΔampR were normalized to PAO1. The following nomenclature will be used to define AmpR-mediated positive and negative regulation. If mRNA expression levels are lower in PAOΔampR compared with PAO1, AmpR positively regulates those genes. Conversely, AmpR negatively regulates the genes if their expression levels are higher in PAOΔampR.
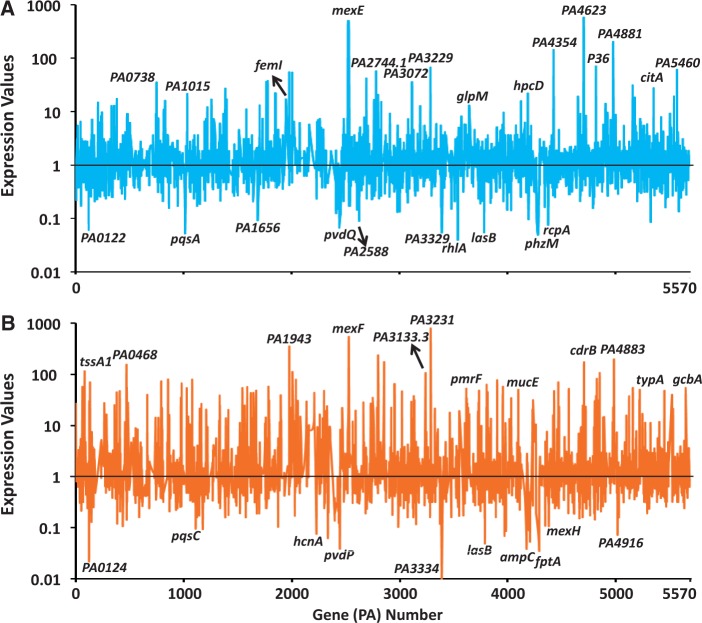
The PAOΔampR data in the uninduced (Panel A, Figure 1) and induced (Panel B, Figure 1) were plotted. The expression profiles were markedly different in PAOΔampR compared with PAO1, both in the absence and presence of antibiotic exposure, attesting to the global regulatory role of AmpR in P. aeruginosa. As expected, the ampC encoding β-lactamase, which is under positive AmpR regulation (21,38), is not significantly activated in the presence β-lactam in PAOΔampR (Panel B, Figure 1). Expression of genes identified in a previous study to be positively regulated by AmpR such as the lasB encoding elastase (21) was also detected here (Panels A, B; Figure 1). The MexEF-OprN efflux pump that provides resistance against fluoroquinolones and chloramphenicol is negatively regulated by AmpR [Panels A, B; Figure 1, (21)]. We had demonstrated AmpR to be a positive regulator of the QS system and in agreement, key QS genes such as pqsA and rhlA are downregulated in the absence of ampR (Panel A, Figure 1). These findings, in addition to others discussed in the following sections, add credence to the current study.
Figure 1.
Gene expression in PAOΔampR. Relative gene expression in PAOΔampR compared with PAO1 (normalized to expression value of 1), based on RNA-Seq data, is shown in the absence (A) and presence (B) of sub-MIC β-lactam stress. Some significantly regulated genes are named. Gene annotations are from the Pseudomonas Genome Database (35).
In addition to the microarray findings, the normalized data identified two transcriptional regulators (PA1015 and PA2588) and one extracytoplasmic function sigma factor (ECF) (femI, PA1912) to be differentially regulated (Panel A, Figure 1). PA2588 is a putative transcriptional regulator of the AraC family (35). Interestingly, it is located downstream of pqsH, which is an important component of the PQS (39). The role of AmpR in regulating the PQS is discussed in a later section. The ECF sigma factor FemI, upregulated in PAOΔampR, is part of a two-gene operon with femR (PA1911). FemR, along with FemA (PA1910), is involved in uptake of the mycobacterial siderophore mycobactin (40). Differential regulation of femI, along with the gene encoding the pyochelin receptor fptA (Panel B in Figure 1), suggests a role for AmpR in iron uptake regulation and is further explored in the later sections of this study. In addition, ncRNAs such as P36, PA2744.1 and PA3133.1 are also differentially regulated (Figure 1).
Identification of AmpR- and AmpR-β-lactam-dependent gene sets
The RNA-Seq data were normalized and pair-wise comparisons were performed to determine differential gene expression (fold change ≥2.0, Bonferroni correction of P ≤0.05). The four pair-wise comparisons performed were PAO1 uninduced versus PAO1 induced (Condition I), PAOΔampR uninduced versus PAOΔampR induced (Condition II), PAO1 uninduced versus PAOΔampR uninduced (Condition III) and PAO1 induced versus PAOΔampR induced (Condition IV). This led to the identification of 2568 genes (Condition I–IV with 384, 672, 532 and 980 genes, respectively) that were differentially expressed across all four pair-wise comparisons. Although these numbers are indicative, they are not a true measure of the AmpR regulatory repertoire due to potential overlaps, i.e. individual genes could be differentially expressed under more than one condition. To address this issue, the pair-wise comparison data were plotted using a four-way Venn diagram (Figure 2). Of the 2568 genes, 865 redundant and 1703 non-redundant (Figure 2) representing 31% of the PAO1 genome were determined. Further, genes that were dependent on AmpR alone, β-lactam alone and AmpR and β-lactam were identified in accordance with the expression criteria listed in Supplementary Table S2. For example, genes in Category B (Figure 2) are regulated in an AmpR-dependent manner, independent of β-lactam exposure.
Figure 2.
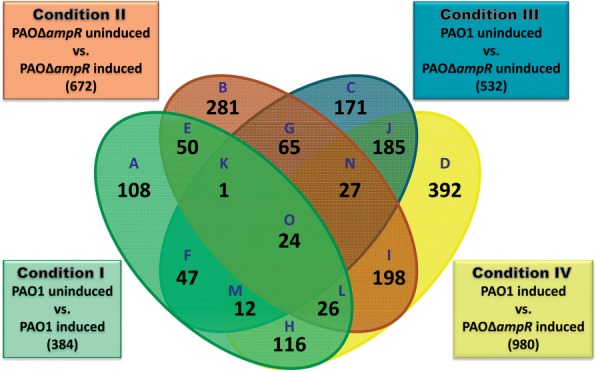
Venn diagram of differentially regulated genes. Distribution of significantly (P ≤ 0.01) regulated genes (≥2.0-fold) in PAO1 and PAOΔampR without (uninduced) and with (induced) sub-MIC β-lactam stress.
Twenty-four genes (Category O) were omitted from further analysis because they were differentially regulated under all conditions, irrespective of AmpR and/or antibiotic exposure. An additional 56 genes could not be assigned unambiguously for analysis [Category H (44 genes), Category K (1 gene), Category M (4 genes) and Category N (7 genes)] were omitted. Thus, of the remaining 1623 genes, we identified 654 AmpR-dependent (Supplementary Table S3), 483 AmpR-dependent only in the presence of β-lactam (Supplementary Table S4) and 486 differentially expressed in response to β-lactam independent of AmpR (Supplementary Table S5).
AmpR-regulated genes are enriched in specific functional categories
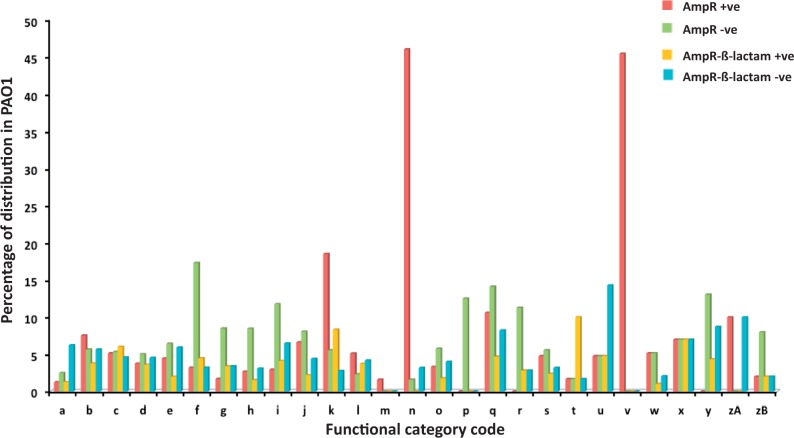
Functional categorization of the AmpR-regulated genes was as per Pseudomonas Genome Database annotation [(35), Figure 3]. The AmpR-regulated genes were expressed as a percentage of the distribution of each functional category in the PAO1 genome (taken as 100%). This revealed a gross upregulation of genes related to phage, transposon or plasmid (45% genes of Category ‘v’; Figure 3), as in the previous transcriptome study (21). Similarly, genes of functional class ‘n’ (secreted factors: toxins, enzymes and alginate) were also shown to be under AmpR positive regulation (46% of genes of the Category ‘n’; Figure 3). This is in agreement with previous study demonstrating that ampR mutant is impaired in production of extracellular enzymes such as LasA and LasB (21). AmpR also positively regulates genes belonging to functional categories ‘k’ (adaptation and protection; 19%), ‘zA’ (antibiotic resistance and susceptibility; 10%) and ‘q’ (central intermediary metabolism; 11%) (Figure 3). This finding agrees with our previous observations from DNA and phenotypic microarrays that AmpR is an important regulator of antibiotic resistance, cell wall recycling enzymes and metabolism in P. aeruginosa (21).
Figure 3.
Functional categorization of AmpR-regulated genes. AmpR differentially regulated genes are functionally categorized and expressed as a percentage of the respective category in the PAO1 genome (35). The functional categories are (a) DNA replication, recombination, modification and repair; (b) fatty acid and phospholipid metabolism; (c) hypothetical; (d) membrane proteins; (e) amino acid biosynthesis, metabolism; (f) translation, post-translational modification, degradation; (g) cell wall/lipopolysaccharide/capsule; (h) transport of small molecules; (i) energy metabolism; (j) biosynthesis of cofactors, prosthetic groups, carriers; (k) adaptation, protection; (l) transcriptional regulators; (m) two-component regulatory systems; (n) secreted factors toxins, enzymes, alginate; (o) putative enzymes; (p) chaperones, heat-shock proteins; (q) central intermediary metabolism; (r) nucleotide biosynthesis and metabolism; (s) carbon compound catabolism; (t) motility and attachment; (u) chemotaxis; (v) related to phage, transposon, plasmid; (w) non-coding RNA genes; (x) protein secretion, export apparatus; (y) cell division; (zA) antibiotic resistance, susceptibility; (zB) transcription, RNA processing, degradation.
In the absence of β-lactam, AmpR negatively regulates genes involved in translation (Category ‘f’ 17%), energy metabolism (Category ‘i’ 12%), nucleotide metabolism (Category ‘r’ 11%), cell division (Category ‘y’ 13%), and chaperones and heat shock (Category ‘p’ 13%). In the presence of β-lactam stress, AmpR positively regulates genes involved in motility and attachment (Category ‘t’ 10%) and negatively regulates genes involved in chemotaxis (Category ‘u’ 15%; Figure 3).
To determine whether the functional categorization in the different gene sets is significant, enrichment analysis was performed using GOEAST (36). Primarily, gene ontology identifications (GOIDs) belonging to biological processes and cellular components were enriched in the AmpR positively and negatively regulated gene sets, respectively (Supplementary Table S6). GOIDs belonging to ribosomal protein biosynthesis and oxidative phosphorylation were statistically significantly enriched in the AmpR-dependent negatively regulated gene set (log odd ratio >1.5; P < 0.05; Supplementary Table S6). There was no enrichment in the AmpR-β-lactam-dependent gene sets.
In the AmpR positively regulated set, many genes involved in pyoverdine biosynthesis were significantly enriched (log odd ratio >2.5, P < 0.03; Supplementary Table S6). This enrichment is, in part, due to the presence of genes encoding the major catalase KatA, and the superoxide dismutase SodA. The physiological effects of enrichment of these genes on iron acquisition and oxidative stress response, and the role of AmpR in their regulation, are discussed in later sections. Furthermore, GOIDs containing genes such as the QS regulator rhlR and the stress-phase sigma factor rpoS, which were identified to be AmpR-regulated previously (21), also showed significant enrichment (Supplementary Table S6).
Regulation of small RNAs by AmpR
RNA-Seq allows detection of expression profiles of small RNAs (sRNAs). Previous microarray studies with PAOΔampR showed dysregulation of the small regulatory RNA rgRsmZ (21). This led us to hypothesize that other sRNAs may also be AmpR-regulated but were not detected in the microarray studies due to technical limitations. As hypothesized, RNA-Seq analysis of the ampR mutant identified many ncRNAs, both in the absence and presence of sub-MIC β-lactam exposure (Supplementary Tables S3 and S4). Some of these were tRNAs, which is expected given their abundance in the cell. Downregulated ncRNAs in PAOΔampR (AmpR positive regulation) include P8 (PA1030.1; uninduced – 30-fold, P 2.14E-06; induced – 92-fold, P 1.03E-14) and prrF1 (PA4704.1; uninduced – 4-fold, P 0.00E+00; induced NS), whereas expression of P7 (PA0887.1; uninduced 6.3-fold, P 0.00E+00; induced 3.1-fold, P 5.35E-14) and amiL (PA3366.1; uninduced 2.5-fold, P 1.82E-13; induced NS) was upregulated (AmpR negative regulation). Interestingly, rgRsmZ, which was detected in the microarray analysis, was not detected in RNA-Seq but dysregulation was confirmed by qPCR (discussed in the following sections).
