Abstract
In its classical form, autophagy is an essential, homeostatic process by which cytoplasmic components are degraded in a double-membrane-bound autophagosome in response to starvation. Paradoxically, although autophagy is primarily a protective process for the cell, it can also play a role in cell death. The roles of autophagy bridge both the innate and adaptive immune systems and autophagic dysfunction is associated with inflammation, infection, neurodegeneration and cancer. In this review, we discuss the contribution of autophagy to inflammatory, infectious and neurodegenerative diseases, as well as cancer.
Keywords: Autophagy, ATG, Beclin 1, neuroinflammation, diseases
1. Introduction
Autophagy is a fundamental cellular homeostatic mechanism, whereby cells autodigest proteins, lipids and organelles of their cytoplasm for removal or turnover [1]. This intracellular recycling process of the cell serves as housekeeping function. During cell injury or accumulation of damaged cellular components, intracellular inclusion bodies may be transferred to the autophagic pathway, and degraded by the lysosome [1]. There are two fates for this catabolic process, a long-term need to prevent tissue damage and disease, and there is also an acute requirement for autophagy to sustain homeostasis in stressful environments.
During autophagy, a double or multi-membrane-bound structure, termed autophagosome or autophagic vacuole, is formed de novo to sequester cytoplasm. The vacuole membrane then fuses with lysosomes to deliver the contents into the organelle lumen, where they are degraded and the resulting macromolecules are recycled [2]. Under normal conditions, autophagy occurs at a low basal level to maintain homeostasis [3]. However, it can be induced under starvation to carry out selective or non-selective bulk degradation to supply a nutrient source, promoting survival. Thus, the regulation of autophagy is important in controlling the level, timing and specificity of cargo elimination [4]. Moreover, multiple additional signals, including endoplasmic reticulum (ER) stress, immune cell activation, oxidative stress and infection, stimulate autophagy [5].
Different studies have shown that autophagy can function as an intracellular pathogen sensing mechanism, and defects in autophagy can lead to increased susceptibility to infection [6]. In this context, several recent studies have implicated autophagy in the removal of pathogens located in phagosomes [7] and the cytosol [8]. For instance, a particle that engages Toll-like receptors (TLRs) on a murine macrophage, while it is phagocytosed, triggers the recruitment of the autophagosome marker LC3 (microtubule-associated protein 1 light chain 3) to the phagosome in an autophagy pathway proteins-dependent manner [9].
The importance of the role of autophagy in innate and adaptive immunity is highlighted in part, by the association of defects in autophagy with neurodegeneration, aging, cancer, metabolic syndrome and inflammatory disorders including Crohn's disease (CD) [2]. An emerging role of autophagy in innate immunity is suggested by findings showing that this process is able to regulates the inflammasome and cell-specific pattern-recognition receptor (PRR) signaling [10–12], as well as the clearance of apoptotic bodies and even the induction of cell death, which was suggested as a potential mechanism to control inflammation [13–14].
About its role in cancer, it has been demonstrated that autophagy may participate in a beneficial or deleterious way of response. It may have a tumour suppressive role through the elimination of oncogenic protein substrates, toxic unfolded proteins and damaged organelles [15]. Alternatively, it may have tumour promoting effects in established cancers through autophagy-mediated intracellular recycling that provides substrates for metabolism and that maintains the functional pool of mitochondrias [15].
In this review we will describe recent advances about autophagy and their roles in the immune responses and diseases.
2. Overview of Autophagy
Autophagy is a general term for pathways by which cytoplasmic materials, including soluble macromolecules and organelles, are delivered to lysosomes for degradation [16]. Autophagy is then, a highly regulated mechanism, as demonstrated by the identification of several autophagy-related (ATG) genes in the yeast Saccharomyces cerevisiae [17–18]. ATG genes encode intracellular machinery that controls the initiation of autophagosome formation, cargo collection and trafficking to the lysosomal compartment. More than 30 of these genes were originally characterized in yeast, and many orthologs have subsequently been identified and confirmed as autophagy regulators in higher eukaryotes [18–19]. There are at least 3 different autophagic mechanisms involved in the lysosomal degradation of de cytoplasm content, including macroautophagy, chaperone-mediated autophagy and microautophagy [16, 20]. This review focused in macroautophagy, usually referred as autophagy. In this pathway, a portion of cytoplasm (usually 0.3–1μm in diameter) is engulfed by an isolation membrane (in mammals) or `phagophore' (in yeast), resulting in the formation of a double-membrane structure known as autophagosome. The outer membrane of the autophagosome fuses with the lysosomal membrane to form an autolysosome, this way, the inner autophagosome membrane and the autophagosome cargo are degraded by lysosomal enzymes [21] (Fig. 1). Following this step, lysosomes can be recycled from autolysosomes allowing the cell to reuse a critical component required for further autophagy, the lysosomal membrane and associated proteins, when there is a scarce resource [22]. In addition, autophagy constitutively and efficiently delivers cytosolic proteins for MHC class II presentation and thus CD4+ T cell stimulation [23].
Fig. 1.
Steps of autophagy pathway activation. The biogenesis of autophagosomes requires the ordered intervention of autophagy-regulated (ATG) proteins that act on different modules. Some of these modules are shown on the figure, including the ULK1complex (ULK1/ATG13/FIP200/ATG101) and PI3K complex (Beclin 1/ATG14/calss III PI3K) in the initiation of phagophore formation. During starvation or rapamycin treatment, mTOR is inhibited, leading to ULK1 dephosphorylation and activation of autophagy. Then, the phagophore nucleation is mediated by a complex involving PI3K. (a) The elongation of phagophore is mediated by two ubiquitin-like conjugation systems that together promote the assembly of the ATG12/ATG5/ATG16L complex and the processing of LC3. This molecule is cleaved by the protease ATG4 to generate LC3-I, which is then activated by ATG7, transferred to ATG3 (a second E2 ubiquitin-like enzyme) and conjugated to PE. The lipidated form of LC3-I (LC3-II) is attached to both faces of the phagophore membrane. This step is characterized by membrane bending and increase in size of the phagophore through addition of membrane, which incorporate cytoplasmic components, such as mithocondria, ER, macromolecules, etc. (b) The elongation step ended off with closure of the autophagosome. In the maturation step, fusion of autophagosomes with lysosomes is required for the formation of autolysomes where the substrates are degraded.
Several hetero-oligomeric protein complexes, that contain ATG proteins, are involved in the initiation and elongation stages of autophagy. Most ATG proteins function primarily at early stages of autophagosome formation; up to the step where the phagophore becomes an autophagosome [24]. When nutrients are abundant, mammalian target of rapamycin (mTOR) interacts with the ULK1–ATG13–FIP200-ATG101 complex and phosphorylates ULK1 and ATG13 to inhibit autophagosome formation. However, autophagy is induced in response to nutrient starvation through the inhibition of mTOR, resulting in dephosphorylation and activation of the ULK1 kinase complex. In turn, activation of this complex causes the initiation of autophagosome nucleation and elongation. In this step, the translocation of the mTOR substrate complex from the cytosol to certain domains of the ER (the site of autophagosome formation), requires a complex containing ATG6 or its mammalian homolog, Beclin 1, that recruits the class III phosphatidylinositol-3-OH kinase (PI(3)K) complex, which includes at least VPS34 kinase (also known as PIK3C3), VPS15 (PIK3R4 and p150), Beclin 1 and ATG14, to the ER [4, 19, 25].
