Abstract
Hereditary hypophosphatemic rickets with hypercalciuria (HHRH) is a rare metabolic disorder, characterized by hypophosphatemia, variable degrees of rickets/osteomalacia, and hypercalciuria secondary to increased serum 1,25-dihydroxyvitamin D [1,25(OH)2D] levels. HHRH is caused by mutations in the SLC34A3 gene, which encodes sodium-phosphate co-transporter type IIc. A 6 ½-year-old female presented with a history of nephrolithiasis. Her metabolic evaluation revealed increased 24- hour urine calcium excretion with high serum calcium, low intact parathyroid hormone (PTH) levels, and elevated 1,25(OH)2D level. In addition, the patient had low to low-normal serum phosphorus with high urine phosphorus. The patient had normal stature; without rachitic or boney deformities or a history of fractures. Genetic analysis of SLC34A3 revealed the patient to be a compound heterozygote for a novel single base pair deletion in exon 12 (c.1304delG) and 30-base pair deletion in intron 6 (g.1440–1469del). The single-base pair mutation causes a frameshift, which results in premature stop codon. The intronic deletion is likely caused by misalignment of the 4-basepair homologous repeats and results in the truncation of an already small intron to 63 bp, which would impair proper RNA splicing of the intron. This is the fourth unique intronic deletion identified in patients with HHRH, suggesting the frequent occurrence of sequence misalignments in SLC34A3 and the importance of screening introns in patients with HHRH.
Keywords: Deletion, HHRH, hypercalciuria, hypophosphatemia, intron, nephrolithiasis
Introduction
Hereditary hypophosphatemic rickets with hypercalciuria (HHRH; OMIM 241530) is a rare genetic disorder of phosphate homeostasis, which was first described in a large Bedouin kindred [1]. Similar to other genetic phosphate wasting disorders such as X-linked hypophosphatemia (XLH) and autosomal dominant hypophosphatemic rickets (ADHR), HHRH is classically characterized by decreased renal phosphate reabsorption, hypophosphatemia, and rickets/osteomalacia [1, 2]. In addition, individuals with HHRH often suffer from bone pain, muscle weakness, and growth retardation [1]. However, HHRH is distinguished from XLH and ADHR by the presence of an appropriately elevated level of serum 1,25- dihydroxyvitamin D [1,25(OH)2D; calcitriol] concentration, which leads to hypercalciuria and suppression of parathyroid hormone (PTH) secretion [1, 2]. The attendant hypercalciuria can lead to the formations of kidney stones and/or the occurrence of nephrocalcinosis in affected patients. HHRH is caused by mutations occurring in the SLC34A3 gene, which encodes the sodium-phosphate co-transporter type IIc (NaPi-IIc or NPT2c) [3, 4]. NaPi-IIc, along with NaPi-IIa encoded by SLC34A1, reabsorb most of filtered phosphate in the renal proximal tubule, and expression of these co-transporters are regulated by two phosphaturic hormones – PTH and fibroblast growth factor 23 (FGF23). Most mutations reported to date are single nucleotide substitutions, leading to changes in encoded amino acids. However, there have been reports of intronic deletions that affect intron sizes rather than splice sites [5]. In this study, we performed mutation analysis of the SLC34A3 gene in a 6 ½-year-old patient presenting with kidney stones and lab/clinic parameters consistent with HHRH.