AmpR-mediated regulation of some of the sRNAs was determined by qPCR. AmpR was found to positively regulate expression of P34 [PA5181.1; Relative Quantity (RQ): uninduced 0.39 ± 0.06, P 0.0003; induced 0.38 ± 0.016, P 0.002]. Expression of P34 requires RpoS (41). Because AmpR positively regulates the expression of rpoS (21), AmpR regulation of P34 is likely via RpoS. Positive regulation of P32 (PA4758.1) by AmpR and its physiological effects on the ampR mutant strain are discussed in the section on heat-shock response. Further, AmpR was also found to positively regulate the antisense RNA asPrrF1 (Supplementary Table S3) and is discussed further in the following section.
AmpR regulates iron uptake positively
Iron is critical in many biological reactions across kingdoms, and P. aeruginosa is no exception. However, freely available iron is in a poorly soluble and biologically unusable ferric (Fe3+) form at neutral pH in aerobic conditions (42). To circumvent this issue, P. aeruginosa has evolved high-affinity iron uptake systems mediated by siderophores. Siderophores are iron chelators that bind extracellular iron and transport it to receptors on the cell surface [reviewed in (42)]. Pseudomonas aeruginosa can produce and take up heme– or iron–siderophore complexes (43). In addition, P. aeruginosa also synthesizes outer membrane receptors for siderophores produced by other bacteria, including pyoverdines produced by other pseudomonads (44), and aerobactin (45) and enterobactin (45,46) produced by Enterobacteriaceae. Pseudomonas aeruginosa produces two main types of siderophores, pyoverdine and pyochelin (42). Pyoverdine is the green-yellow fluorescent pigment that is produced typically under conditions of iron limitation (47). The pvd genes involved in the synthesis of pyoverdine are clustered (PA2385–PA2426) on the PAO1 genome (35). RNA-Seq analysis of the ampR mutant revealed downregulation (3- to 103-fold) of many pvd genes (Supplementary Table S3). This includes pvdS (PA2426), the ECF sigma factor (uninduced – 7.6, P 3.49E-09; induced – 38.2, P 1.93E-08) that is known to regulate expression of the pvd genes (48). Genes encoding the second siderophore system, pyochelin, are part of a gene cluster (PA4220–PA4231) and consist of three operons (PA4220–PA4221, PA4222–PA4226 and PA4228–PA4231). Genes of these operons (PA4224–PA4226, PA4228–PA4231) are significantly downregulated (14-fold to 193-fold) in PAOΔampR in a β-lactam-independent manner (Supplementary Table S3). This is also reflected in the GOEAST analysis of the AmpR positively regulated gene set revealing a significant enrichment (log odds ratio ≥1.0, P ≤ 0.05) of the pvd genes (Supplementary Table S6). These findings suggest that AmpR is potentially involved in iron uptake.
Further, comparison of AmpR-regulated genes from this study with the iron-related genes identified as part of a previous transcriptome meta-analysis (49) revealed overlaps (Figure 4A). Genes involved in pyochelin biosynthesis are part of the 19 that are shared between the iron-regulated and AmpR positively regulated datasets (Figure 4A). The overlapping set also includes prpL and pfeR encoding a PvdS-regulated protease and a transcriptional regulator, respectively. PrpL has been implicated in virulence (50). PfeR, a two-component response regulator, positively regulates pfeA, encoding the enterobactin outer membrane receptor (51). Moreover, positive regulator of iron uptake, asPrrF1 is also downregulated in PAOΔampR in the RNA-Seq (uninduced: 4-fold, P < 0.001; induced: NS) as well as qPCR (uninduced: 0.47 ± 0.04, P 0.004; induced: NS) assays. However, expression of the master repressor of iron uptake Fur (52) is not significantly differentially regulated in the ampR RNA-Seq analyses. Downregulation of genes involved in siderophore biosynthesis, and comparison with previous meta-analysis led us to hypothesize that AmpR plays a positive regulatory role in iron uptake.
Figure 4.
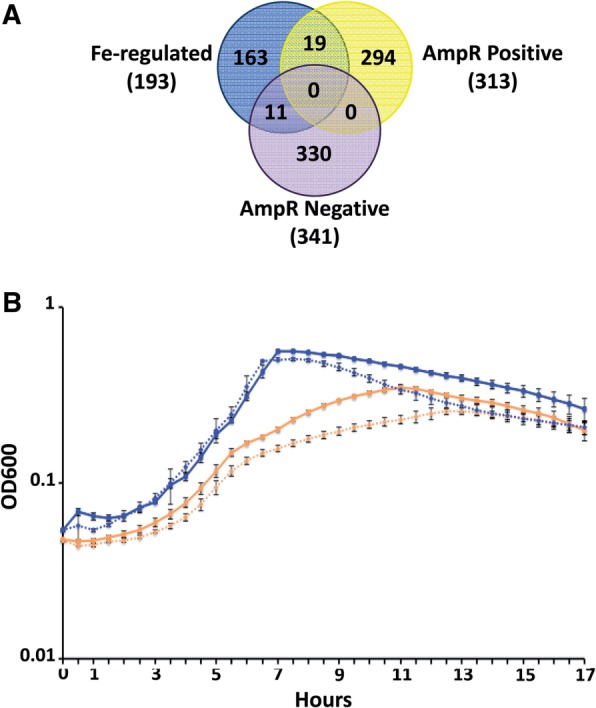
AmpR regulates iron uptake. (A) Comparing the AmpR positively and negatively regulated genes against the iron-regulated gene set shows overlaps between the datasets. (B) Growth in iron-limited media of PAO1 (solid lines) and PAOΔampR (dashed lines) in the absence (orange lines) and presence (blue lines) of exogenously added FeCl3.
To confirm the role of AmpR in iron uptake, growth curves were performed in iron-limited SSM (27). Loss of ampR resulted in impaired growth in SSM compared with PAO1 (orange lines; Figure 4B). Maximum growth difference between these strains was seen in the log and early stationary phases (P < 0.0001 at all time points between 4 and 14 h) of growth (orange lines; Figure 4B). The reduced growth of PAOΔampR in iron-deficient media can be a result of impairment in either uptake or utilization of iron. To address this, growth curves were performed with exogenously added FeCl3, making the media iron-replete (blue lines; Figure 4B). Addition of iron to the media enhanced the growth rate of both PAO1 and PAOΔampR (blue lines; Figure 4B), compared with growth in SSM alone (orange lines; Figure 4B). Moreover, under iron-replete conditions, growth of the two strains is very similar till about 10 h, after which the ampR mutant shows significantly accelerated death (blue lines; Figure 4B). Thus, in the presence of excess iron in the log phase, PAOΔampR shows no growth deficiency. This observation, and the transcriptome data, strongly suggests that AmpR plays a role in iron uptake, and not in iron utilization. The accelerated death phase seen under iron-replete conditions with PAOΔampR between 10 and 16 h is significant (blue lines, Figure 4B; P ≤ 0.0003 at all points). This is possibly due to the fact that PAOΔampR uses up all the freely available iron to maintain growth rates similar to PAO1 for the first 10 h of the experiment. When conditions start to become iron limiting (after 10 h, Figure 4B), continued growth of PAOΔampR is hampered due to impaired iron uptake. Moreover, under iron-replete conditions, neither strain produced pyoverdine, seen visually as a lack of yellow-green color of the cultures (data not shown), suggesting pyoverdine-independent iron uptake. Pseudomonas aeruginosa also has an uncharacterized low-affinity iron uptake system that functions under iron-replete conditions (P. Cornelis, personal communication) and a citrate-mediated iron uptake system (53), potentially explaining growth.
In addition to the siderophore-mediated uptake, expression of the gene encoding heme acquisition protein HasAp (PA3407) is downregulated in an AmpR-β-lactam-dependent manner (34.9-fold, P 8.5E-08; Supplementary Table S4). This further attests to the role of AmpR in iron uptake in P. aeruginosa. The pH of the media is known to influence growth in SSM (27), but there was no difference in the pH of the media between the strains (data not shown). Thus, the gene expression and phenotypic data clearly indicate a positive regulatory role for AmpR in iron uptake in P. aeruginosa.
Pseudomonas aeruginosa AmpR regulates heat-shock response by modulating rgP32 expression
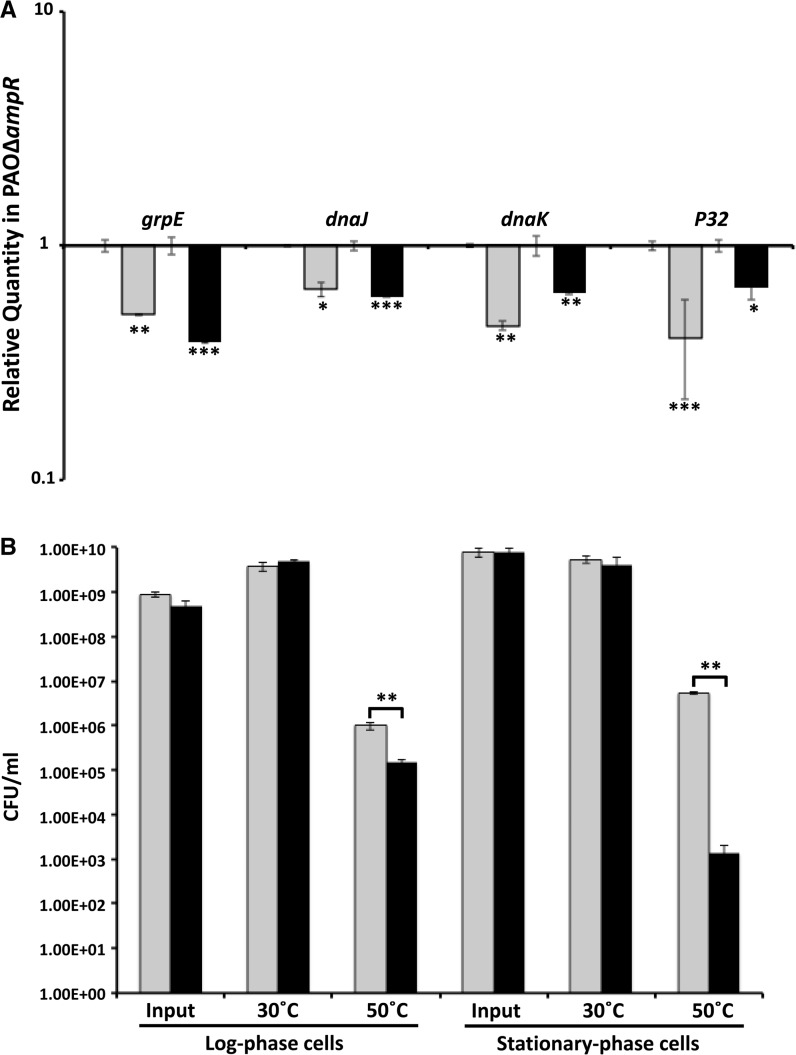
RNA-Seq analysis revealed that AmpR positively regulates the sRNA rgP32 (20.8-fold downregulated in PAOΔampR, P 7.77E-14; Supplementary Table S3). P32 is the last gene of a three-gene operon with dnaJ and dapB (35). DnaJ is part of the Hsp70 heat-shock response system (54). The DnaJ–DnaK–GrpE (PA4760–PA4762) chaperone prevents premature folding of nascent polypeptides and, along with the GroEL (Hsp60) system, helps in the heat-shock response in bacteria (54–56). DnaK is the P. aeruginosa homolog of E. coli Hsp70 (35). Conversion between the ATP- or ADP-bound forms of DnaK is controlled by DnaJ and GrpE, which function as a co-chaperone and a nucleotide exchange factor, respectively (57). Given that the sRNA is part of the operon encoding genes involved in the Hsp70 system, we hypothesized differential regulation of the Hsp70 heat-shock system in PAOΔampR compared with PAO1. qPCR analysis showed that the expression of grpE (RQ: uninduced 0.51 ± 0.004, P 0.0017; induced 0.38 ± 0.003, P 0.0003), dnaJ (RQ: uninduced 0.65 ± 0.045, P 0.015; induced 0.6 ± 0.005, P 0.0004) and dnaK (RQ: uninduced 0.45 ± 0.02, P 0.0019; induced 0.62 ± 0.007, P 0.0028) is downregulated in PAOΔampR (Figure 5A). The positive regulation is independent of sub-MIC β-lactam in the system (Figure 5A).
Figure 5.