The PI(3)K complex can also be activated by proteins that interact with Beclin 1, including UV radiation resistance–associated gene (UVRAG) and activating molecule in Beclin 1-regulated autophagy protein 1(AMBRA1) [17]. Once activated, this complex produces phosphatidylinositol-3-phosphate (PtdIns(3)P), which recruits effectors molecules such as double FYVE-containing protein 1 (DFCP1) and WD-repeat domain phosphoinositide-interacting (WIPI) family proteins WIPI1 andWIPI2, which are orthologs of the yeast PI3P-binding protein ATG18 [25]. DFCP1 translocates to the site of autophagosome formation in a PI3P-dependent manner and is essential for the generation of membranous structure, closely associated with both the autophagosome and the ER, termed omegasomes [26]. In addition, in yeast, ATG14L mediates the localization of the autophagy specific PI3-kinase complex to the ER, and its recruitment is a critical determinant of the omegasomes formation, possibly ensuring a constant supply of PI3P at sites of omegasome/autophagosome formation, however, detailed function of ATG14L in mammalian autophagy has not yet been characterized [27].
At the final step of autophagosome formation, elongation of the isolation membrane and/or completion of enclosure require two ubiquitin-like conjugates. The first system involves conjugation of the ATG12-ATG5 by the E1-like ATG7 and E2-like ATG10 enzymes, which together bind ATG16L1 to form pre-autophagosomal structures [28]. In the second ubiquitin-like reaction, LC3 and GABARAPs (mammalian ATG8 homologs) are cleaved by the protease ATG4. Phosphatidylethanolamine is conjugated to cleaved LC3 (LC3-II) and GABARAPs by ATG7 and a second E2-like enzyme, ATG3, then, this lapidated LC3-II associates with newly forming autophagosome membranes and is found on the inner and outer surfaces of the autophagosome. ATG8s are degraded along with the inner membrane upon formation of the autolysosome, but LC3-II remains on mature autophagosomes until fusion with lysosomes and is commonly used to monitor autophagy [29–33]
In addition to the ER, other membranes may be involved in autophagosome formation. ATG9, another transmembrane protein in mammals, is essential for autophagy and traffics between the trans-Golgi network, late-endosomes and autophagosome precursors [34]. A recent investigation suggests that mitochondria, the plasma membrane and the nuclear membrane could also be membrane sources for autophagosome formation [35–36]. However, further research is needed to develop specific protein markers for these structures on the autophagosomal membrane. It is possible that cells may use different membrane sources, to form the autophagosome, in different situations.
During maduration stage, autophagosomes are transported along microtubules towards the microtubule-organizing center, where lysosomes are abundant [37]. The autophagosomal membranes assemble around cargo, encapsulating it in a vesicle that subsequently fuses with a lysosome, generating an auto-lysosome. A number of SNARE proteins, including VAMP8 and Vti1B, are believed to be involved in regulating heterotypic fusion between autophagosomes and the lysosomal compartment [38]. The contents are then degraded by lysosomal enzymes and the lysosomal permeases release the breakdown products into the cytosol, where they are available for synthetic and metabolic pathways [4].
2.1. Autophagy: a selective degradation process
For a long time macroautophagy has been considered a rather unselective process for bulk degradation of long-lived proteins and organelles that during nutrition deprivation can recycle building blocks and help restore the energy balance of the cell. [39]. Later, it was observed that autophagosomes preferentially degrade particular macromolecular constituents within the cytosol, thus, the idea of “targeted” or specific autophagy began [40]. Recent reports show evidences of selective autophagic degradation of protein inclusions caused by aggregate-prone or misfolded proteins (aggrephagy) [41–44], of organelles such as peroxisomes (pexophagy) [45], mitochondria (mitophagy) [46], bacteria and virus (xenophagy) [47–48], surplus ER (reticulophagy) [49], and ribosomes (ribophagy) [50]. As suggested by Johansen et al. [51] selective autophagy depends on binding of substrates to the inner surface of the growing phagophore, and this can be achieved by cargo receptors that are associated both with the substrate and with lipidated ATG8 family proteins anchored to the phagophore. Aggregation of the substrate and/or cargo receptor is required for efficient sequestration. Closure results in the formation of a double-membrane autophagosome. Fusion of autophagosomes with late endosomes or lysosomes (maturation step) is then required for the formation of autolysomes where the substrates are degraded.
The autophagic adaptors p62/SQSTM1 (sequestosome 1) and NBR1 (neighbor of Brca1 gene) are both selectively degraded by autophagy and able to act as cargo receptors for degradation of ubiquitinated substrates [51–52]. A direct interaction between these autophagic adapters and LC3, mediated by a LIR (LC3-interacting region) motif, their inherent ability to polymerize or aggregate as well as their ability to specifically recognize substrates, are all required for efficient selective autophagy [51–52].
3. Autophagy and inflammation
Inflammation is a vital host response to the loss of cellular and tissue homeostasis with many important roles, such as host defence, tissue remodeling and repair, and the regulation of metabolism [53] During infections or tissue damage, a cascade of signals leads to the recruitment of inflammatory cells, particularly phagocytes such as neutrophils and macrophages. These cells phagocytose infectious agents and damaged tissue components and produce additional cytokines and chemokines that lead to the activation of adaptive immune responses. Sterile inflammation occurs in the absence of any microorganisms during trauma, ischaemia-reperfusion injury or chemically induced injury. During both, microbially-induced and sterile inflammation, the recruitment of phagocytes and the production of pro-inflammatory cytokines and chemokines, notably tumour necrosis factor (TNF) and interleukin1 (IL1)[54].
The pro-inflammatory cytokines IL-1β and IL-18 are synthesized as precursors, which can be processed by caspase-1 to become bioactive forms that are then secreted. This process occurs in specialized protein platforms, referred to as inflammasomes that are assembled after pro-inflammatory stimulation of the cells [55].
3.1. Autophagy, TLRs and the inflammasome
There are multiple crossroads between autophagy and inflammation [52]. The autophagy pathways and/or proteins are suggested to play a role in the control of inflammatory signaling [56]. Autophagy contributes to host defense responses by promoting the elimination of pathogens and the induction of acquired immunity. In this scenario, several TLRs including TLR2, TLR4, and TLR9 are able to induce autophagy. We have demonstrated that activation of TLR2 with peptidoglycan (PGN) from S. aureus induces autophagic cell death in phagocytes [14] (Fig. 2). Other investigators have reported that TLR4 adaptor protein TRAF ubiquitinates Beclin 1 to promote autophagy and to stimulate NF-κB signaling [57]. A recent study showed that DNA containing immune complexes activates TLR9 and induces secretion of type I interferons (IFNs) from plasmacytoid dendritic cells, by a mechanism that involved the convergence of the phagocytic and autophagic pathways [58].
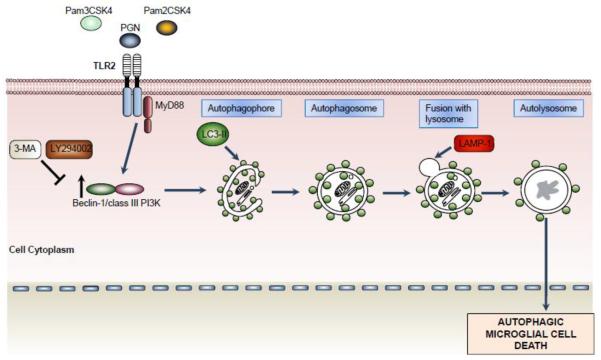
Fig. 2.
TLR2 activation induces microglial cell death by inducing autophagy. The stimulation of TLR2 with PGN, Pam2CSK4 and Pam3CSK4 induce activation of autophagy revealed by increased protein levels of Beclin 1 and LC3-II. Moreover, the co-localization of LAMP-1 and LC3-II indicated fusion of autophagosomes with lysosomes. Persistent induction of autophagy pathway promotes microglial cell death with a significantly reduction of microglial cells in the brain parenchyma. The microglial cell death was reverted in the presence of autophagy inhibitors (3-MA and LY294002).