Subjects and Methods
Case
Our patient is an 11–1/2 year old female that presented at 6 years of age with gross hematuria, right flank pain, nausea, dysuria, and urinary urgency. She was diagnosed with a right lower-pole renal calculus via abdominal CT which otherwise revealed normal size and appearance of both kidneys. The patient was well developed without rachitic or boney deformities. Height was at the 20th percentile. The patient had no history of fractures. There was neither a family history of rickets nor kidney stone disease. As shown in Table 1, her initial metabolic evaluation revealed a 24-hour urine calcium excretion of 17.8 mg/kg/day (normal < 4 mg/kg/day) with a serum calcium level of 9.6 mg/dL and intact PTH of 12.6 pg/mL. The patient had a serum phosphorus of 3.0 mg/dL (normal 3.7 – 5.6), 25-hydroxyvitamin D [25(OH)D] level was 42.4 ng/mL with a 1,25(OH)2D of 127 pg/mL (normal 15 – 90). FGF23 level was 56 pg/mL (normal, 29.7 ± 20.7) [6]. The patient had normal renal function with an estimated GFR of 95 ml/min/1.73m2. Her urinalysis was unremarkable without glucosuria or proteinuria. Her urine beta-2 microglobulin excretion was normal (34 μg/gram creatinine). Repeat labs revealed serum phosphorus of 4.0 mg/dL with a low tubular threshold for phosphate reabsorption (TmP/GFR) of 3.75 mg/dL and inappropriately high phosphorus excretion of 105 mg/kg/day. A chest x-ray and erythrocyte sedimentation rate were both normal. A renal ultrasound showed bilateral nephrocalcinosis. Her biologic parent’s urine calcium to creatinine ratios were in the normal range. The patient underwent dual x-ray absorptiometry scanning which revealed an L-spine z-score of −1.9. The patient was supplemented with 2 mmol/kg/day of oral phosphorus. Within a month of treatment, the urine calcium to creatinine ratio was reduced to 0.08 despite persistent elevations in 1,25(OH)2D. An MRI of the lower extremities did not show rickets. She did pass a 6 mm stone when she first presented in 2007, and in spite of generous fluid intake and phosphorus supplementation, she had another episode of renal colic at age 9 years, secondary to another passing kidney stone. A renal ultrasound at that time showed a 7 mm stone in the right UVJ with hydronephrosis and pelviectasis. At her follow up visit in August 2011, a timed urine collection showed a high urinary calcium (10.7 mg/Kg/day), bone age films did not show rickets and her biochemical profile revealed a serum creatinine of 0.7 mg/dl (eGFR 107 ml/min.1.73 m2), serum calcium of 10.8 mg/dl (normal range 8.6–10.2), serum phosphorus 4.0 (normal range 4.1–5.4), intact PTH 6 pg/ml (normal range 12–65), and 1,25(OH)2D of 78 pg/ml. Her treatment consisted of phosphorus supplementation (K-Phos neutral) 2,750 mg/day in three divided doses.
Table 1.
Demographic and clinical characteristics
| 9/19/08 | 8/24/09 | 12/10/10 | 7/18/11 | 7/30/12 | Reference Values | |
|---|---|---|---|---|---|---|
| Age (year) | 6.9 | 7.8 | 9.1 | 9.7 | 10.7 | |
| Height (cm) | 116.1 | 124 | 134.1 | 135.2 | 141.2 | |
| Percentile (%) | 20 | 25 | 39 | 43 | 45 | |
| Weight (kg) | 19.3 | 23.8 | 28 | 30.3 | 31.5 | |
| Percentile (%) | 25 | 25 | 39 | 41 | 24 | |
| Calcium (mg/dL) | 9.6 | 10.2 | 9.8 | 10.8 | 10.8 | 8.6–10.2 |
| Phosphorus (mg/dL) | 3.0 | 4.0 | 3.8 | 4.0* | 3.0 | 3.7–5.6, 4.1–5.4* |
| Creatinine (mg/dL) | 0.5 | 0.6 | 0.6 | 0.2–0.73 | ||
| Intact PTH (pg/mL) | 12.6 | 14.5 | 6 | 5 | 12–65 | |
| 25-hydroxyvitamin D (ng/mL) | 42.4 | 46.2 | 25–80 | |||
| 1,25-dihyroxyvitamin D (pg/mL) | 127 | 143 | 78 | 94* | 15–90, 30–83* | |
| 24-hour urine calcium (mg/kg/day) | 17.8 | 4.5 | 10.7 | Mean 2.4 SD 0.7 | ||
| 24-hour urine phosphorus (mg/kg/day) | 24 | 105 | 50 | Mean 14.3 SD 7.2 | ||
| TmP/GFR (mg/100 mL GFR) | 3.75 |
SI unit conversion: Multiply by 0.25 (calcium, mmol/L), 0.323 (phosphorus, mmol/L), 88.4 (creatinine, μmol/L), 1.0 (PTH, ng/L), 2.496 (25-hydroxyvitamin D, nmol/L), 2.6 (1,25-dihyroxyvitamin D, pmol/L).
The study was approved by the Institutional Review Board of Indiana University-Purdue University Indianapolis. Written informed consent was obtained from the patient’s legal guardians prior to participation in the study.