Regulation of heat-shock response by AmpR. (A) qPCR of hsp70 genes: RNA was isolated from PAO1 and PAOΔampR cells, without and with sub-MIC β-lactam stress, reverse transcribed to cDNA and tested by qPCR with gene-specific primers, as described in the text. Relative gene expression in PAOΔampR is shown without (gray bars) and with (black bars) sub-MIC β-lactam exposure. Values have been normalized to expression in PAO1 under the same conditions (log10 RQ = 1) and bars above and below the threshold represent up- and downregulation, respectively. (B) CFU counts of heat-shock exposed and unexposed cells in the log and stationary growth phases: Cells grown at 3°C were OD600-normalized, split into two aliquots and maintained at 30° and 50°C for varying periods (3 h for log phase, 1 h for stationary phase) before enumeration. Data of PAO1 (gray bars) and PAOΔampR (black bars) are represented. *P < 0.02, **P < 0.003, ***P < 0.0005.
Downregulation of the Hsp70 heat-shock system genes in the ampR mutant led us to hypothesize that, compared with PAO1, PAOΔampR would behave differently at higher temperatures. However, no difference in growth pattern was observed between PAO1 and PAOΔampR at 30°C, 37°C or 43°C (data not shown). The heat tolerance of the two strains was then examined by enumerating CFUs after exposure of both log- and stationary-phase cells to 50°C, inducing the cellular heat-shock response. There was no significant difference in CFU for either log- or stationary-phase cells grown at 30°C (Figure 5B). However, PAOΔampR log-phase cells were more sensitive compared with PAO1, with >90% of cells killed after 3 h of 50°C exposure (P 0.0018; Figure 5B). The stationary-phase PAOΔampR cells also followed a similar trend. After a one hour exposure, loss of ampR led to a 99.9% drop in cell viability compared with PAO1, when stationary-phase cells were exposed to 50°C (P 0.0014; Figure 5B).
Compared with the log phase, the stationary-phase cells were more sensitive to the elevated temperature and show a 2-log greater drop in CFU even after a brief exposure (Figure 5B). This is counterintuitive, as at stationary phase, the cells are thought to be more resistant to changing conditions as compared with log phase. Downregulation of rpoS encoding the stationary-phase sigma factor in PAOΔampR (21) potentially plays a role in the enhanced sensitivity to temperature. Regulation of rpoH, encoding the heat-shock sigma factor, was also not significantly different between the strains in the RNA-Seq analysis (data not shown). This, however, is not surprising because expression of many sigma factors, including RpoH, is regulated post-transcriptionally (58,59). Dysregulation of the hsp70 genes, confirmed by reduced temperature tolerance of PAOΔampR, suggests a positive regulatory role for AmpR in the heat-shock response of P. aeruginosa.
AmpR positively regulates P. aeruginosa oxidative stress response
H2O2 is a byproduct of O2 metabolism whose deleterious effects on cells include altered membrane potential (60) and DNA mutation caused by single-stranded nicks (61). Intracellular H2O2 detoxification is achieved by the enzyme catalase and P. aeruginosa has four homologs: KatA (PA4236), KatB (PA4613), KatE (PA2147) and KatN [PA2185; (62)]. Of these, KatA is the major catalase and is expressed in all stages of cell growth but is produced more in the stationary phase (62). RNA-Seq analysis of PAOΔampR revealed downregulation of katA expression (−2.1-fold, P 1.6E-09) compared with PAO1 in the absence of antibiotic exposure, suggesting AmpR-dependent expression (Supplementary Table S3). Differential expression of katA was validated using qPCR (RQ uninduced: 0.12 ± 0.01, P 0.0012). The sRNA rgRgsA (PA2958.1), which requires GacA and RpoS for its expression, contributes to H2O2 resistance (63). The expression of rgRgsA is also downregulated >2-fold in PAOΔampR (Supplementary Table S3), indicating positive AmpR regulation.
Previous meta-analysis studies of P. aeruginosa transcriptomes led to the identification of genes that were specifically differentially regulated under oxidative stress conditions (49). The differential regulation of katA and rgRgsA in PAOΔampR prompted comparison of the AmpR-regulated genes with the oxidative stress gene set. Seventy genes were shared between the two conditions, 48 and 22 of which are positively and negatively regulated by AmpR, respectively (Figure 6A). The 48 positively regulated genes include the major P. aeruginosa catalase katA, and four genes involved in PQS signal biogenesis [pqsA (PA0996), pqsE (PA1000), phnA (PA1001) and phnB (PA1002)]. Interestingly, 22 of the 48 genes are clustered in a single locus on the genome that is involved in the production of R- and F-type pyocins (64,65), and are located in regions of genome plasticity (RGPs), RGP03 and RGP04 (66). These genes were also identified in a previous transcriptome study to be AmpR-regulated (21). Most of these 22 AmpR-downregulated genes that are shared with the oxidative stress gene set are involved in metabolism, including six of the nuo genes, which synthesize components of nicotinamide adenine dinucleotide dehydrogenase I (67). Another member of the LTTR family of transcriptional regulators, OxyR regulates katA expression in response to oxidative stress (68). Although ampR deletion in PAO1 did not affect oxyR expression in the RNA-Seq analysis (data not shown), qPCR analysis revealed that AmpR positively regulates oxyR expression (RQ uninduced: 0.31 ± 0.012, P 0.001).
Figure 6.
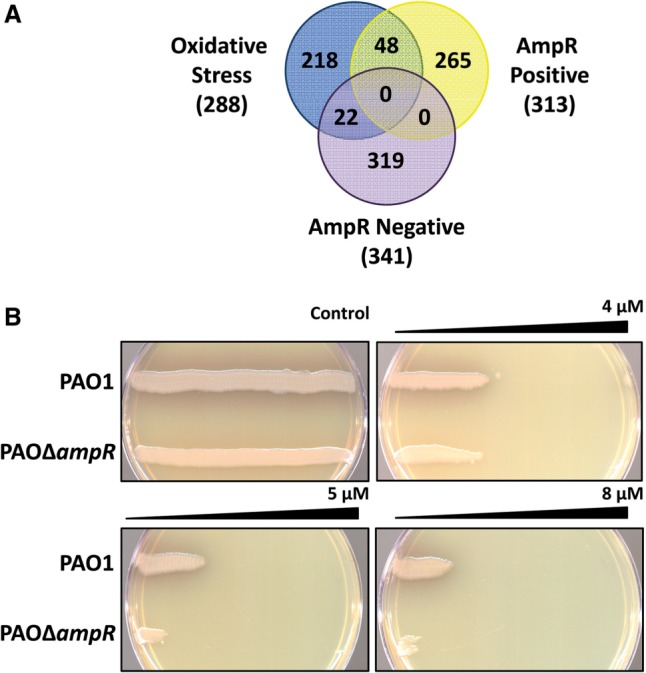
AmpR regulates resistance to oxidative stress. (A) Comparing the AmpR positively and negatively regulated genes against the oxidative stress gene set shows overlaps between the datasets. Overlapping genes include those identified previously to play a role in the oxidative stress response. (B) Gradient plates demonstrate decreased resistance of PAOΔampR to H2O2, compared with the control plate without H2O2. Representative data from four independent experiments are shown.
To determine whether reduced expression of katA and other oxidative stress-response genes translates into an observable phenotype, the H2O2 sensitivity of PAO1 and PAOΔampR was compared using the gradient plate method (37). PAOΔampR demonstrates a concentration-dependent reduced growth compared with PAO1 on the H2O2 gradient (Figure 6B), suggesting an impaired resistance to oxidative stress. This finding is in agreement with downregulation of oxidative stress response genes in PAOΔampR.
Thus, the transcriptomic and phenotypic data demonstrate a role for AmpR in positively regulating oxidative stress response in P. aeruginosa.
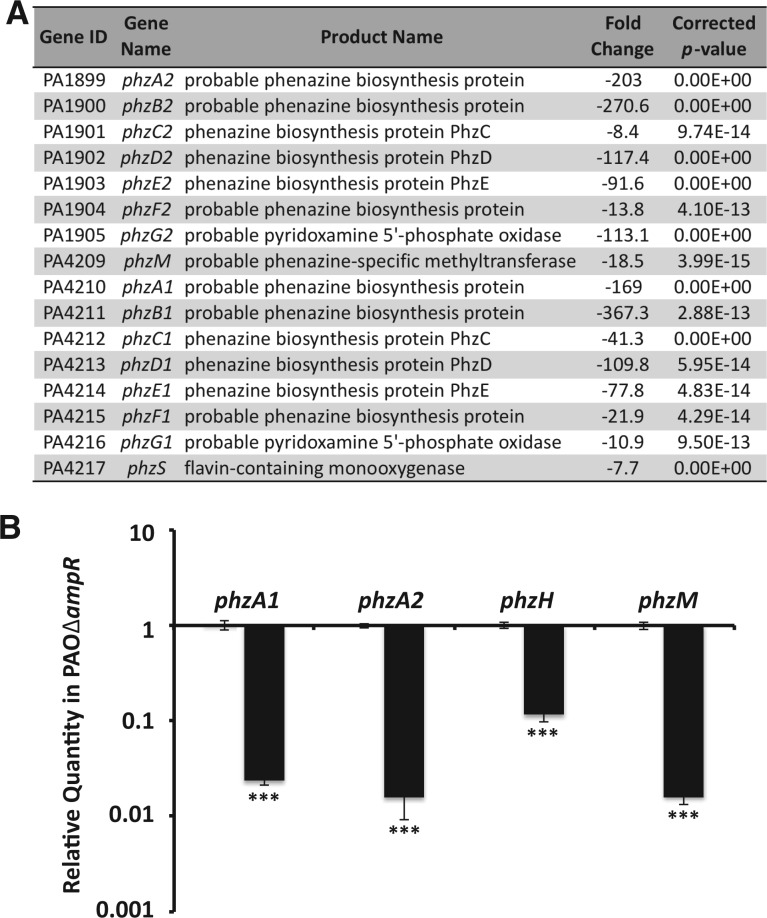
AmpR regulates phenazine production by modulating expression of phzA1-G1 and phzA2-G2 operons
The PAOΔampR strain is impaired in producing pyocyanin (21). Pseudomonas aeruginosa PAO1 and PA14 have two redundant operons phzA1-G1 (PA4210–PA4216; phz1 operon) and phzA2-G2 (PA1899–PA1905; phz2 operon) that are involved in biosynthesis of the phenazine precursor, phenazine-1-carboxylic acid (35). This precursor is then sequentially modified by a methyltransferase (PhzM, PA4209) and a monooxygenase (PhzS, PA4217) to form pyocyanin (69). The expression of all the genes involved in pyocyanin biosynthesis and export (phz1 and phz2 operons, phzM, phzS, mexGHI-opmD) was significantly reduced in the ampR mutant (positive AmpR regulation) in the RNA-Seq analysis (Figure 7A and Supplementary Table S3). The positive AmpR regulation of phzA1, phzA2 (the first genes of phz operons), phzH and phzM was also confirmed by qPCR using gene-specific primers (Figure 7B). The GOIDs under which phzM and phzS are classified (antibiotic metabolic processes, antibiotic biosynthetic processes and drug metabolic processes) also showed functional enrichment (log-odd ratio 3.46, P 0.03; Supplementary Table S6).
Figure 7.
Regulation of phenazine genes by AmpR. (A) Genes involved in phenazine biosynthesis are significantly downregulated in PAOΔampR as seen in RNA-Seq analysis. (B) Differential regulation of the first genes of the phenazine biosynthetic operons phzA1 and phzA2, and the modifying enzymes phzH and phzM, was validated by qPCR.***P < 0.0001.
The phz1 operon is under QS control, whereas phz2 operon regulators have not been identified (69,70). AmpR-mediated positive regulation of the phz2 operon is a novel finding.
The PQS is positively regulated by AmpR
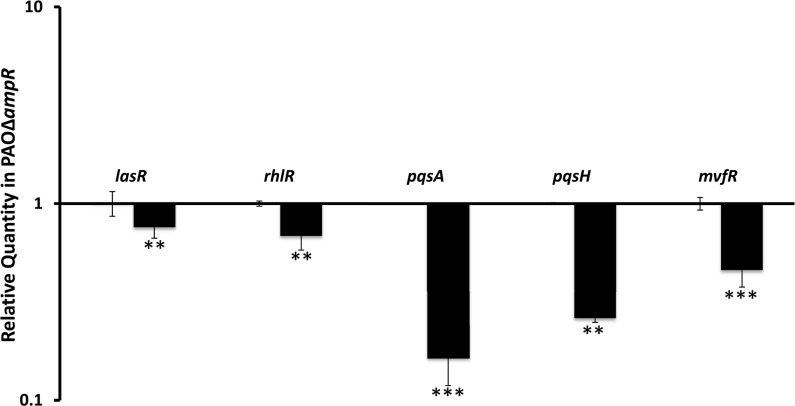
Of the 654 AmpR-dependent genes, 313 and 341 are positively and negatively regulated, respectively. Genes that are positively regulated by AmpR include the QS-controlled genes lasA, lasB, rhlAB, rhlR and hcnABC operon (Supplementary Table S3). The positive regulation of lasR and rhlR expression by AmpR was confirmed by qPCR (Figure 8). Furthermore, even under sub-MIC β-lactam exposure, AmpR positively regulated lasR (RQ induced: 0.84 ± 0.03, P 0.001) and rhlR (RQ induced: 0.71 ± 0.007, P 0.0001) expression. These concur with our previous findings that AmpR is a positive regulator of some QS phenotypes (21).