Another important effect of autophagy proteins on inflammatory signalling is related to the regulation of the inflammasome-dependent responses. Autophagy regulates these responses by controlling the levels of pro-inflammatory cytokine secretion, such as interleukin-1β (IL-1β) and IL-18. The inflammasome complex contains NOD-like receptor (NLR) cryopyrin proteins, the adaptor protein ASC and caspase 1, and is activated by cellular infection or other stress stimuli, to promote the maturation of IL-1β and IL-18. The involvement of autophagy in the regulation of pro-inflammatory cytokine secretion was recently demonstrated in ATG-16 deficient mice, which produce exaggerated amounts of IL-1β and IL-18 in response to LPS and other pathogen associated molecular patterns (PAMPs) [59]. ATG16L1-deficiency disrupts the recruitment of the ATG12-ATG5 conjugate to the isolation membrane, resulting in a loss of microtubule LC3 conjugation to phosphatidylethanolamine. Consequently, both autophagosome formation and degradation of long-lived proteins are severely impaired in ATG16L1-deficient cells. Mice lacking ATG16L1 in haematopoietic cells are highly susceptible to dextran sulphate sodium-induced acute colitis [59]. Although in this study, the nature of the inflammasome scaffold was not determined, LPS-induced inflammasome activation in ATG16L1-deficient cells is dependent on K+ efflux and ROS, suggestive of NLRP3 involvement.
The NLRP3 inflammasome consists of the NLRP3 scaffold, the ASC (PYCARD) adaptor, and caspase-1. NLRP3 is activated upon exposure to whole pathogens, as well as a number of structurally diverse PAMPs, damage-associated molecular patterns (DAMPs), and environmental irritants [60]. Recent reports reveal that NLRP3 inflammasome activity is suppressed by ROS blockade [59, 61] and inhibition of mitophagy/autophagy with 3-methiladenine (3-MA) in THP1 macrophages, resulted in the accumulation of ROS-producing damaged mitochondria, as a consequence of the inflammasome activation [62]. Additionally, the induction of AIM2 or NLRP3 inflammasomes in macrophages triggered activation of the small G protein RalB and autophagosome formation. Assembled inflammasomes underwent ubiquitination and recruited the autophagic adaptor p62, which assisted their delivery to autophagosomes [63]. A recent study found that Map1lc3b−/− and Becn1+/− macrophages released mitochondrial DNA in excess quantities into the cytosol in response to LPS and ATP treatment, explaining the increased caspase-1 activation and IL-1β and IL-18 secretion observed in these cells [12]. Further research is required to fully understand the complex reciprocal regulation of inflammasome and autophagic pathways. Dying, stressed or injured cells release or expose molecules on their surface that can function as either adjuvant or danger signals for the innate immune system [64]. These immunogenic endogenous molecules are called DAMPs and they can stimulate the immune system through a broad family of membrane-bound or cytoplasmic pattern-recognition receptors, which include TLRs, NLRs and RIG-I-like receptors [65–66].
Some DAMPs, such as ATP and high mobility group protein B1 (HMGB1), are secreted or released and others, such as calreticulin and heat shock protein 90 (HSP90), are exposed de novo or become enriched on the outer leaflet of the plasma membrane [66]. In addition, other DAMPs, such as uric acid, are produced as end-stage degradation products during the course of cell death [66]. In the past few years, the concept that autophagy regulates release and degradation of DAMPs such as HMGB1, ATP, IL1β, and DNA in several cell types has emerged [67]. For instance, recent evidences suggest that autophagy can regulate HMGB1 release in a ROS-dependent manner in fibroblasts, macrophages and cancer cells, and neutrophil extracellular trap-mediated HMGB1 release in neutrophils [67–69]. Autophagy can also induce the degradation of endocytosed exogenous HMGB1 in macrophages [70]. In addition, autophagy is required for the release of ATP by cancer cells, which may stimulate antitumor immune responses [71].
Multiple signals and molecules can participate in the regulation of autophagy, and it is suggested that DAMPs, such as HMGB1 and ATP, are powerful autophagic stimuli and regulators [67–68, 72].
ATP induces P2RX7-dependent autophagy in human monocytes/macrophages, which is associated with rapid killing of intracellular mycobacteria [73] and stimulates in microglial cells the release of autophagolysosomes to the extracellular space [74]. The induction of autophagy by HMGB1 has been well studied and both, intracellular and extracellular HMGB1-mediated autophagy promote chemoresistance in colon cancer, pancreatic cancer and leukemia [67, 75–76].
3.2. Autophagy and inflammatory pathology: Crohn'disease
Crohn'disease is a common inflammatory bowel disorder thought to result from a breakdown in self-recognition of commensal gut flora together with defects in mucosal barrier function [77]. The etiology of CD remains a controversial topic, but recent studies have revealed three CD susceptibility genes, ATG16L1, immunity-related GTPase family M (IRGM) and NOD2 [78–79]. As we described earlier, ATG16L1 is not only present but also is essential for proper elongation of the isolation membrane. IRGM induces autophagy in response to IFN-γ, leading to increased clearance of bacterial pathogens [80]. Finally, NOD2 is an intracellular PRR of the NLR family expressed in a limited number of tissues and cells that includes Paneth cells and monocyte-derived cells of the immune system. This molecule recruits ATG16L1 to bacterial entry sites, targeting bacteria for autophagic degradation [81].
Interestingly, recent studies showed that NOD2 activation by its bacterial ligand, muramyldipeptide (MDP), was capable of inducing autophagy in primary human antigen-presenting cells, monocyte-derived dendritic cells (DCs). This phenomenon required NOD2 and the NOD2 signaling mediator RIPK-2, but not NALP3, a PRR that also recognizes MDP. In addition, other studies also reported that NOD2-induced autophagy requires autophagy proteins including PI3K, ATG5, ATG7 and ATG16L [77]. Moreover, in human DCs, NOD2-mediated antigen presentation requires autophagy, and DCs expressing CD variant NOD2 (1007fsinsC, R702W or G908R) and ATG16L1 T300A display reduced surface MHC class II and are less capable of inducing antigen-specific CD4+ T cell responses [82–83]. Additionally, some studies have suggested that the ATG16L1 T300A variant has reduced autophagic clearance of enteric pathogens such as adherent-invasive Escherichia coli or S. typhimurium [77].
Another study has demonstrated that ATG16L1 mutation (null or hypomorphic alleles) in mice results in abnormalities relevant to CD pathogenesis [84]. Here, the author reports a striking genetic interaction between ATG16L1 mutation and a specific strain of an enteric virus, murine norovirus. This virus-plus-susceptibility gene interaction alters the transcriptional signature of Paneth cells and the nature of the inflammatory response in mice treated with the toxic substance dextran sodium sulfate by a mechanism that involved TNFα and IFNγ secretion as well as commensal bacteria [83]. In addition, macrophages from chimeric mice lacking ATG16L1 in hematopoietic cells have increased IL-1β production in response to LPS stimulation or infection with noninvasive enteric bacteria. These mice are highly sensitive to sodium sulfate-induced colitis, suggesting that increased pro-inflammatory cytokine production by macrophages may also promote intestinal damage in ATG16L1-dependent CD [59]. Overall, defects in autophagy may alter xenophagic bacterial clearance, pro-inflammatory cytokine secretion, and extracellular secretion pathways, promoting CD pathogenesis.