Mutation Analysis
Genomic DNA was extracted from blood obtained from the patient. All 13 exons (one non-coding and 12 coding), as well as adjacent intronic sequences, were amplified in eight fragments using Multiplex PCR Kit (QIAGEN Inc., Valencia, CA). Primer sequences and PCR conditions are available upon request. PCR products were electrophoresed in a 2% agarose gel and purified using DNA Gel Extraction Kit (QIAGEN). Approximately 100 ng of PCR products were directly sequenced from forward and/or reverse PCR primer, using Big-Dye Terminator Cycle Sequencing Kit and the ABI PRISM 3100 Genetic Analyzer (Applied Biosystems).
Sequence variations identified by DNA sequencing were tested in race-matched control DNA samples. Single nucleotide deletion in exon 12 was analyzed by PCR-RFLP analysis. Genomic DNA fragments were amplified by PCR (forward: 5’-ACGTTGGATGTGCTGGCTGCCCTGGTCA-3’ and reverse: 5’- ACGTTGGATGGTGGGACTCGGCCTTTGCAG-3’). The 10-bp tails (italicized) were added to each primer to improve separation between normal and mutant alleles during agarose gel electrophoresis. The mismatch nucleotide (underlined) was introduced in the forward primer to create BstEII (New England Biolabs, Beverly, MA) restriction site only in the mutant allele. The digested PCR products were electrophoresed in an agarose gel and visualized under the UV light. Deletion in intron 6 was tested by using the same PCR primers originally used for sequencing the patient, and the PCR products were directly analyzed by agarose gel electrophoresis.
Results
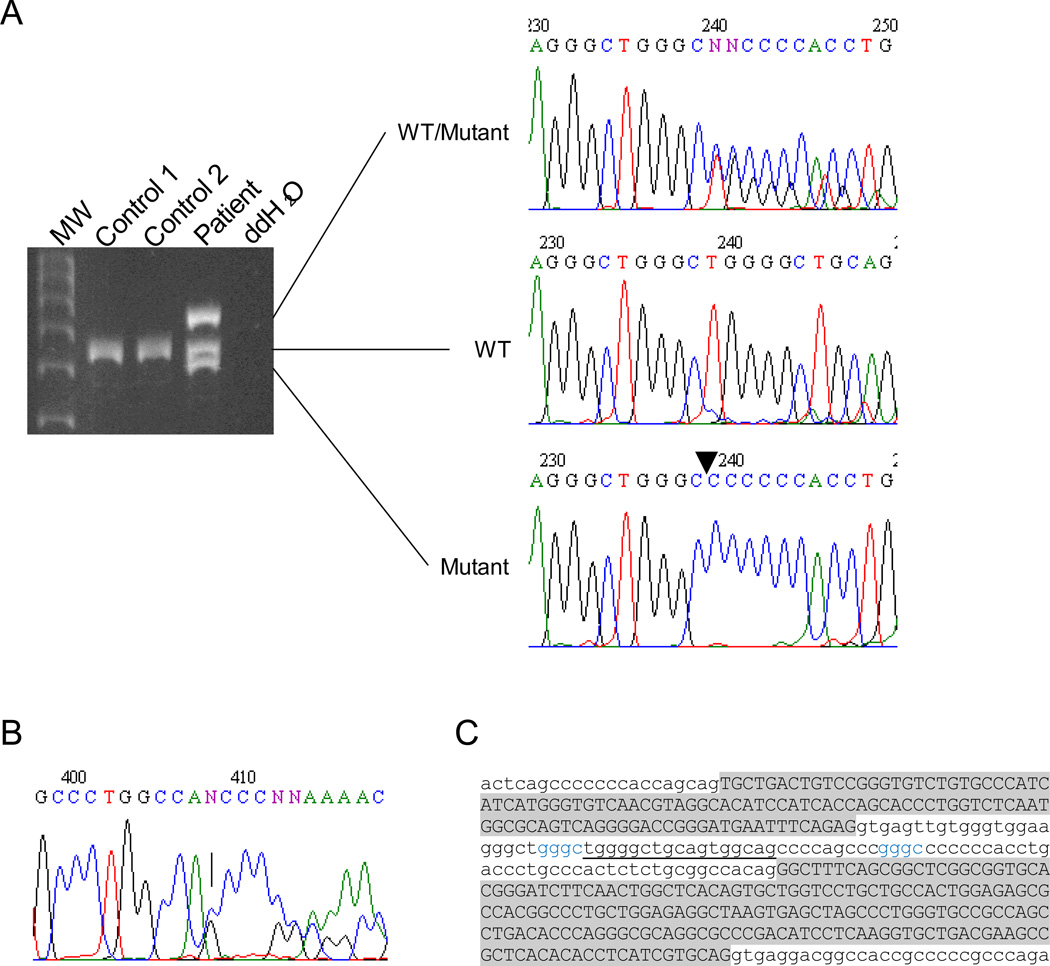
Genetic analysis of the SLC34A3 gene revealed that the patient is a compound heterozygote for a single base pair deletion in exon 12 (c.1304delG) and 30-basepair deletion in intron 6 (g.1440–1469del) (Figure 1A-B). While the intronic deletion was not found in over 180 unrelated healthy white controls, the deletion in exon 12 was found in one of 200 controls.