Figure 8.
AmpR positively regulates critical QS regulators of the Las system, Rhl system and PQS. Expression of the genes was determined by qPCR. The expression levels in PAOΔampR are shown, normalized to expression in PAO1. *P < 0.02, **P < 0.003, ***P < 0.0005.
The PQS is a critical part of QS signaling in P. aeruginosa, and complements the las and rhl systems (71,72). Genes of the two operons, pqsABCDE (PA0996–PA1000) and phnAB (PA1001–PA1002), and pqsH (PA2587) are involved in PQS biosynthesis (73). PhnAB converts the PQS precursor chorismate to anthralinate, which is further converted to the signaling molecule PQS by PqsA-D and PqsH (39,73). The RNA-Seq data show that AmpR positively regulates all these genes except for pqsH (Supplementary Table S3). We validated the RNA-Seq data using qPCR of the first gene of the pqs operon (pqsA) and pqsH (Figure 8). MvfR (PA1003) lies downstream of the pqs genes and positively regulates genes in this cluster (74,75). qPCR analysis revealed that AmpR positively regulates mvfR expression (Figure 8). Thus, it is likely that AmpR-mediated regulation of the PQS in P. aeruginosa is via MvfR.
Pseudomonas aeruginosa qscR (PA1898) encoding another QS regulator is located in the same locus as the phzA2 operon (76). qPCR analysis showed that the expression of qscR is significantly reduced in PAOΔampR (RQ: uninduced– 0.38 ± 0.17, P 0.0057). This further supports the role of AmpR as a QS regulator.
V5-tagged AmpR is functional in vivo
The previous study and current study suggest that the AmpR regulon in P. aeruginosa is extensive (21,22). However, it is highly unlikely that all the genes are under direct AmpR regulation. It is possible that AmpR indirectly controls a subset of genes via other regulators. Accordingly, we identified transcriptional regulators that could be potential targets of AmpR [Supplementary Tables S3 and S4, (21)]. Moreover, using the putative binding site (77), in silico analysis of the P. aeruginosa PAO1 genome identified genes that may be direct targets of AmpR (21). Some of these targets were confirmed to be differentially regulated using DNA microarrays (21). However, direct AmpR targets have not been demonstrated as yet.
To identify the direct targets of AmpR, ChIP-Seq studies were performed using a 3x-V5-tagged AmpR. As AmpR has a positive regulatory role in β-lactam resistance, the functionality of the tagged AmpR was verified by determining the MIC. Amoxicillin had an MIC of 4 µg/ml for PAOΔampR, whereas the wild-type PAO1 is resistant (>256 µg/ml). The MIC of amoxicillin on PAOΔampR::ampR-V5 was >256 µg/ml, similar to PAO1, indicating that C-terminus tagging did not inhibit AmpR function. ChIP was then performed both in the presence and absence of sub-MIC β-lactam exposure. Before proceeding with the high-throughput sequencing, validity of the pull-down assay was tested by qPCR for PampC. AmpR occupancy data for the ampC promoter revealed that compared with the input DNA, the ChIP DNA showed a 17.1 ± 1.2-fold and 21.5 ± 4.3-fold higher occupancy in the absence and presence of β-lactam stress, respectively. This demonstrated that the tagged AmpR was able to effectively bind the ampC promoter in vivo. Thus, the MIC and ChIP–qPCR studies confirmed in vivo functionality of the V5-tagged AmpR protein.
Identifying direct AmpR targets by ChIP-Seq
After confirming functionality of the tagged AmpR protein in vivo, ChIP and input DNA samples were processed on the Helicos sequencer. Data analysis was performed on the CLC Genomics Workbench as described in the Methods section. The target regions on the PAO1 genome that AmpR binds are shown in Table 2. All the loci in the table have significant Wilcoxon P < 3.0E-05. In agreement with the ChIP–qPCR data (previous section), ChIP-Seq data showed that AmpR binds to promoter DNA upstream of ampC under both induced and uninduced conditions (Table 2). This is typical of LTTRs, which bind their target sequences irrespective of effector binding (78,79), and is also seen in the PampC ChIP–qPCR (previous section).
Table 2.
AmpR ChIP peaks
| Chromosomal locus | Average read length | % Reads mapping to locus | Strand | FDR (%) | Flanking genes |
FC in RNA-Seq (AmpR Regulation) | |
|---|---|---|---|---|---|---|---|
| 5′ | 3′ | ||||||
| Uninduced | |||||||
| 586888–586930 | 42 | 88 | + | 2.0E–01 | rsmY | PA0528 | ND |
| 797294–797374 | 80 | 53 | + | 7.5E–01 | PA0728 | PA0729 | ND |
| 901840–901853 | 13 | 95 | – | 1.9E–01 | PA0826 | ssrA | ND |
| 1668963–1669003 | 40 | 95 | + | 1.0E+00 | ffs | PA1531 | ND |
| 1921430–1921543 | 113 | 95 | + | 1.2E+00 | oprF | cobA | ND |
| 3123393–3123411 | 18 | 83 | – | 5.3E–02 | PA2763 | PA2764 | ND |
| 4057616–4057641 | 25 | 89 | – | 1.6E–09 | fdxA | rsmZ | 2.0 (–ve) |
| 4592895–4593005 | 110 | 97 | – | 1.3E–04 | PA4108 | ampR | ND |
| 4362457–4362649 | 192 | 88 | + | 1.6E+00 | PA4140 | PA4141 | ND |
| 4782725–4783036 | 311 | 89 | – | 3.4E+00 | rplA | rplK | 2.1 (–ve) |
| 4956459–4956671 | 212 | 91 | – | 1.9E+00 | rnpB | PA4422 | ND |
| 5387789–5387840 | 51 | 95 | – | 6.8E–02 | PA4802 | PA4802.1 | ND |
| 5884393-5884467 | 74 | 94 | + | 1.3E+00 | ssrS | PA5228 | ND |
| 5986032–5986133 | 101 | 90 | – | 1.7E+00 | rpmG | rpmB | 2.2 (+ve) |
| 6183549–6183590 | 41 | 88 | – | 2.6E+00 | PA5492 | polA | ND |
| Induced | |||||||
| 586884–586929 | 45 | 96 | + | 2.3E–03 | rsmY | PA0528 | ND |
| 901794–901852 | 58 | 92 | – | 3.1E–03 | PA0826 | ssrA | ND |
| 1668962–1669008 | 46 | 98 | + | 6.3E–02 | ffs | PA1531 | 41.3 (+ve) |
| 3206872–3207029 | 157 | 96 | + | 1.0E–02 | PA2852.1 | oprI | ND |
| 4057616–4057641 | 25 | 88 | – | 3.2E–05 | fdxA | rsmZ | ND |
| 4592896–4593007 | 111 | 73 | – | 2.5E–03 | PA4108 | ampR | ND |
| 4956491–4956668 | 177 | 91 | – | 1.9E–03 | rnpB | PA4422 | ND |
| 5308608–5308852 | 244 | 93 | + | 3.8E–02 | crcZ | PA4726.2 | ND |
| 5387787–5387840 | 53 | 94 | – | 7.2E-02 | PA4802 | PA4802.1 | ND |
| 5884382–5884467 | 85 | 93 | + | 3.7E–01 | ssrS | PA5228 | ND |
| 6183549–6183593 | 44 | 90 | – | 1.3E+00 | PA5492 | polA | 2.4 (–ve) |
ChIP-Seq studies were performed on P. aeruginosa PAOΔampR strain harboring V5-tagged AmpR in the absence (uninduced) and presence (induced) of sub-MIC β-lactam stress. Regions on the chromosome that were enriched in the ChIP DNA samples compared with the input DNA are shown. All readings have a Wilcoxon Filter P ≤ 3.0E-05. FDR, False discovery rate; FC, Fold change; ND, Not detected.
The region with the least % false discovery rate value, irrespective of inducer presence, is within rgRsmZ (PA3621.1). The locus that was pulled down in ChIP is on the negative strand (Table 2) and corresponds to a 25-bp region in the rsmZ gene. AmpR-dependent regulation of the rsmZ gene was also seen previously in transcriptome studies using microarrays (21) and in the current RNA-Seq analysis (Supplementary Table S3). The rgRNAs rsmY and rsmZ are thought to be functionally redundant (80) and play a major role in the acute to chronic lifestyle transition of P. aeruginosa (81–83). Transcription of rsmY and rsmZ is repressed by NarL [PA3879, (84)], CafA [PA4477, (85)] and MvaT/U [PA4315, (83)], and is activated by GacA [PA2586, (86,87)].
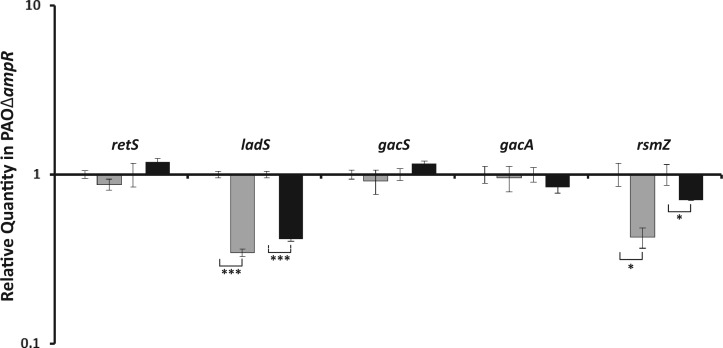
In an attempt to understand the role of AmpR in this process, qPCR assays were performed with the other players of this well-established regulatory cascade (Figure 9). Two hybrid sensor kinases LadS (PA3974) and RetS (PA4856) have a positive and negative effect, respectively, on the sensor kinase GacS (81,82). GacS activates transcription of rgRsmZ and rgRsmY indirectly through GacA (83). The rgRNAs rsmY and rsmZ sequester the RNA-binding protein RsmA (PA0905), which plays a key role in regulating > 500 genes, controlling the acute versus chronic lifestyle transition (88). qPCR analysis revealed that in the ampR mutant, expression of ladS (RQ: uninduced 0.35 ± 0.018, P < 0.0001; induced 0.42 ± 0.012, P < 0.0001) and rgRsmZ (RQ: uninduced 0.43 ± 0.059, P = 0.01; induced 0.71 ± 0.006, P = 0.02) was reduced, indicating that AmpR is required for their expression (Figure 9). No dysregulation of retS, gacS or gacA was observed (Figure 9). Our previous microarray studies (21) and our current RNA-Seq, qPCR and ChIP-Seq studies show that AmpR activates transcription of rsmZ. Furthermore, P. aeruginosa AmpR is a positive regulator of acute virulence factors, many of which are QS-regulated, while negatively regulating chronic infection phenotypes such as biofilm formation (21). The current ChIP-Seq data seem to suggest that this regulation by AmpR is mediated by the regulatory RNA rgRsmZ.
Figure 9.
Quantitative PCR analysis of Gac-Rsm pathway genes. Relative expression of genes of the RetS-LadS-GacSA-Rsm pathway in PAOΔampR compared with PAO1 was analyzed in the absence (gray bars) and presence (black bars) of sub-MIC β-lactam stress. Gene expression in the ampR mutant has been normalized to the corresponding condition in the wild-type strain and expressed as relative numbers of gene-specific transcripts. *P ≤ 0.02, ***P < 0.0001.
AmpR-binding site analysis
The ChIP data were used to identify the AmpR-binding site. A region of about 200 bp upstream of the genes that were identified to be AmpR-bound (Table 2) was used as input for regulatory sequence analysis tools [RSAT; rsat.ulb.ac.be; (89)]. These regions were queried using the putative AmpR-binding site identified earlier (77) and the matrix derived as part of genome-wide analysis (21). Using the binding sites identified in the promoter regions of the AmpR-regulated genes as input for RSAT, WebLogos were generated for both uninduced and induced datasets (Figure 10). The DNA motif that AmpR seems to bind, both in the presence and absence of effectors, is almost identical, except for minor changes at positions 1, 2, 3 and 5 (Panels A and B, Figure 10). The AmpR motif, like the LTTR box (79), is AT-rich and the conserved consensus binding sequence is As and Ts at positions 1, 6, 9, 10, 13 and 14 (Figure 10).
Figure 10.
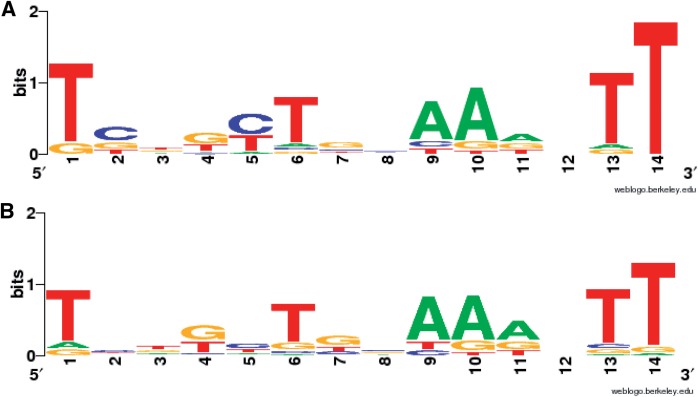
ChIP-Seq data-based AmpR-binding site analysis. Promoters of the genes that were identified by ChIP-Seq to be AmpR-regulated, either in the absence or presence of β-lactam stress, were scanned for the presence of the putative AmpR-binding motif using RSAT. The binding sites upstream of each gene were then used as input to generate a WebLogo for the uninduced (A) and induced (B). P 0.001.