4. Autophagy and infection
Xenophagy protects against infectious disease by degrading intracellular bacteria such as Streptococcus pyogenes, Mycobacterium tuberculosis, Shigella flexneri, Salmonella enterica, Listeria monocytogenes, viruses such as herpes simplex virus type 1 (HSV-1) and protozoan pathogens including Toxoplasma gondii [85–86]. In addition, autophagy induction, along with activation of other innate immune responses, represents the first line of defense during pathogen infections. However, autophagy-mediated responses have different functions depending on the microorganism and host cell type involved. Indeed, the efficacy of intracellular microbe sensing may have driven the evolution of pathogen mechanisms aimed to evade, inhibit, and usurp host cell autophagy to promote survival, replication, and pathogenesis [52].
Multiple studies have confirmed the important role of autophagy during infection. For example, group A Streptococcus (GAS) is an extracellular pathogen that invades the cytoplasm of epithelial cells but is rapidly degraded by an autophagy-dependent pathway [7]. The diameter of GAS-containing autophagosome-like vacuoles can be as large as 10 μm compared with 0.3–1.0 μm for non-selective autophagosomes. Several reports demonstrated that this process required not only the common machinery of autophagy, but also Rab7 [87–88], Rab9 and Rab23 [89] as additional components.
Salmonella enterica serotype typhimurium infection represents another situation in which a pathogen is targeted inside a vesicular structure. Upon infection of epithelial cells, S. typhimurium resides in Salmonella-containing vacuoles (SCVs), which promote bacterial survival and replication. In the process of bacterial internalization, autophagy adaptors have a crucial role in targeting pathogens to autophagosomes. These adaptors include p62/sequestosome 1 [90], nuclear dot protein 52 kDa (NDP52) [47, 91], neighbor of BRCA1 gene 1 (NBR1) [92], and Optineurin (OPTN) [93], all of which bind ubiquitin, LC3 and GABARAP-1, mediating interaction between LC3-positive isolation membranes and ubiquitinated targets. Finally, LIR domains of the autophagic adapter proteins allow for the delivery of cytosolic S. typhimurium into autophagosomes [51]. Autophagic degradation of several other pathogens, including L. monocytogenes [94] and S. flexneri [95], is also mediated by p62.
Autophagy mediated by NDP52 and OPTN involves TANK-binding kinase (TBK1) and this kinase is known to interact with OPTN. The LPS/TLR4-mediated activation of TBK1 likely promotes phosphorylation of S177 in the LIR domain of OPTN, facilitating LC3 binding and subsequent targeting of S. typhimurium to autophagosomes [93]. An ubiquitin-independent pathway, involving the lipid second messenger diacyl-glycerol, that does not involve p62 or NDP52, could also function in targeting damaged SCVs to the autophagosome. In this pathway, diacyl-glycerol acts as a signal for the co-localization of SCVs with LC3-positive autophagosomes by a mechanism that involves protein kinase C and its downstream targets, JNK and NADPH oxidase [96].
Numerous studies have shown a crucial role for autophagy in defense against mycobacterial infection in human cells, and a genome-wide siRNA screen of host genes that regulate Mycobacterium tuberculosis (M. tuberculosis) replication in human macrophage-like THP-1 cells, identified host proteins that influence maintenance of intracellular pathogen load [97]. Most of the identified M. tuberculosis-strain-independent host factors function in autophagy, indicating a central role for this process in controlling M. tuberculosis infections. Previous studies focused on the role of autophagy in mycobacterial clearance were performed using Mycobacterium bovis Bacille Calmette-Guérin (BCG), the attenuated vaccine strain [98] that lack of several virulence factors, including the type VII secretion system ESX-1 [99–100], that make this strain defective for replication within macrophages and fails to activate innate responses of macrophages. In these studies, targeting of LC3 to BCG-containing vacuoles required exogenous stimulation of autophagy. Recently, it was reported that wild-type (WT) M. tuberculosis cells elicit ubiquitin-mediated targeting to autophagy in resting macrophages, resulting in the delivery of bacilli to lysosomes. Targeting requires both the bacterial ESX-1 system and host cytosolic DNA sensing pathway, and the delivery of bacilli to autophagosomes requires the ubiquitin-autophagy receptors p62 and NDP52 and the DNA-responsive kinase TBK1 [100].
Interestingly, the IRGM leads to enhanced autophagic clearance of vacuolar M. tuberculosis in human myeloid cells [8]. In addition to the delivery of the bacterial pathogen to lysosomes, autophagy contributes to the elimination of M. tuberculosis through the production of antimicrobial peptides by a mechanism p62-dependet [101–102]. Moreover two members of the guanylate-binding protein (Gbp) family of GTPases (Gbp1 y Gbp7) were recently found to be involved in the production of antimicrobial peptides in autolysosomes [103]. However, M. tuberculosis may also block phagosomal maturation in order to evade degradation [98].
In a similar fashion to the M. tuberculosis-containing vesicles, conditioned phagosomes are also used by the parasite Toxoplasma gondii for its replication. Nevertheless, there are two mechanisms of macrophage activation that result in killing and clearance of T. gondii in cultured cells, one dependent on IFNγ/LPS signaling and the other one dependent on ligation of CD40, being the IFNγ-mediated pathway very important for the control of acute infection. Recent studies have shown that expression of the Atg5 in granulocytes and macrophages is required for in vivo resistance to infection with T. gondii and recruitment of the IFNγ-inducible p47 GTPase IIGP1 (Irga6) to the vacuole membrane [104–105]. These studies suggest that pathogen-conditioned phagosomes might need processing for efficient clearance of the contained parasites. In addition to IFNγ, activation of macrophages by CD40-CD40L ligation increases the levels of Beclin 1 and stimulates autophagy to kill T. gondii [106].
During viral infections, autophagy-mediated responses have different outcomes depending on the virus and host cell type. The viruses have developed strategies to both target autophagosome generation and autophagosome maturation. Several viral proteins target the core autophagy protein Beclin 1. In fact, autophagosome initiation is blocked by interactions between Beclin 1 and the α-herpes simplex virus 1 (HSV-1) neurovirulence factor ICP34.5 [107] or the oncogenic γ-herpes virus-encoded viral BCL2-like proteins [108]. The factor ICP34.5 also interacts with other host proteins, including protein phosphatase 1α (PP1α) to promote neuropathogenesis. The GADD34 homology domain of ICP34.5 interacts with PP1α to facilitate eIF2α dephosphorylation and enable HSV-1 replication [107, 109].
Interestingly, the Beclin 1-binding domain (BBD) of ICP34.5 has been demonstrated to be necessary to inhibit starvation-induced autophagy in the breast cancer cell line MCF7. Mutant HSV-1 expressing a version of ICP34.5 that lacks the BBD was unable to inhibit autophagy in infected primary neurons and this effect finally leads to reduction of replication in vivo [109]. Moreover, a mouse γ-herpes virus that encodes a mutant viral BCL2 inhibits autophagy maintaining latent infections [110]. Thus, herpes viruses sequester Beclin 1 to inhibit autophagosome formation to enhance their neurovirulence and increase their persistence in vivo.
RNA viruses seem to stabilize autophagosomes by preventing their degradation. The human immunodeficiency virus type-1 (HIV) blocks autophagosome maturation in infected macrophages [111]. HIV relies on several components of autophagy for its replication since silencing of autophagy proteins inhibits HIV replication [112]. In macrophages, HIV group-specific antigen (Gag)-derived proteins accumulate in the stabilized autophagosomes enhancing viral replication. The Gag proteins co-localize and interact with LC3B, and are accumulated at LC3B-II enriched membranes via its nef protein, which co-localizes with autophagosomes and binds to Beclin 1 via its DD motif (aa 174–175). This fusion block is also established in DCs upon HIV infection, preventing the formation of so-called immunoamphisomes [111, 113]. Moreover, HIV-1 blocks autophagy in DCs by activating the mTOR pathway through the interaction of HIV-1 envelope glycoproteins (Env) with the CD4 receptor [113]. The inhibition of autophagy in DCs leads to increased cellular viral content, increased transfer of HIV-1 to CD4+ T cells and impaired MHC class II presentation of HIV-specific antigens to HIV-specific CD4+ T cells [113–114]. In addition, HIV-1 inhibits autophagy in infected CD4+ T cells by downregulating Beclin 1 at the transcriptional level [115]. Thus this manner, HIV-1 may manipulate autophagy to evade degradation.