Figure 1.
SLC34A3 mutations in the HHRH patient. A. Deletion in intron 6. Agarose gel electrophoresis shows three distinct PCR products from the patient. DNA sequencing revealed that top, middle and bottom products represent wild-type/mutant heteroduplex, wild-type, and mutant, respectively. WT, wild type. B. Electropherogram of exon 12. The arrow denotes a deletion of nucleotide “G”. C. Sequence analysis near the intron 6 deletion. The deletion (underline) likely involves four base pair repeats (in blue), which are separated by 26 bp. These short direct repeats are predicted to promote sequence misalignment during meiosis, leading to deletion of one of the repeats along with internal sequence between the repeats. Upper case, exon; lower case, intron.
The c.1304delG mutation results in a shift in the open reading frame, leading to the premature termination of the encoded NaPi-IIc protein – 599 amino acids to 479 amino acids. The other deletion is located in intron 6 and does not remove any apparent cis-acting elements, including conserved donor and acceptor splice sites. Instead, it only reduces the intron size from 93 base pairs to 63. The sequence analysis of the deleted region showed the presence of four base pair direct repeats (GGGC) in the middle of intron 6 – g.1436–1439 and g.1466–1469 (Figure 1C). These repeats likely promote misalignment during meiosis, leading to deletion of one of the repeats and the sequence flanked by the repeats.
Discussion
In the present study, we describe an 11 1/2-year-old girl with increased urinary excretion of calcium and phosphorus and recurrent nephrolithiasis. Despite persistent hypophosphatemia and low bone mineral density, the patient did not have any apparent bone deformities or rachitic changes. However, the clinical phenotype of HHRH can be quite variable even in the same family. Two siblings in the previously described HHRH family carried the identical mutations; however, only one developed bone abnormalities without kidney stones and the other had no bone defects, but had recurrent nephrolithiasis [5]. We cannot rule out the possibility that the patient described in this study may develop bone abnormalities in the future.
The patient carries compound heterozygous mutations in the SLC34A3 gene. The c.1304delG mutation is a novel mutation that causes frameshift in the reading frame. The other mutation is a deletion in intron 6, which was also recently found in an unrelated family with HHRH [7]. As in our previous study of similar intronic deletions in this gene [5], this deletion does not remove any apparent cis-acting elements, but only reduces intron size to 63 bp – below the minimally required size of approximately 87–92 bp in humans [8, 9]. Therefore, the deletion likely prevents proper RNA processing, leading to retention of the truncated intron 6.