LasR is a direct target of AmpR
The regulator LasR is at the top of the QS regulatory hierarchy in P. aeruginosa (90–92). Other regulators, in addition to LasR, regulate the Rhl system and PQS [reviewed in (11)]. Our findings have identified that AmpR is one of these regulators that positively regulates the Las and Rhl systems transcriptionally (PQS section earlier) and phenotypically [Figure 8; (21)]. ChIP-Seq data suggested that AmpR regulates the QS master regulator LasR directly (Table 2). This finding is further strengthened by the presence of a putative AmpR-binding motif upstream of the lasR gene. To validate the ChIP-Seq data, ChIP–qPCR was performed for selected targets using the V5-tagged AmpR strain.
As expected, the ampC promoter showed 79-fold enrichment in the ChIP DNA compared with the control DNA, demonstrating strong AmpR binding. AmpR pull-down of PampC was also confirmed using a VSVG-tagged AmpR (data not shown). ChIP–qPCR data showed that the lasR promoter was enriched 3-fold compared with the input DNA, indicating direct binding of AmpR. This binding, although not as strong as AmpR binding at PampC, is significant. Transcriptome and phenotypic data show that even at this reduced binding, AmpR is able to bring about significant changes in the QS system [Figure 8, (21)].
Comparison of microarray and RNA-Seq
To get a comprehensive picture of the AmpR regulon in P. aeruginosa, we have performed transcriptomics studies using DNA microarrays (21) and RNA-Seq (this study). Both the transcriptomics studies were performed using the same two strains (PAO1 and PAOΔampR), under identical conditions (without and with sub-MIC β-lactam exposure).
The normalized data from the microarrays and RNA-Seq were compared to compute the extent of correlation. The Pearson correlation coefficients ranged from 0.31 to 0.70 for the four different conditions. Further, comparing the differentially expressed genes under the same four conditions showed correlation coefficients ranging from 0.66 to 0.99. This degree of correlation agrees with work done previously by ‘t Hoen et al. (93).
Overlaps between the microarray and RNA-Seq datasets were determined for all the significantly differentially regulated genes in both PAO1 and PAOΔampR (Figure 11A). This revealed that deep sequencing identified 50% of the genes detected in the microarray (Figure 11A), with a correlation coefficient of 0.56 between the overlapping datasets. Examples of the overlapping genes have been discussed in the previous sections and include both positively regulated (numerous QS-regulated genes, ampC) and negatively regulated (mexEF-oprN, alginate regulators) genes. Moreover, no discordance was observed in the direction of fold change between the two techniques, and is in agreement with previous studies (94–96).
Figure 11.

Comparative analyses of AmpR microarray and RNA-Seq datasets. All the differentially expressed genes (A) and the AmpR-regulated genes (B) from our previous AmpR microarray analysis (21) and the RNA-Seq data from the current study were compared.
Comparing the AmpR-regulated genes between the microarray (21), and RNA-Seq datasets (this study) also revealed overlaps (Figure 11B). The genes identified in both the transcriptome studies to be AmpR-regulated include ampC (the primary target of AmpR) and genes of the type VI secretion system (tssJ1, tssM1 and the tse2-tsi2 operon). AmpR positively regulates the QS system [(21), this study]. Accordingly, the transcriptome studies identified AmpR-regulated QS genes such as pqsE, hcnABC (the hydrogen cyanide biosynthetic operon), phz (phenazine biosynthetic genes) and mexGHI-opmD (MexGHI-OpmD efflux pump).
Genes of the mexEF-oprN efflux pump are also downregulated in both of the transcriptome analyses (Figure 11B), and concurs with data from MIC studies (21). AmpR regulates genes present in RGPs by modulating expression of other transcriptional regulators (21). Accordingly, both microarray and transcriptome studies identified 13 genes in RGP03 and RGP04 to be differentially expressed in an AmpR-dependent manner.
Another interesting gene identified in both the microarray and the RNA-Seq analyses is PA4378. The PA4378 gene is part of a three-gene operon (PA4377–PA4379). Both the array and RNA-Seq data showed increased expression of PA4378 and PA4379 in the absence of ampR. PA4378 has a 61% similarity to E. coli InaA (35). The expression of inaA is induced under stress conditions such as pH change in E. coli (35) and ice-nucleation in Erwinia ananas (97). In E. coli, inaA expression is regulated by SoxRS, the superoxide stress response system (98). The opposing regulatory effect on inaA expression (negative) and oxidative stress (positive) suggests that AmpR might regulate these two via SoxRS. However, there was no differential regulation of soxRS in the transcriptome studies. This interesting co-regulation of antibiotic resistance with oxidative stress response and the role of AmpR in this process need further investigation.
DISCUSSION
In the opportunistic human pathogen P. aeruginosa, gene expression is a tightly controlled process with many regulators acting in concert to control virulence traits (11,12). In silico analyses and empirical evidence have identified critical regulators such as Vfr, the P. aeruginosa homolog of the E. coli cAMP receptor protein, to be central to P. aeruginosa pathogenicity (99–102). This study establishes the role of P. aeruginosa AmpR as not only a regulator of virulence factors, but also of important physiological processes such as response to oxidative stress and heat shock. In this study, we also identified sRNAs to be targets of AmpR regulation.
In the CF lung, chronic exposure to H2O2 released by polymorphonuclear leukocytes leads to P. aeruginosa overproducing the extracellular polysaccharide alginate (103). It has been established that the ECF sigma factor AlgT/U is the master regulator of alginate production by turning on constitutive expression of the algD operon during chronic infection (104). AmpR positively regulates the H2O2-mediated oxidative stress response, whereas negatively regulates algT/U expression in response to unidentified signals (22). Although it appears contradictory, AmpR is required for acute but not chronic infection, and thus it is expected to negatively regulate algT/U expression. Even though loss of ampR leads to enhanced-algT/U transcription, it does not translate into alginate production. This is due to posttranslational control of AlgT/U by its cognate anti-sigma factor MucA preventing AlgT/U-mediated algD transcription (105). Moreover, gene regulation is a complex interlinked process in P. aeruginosa, and multiple tiers of regulation for critical pathways are common (11,99,106).
AmpR is required for bacterial survival upon heat shock. However, AmpR did not regulate expression of rpoH, encoding the heat-shock sigma factor (data not shown). This is not surprising because expression of many sigma factors, including RpoH, is regulated post-transcriptionally (58,59). However, high-temperature survival may be mediated by small regulatory RNA rgP32, DnaJ and DapB (35), the members of Hsp70 heat-shock response system (54) that are positively regulated by AmpR (Figure 12).
Figure 12.
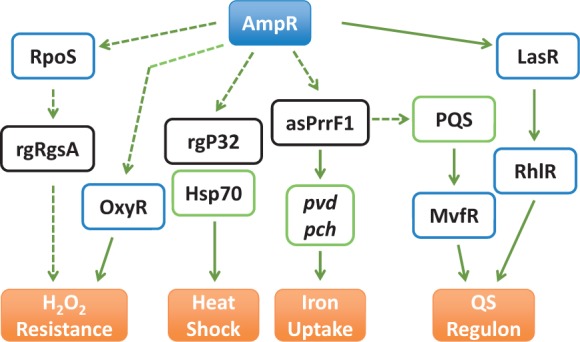
AmpR-mediated regulation of virulence and physiological processes in P. aeruginosa. AmpR affects expression of QS genes by directly binding to PlasR and modulating lasR expression. AmpR also positively regulates the PQS by modulating levels of the antisense RNA asPrrF1, thus also affecting iron uptake. By regulating expression of the stationary-phase sigma factor RpoS (21), oxyR and the small RNA rgRgsA, AmpR positively regulates the oxidative stress response. The genes encoding the Hsp70 heat-shock response system are also positively regulated by AmpR. This and previous findings (21) demonstrate that AmpR is a major regulator of virulence and physiological processes in P. aeruginosa.
Iron acquisition is a critical determinant of P. aeruginosa pathogenicity and has been proposed as a potential target to counter infections (107). In healthy lungs, free iron is available only in very low quantities, as it is typically bound by ferritin and transferrin (108). In the CF lungs, however, higher free iron concentration is attributed to successful colonization by bacteria such as P. aeruginosa (108). Pseudomonas aeruginosa uses siderophores pyoverdine and pyochelin to sequester extracellular iron under limiting conditions (109,110). In addition, the P. aeruginosa PQS helps in iron chelation, facilitating the activity of pyoverdine and pyochelin (111–113). AmpR positively regulates expression of both siderophores and PQS genes, and the loss of ampR results in growth impairment under iron-limited conditions (Figure 4). However, iron is not a limiting factor in the CF lung (108,114). Thus, the role of AmpR in iron uptake in the CF lungs needs further investigation.
AmpR positively regulates the stress-specific sigma factor RpoS and RpoS-regulated virulence factors in a growth-phase dependent manner (21). So, a subset of the AmpR-regulated genes is possibly regulated via RpoS. As the link between QS and RpoS is well established (115,116), AmpR regulation of both these processes is not surprising (Figure 12). RpoS positively regulates expression of the sRNA rgsA (PA2958.1) that contributes to H2O2 resistance in P. aeruginosa (63). GacA positively regulates rgRgsA (63). In this study, AmpR positively regulates RpoS, GacA and rgRgsA (Figure 12), as well as OxyR, the major regulator of the catalase KatA that ultimately affects resistance to H2O2 (Figure 12). GacA affects QS signaling by controlling the expression of RsmA, a negative regulator of the las QS system (83,88,117,118).
Iron uptake, QS and oxidative stress are positively regulated by AmpR. However, these phenotypes appear disparate but they are, in fact, interlinked [Figure 12, (11)]. The relationship between iron uptake and QS is complex, and some of the transcriptional regulators (such as MvfR) involved in QS regulation also modulate iron response (119–121). The iron-responsive sigma factor PvdS (PA2426) turns on mvfR transcription in response to iron starvation (122,123). In addition, the antisense RNAs, asPrrF1 and asPrrF2, positively regulate production of the PQS signaling molecule (124). Our data demonstrate that AmpR positively regulates expression of pvd (pyoverdin genes), pch (pyochelin genes), mvfR and prrF1 but not prrF2, thus affecting iron uptake and PQS synthesis. Of the 15 genes in the rgPrrF1 regulon (125), AmpR regulates positively PA2514 and PA4236, whereas negatively PA1581 and PA3531. Both PA2514 and PA1581 are part of two distinct operons (35). So AmpR could also potentially regulate other genes in the operon, but were not detected in our assay. However, it is tempting to speculate that these genes are regulated in an AmpR-independent manner. This manner of multiple tiers of gene regulation is not surprising in P. aeruginosa (11,12).
CF lung isolates are genotypically and phenotypically heterogeneous [reviewed in (126)]. Higher mutation rates are the driving force behind the P. aeruginosa population heterogeneity (127–129). Specifically, mutations in mucA and lasR arise early in the colonization process, followed by mutations in anti-mutator genes such as mutS, mutT, mutY and mutM (130). MucA mutations trigger the regulated intramembrane proteolytic cascade, freeing AlgT/U and allowing for overexpression of alginate [reviewed in (131)]. Mutations in lasR would abolish expression of QS-regulated acute virulence factors [reviewed in (132)]. In the CF lung, P. aeruginosa loses the ability to produce acute virulence phenotypes after initial colonization and starts to overexpress chronic infection traits (133). AmpR mutant strains display some characteristics reminiscent of CF isolates, including acquisition of fluoroquinolone resistance, reduced production of QS-regulated virulence factors such as proteases and pyocyanin and enhanced biofilm formation [this study, (21)]. Furthermore, loss of ampR leads to increased expression of mutY (PA5147; 2.4-fold, P 9.25E-06) and mutM (PA0357; 2.6-fold, P 5.28E-06) when exposed to β-lactam (Supplementary Table S4), indicating negative AmpR regulation. So, one would expect the mutation frequencies to decrease in the absence of ampR, given that some anti-mutators are overexpressed. However, loss of ampR did not result in significant change in mutation frequencies for rifampicin and streptomycin (data not shown). This seeming contradiction is not surprising because MutY and MutM are known to be weak anti-mutators in P. aeruginosa, unlike MutS (130). These data suggest that inactivating ampR in the CF lung, in addition to other mutations, will help P. aeruginosa colonize better. However, the occurrence and frequency of ampR mutations in CF isolates needs to be determined.
Our previous and current analyses showed that AmpR positively modulates LasR, affecting QS phenotypes [Figure 8, (21)]. ChIP-Seq and complementary data demonstrated that AmpR directly binds to PlasR. AmpR-binding site analysis using the ChIP-Seq data revealed a motif that was very similar to that identified in our previous studies using microarrays (21). Minor motif variations from the consensus sequence are expected. A consensus sequence arrived using data from multiple promoters is likely to be more accurate as compared with footprinting studies that look at individual promoters. The AmpR motif identified in this study (Figure 10) using ChIP-Seq data appears to be more refined as compared with previous analysis (21). This motif is likely to closely resemble the AmpR-binding site, as it was determined based on multiple promoters.