5. Autophagy, neuroinflammation and neurodegeneration
5.1. Microglial cells and neuroinflammation
The central nervous system (CNS) contains sensitive tissues with poor regenerative capacity; therefore the `immune privilege' is indispensable for damage limitation during inflammation [116]. It is now clear that while peripheral immune components access to the CNS is restricted and tightly controlled, the CNS is capable of mounting dynamic immunologic and inflammatory responses to a variety of insults [117–119]. Several stimuli such as trauma, infections, toxins and systemic pro-inflammatory cytokines are capable of eliciting an immediate and short lived activation of the innate immune system within the CNS [118, 120]. This acute neuroinflammatory response includes activation of microglia (resident immune cells) resulting in their morphological and phenotypical changes, and the release of inflammatory mediators such as cytokines and chemokines by these cells [119]. Under physiological conditions, microglia exhibit a quiescent phenotype which is associated with the production of anti-inflammatory and neurotrophic factors [121]. Activated microglia, however, promote an inflammatory response that serves to further engage the immune system and initiate tissue repair [122]. This response is frequently self-limiting, resolving once infection has been eradicated or the tissue damage has been repaired. However, persistence of inflammatory stimulation, by exogenous or endogenous factors, or a failure in normal resolution mechanisms caused by overwhelming inflammatory cycles, may result in pathological consequences [122]. Activated microglial cells and macrophages are essential for the clearance of invading microorganisms and injured tissue. They can be stimulated to express a variety of pro-inflammatory cytokines such as IL-1, TNFα and IL-6, as well as superoxide and nitric oxide, which are neurotoxic and may amplify underlying disease states [123].
The current knowledge about the role of autophagy in the CNS is still patchy [124]. Autophagosomes accumulate in several brain disorders [125–126] and autophagy seems to be essential for neuronal homeostasis, plasticity and protein quality control in neurons [127–128]. Most of the existing literature related to autophagy in the CNS focuses on neurons and little is known about the effects of the autophagic process and its regulation in microglial cells. Our recent in vitro and in vivo studies show that activation of microglial cells with bacterial PGN and other TLR2 ligands results initially in microglial cell activation and later in the induction of microglial cell death by mechanisms involving autophagy [14]. Thus, PGN might act as a regulator of microglial cell survival through the induction of autophagic cell death in pathological conditions where PGN, or other TLR2 ligands, are present in the CNS. Our data revealed that stimulation of TLR2 in microglial cells initially activates the cells followed by the induction of autophagic cell death [14]. Therefore, TLR2, an important PRR involved in host defense and neurodegeneration [116], has the potential to control microglial cell population, and this capacity might be exploited by pathogens to evade innate host immune responses. On the other hand, recent evidences support the notion that mTOR is involved in microglial pro-inflammatory activation, thus making this kinase a possible target for therapeutic intervention to reduce brain inflammatory responses [129].
As we mentioned before, following activation, microglia become capable of numerous functions depending on the stimuli in the surrounding environment. One such function is phagocytosis, which facilitates brain homeostasis via the clearance of cellular debris and possibly the pruning of synapses [130–131]. Interestingly, Lucin et al. recently show that Beclin 1, together with its phosphatidylinositol 3-kinase (PI3K) binding partner, Vps34, participate in microglial receptor-mediated phagocytosis by regulating the retromer complex, which is involved in sorting cellular components to the lysosome or recycling the components back to defined compartments such as the cell surface [131]. Therefore, reduction of Beclin 1 results in decreased retromer levels, phagocytic receptor recycling, and phagocytosis of latex beads and Alzheimer's disease (AD) –associated β-amyloid (Aβ) peptide. In addition, they showed that Beclin 1 and retromer are reduced in microglia isolated from postmortem human AD brains [131].
Together these findings suggest that autophagy may participate in different microglial cell functions that play important roles in neuroinflammation; however, further investigation is required in order to dissect in detail the molecules involved in these processes.
Although the causes of several pathological conditions in the CNS, such as AD, Parkinson's disease (PD) and multiple sclerosis (MS), are complex and may involve multiple factors, an active role of the innate host defense mediated by mononuclear phagocytes has been clearly demonstrated [116]. Despite the fact that inflammation may not typically represent an initiating factor in neurodegenerative diseases, it is clear that a balance between pro- and anti-inflammatory signals, determining sustained inflammatory responses in the CNS involving microglia and astrocytes contribute to disease progression [122].
Neurodegenerative diseases are pathological conditions in the nervous system characterized by progressive neuron loss, normally accompanied by accumulation of abnormal protein aggregates in the affected regions. Autophagy is essential for proper neuron function and generally plays a cytoprotective role against the onset of neurodegeneration preventing accumulation of aggregate-prone proteins and damaged mitochondria [132]. Pharmacological induction of autophagy can enhance the clearance of intracytoplasmic protein aggregates, such as mutant forms of huntington, and ameliorate pathology in cell and animal models of neurodegenerative diseases such as AD, PD and forms of motor neuron disease [133]. Interestingly, Beclin 1 expression decreases with age in the human brain, suggesting that decreased autophagy may underlie the observed association between advanced age and increased incidence of neurodegenerative diseases [134]. Next, we will describe the role of autophagic dysfunction in the etiology of some neurodegenerative diseases, such as AD and PD and the different ways in which autophagic pathways might be manipulated for the therapeutic benefit of patients with those neurodegenerative disorders.
5.2. Alzheimer's disease
Alzheimer's disease is a late-onset, neurodegenerative disorder characterized by progressive accumulation of hyperphosphorylated tau protein in intracellular neurofibrillary tangles and deposition of extracellular Aβ plaques [84]. Previous studies in well-preserved biopsied AD neocortex have distinguished dense lysosomes and also various types of autophagic vacuoles including autophagosomes, amphisomes, multilamellar bodies, and autolysosomes, representing “intermediate” stages in the progression of autophagy [135]. These observations suggested that defects in autophagic maturation may be a general feature of AD pathology.
For instance, in AD, autophagy may be impaired at both levels, autophagosome degradation [136] and autophagosome formation [137], although these effects may vary accordingly to the patient's genotype or the stage of the disease. Genetic studies have identified several mutations that cause rare familial forms of AD, such as mutations in amyloid precursor protein (APP) and in presenilin (PS) 1 and 2 [138–139].
PS1 mutations are the most common cause of early-onset familial AD and this transmembrane protein has a critical role in lysosome acidification. Furthermore, PS1 is essential for the activation of lysosomal proteases during autophagy since mutations in PS1 lead to impaired targeting of α subunit of V0-ATPase to the lysosome resulting in deficient lysosomal acidification [136].
Other evidence suggests that autophagy might be disrupted at the level of autophagosome formation in patients with AD. Compared with healthy individuals, the brains of patients with AD show reduced expression of Beclin 1, which could lead to impairment in autophagic activity [137]. Heterozygous deletion of Beclin 1 in mice that express the AD-associated mutant human APP have increased APP and Aβ aggregation and present a more severe neurodegeneration compared to Beclin 1 WT mice expressing mutant human APP [137]. Moreover, not only pathogenic APP but also pathogenic tau is degraded by autophagy. Supporting these data, 3-MA (autophagy inhibitor) increases tau aggregation and toxicity, and rapamycin (autophagy inductor) decreases toxicity in cells that overexpress mutant tau [128, 140]. However, another study indicates that both treatment with 3-MA or Beclin 1 knockdown decreases Aβ toxicity in human neuroblastoma and glioma cell lines [141]. This controversy about the cytoprotective vs. cytotoxic roles of autophagy in AD models may be addressed by evaluating the autophagic flux and the degree of lysosomal defect in each case. Further research is required to understand the participation of the autophagic response in AD.