This is the fourth unique intronic deletion identified in HHRH kindreds: 30-bp deletion in intron 6 (g.1440–1469del) (this study, [7]), 101-bp deletion in intron 9 (g.2259_2359del) [3, 5, 10], 85-bp deletion in intron 10 (g.2615_2699del) [5], and 26-bp deletion in intron 12 (g.4225_50del) [11]. The deletions found in introns 6, 9, and 10 all appear to affect the minimal intron size required for proper splicing, whereas the deletion in intron 12 likely removes cis-acting elements at acceptor splice site [11]. Frequent intronic deletions in the small number of reported HHRH cases are intriguing and are also important from the genetic analysis perspective. All four intronic mutations described to date involve homologous sequence repeats (Figure 2), suggesting the same molecular mechanism for the deletion. These repeats likely facilitate misalignment during meiosis and remove the internal sequences between the repeats (along with one of the repeats). Furthermore, the SLC34A3 gene (variant 1, NM_001177316.1) is a relatively small gene (5,798 bp), which harbors 13 exons and has high GC content of 66.9%. Since GC content is inversely related to intron size [9], it is not surprising that many of the SLC34A3 introns are significantly smaller than average intron size – mean 3,749 bp, median 1,334 bp [12] (Table 2). However, as we postulated previously [5], the existence of homologous repeats in or near these small introns likely make the SLC34A3 gene particularly vulnerable to unequal crossing over events affecting intron sizes. Since HHRH is an autosomal recessive disease, biallelic mutations are required for full-scale disease manifestations. However, clinical phenotypes are sometimes seen in carriers of single SLC34A3 mutations [3–5, 13, 14], some of which could be explained by the inability to find second mutations. In heterozygous individuals, deletions could be easily missed if PCR primers were designed within the deleted sequences because one normal allele would allow amplification and mask the presence of the deletion. In addition, routine genetic testing only screens exons and adjacent intron sequences, and most intronic sequences go unscreened. Thus, screening of SLC34A3 introns should be considered part of genetic analysis for HHRH, particularly when only one mutation is found in the affected patients. In this regard, we screen 13 SLC34A3 exons in eight fragments, which cover most of the small introns [5]. Alternatively, PCR amplification and sequencing of the entire gene in four overlapping fragments – the approach used by Bergwitz and colleagues [3] – ensures screening of all exons and introns in this gene. In conclusion, we identified two SLC34A3 mutations in a patient with HHRH. One of the mutations was a disease-causing deletion in a short intron, likely leading to retention of the truncated intron. The frequent deletion events in SLC34A3 suggest that the gene is particularly susceptible to unequal crossovers due to the existence of intragenic homologous sequences in or near small introns. More importantly, prevalent intronic deletions indicate the critical importance of including introns as a part of genetic analysis for HHRH.
Figure 2.
Homologous sequences involved in intronic deletions in the SLC34A3 gene. Direct repeats of homologous sequences (gray highlights) likely misalign during meiosis, resulting in deletions of one of the repeats and the sequence flanked by the repeats. Uppercase and lower cases denote exonic and intronic sequences, respectively. Underlines denote deleted sequences. The most 3’ repeats are arbitrarily assigned to be deleted.
Table 2.
Characteristics of introns in the SLC34A3 gene
| Intron | Length (bp)1 | GC (%)1 |
|---|---|---|
| 1 | 725 | 64.7 |
| 2 | 284 | 65.5 |
| 3 | 413 | 67.6 |
| 4 | 80 | 71.3 |
| 5 | 76 | 76.3 |
| 6 | 93 | 73.1 |
| 7 | 228 | 74.1 |
| 8 | 140 | 67.1 |
| 9 | 167 | 76.0 |
| 10 | 139 | 70.5 |
| 11 | 74 | 70.4 |
| 12 | 1220 | 65.6 |
Based on transcript variant 1 (RefSeq NM_001177316.1)
Highlights.
Compound heterozygous mutations, c.1304delG and g.1440–1469del, were identified in a patient with HHRH.
The deletion in intron 6 (g.1440–1469del) is the fourth intronic deletion found in the SLC34A3 gene.
The intronic deletion is likely mediated by sequence misalignments of homologous repeats and affects minimal intron size required for splicing.
Frequent deletions in the SLC34A3 introns suggest the importance of screening introns in genetic analysis of HHRH.
Acknowledgements
This study was supported by National Institutes of Health grants R01 AR42228 (to MJE) and KL2 career development award (to SI) from the Indiana Clinical and Translational Sciences Institute funded in part by the National Institutes of Health grant RR025760.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure statement: The authors have nothing to disclose.