In conclusion, the data presented here, and previously by our laboratory, demonstrate that P. aeruginosa AmpR is a critical component in regulating virulence, metabolism and physiological processes. The clinical significance is highlighted in a recent study on extremely drug-resistant high-risk P. aeruginosa hospital isolates that harbor constitutive AmpR-activating mutations leading to AmpC overproduction (134). Moreover, given the global regulatory effect, it is likely that other virulence traits exhibited by high-risk clinical isolates (134) are also AmpR-mediated. Small molecule inhibitors targeting specific proteins such as P. aeruginosa NagZ (135) and Vibrio cholera LuxO (136) show therapeutic promise. Inhibitors of AmpR function will render the strain sensitive to β-lactam antibiotics and reduce the production of acute virulence factors. Thus, combination therapies using AmpR inhibitors and antibiotics will potentially provide us with means to counter P. aeruginosa infections, and warrant further investigation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health-Minority Biomedical Research Support SCORE grants [S06 GM08205, 5SC1AI081376 to K.M.]; Florida Department of Health [09KW-10 to G.N. and K.M.]; Florida International University (FIU) Research Assistantship (Herbert Werthiem College of Medicine, to D.B.); FIU College of Computing and Information Science Post Doctoral Fellowship (to D.B.); and FIU University Graduate School Dissertation Year Fellowship (to D.B.). Funding for open access charge: College of Engineering and Computing, Florida International University (to G.N.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Deborah Yoder-Himes (University of Louisville), Bryan Davies and William Robins (Mekalanos lab, Harvard Medical School) for guidance with RNA-Seq and ChIP-Seq experiments. They are also grateful to Lisa Schneper (Mathee lab) for helpful discussions.
REFERENCES
- 1.Morrison AJ, Jr, Wenzel RP. Epidemiology of infections due to Pseudomonas aeruginosa. Rev. Infect. Dis. 1984;6:S627–S642. doi: 10.1093/clinids/6.supplement_3.s627. [DOI] [PubMed] [Google Scholar]
- 2.Pollack M. Pseudomonas aeruginosa. In: Mandell GL, Dolan R, Bennett JE, editors. Principles and Practices of Infectious Diseases. New York: Churchill Livingston; 1995. pp. 1820–2003. [Google Scholar]
- 3.Cystic Fibrosis Foundation Annual Report. Cystic Fibrosis Foundation. Bethesda, MD; 2011. [Google Scholar]
- 4.Pier GB. Pseudomonas aeruginosa lipopolysaccharide: a major virulence factor, initiator of inflammation and target for effective immunity. Int. J. Med. Microbiol. 2007;297:277–295. doi: 10.1016/j.ijmm.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Craig L, Pique ME, Tainer JA. Type IV pilus structure and bacterial pathogenicity. Nat. Rev. Microbiol. 2004;2:363–378. doi: 10.1038/nrmicro885. [DOI] [PubMed] [Google Scholar]
- 6.Kuang Z, Hao Y, Hwang S, Zhang S, Kim E, Akinbi HT, Schurr MJ, Irvin RT, Hassett DJ, Lau GW. The Pseudomonas aeruginosa flagellum confers resistance to pulmonary surfactant protein-A by impacting the production of exoproteases through quorum-sensing. Mol. Microbiol. 2011;79:1220–1235. doi: 10.1111/j.1365-2958.2010.07516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 8.Ng WL, Bassler BL. Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riedel K, Hentzer M, Geisenberger O, Huber B, Steidle A, Wu H, Hoiby N, Givskov M, Molin S, Eberl L. N-acylhomoserine-lactone-mediated communication between Pseudomonas aeruginosa and Burkholderia cepacia in mixed biofilms. Microbiology. 2001;147:3249–3262. doi: 10.1099/00221287-147-12-3249. [DOI] [PubMed] [Google Scholar]
- 10.Bassler BL, Greenberg EP, Stevens AM. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J. Bacteriol. 1997;179:4043–4045. doi: 10.1128/jb.179.12.4043-4045.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balasubramanian D, Schneper L, Kumari H, Mathee K. A dynamic and intricate regulatory network determines Pseudomonas aeruginosa virulence. Nucleic Acids Res. 2013;41:1–20. doi: 10.1093/nar/gks1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jimenez PN, Koch G, Thompson JA, Xavier KB, Cool RH, Quax WJ. The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 2012;76:46–65. doi: 10.1128/MMBR.05007-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harmsen M, Yang L, Pamp SJ, Tolker-Nielsen T. An update on Pseudomonas aeruginosa biofilm formation, tolerance, and dispersal. FEMS Immunol. Med. Microbiol. 2010;59:253–268. doi: 10.1111/j.1574-695X.2010.00690.x. [DOI] [PubMed] [Google Scholar]
- 14.Aendekerk S, Diggle SP, Song Z, Hoiby N, Cornelis P, Williams P, Camara M. The MexGHI-OpmD multidrug efflux pump controls growth, antibiotic susceptibility and virulence in Pseudomonas aeruginosa via 4-quinolone-dependent cell-to-cell communication. Microbiology. 2005;151:1113–1125. doi: 10.1099/mic.0.27631-0. [DOI] [PubMed] [Google Scholar]
- 15.Piddock LJ. Multidrug-resistance efflux pumps - not just for resistance. Nat. Rev. Microbiol. 2006;4:629–636. doi: 10.1038/nrmicro1464. [DOI] [PubMed] [Google Scholar]
- 16.Lister PD, Wolter DJ, Hanson ND. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin. Microbiol. Rev. 2009;22:582–610. doi: 10.1128/CMR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fernandez L, Hancock RE. Adaptive and mutational resistance: role of porins and efflux pumps in drug resistance. Clin. Microbiol. Rev. 2012;25:661–681. doi: 10.1128/CMR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fothergill JL, Winstanley C, James CE. Novel therapeutic strategies to counter Pseudomonas aeruginosa infections. Expert Rev. Anti. Infect. Ther. 2012;10:219–235. doi: 10.1586/eri.11.168. [DOI] [PubMed] [Google Scholar]
- 19.National Nosocomial Infections Surveillance System. National Nosocomial Infections Surveillance (NNIS) System Report, data summary from January 1992 through June 2004, issued October 2004. Am. J. Infect. Control. 2004;32:470–485. doi: 10.1016/S0196655304005425. [DOI] [PubMed] [Google Scholar]
- 20.Kohler T, Michea-Hamzehpour M, Henze U, Gotoh N, Curty LK, Pechere JC. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol. Microbiol. 1997;23:345–354. doi: 10.1046/j.1365-2958.1997.2281594.x. [DOI] [PubMed] [Google Scholar]
- 21.Balasubramanian D, Schneper L, Merighi M, Smith R, Narasimhan G, Lory S, Mathee K. The regulatory repertoire of Pseudomonas aeruginosa AmpC ß-lactamase regulator AmpR includes virulence genes. PLoS One. 2012;7:e34067. doi: 10.1371/journal.pone.0034067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balasubramanian D, Kong KF, Jayawardena SR, Leal SM, Sautter RT, Mathee K. Co-regulation of {beta}-lactam resistance, alginate production and quorum sensing in Pseudomonas aeruginosa. J. Med. Microbiol. 2011;60:147–156. doi: 10.1099/jmm.0.021600-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, et al. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 24.Moqtaderi Z, Struhl K. Expanding the repertoire of plasmids for PCR-mediated epitope tagging in yeast. Yeast. 2008;25:287–292. doi: 10.1002/yea.1581. [DOI] [PubMed] [Google Scholar]
- 25.Hoang TT, Kutchma AJ, Becher A, Schweizer HP. Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid. 2000;43:59–72. doi: 10.1006/plas.1999.1441. [DOI] [PubMed] [Google Scholar]
- 26.Choi KH, Kumar A, Schweizer HP. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods. 2006;64:391–397. doi: 10.1016/j.mimet.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 27.Meyer JM, Abdallah MA. The fluorescent pigment of Pseudomonas fluorescens: biosynthesis, purification and physicochemical properties. J. Gen. Microbiol. 1978;107:319–328. [Google Scholar]
- 28.Yoder-Himes DR, Chain PS, Zhu Y, Wurtzel O, Rubin EM, Tiedje JM, Sorek R. Mapping the Burkholderia cenocepacia niche response via high-throughput sequencing. Proc. Natl Acad. Sci. USA. 2009;106:3976–3981. doi: 10.1073/pnas.0813403106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 30.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davies BW, Bogard RW, Mekalanos JJ. Mapping the regulon of Vibrio cholerae ferric uptake regulator expands its known network of gene regulation. Proc. Natl Acad. Sci. USA. 2011;108:12467–12472. doi: 10.1073/pnas.1107894108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mandlik A, Livny J, Robins WP, Ritchie JM, Mekalanos JJ, Waldor MK. RNA-Seq-based monitoring of infection-linked changes in Vibrio cholerae gene expression. Cell Host Microbe. 2011;10:165–174. doi: 10.1016/j.chom.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davies BW, Bogard RW, Young TS, Mekalanos JJ. Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell. 2012;149:358–370. doi: 10.1016/j.cell.2012.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wurtzel O, Yoder-Himes DR, Han K, Dandekar AA, Edelheit S, Greenberg EP, Sorek R, Lory S. The single-nucleotide resolution transcriptome of Pseudomonas aeruginosa grown in body temperature. PLoS Pathog. 2012;8:e1002945. doi: 10.1371/journal.ppat.1002945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winsor GL, Lam DK, Fleming L, Lo R, Whiteside MD, Yu NY, Hancock RE, Brinkman FS. Pseudomonas Genome Database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res. 2011;39:D596–D600. doi: 10.1093/nar/gkq869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng Q, Wang XJ. GOEAST: a web-based software toolkit for Gene Ontology enrichment analysis. Nucleic Acids Res. 2008;36:W358–W363. doi: 10.1093/nar/gkn276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown SM, Howell ML, Vasil ML, Anderson AJ, Hassett DJ. Cloning and characterization of the katB gene of Pseudomonas aeruginosa encoding a hydrogen peroxide-inducible catalase: purification of KatB, cellular localization, and demonstration that it is essential for optimal resistance to hydrogen peroxide. J. Bacteriol. 1995;177:6536–6544. doi: 10.1128/jb.177.22.6536-6544.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kong KF, Jayawardena SR, Indulkar SD, Del Puerto A, Koh CL, Hoiby N, Mathee K. Pseudomonas aeruginosa AmpR is a global transcriptional factor that regulates expression of AmpC and PoxB beta-lactamases, proteases, quorum sensing, and other virulence factors. Antimicrob. Agents Chemother. 2005;49:4567–4575. doi: 10.1128/AAC.49.11.4567-4575.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gallagher LA, McKnight SL, Kuznetsova MS, Pesci EC, Manoil C. Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J. Bacteriol. 2002;184:6472–6480. doi: 10.1128/JB.184.23.6472-6480.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Llamas MA, Mooij MJ, Sparrius M, Vandenbroucke-Grauls CM, Ratledge C, Bitter W. Characterization of five novel Pseudomonas aeruginosa cell-surface signalling systems. Mol. Microbiol. 2008;67:458–472. doi: 10.1111/j.1365-2958.2007.06061.x. [DOI] [PubMed] [Google Scholar]
- 41.Livny J, Brencic A, Lory S, Waldor MK. Identification of 17 Pseudomonas aeruginosa sRNAs and prediction of sRNA-encoding genes in 10 diverse pathogens using the bioinformatic tool sRNAPredict2. Nucleic Acids Res. 2006;34:3484–3493. doi: 10.1093/nar/gkl453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vasil ML. How we learnt about iron acquisition in Pseudomonas aeruginosa: a series of very fortunate events. Biometals. 2007;20:587–601. doi: 10.1007/s10534-006-9067-2. [DOI] [PubMed] [Google Scholar]
- 43.Cornelis P. Iron uptake and metabolism in pseudomonads. Appl. Environ. Microbiol. 2010;86:1637–1645. doi: 10.1007/s00253-010-2550-2. [DOI] [PubMed] [Google Scholar]