5.3. Parkinson's disease
Parkinson's disease is a late-onset neurodegenerative disorder caused by degeneration of dopaminergic neurons in the substantia nigra. This pathology is characterized by the presence of intracellular inclusions named Lewy bodies containing the proteins α-synuclein and ubiquitin and accumulation of autophagic vacuoles and damaged mitochondria. The most frequently type of PD is sporadic, although familial forms do exist [142].
Mutations in two genes: PINK1 and PARK2 have been associated with autosomal recessive PD. PINK1 is a gene that leads to the ubiquitination of outer mitochondrial membrane proteins, AP recruitment, and mitophagic degradation and PARK2 encodes Parkin [143–144], which is selectively recruited by PINK. Both, WT and mutant A53T α-synuclein are degraded by autophagy and the overexpression of mutant A53T α-synuclein in neurons may lead to PD. Mutant A53T-expressing neurons induce mitophagy, accompanied by depletion of cellular ATP and cell death[145]. In addition, treatment with 3-MA or knockdown of Parkin or Beclin 1 partially protects against A53T-mediated cytotoxicity.
Moreover, mutations that cause gene duplications of α-synuclein, are sufficient to cause familial PD [146]. Interestingly, whereas autophagy is inhibited by overexpression of mutant A53T, excess of intracellular levels of α-synuclein impairs autophagy by causing inhibition of the small GTPase Rab-1A [147]. The accumulation of α-synuclein might increase the level of protein aggregation and reduce autophagy, preventing effective clearance of dysfunctional mitochondria, and enhancing neuronal susceptibility to pro-apoptotic insults [147].
6. Autophagy and cancer
The autophagic machinery seems to be involved in some type of cancers. There are really two links, one at the level of cancer development and the second one at the level of cancer treatment. Several studies revealed that allelic deletion of Beclin 1+/− has been associated with enhanced susceptibility to ovarian, prostate and breast cancer in humans [148] and heterozygous Beclin 1+/− mice increased spontaneous tumorigenesis [148]. In addition, spontaneous tumorigenesis was also reported later upon deletion of others ATGs, such as UVRAG, LC3 and ATG4c [149–151]. Furthermore, inactivation of autophagy-specific genes may be used like a tumour-suppressor pathway, and its decreased activity may be involved in the development of human cancer.
Autophagy was initially classified as an anti-oncogenic mechanism. This conclusion was based on evidence that demonstrate a tumor-suppressive role for autophagy when considering genetic inactivation of it, either indirectly by constitutive activation of the PI3K/AKT pathway via activating PI3K mutations, AKT amplifications, or PTEN (tumor suppressor gene) loss or directly by allelic loss of Beclin 1 or deficiency in Atg5 that in increased tumorigenesis [152]. Paradoxically, this concept has been challenged by some evidence suggesting that autophagy can also be pro-oncogenic because it might help to maintain tumor cell survival [153].
Regulatory mechanisms of autophagy overlap with signaling pathways that regulate tumorigenesis. Consistently, tumour-suppressor genes involved in the upstream inhibition of mTOR signaling (PTEN, TSC1, and TSC2) turn autophagy on, and conversely, mTOR activating oncogene products such as class I PI3K and Akt genes turn it off [154–156]. Moreover, p53 and DAPK, that are frequently mutated in human cancer, positively regulates autophagy [2, 157–158]. The cellular proto-oncoproteins, Bcl-2 and Bcl-XL, which are often overexpressed in human cancers, inhibit autophagy by binding to Beclin 1 [2].
The overexpression of p62 (normally degraded by autophagy) as a result of autophagy inhibition, was shown to be important in the promotion of tumorigenesis through deregulation of NF-κB signaling, activation of NF-E2 related factor 2 (Nrf-2), accumulation of ROS, and increased DNA damage.
The conflicting pro-survival and pro-death functions of autophagy make the connection to cancer treatment more complex. However, it has been proposed that in the early stages of cancer development, quality control by autophagy, particularly over genome maintenance, inhibits tumorigenesis, conferring anti-oncogenic functions upon this pathway. In fact, autophagy could also coordinate the maintenance or entry of cells into the G0 phase and consequently, prevent spontaneous hyperproliferation of cells. In contrast, in the late stages of oncogenesis, autophagy may help tumour cells endure metabolic stress and resist death triggered by chemotherapeutic [159].
6.1. Autophagy and cancer therapy
The particular ability of autophagy to promote cell survival during metabolic stress or cell death as a result of an imbalance in cell metabolism, where autophagic cellular consumption exceeds the cellular capacity for synthesis, is a promising avenue for cancer therapy.
In a study using mice harboring c-Myc-induced lymphomas, it was reported that chloroquine, an alkalinizing lysosomotropic drug, impaired autophagic degradation, by a mechanism that enhanced the ability of either p53 or a DNA alkylating agent to induce tumor cell death and tumor regression [160]. This suggests that inhibition of autophagy might be beneficial in cancer chemotherapy. In addition, some recent studies in glioma cells demonstrated that shRNA-mediated knockdown of BECN1 and ATG5 protected this cell against temozolomide (a DNA alkylating agent)-induced death [161]. Furthermore, numerous assays using autophagy inhibitors (3-MA or chloroquine treatment) or genetic knockdown of autophagy genes have demonstrated that inhibition of autophagy may sensitize tumour cells to cell death induced by diverse cytotoxic agents [86, 159, 162–163]. In addition, there are several Phase I/II clinical trials in progress using the chloroquine or hydroxychloroquine in combination with chemotherapy for the treatment of a range of haematological and solid tumours [162, 164].
Proteasome inhibitors are also known as autophagy inducers. Protein turnover by lysosomal degradation through the autophagy pathway is functionally coupled to, and compensatory with, the ubiquitin-mediated proteasome protein degradation pathway. In this scenario, the proteasome inhibitor Bortezomib was shown to induce autophagy in colorectal cancer cells and this drug was approved by US Food and Drug Administration for the treatment of relapsed multiple myeloma [165–166]. Similar findings were observed in another study, which demonstrated that the inhibition of proteasome in prostate cancer cells by NPI-0052 could facilitate autophagy through an eIF2α-dependent mechanism that up-regulated transcription of ATG genes [167]. Thus, simultaneous inhibition of both mechanisms proteasome- and autophagy-mediated protein degradation could result in more efficient tumour cell elimination than the inhibition of either pathway alone, which is worth testing therapeutically.
7. Perspectives
Autophagy has been shown to play essential roles in infection, inflammatory diseases and cancer. Better understanding of the relevance of autophagy contribution to diseases has great clinical potential. While the study of autophagy is still in progress, it is clear that autophagy is deeply integrated into metabolism, stress response and cell death pathways. Thus, this process and their associated responses might provide information about the capacity of the host to interact with exogenous pathogens and endogenous molecules produced under stress conditions, but these events could be conditioned by the type of stress and cell type. In the meanwhile, how autophagy is controlled and regulated, and the specificity that is associated with cellular consumption, requires further investigation. It will be important to define and characterize molecular and biochemical events involved in the complex interplay between autophagy and diverse pathologies, for the development of novel therapeutic strategies for patients suffering either neurodegenerative and autoimmune diseases or cancer. Finally, further pre-clinical and clinical studies are also warranted to explore the role of autophagy up-regulation in cancer prevention.