References
- 1.Tieder M, Modai D, Samuel R, Arie R, Halabe A, Bab I, Gabizon D, Liberman UA. Hereditary hypophosphatemic rickets with hypercalciuria. N Engl J Med. 1985;312:611–617. doi: 10.1056/NEJM198503073121003. [DOI] [PubMed] [Google Scholar]
- 2.Drezner MK. Clinical disorders of phosphate homeostasis. In: Feldman D, Pike JW, Glorieux F, editors. Vitamin D. 2nd ed. Burlington, MA: Elsevier Academic Press; 2005. pp. 1159–1187. [Google Scholar]
- 3.Bergwitz C, Roslin NM, Tieder M, Loredo-Osti JC, Bastepe M, Abu-Zahra H, Frappier D, Burkett K, Carpenter TO, Anderson D, Garabédian M, Sermet I, Fujiwara TM, Morgan K, Tenenhouse HS, Jüppner H. SLC34A3 Mutations in Patients with Hereditary Hypophosphatemic Rickets with Hypercalciuria Predict a Key Role for the Sodium-Phosphate Cotransporter NaPi-IIc in Maintaining Phosphate Homeostasis. Am J Hum Genet. 2006;78:179–192. doi: 10.1086/499409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lorenz-Depiereux B, Benet-Pages A, Eckstein G, Tenenbaum-Rakover Y, Wagenstaller J, Tiosano D, Gershoni-Baruch R, Albers N, Lichtner P, Schnabel D, Hochberg Z, Strom TM. Hereditary Hypophosphatemic Rickets with Hypercalciuria Is Caused by Mutations in the Sodium-Phosphate Cotransporter Gene SLC34A3. Am J Hum Genet. 2006;78:193–201. doi: 10.1086/499410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ichikawa S, Sorenson AH, Imel EA, Friedman NE, Gertner JM, Econs MJ. Intronic deletions in the SLC34A3 gene cause hereditary hypophosphatemic rickets with hypercalciuria. J Clin Endocrinol Metab. 2006;91:4022–4027. doi: 10.1210/jc.2005-2840. [DOI] [PubMed] [Google Scholar]
- 6.Imel EA, Peacock M, Pitukcheewanont P, Heller HJ, Ward LM, Shulman D, Kassem M, Rackoff P, Zimering M, Dalkin A, Drobny E, Colussi G, Shaker JL, Hoogendoorn EH, Hui SL, Econs MJ. Sensitivity of fibroblast growth factor 23 measurements in tumor-induced osteomalacia. J Clin Endocrinol Metab. 2006;91:2055–2061. doi: 10.1210/jc.2005-2105. [DOI] [PubMed] [Google Scholar]
- 7.Yu Y, Sanderson SR, Reyes M, Sharma A, Dunbar N, Srivastava T, Juppner H, Bergwitz C. Novel NaPi-IIc mutations causing HHRH and idiopathic hypercalciuria in several unrelated families: long-term follow-up in one kindred. Bone. 2012;50:1100–1106. doi: 10.1016/j.bone.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu J, Yang Z, Kibukawa M, Paddock M, Passey DA, Wong GK. Minimal introns are not "junk". Genome Res. 2002;12:1185–119. doi: 10.1101/gr.224602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowski J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 10.Hasani-Ranjbar S, Amoli MM, Ebrahim-Habibi A, Dehghan E, Soltani A, Amiri P, Larijani B. SLC34A3 intronic deletion in a new kindred with hereditary hypophosphatemic rickets with hypercalciuria. J Clin Res Pediatr Endocrinol. 2012;4:89–93. doi: 10.4274/jcrpe.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jaureguiberry G, Carpenter TO, Forman S, Juppner H, Bergwitz C. A novel missense mutation in SLC34A3 that causes hereditary hypophosphatemic rickets with hypercalciuria in humans identifies threonine 137 as an important determinant of sodium-phosphate cotransport in NaPi-IIc. Am J Physiol Renal Physiol. 2008;295:F371–F379. doi: 10.1152/ajprenal.00090.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hong X, Scofield DG, Lynch M. Intron size, abundance, and distribution within untranslated regions of genes. Mol Biol Evol. 2006;23:2392–2404. doi: 10.1093/molbev/msl111. [DOI] [PubMed] [Google Scholar]
- 13.Mejia-Gaviria N, Gil-Pena H, Coto E, Perez-Menendez TM, Santos F. Genetic and clinical peculiarities in a new family with hereditary hypophosphatemic rickets with hypercalciuria: a case report. Orphanet J Rare Dis. 2010;5:1. doi: 10.1186/1750-1172-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamamoto T, Michigami T, Aranami F, Segawa H, Yoh K, Nakajima S, Miyamoto K, Ozono K. Hereditary hypophosphatemic rickets with hypercalciuria: a study for the phosphate transporter gene type IIc and osteoblastic function. J Bone Miner Metab. 2007;25:407–413. doi: 10.1007/s00774-007-0776-6. [DOI] [PubMed] [Google Scholar]