- 44.Hohnadel D, Meyer JM. Specificity of pyoverdine-mediated iron uptake among fluorescent Pseudomonas strains. J. Bacteriol. 1988;170:4865–4873. doi: 10.1128/jb.170.10.4865-4873.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu PV, Shokrani F. Biological activities of pyochelins: iron-chelating agents of Pseudomonas aeruginosa. Infect. Immun. 1978;22:878–890. doi: 10.1128/iai.22.3.878-890.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poole K, Young L, Neshat S. Enterobactin-mediated iron transport in Pseudomonas aeruginosa. J. Bacteriol. 1990;172:6991–6996. doi: 10.1128/jb.172.12.6991-6996.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meyer JM. Pyoverdines: pigments, siderophores and potential taxonomic markers of fluorescent Pseudomonas species. Arch. Microbiol. 2000;174:135–142. doi: 10.1007/s002030000188. [DOI] [PubMed] [Google Scholar]
- 48.Miyazaki H, Kato H, Nakazawa T, Tsuda M. A positive regulatory gene, pvdS, for expression of pyoverdin biosynthetic genes in Pseudomonas aeruginosa PAO. Mol. Gen. Genet. 1995;248:17–24. doi: 10.1007/BF02456609. [DOI] [PubMed] [Google Scholar]
- 49.Balasubramanian D, Mathee K. Comparative transcriptome analyses of Pseudomonas aeruginosa. Hum. Genomics. 2009;3:349–361. doi: 10.1186/1479-7364-3-4-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilderman PJ, Vasil AI, Johnson Z, Wilson MJ, Cunliffe HE, Lamont IL, Vasil ML. Characterization of an endoprotease (PrpL) encoded by a PvdS-regulated gene in Pseudomonas aeruginosa. Infect. Immun. 2001;69:5385–5394. doi: 10.1128/IAI.69.9.5385-5394.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dean CR, Neshat S, Poole K. PfeR, an enterobactin-responsive activator of ferric enterobactin receptor gene expression in Pseudomonas aeruginosa. J. Bacteriol. 1996;178:5361–5369. doi: 10.1128/jb.178.18.5361-5369.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ochsner UA, Vasil AI, Vasil ML. Role of the ferric uptake regulator of Pseudomonas aeruginosa in the regulation of siderophores and exotoxin A expression: purification and activity on iron-regulated promoters. J. Bacteriol. 1995;177:7194–7201. doi: 10.1128/jb.177.24.7194-7201.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marshall B, Stintzi A, Gilmour C, Meyer JM, Poole K. Citrate-mediated iron uptake in Pseudomonas aeruginosa: involvement of the citrate-inducible FecA receptor and the FeoB ferrous iron transporter. Microbiology. 2009;155:305–315. doi: 10.1099/mic.0.023531-0. [DOI] [PubMed] [Google Scholar]
- 54.Genevaux P, Georgopoulos C, Kelley WL. The Hsp70 chaperone machines of Escherichia coli: a paradigm for the repartition of chaperone functions. Mol. Microbiol. 2007;66:840–857. doi: 10.1111/j.1365-2958.2007.05961.x. [DOI] [PubMed] [Google Scholar]
- 55.Meyer AS, Baker TA. Proteolysis in the Escherichia coli heat shock response: a player at many levels. Curr. Opin. Microbiol. 2011;14:194–199. doi: 10.1016/j.mib.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hughes KT, Mathee K. The anti-sigma factors. Annu. Rev. Microbiol. 1998;52:231–286. doi: 10.1146/annurev.micro.52.1.231. [DOI] [PubMed] [Google Scholar]
- 57.Harrison C. GrpE, a nucleotide exchange factor for DnaK. Cell Stress Chaperones. 2003;8:218–224. doi: 10.1379/1466-1268(2003)008<0218:ganeff>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Osterberg S, del Peso-Santos T, Shingler V. Regulation of alternative sigma factor use. Ann. Rev. Microbiol. 2011;65:37–55. doi: 10.1146/annurev.micro.112408.134219. [DOI] [PubMed] [Google Scholar]
- 59.Potvin E, Sanschagrin F, Levesque RC. Sigma factors in Pseudomonas aeruginosa. FEMS Microbiol. Rev. 2008;32:38–55. doi: 10.1111/j.1574-6976.2007.00092.x. [DOI] [PubMed] [Google Scholar]
- 60.Farr SB, Touati D, Kogoma T. Effects of oxygen stress on membrane functions in Escherichia coli: role of HPI catalase. J. Bacteriol. 1988;170:1837–1842. doi: 10.1128/jb.170.4.1837-1842.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moody CS, Hassan HM. Mutagenicity of oxygen free radicals. Proc. Natl Acad. Sci. USA. 1982;79:2855–2859. doi: 10.1073/pnas.79.9.2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee JS, Heo YJ, Lee JK, Cho YH. KatA, the major catalase, is critical for osmoprotection and virulence in Pseudomonas aeruginosa PA14. Infect. Immun. 2005;73:4399–4403. doi: 10.1128/IAI.73.7.4399-4403.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gonzalez N, Heeb S, Valverde C, Kay E, Reimmann C, Junier T, Haas D. Genome-wide search reveals a novel GacA-regulated small RNA in Pseudomonas species. BMC Genomics. 2008;9:167. doi: 10.1186/1471-2164-9-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Michel-Briand Y, Baysse C. The pyocins of Pseudomonas aeruginosa. Biochimie. 2002;84:499–510. doi: 10.1016/s0300-9084(02)01422-0. [DOI] [PubMed] [Google Scholar]
- 65.Nakayama K, Takashima K, Ishihara H, Shinomiya T, Kageyama M, Kanaya S, Ohnishi M, Murata T, Mori H, Hayashi T. The R-type pyocin of Pseudomonas aeruginosa is related to P2 phage, and the F-type is related to lambda phage. Mol. Microbiol. 2000;38:213–231. doi: 10.1046/j.1365-2958.2000.02135.x. [DOI] [PubMed] [Google Scholar]
- 66.Mathee K, Narasimhan G, Valdes C, Qiu X, Matewish JM, Koehrsen M, Rokas A, Yandava CN, Engels R, Zeng E, et al. Dynamics of Pseudomonas aeruginosa genome evolution. Proc. Natl Acad. Sci. USA. 2008;105:3100–3105. doi: 10.1073/pnas.0711982105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weidner U, Geier S, Ptock A, Friedrich T, Leif H, Weiss H. The gene locus of the proton-translocating NADH: ubiquinone oxidoreductase in Escherichia coli. Organization of the 14 genes and relationship between the derived proteins and subunits of mitochondrial complex I. J. Mol. Biol. 1993;233:109–122. doi: 10.1006/jmbi.1993.1488. [DOI] [PubMed] [Google Scholar]
- 68.Heo YJ, Chung IY, Cho WJ, Lee BY, Kim JH, Choi KH, Lee JW, Hassett DJ, Cho YH. The major catalase gene (katA) of Pseudomonas aeruginosa PA14 is under both positive and negative control of the global transactivator OxyR in response to hydrogen peroxide. J. Bacteriol. 2010;192:381–390. doi: 10.1128/JB.00980-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Recinos DA, Sekedat MD, Hernandez A, Cohen TS, Sakhtah H, Prince AS, Price-Whelan A, Dietrich LE. Redundant phenazine operons in Pseudomonas aeruginosa exhibit environment-dependent expression and differential roles in pathogenicity. Proc. Natl Acad. Sci. USA. 2012;109:19420–19425. doi: 10.1073/pnas.1213901109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liang H, Li L, Kong W, Shen L, Duan K. Identification of a novel regulator of the quorum-sensing systems in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 2009;293:196–204. doi: 10.1111/j.1574-6968.2009.01544.x. [DOI] [PubMed] [Google Scholar]
- 71.Pesci EC, Milbank JB, Pearson JP, McKnight S, Kende AS, Greenberg EP, Iglewski BH. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc. Natl Acad. Sci USA. 1999;96:11229–11234. doi: 10.1073/pnas.96.20.11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McKnight SL, Iglewski BH, Pesci EC. The Pseudomonas quinolone signal regulates rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 2000;182:2702–2708. doi: 10.1128/jb.182.10.2702-2708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bredenbruch F, Nimtz M, Wray V, Morr M, Muller R, Haussler S. Biosynthetic pathway of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines. J. Bacteriol. 2005;187:3630–3635. doi: 10.1128/JB.187.11.3630-3635.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xiao G, He J, Rahme LG. Mutation analysis of the Pseudomonas aeruginosa mvfR and pqsABCDE gene promoters demonstrates complex quorum-sensing circuitry. Microbiology. 2006;152:1679–1686. doi: 10.1099/mic.0.28605-0. [DOI] [PubMed] [Google Scholar]
- 75.Deziel E, Gopalan S, Tampakaki AP, Lepine F, Padfield KE, Saucier M, Xiao G, Rahme LG. The contribution of MvfR to Pseudomonas aeruginosa pathogenesis and quorum sensing circuitry regulation: multiple quorum sensing-regulated genes are modulated without affecting lasRI, rhlRI or the production of N-acyl-L-homoserine lactones. Mol. Microbiol. 2005;55:998–1014. doi: 10.1111/j.1365-2958.2004.04448.x. [DOI] [PubMed] [Google Scholar]
- 76.Chugani SA, Whiteley M, Lee KM, D'Argenio D, Manoil C, Greenberg EP. QscR, a modulator of quorum-sensing signal synthesis and virulence in Pseudomonas aeruginosa. Proc. Natl Acad. Sci. USA. 2001;98:2752–2757. doi: 10.1073/pnas.051624298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zeng E, Mathee K, Narasimhan G. IEM: an algorithm for iterative enhancement of motifs using comparative genomics data. Comput. Syst. Bioinform. Conf. 2007;6:227–235. [PubMed] [Google Scholar]
- 78.Schell MA. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 1993;47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- 79.Maddocks SE, Oyston PC. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology. 2008;154:3609–3623. doi: 10.1099/mic.0.2008/022772-0. [DOI] [PubMed] [Google Scholar]
- 80.Kay E, Humair B, Denervaud V, Riedel K, Spahr S, Eberl L, Valverde C, Haas D. Two GacA-dependent small RNAs modulate the quorum-sensing response in Pseudomonas aeruginosa. J. Bacteriol. 2006;188:6026–6033. doi: 10.1128/JB.00409-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ventre I, Goodman AL, Vallet-Gely I, Vasseur P, Soscia C, Molin S, Bleves S, Lazdunski A, Lory S, Filloux A. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc. Natl Acad. Sci. USA. 2006;103:171–176. doi: 10.1073/pnas.0507407103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Goodman AL, Kulasekara B, Rietsch A, Boyd D, Smith RS, Lory S. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev.Cell. 2004;7:745–754. doi: 10.1016/j.devcel.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 83.Brencic A, McFarland KA, McManus HR, Castang S, Mogno I, Dove SL, Lory S. The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol. Microbiol. 2009;73:434–445. doi: 10.1111/j.1365-2958.2009.06782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.O'Callaghan J, Reen FJ, Adams C, O'Gara F. Low oxygen induces the type III secretion system in Pseudomonas aeruginosa via modulation of the small RNAs rsmZ and rsmY. Microbiology. 2011;157:3417–3428. doi: 10.1099/mic.0.052050-0. [DOI] [PubMed] [Google Scholar]
- 85.Petrova OE, Sauer K. The novel two-component regulatory system BfiSR regulates biofilm development by controlling the small RNA rsmZ through CafA. J. Bacteriol. 2010;192:5275–5288. doi: 10.1128/JB.00387-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Heeb S, Blumer C, Haas D. Regulatory RNA as mediator in GacA/RsmA-dependent global control of exoproduct formation in Pseudomonas fluorescens CHA0. J. Bacteriol. 2002;184:1046–1056. doi: 10.1128/jb.184.4.1046-1056.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Valverde C, Heeb S, Keel C, Haas D. RsmY, a small regulatory RNA, is required in concert with RsmZ for GacA-dependent expression of biocontrol traits in Pseudomonas fluorescens CHA0. Mol. Microbiol. 2003;50:1361–1379. doi: 10.1046/j.1365-2958.2003.03774.x. [DOI] [PubMed] [Google Scholar]
- 88.Brencic A, Lory S. Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol. Microbiol. 2009;72:612–632. doi: 10.1111/j.1365-2958.2009.06670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Turatsinze JV, Thomas-Chollier M, Defrance M, van Helden J. Using RSAT to scan genome sequences for transcription factor binding sites and cis-regulatory modules. Nat. Protoc. 2008;3:1578–1588. doi: 10.1038/nprot.2008.97. [DOI] [PubMed] [Google Scholar]
- 90.Pesci EC, Pearson JP, Seed PC, Iglewski BH. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 1997;179:3127–3132. doi: 10.1128/jb.179.10.3127-3132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Latifi A, Foglino M, Tanaka K, Williams P, Lazdunski A. A hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhIR (VsmR) to expression of the stationary-phase sigma factor RpoS. Mol. Microbiol. 1996;21:1137–1146. doi: 10.1046/j.1365-2958.1996.00063.x. [DOI] [PubMed] [Google Scholar]
- 92.Medina G, Juarez K, Diaz R, Soberon-Chavez G. Transcriptional regulation of Pseudomonas aeruginosa rhlR, encoding a quorum-sensing regulatory protein. Microbiology. 2003;149:3073–3081. doi: 10.1099/mic.0.26282-0. [DOI] [PubMed] [Google Scholar]