Highlights
-
>
Autophagy is a homeostatic process by which cell components are degraded.
-
>
Autophagy can also play a role in cell death.
-
>
Autophagy contributes to host defense and inflammation.
-
>
Autophagic dysfunction is associated with cancer
Acknowledgements
This work was supported by Grant Number 1R01TW007621-01A4 from Fogarty International Center, NIH, USA, CONICET and SECyT-UNC, Argentina. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the Fogarty International Center (FIC), NIH, USA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Mehrpour M, Esclatine A, Beau I, Codogno P. Overview of macroautophagy regulation in mammalian cells. Cell Res. 2010;20:748–62. doi: 10.1038/cr.2010.82. [DOI] [PubMed] [Google Scholar]
- [4].Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344–8. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kouroku Y, Fujita E, Tanida I, Ueno T, Isoai A, Kumagai H, et al. ER stress (PERK/eIF2alpha phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 2007;14:230–9. doi: 10.1038/sj.cdd.4401984. [DOI] [PubMed] [Google Scholar]
- [6].Shin DM, Jeon BY, Lee HM, Jin HS, Yuk JM, Song CH, et al. Mycobacterium tuberculosis eis regulates autophagy, inflammation, and cell death through redox-dependent signaling. PLoS Pathog. 2010;6:e1001230. doi: 10.1371/journal.ppat.1001230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, et al. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037–40. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- [8].Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438–41. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- [9].Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Connell S, et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–7. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- [10].Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315:1398–401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- [11].Franchi L, Eigenbrod T, Munoz-Planillo R, Nunez G. The inflammasome: a caspase-1-activation platform that regulates immune responses and disease pathogenesis. Nat Immunol. 2009;10:241–7. doi: 10.1038/ni.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12:222–30. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Qu X, Zou Z, Sun Q, Luby-Phelps K, Cheng P, Hogan RN, et al. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007;128:931–46. doi: 10.1016/j.cell.2006.12.044. [DOI] [PubMed] [Google Scholar]
- [14].Arroyo DS, Soria JA, Gaviglio EA, Garcia-Keller C, Cancela LM, Rodriguez-Galan MC, et al. Toll-like receptor 2 ligands promote microglial cell death by inducing autophagy. FASEB J. 2013;27:299–312. doi: 10.1096/fj.12-214312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12:401–10. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Munz C. Enhancing immunity through autophagy. Annu Rev Immunol. 2009;27:423–49. doi: 10.1146/annurev.immunol.021908.132537. [DOI] [PubMed] [Google Scholar]
- [17].Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–5. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- [18].Virgin HW, Levine B. Autophagy genes in immunity. Nat Immunol. 2009;10:461–70. doi: 10.1038/ni.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol. 2010;22:132–9. doi: 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- [20].Harris H, Rubinsztein DC. Control of autophagy as a therapy for neurodegenerative disease. Nat Rev Neurol. 2012;8:108–17. doi: 10.1038/nrneurol.2011.200. [DOI] [PubMed] [Google Scholar]
- [21].Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–26. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Yu L, McPhee CK, Zheng L, Mardones GA, Rong Y, Peng J, et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 2010;465:942–6. doi: 10.1038/nature09076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Schmid D, Pypaert M, Munz C. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity. 2007;26:79–92. doi: 10.1016/j.immuni.2006.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Tooze SA, Yoshimori T. The origin of the autophagosomal membrane. Nat Cell Biol. 2010;12:831–5. doi: 10.1038/ncb0910-831. [DOI] [PubMed] [Google Scholar]
- [25].Kuballa P, Nolte WM, Castoreno AB, Xavier RJ. Autophagy and the immune system. Annu Rev Immunol. 2012;30:611–46. doi: 10.1146/annurev-immunol-020711-074948. [DOI] [PubMed] [Google Scholar]
- [26].Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Matsunaga K, Morita E, Saitoh T, Akira S, Ktistakis NT, Izumi T, et al. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J Cell Biol. 2010;190:511–21. doi: 10.1083/jcb.200911141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Mizushima N, Kuma A, Kobayashi Y, Yamamoto A, Matsubae M, Takao T, et al. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J Cell Sci. 2003;116:1679–88. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- [29].Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–8. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci. 2004;117:2805–12. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- [31].Satoo K, Noda NN, Kumeta H, Fujioka Y, Mizushima N, Ohsumi Y, et al. The structure of Atg4B-LC3 complex reveals the mechanism of LC3 processing and delipidation during autophagy. EMBO J. 2009;28:1341–50. doi: 10.1038/emboj.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Radoshevich L, Murrow L, Chen N, Fernandez E, Roy S, Fung C, et al. ATG12 conjugation to ATG3 regulates mitochondrial homeostasis and cell death. Cell. 2010;142:590–600. doi: 10.1016/j.cell.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Weidberg H, Shvets E, Elazar Z. Biogenesis and cargo selectivity of autophagosomes. Annu Rev Biochem. 2011;80:125–56. doi: 10.1146/annurev-biochem-052709-094552. [DOI] [PubMed] [Google Scholar]
- [35].Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, et al. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–67. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol. 2010;12:747–57. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Luzio JP, Pryor PR, Bright NA. Lysosomes: fusion and function. Nat Rev Mol Cell Biol. 2007;8:622–32. doi: 10.1038/nrm2217. [DOI] [PubMed] [Google Scholar]
- [38].Furuta N, Fujita N, Noda T, Yoshimori T, Amano A. Combinational soluble N-ethylmaleimide-sensitive factor attachment protein receptor proteins VAMP8 and Vti1b mediate fusion of antimicrobial and canonical autophagosomes with lysosomes. Mol Biol Cell. 2010;21:1001–10. doi: 10.1091/mbc.E09-08-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Deter RL, Baudhuin P, De Duve C. Participation of lysosomes in cellular autophagy induced in rat liver by glucagon. J Cell Biol. 1967;35:C11–6. doi: 10.1083/jcb.35.2.c11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Baba M, Osumi M, Scott SV, Klionsky DJ, Ohsumi Y. Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J Cell Biol. 1997;139:1687–95. doi: 10.1083/jcb.139.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Szeto J, Kaniuk NA, Canadien V, Nisman R, Mizushima N, Yoshimori T, et al. ALIS are stress-induced protein storage compartments for substrates of the proteasome and autophagy. Autophagy. 2006;2:189–99. doi: 10.4161/auto.2731. [DOI] [PubMed] [Google Scholar]
- [42].Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–14. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Klionsky DJ, Cuervo AM, Dunn WA, Jr, Levine B, van der Klei I, Seglen PO. How shall I eat thee? Autophagy. 2007;3:413–6. doi: 10.4161/auto.4377. [DOI] [PubMed] [Google Scholar]
- [44].Knaevelsrud H, Simonsen A. Fighting disease by selective autophagy of aggregate-prone proteins. FEBS Lett. 2010;584:2635–45. doi: 10.1016/j.febslet.2010.04.041. [DOI] [PubMed] [Google Scholar]