- 93.'t Hoen PA, Ariyurek Y, Thygesen HH, Vreugdenhil E, Vossen RH, de Menezes RX, Boer JM, van Ommen GJ, den Dunnen JT. Deep sequencing-based expression analysis shows major advances in robustness, resolution and inter-lab portability over five microarray platforms. Nucleic Acids Res. 2008;36:e141. doi: 10.1093/nar/gkn705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sirbu A, Kerr G, Crane M, Ruskin HJ. RNA-Seq vs dual- and single-channel microarray data: sensitivity analysis for differential expression and clustering. PLoS One. 2012;7:e50986. doi: 10.1371/journal.pone.0050986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kogenaru S, Qing Y, Guo Y, Wang N. RNA-seq and microarray complement each other in transcriptome profiling. BMC Genomics. 2012;13:629. doi: 10.1186/1471-2164-13-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nookaew I, Papini M, Pornputtapong N, Scalcinati G, Fagerberg L, Uhlen M, Nielsen J. A comprehensive comparison of RNA-Seq-based transcriptome analysis from reads to differential gene expression and cross-comparison with microarrays: a case study in Saccharomyces cerevisiae. Nucleic Acids Res. 2012;40:10084–10097. doi: 10.1093/nar/gks804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Abe K, Watabe S, Emori Y, Watanabe M, Arai S. An ice nucleation active gene of Erwinia ananas. Sequence similarity to those of Pseudomonas species and regions required for ice nucleation activity. FEBS Lett. 1989;258:297–300. doi: 10.1016/0014-5793(89)81678-3. [DOI] [PubMed] [Google Scholar]
- 98.Rosner JL, Slonczewski JL. Dual regulation of inaA by the multiple antibiotic resistance (mar) and superoxide (soxRS) stress response systems of Escherichia coli. J. Bacteriol. 1994;176:6262–6269. doi: 10.1128/jb.176.20.6262-6269.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Balasubramanian D, Murugapiran SK, Silva-Herzog E, Schneper L, Yang X, Tatke G, Narasimhan G, Mathee K. In: Bacterial Gene Regulation and Transcriptional Networks. Madan Babu MB, editor. Poole, UK: Caiser Academic Press; 2013. [Google Scholar]
- 100.Fuchs EL, Brutinel ED, Jones AK, Fulcher NB, Urbanowski ML, Yahr TL, Wolfgang MC. The Pseudomonas aeruginosa Vfr regulator controls global virulence factor expression through cyclic AMP-dependent and -independent mechanisms. J. Bacteriol. 2010;192:3553–3564. doi: 10.1128/JB.00363-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Davinic M, Carty NL, Colmer-Hamood JA, San Francisco M, Hamood AN. Role of Vfr in regulating exotoxin A production by Pseudomonas aeruginosa. Microbiology. 2009;155:2265–2273. doi: 10.1099/mic.0.028373-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ferrell E, Carty NL, Colmer-Hamood JA, Hamood AN, West SE. Regulation of Pseudomonas aeruginosa ptxR by Vfr. Microbiology. 2008;154:431–439. doi: 10.1099/mic.0.2007/011577-0. [DOI] [PubMed] [Google Scholar]
- 103.Mathee K, Ciofu O, Sternberg C, Lindum PW, Campbell JI, Jensen P, Johnsen AH, Givskov M, Ohman DE, Molin S, et al. Mucoid conversion of Pseudomonas aeruginosa by hydrogen peroxide: a mechanism for virulence activation in the cystic fibrosis lung. Microbiology. 1999;145:1349–1357. doi: 10.1099/13500872-145-6-1349. [DOI] [PubMed] [Google Scholar]
- 104.Wozniak DJ, Ohman DE. Transcriptional analysis of the Pseudomonas aeruginosa genes algR, algB, and algD reveals a hierarchy of alginate gene expression which is modulated by algT. J. Bacteriol. 1994;176:6007–6014. doi: 10.1128/jb.176.19.6007-6014.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mathee K, McPherson CJ, Ohman DE. Posttranslational control of the algT (algU)-encoded sigma22 for expression of the alginate regulon in Pseudomonas aeruginosa and localization of its antagonist proteins MucA and MucB (AlgN) J. Bacteriol. 1997;179:3711–3720. doi: 10.1128/jb.179.11.3711-3720.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Coggan KA, Wolfgang MC. Global regulatory pathways and cross-talk control Pseudomonas aeruginosa environmental lifestyle and virulence phenotype. Curr. Issues Mol. Biol. 2012;14:47–70. [PubMed] [Google Scholar]
- 107.Smith DJ, Lamont IL, Anderson GJ, Reid DW. Targeting iron uptake to control Pseudomonas aeruginosa infections in cystic fibrosis. Eur. Respir J. 2012;42:1723–1736. doi: 10.1183/09031936.00124012. [DOI] [PubMed] [Google Scholar]
- 108.Reid DW, Carroll V, O'May C, Champion A, Kirov SM. Increased airway iron as a potential factor in the persistence of Pseudomonas aeruginosa infection in cystic fibrosis. Eur. Respir J. 2007;30:286–292. doi: 10.1183/09031936.00154006. [DOI] [PubMed] [Google Scholar]
- 109.Meyer JM, Neely A, Stintzi A, Georges C, Holder IA. Pyoverdin is essential for virulence of Pseudomonas aeruginosa. Infect. Immun. 1996;64:518–523. doi: 10.1128/iai.64.2.518-523.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Martin LW, Reid DW, Sharples KJ, Lamont IL. Pseudomonas siderophores in the sputum of patients with cystic fibrosis. Biometals. 2011;24:1059–1067. doi: 10.1007/s10534-011-9464-z. [DOI] [PubMed] [Google Scholar]
- 111.Diggle SP, Matthijs S, Wright VJ, Fletcher MP, Chhabra SR, Lamont IL, Kong X, Hider RC, Cornelis P, Camara M, et al. The Pseudomonas aeruginosa 4-quinolone signal molecules HHQ and PQS play multifunctional roles in quorum sensing and iron entrapment. Chem. Biol. 2007;14:87–96. doi: 10.1016/j.chembiol.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 112.Bredenbruch F, Geffers R, Nimtz M, Buer J, Haussler S. The Pseudomonas aeruginosa quinolone signal (PQS) has an iron-chelating activity. Environ. Microbiol. 2006;8:1318–1329. doi: 10.1111/j.1462-2920.2006.01025.x. [DOI] [PubMed] [Google Scholar]
- 113.Mashburn LM, Whiteley M. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature. 2005;437:422–425. doi: 10.1038/nature03925. [DOI] [PubMed] [Google Scholar]
- 114.Stites SW, Walters B, O'Brien-Ladner AR, Bailey K, Wesselius LJ. Increased iron and ferritin content of sputum from patients with cystic fibrosis or chronic bronchitis. Chest. 1998;114:814–819. doi: 10.1378/chest.114.3.814. [DOI] [PubMed] [Google Scholar]
- 115.Schuster M, Greenberg EP. Early activation of quorum sensing in Pseudomonas aeruginosa reveals the architecture of a complex regulon. BMC Genomics. 2007;8:287. doi: 10.1186/1471-2164-8-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Schuster M, Hawkins AC, Harwood CS, Greenberg EP. The Pseudomonas aeruginosa RpoS regulon and its relationship to quorum sensing. Mol. Microbiol. 2004;51:973–985. doi: 10.1046/j.1365-2958.2003.03886.x. [DOI] [PubMed] [Google Scholar]
- 117.Heeb S, Haas D. Regulatory roles of the GacS/GacA two-component system in plant-associated and other Gram-negative bacteria. Mol. Plant Microbe. Interact. 2001;14:1351–1363. doi: 10.1094/MPMI.2001.14.12.1351. [DOI] [PubMed] [Google Scholar]
- 118.Wolfgang MC, Lee VT, Gilmore ME, Lory S. Coordinate regulation of bacterial virulence genes by a novel adenylate cyclase-dependent signaling pathway. Dev. Cell. 2003;4:253–263. doi: 10.1016/s1534-5807(03)00019-4. [DOI] [PubMed] [Google Scholar]
- 119.Lamb JR, Patel H, Montminy T, Wagner VE, Iglewski BH. Functional domains of the RhlR transcriptional regulator of Pseudomonas aeruginosa. J. Bacteriol. 2003;185:7129–7139. doi: 10.1128/JB.185.24.7129-7139.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Schuster M, Lostroh CP, Ogi T, Greenberg EP. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 2003;185:2066–2079. doi: 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Juhas M, Wiehlmann L, Salunkhe P, Lauber J, Buer J, Tummler B. GeneChip expression analysis of the VqsR regulon of Pseudomonas aeruginosa TB. FEMS Microbiol. Lett. 2005;242:287–295. doi: 10.1016/j.femsle.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 122.Visca P, Leoni L, Wilson MJ, Lamont IL. Iron transport and regulation, cell signalling and genomics: lessons from Escherichia coli and Pseudomonas. Mol. Microbiol. 2002;45:1177–1190. doi: 10.1046/j.1365-2958.2002.03088.x. [DOI] [PubMed] [Google Scholar]
- 123.Ochsner UA, Wilderman PJ, Vasil AI, Vasil ML. GeneChip expression analysis of the iron starvation response in Pseudomonas aeruginosa: identification of novel pyoverdine biosynthesis genes. Mol. Microbiol. 2002;45:1277–1287. doi: 10.1046/j.1365-2958.2002.03084.x. [DOI] [PubMed] [Google Scholar]
- 124.Oglesby AG, Farrow JM, 3rd, Lee JH, Tomaras AP, Greenberg EP, Pesci EC, Vasil ML. The influence of iron on Pseudomonas aeruginosa physiology: a regulatory link between iron and quorum sensing. J. Biol. Chem. 2008;283:15558–15567. doi: 10.1074/jbc.M707840200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wilderman PJ, Sowa NA, FitzGerald DJ, FitzGerald PC, Gottesman S, Ochsner UA, Vasil ML. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc. Natl Acad. Sci. USA. 2004;101:9792–9797. doi: 10.1073/pnas.0403423101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Folkesson A, Jelsbak L, Yang L, Johansen HK, Ciofu O, Hoiby N, Molin S. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: an evolutionary perspective. Nat. Rev. Microbiol. 2012;10:841–851. doi: 10.1038/nrmicro2907. [DOI] [PubMed] [Google Scholar]
- 127.Feliziani S, Lujan AM, Moyano AJ, Sola C, Bocco JL, Montanaro P, Canigia LF, Argarana CE, Smania AM. Mucoidy, quorum sensing, mismatch repair and antibiotic resistance in Pseudomonas aeruginosa from cystic fibrosis chronic airways infections. PLoS One. 2010;5:e12669. doi: 10.1371/journal.pone.0012669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Oliver A, Mena A. Bacterial hypermutation in cystic fibrosis, not only for antibiotic resistance. Clin. Microbiol. Infect. 2010;16:798–808. doi: 10.1111/j.1469-0691.2010.03250.x. [DOI] [PubMed] [Google Scholar]
- 129.Mena A, Smith EE, Burns JL, Speert DP, Moskowitz SM, Perez JL, Oliver A. Genetic adaptation of Pseudomonas aeruginosa to the airways of cystic fibrosis patients is catalyzed by hypermutation. J. Bacteriol. 2008;190:7910–7917. doi: 10.1128/JB.01147-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ciofu O, Mandsberg LF, Bjarnsholt T, Wassermann T, Hoiby N. Genetic adaptation of Pseudomonas aeruginosa during chronic lung infection of patients with cystic fibrosis: strong and weak mutators with heterogeneous genetic backgrounds emerge in mucA and/or lasR mutants. Microbiology. 2010;156:1108–1119. doi: 10.1099/mic.0.033993-0. [DOI] [PubMed] [Google Scholar]
- 131.Damron FH, Goldberg JB. Proteolytic regulation of alginate overproduction in Pseudomonas aeruginosa. Mol. Microbiol. 2012;84:595–607. doi: 10.1111/j.1365-2958.2012.08049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Venturi V. Regulation of quorum sensing in Pseudomonas. FEMS Microbiol. Rev. 2006;30:274–291. doi: 10.1111/j.1574-6976.2005.00012.x. [DOI] [PubMed] [Google Scholar]
- 133.Ciofu O, Mandsberg LF, Wang H, Hoiby N. Phenotypes selected during chronic lung infection in cystic fibrosis patients: implications for the treatment of Pseudomonas aeruginosa biofilm infections. FEMS Immunol. Med. Microbiol. 2012;65:215–225. doi: 10.1111/j.1574-695X.2012.00983.x. [DOI] [PubMed] [Google Scholar]
- 134.Cabot G, Ocampo-Sosa AA, Dominguez MA, Gago JF, Juan C, Tubau F, Rodriguez C, Moya B, Pena C, Martinez-Martinez L, et al. Genetic markers of widespread extensively drug-resistant Pseudomonas aeruginosa high-risk clones. Antimicrob. Agents Chemother. 2012;56:6349–6357. doi: 10.1128/AAC.01388-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Asgarali A, Stubbs KA, Oliver A, Vocadlo DJ, Mark BL. Inactivation of the glycoside hydrolase NagZ attenuates antipseudomonal beta-lactam resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2009;53:2274–2282. doi: 10.1128/AAC.01617-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ng WL, Perez L, Cong J, Semmelhack MF, Bassler BL. Broad spectrum pro-quorum-sensing molecules as inhibitors of virulence in vibrios. PLoS Pathog. 2012;8:e1002767. doi: 10.1371/journal.ppat.1002767. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.