- [45].Kim PK, Hailey DW, Mullen RT, Lippincott-Schwartz J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc Natl Acad Sci U S A. 2008;105:20567–74. doi: 10.1073/pnas.0810611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–31. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- [47].Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol. 2009;10:1215–21. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- [48].Orvedahl A, MacPherson S, Sumpter R, Jr, Talloczy Z, Zou Z, Levine B. Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe. 2010;7:115–27. doi: 10.1016/j.chom.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Bernales S, McDonald KL, Walter P. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 2006;4:e423. doi: 10.1371/journal.pbio.0040423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol. 2008;10:602–10. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- [51].Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–96. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol. 2013;13:722–37. doi: 10.1038/nri3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Medzhitov R, Horng T. Transcriptional control of the inflammatory response. Nat Rev Immunol. 2009;9:692–703. doi: 10.1038/nri2634. [DOI] [PubMed] [Google Scholar]
- [54].Chen GY, Nunez G. Sterile inflammation: sensing and reacting to damage. Nat Rev Immunol. 2010;10:826–37. doi: 10.1038/nri2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Stehlik C, Dorfleutner A. COPs and POPs: modulators of inflammasome activity. J Immunol. 2007;179:7993–8. doi: 10.4049/jimmunol.179.12.7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–35. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Shi CS, Kehrl JH. TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci Signal. 2010;3:ra42. doi: 10.1126/scisignal.2000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Henault J, Martinez J, Riggs JM, Tian J, Mehta P, Clarke L, et al. Noncanonical autophagy is required for type I interferon secretion in response to DNA-immune complexes. Immunity. 2012;37:986–97. doi: 10.1016/j.immuni.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–8. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- [60].Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140:821–32. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- [61].Bensaad K, Cheung EC, Vousden KH. Modulation of intracellular ROS levels by TIGAR controls autophagy. EMBO J. 2009;28:3015–26. doi: 10.1038/emboj.2009.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–5. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- [63].Shi CS, Shenderov K, Huang NN, Kabat J, Abu-Asab M, Fitzgerald KA, et al. Activation of autophagy by inflammatory signals limits IL-1beta production by targeting ubiquitinated inflammasomes for destruction. Nat Immunol. 2012;13:255–63. doi: 10.1038/ni.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Matzinger P. Tolerance, danger, and the extended family. Annu Rev Immunol. 1994;12:991–1045. doi: 10.1146/annurev.iy.12.040194.005015. [DOI] [PubMed] [Google Scholar]
- [65].Garg AD, Nowis D, Golab J, Vandenabeele P, Krysko DV, Agostinis P. Immunogenic cell death, DAMPs and anticancer therapeutics: an emerging amalgamation. Biochim Biophys Acta. 2010;1805:53–71. doi: 10.1016/j.bbcan.2009.08.003. [DOI] [PubMed] [Google Scholar]
- [66].Krysko DV, Garg AD, Kaczmarek A, Krysko O, Agostinis P, Vandenabeele P. Immunogenic cell death and DAMPs in cancer therapy. Nat Rev Cancer. 2012;12:860–75. doi: 10.1038/nrc3380. [DOI] [PubMed] [Google Scholar]
- [67].Zhang Q, Kang R, Zeh HJ, 3rd, Lotze MT, Tang D. DAMPs and autophagy: cellular adaptation to injury and unscheduled cell death. Autophagy. 2013;9:451–8. doi: 10.4161/auto.23691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Tang D, Kang R, Livesey KM, Zeh HJ, 3rd, Lotze MT. High mobility group box 1 (HMGB1) activates an autophagic response to oxidative stress. Antioxid Redox Signal. 2011;15:2185–95. doi: 10.1089/ars.2010.3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Thorburn J, Horita H, Redzic J, Hansen K, Frankel AE, Thorburn A. Autophagy regulates selective HMGB1 release in tumor cells that are destined to die. Cell Death Differ. 2009;16:175–83. doi: 10.1038/cdd.2008.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Li W, Zhu S, Li J, Assa A, Jundoria A, Xu J, et al. EGCG stimulates autophagy and reduces cytoplasmic HMGB1 levels in endotoxin-stimulated macrophages. Biochem Pharmacol. 2011;81:1152–63. doi: 10.1016/j.bcp.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Michaud M, Martins I, Sukkurwala AQ, Adjemian S, Ma Y, Pellegatti P, et al. Autophagy-dependent anticancer immune responses induced by chemotherapeutic agents in mice. Science. 2011;334:1573–7. doi: 10.1126/science.1208347. [DOI] [PubMed] [Google Scholar]
- [72].Tang D, Kang R, Coyne CB, Zeh HJ, Lotze MT. PAMPs and DAMPs: signal 0s that spur autophagy and immunity. Immunol Rev. 2012;249:158–75. doi: 10.1111/j.1600-065X.2012.01146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Biswas D, Qureshi OS, Lee WY, Croudace JE, Mura M, Lammas DA. ATP-induced autophagy is associated with rapid killing of intracellular mycobacteria within human monocytes/macrophages. BMC Immunol. 2008;9:35. doi: 10.1186/1471-2172-9-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Takenouchi T, Nakai M, Iwamaru Y, Sugama S, Tsukimoto M, Fujita M, et al. The activation of P2X7 receptor impairs lysosomal functions and stimulates the release of autophagolysosomes in microglial cells. J Immunol. 2009;182:2051–62. doi: 10.4049/jimmunol.0802577. [DOI] [PubMed] [Google Scholar]
- [75].Livesey KM, Kang R, Vernon P, Buchser W, Loughran P, Watkins SC, et al. p53/HMGB1 complexes regulate autophagy and apoptosis. Cancer Res. 2012;72:1996–2005. doi: 10.1158/0008-5472.CAN-11-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Huang J, Ni J, Liu K, Yu Y, Xie M, Kang R, et al. HMGB1 promotes drug resistance in osteosarcoma. Cancer Res. 2012;72:230–8. doi: 10.1158/0008-5472.CAN-11-2001. [DOI] [PubMed] [Google Scholar]
- [77].Cooney R, Baker J, Brain O, Danis B, Pichulik T, Allan P, et al. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med. 2010;16:90–7. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- [78].Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet. 2010;42:1118–25. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008;40:955–62. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Feng CG, Zheng L, Jankovic D, Bafica A, Cannons JL, Watford WT, et al. The immunity-related GTPase Irgm1 promotes the expansion of activated CD4+ T cell populations by preventing interferon-gamma-induced cell death. Nat Immunol. 2008;9:1279–87. doi: 10.1038/ni.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhaes JG, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- [82].Brain O, Allan P, Simmons A. NOD2-mediated autophagy and Crohn disease. Autophagy. 2010;6:412–4. doi: 10.4161/auto.6.3.11389. [DOI] [PubMed] [Google Scholar]
- [83].Cadwell K, Patel KK, Maloney NS, Liu TC, Ng AC, Storer CE, et al. Virus-plus-susceptibility gene interaction determines Crohn's disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–45. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Murrow L, Debnath J. Autophagy as a stress-response and quality-control mechanism: implications for cell injury and human disease. Annu Rev Pathol. 2013;8:105–37. doi: 10.1146/annurev-pathol-020712-163918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Schmid D, Munz C. Innate and adaptive immunity through autophagy. Immunity. 2007;27:11–21. doi: 10.1016/j.immuni.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Rubinsztein DC, Codogno P, Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat Rev Drug Discov. 2012;11:709–30. doi: 10.1038/nrd3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Sakurai A, Maruyama F, Funao J, Nozawa T, Aikawa C, Okahashi N, et al. Specific behavior of intracellular Streptococcus pyogenes that has undergone autophagic degradation is associated with bacterial streptolysin O and host small G proteins Rab5 and Rab7. J Biol Chem. 2010;285:22666–75. doi: 10.1074/jbc.M109.100131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Yamaguchi H, Nakagawa I, Yamamoto A, Amano A, Noda T, Yoshimori T. An initial step of GAS-containing autophagosome-like vacuoles formation requires Rab7. PLoS Pathog. 2009;5:e1000670. doi: 10.1371/journal.ppat.1000670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Nozawa T, Aikawa C, Goda A, Maruyama F, Hamada S, Nakagawa I. The small GTPases Rab9A and Rab23 function at distinct steps in autophagy during Group A Streptococcus infection. Cell Microbiol. 2012;14:1149–65. doi: 10.1111/j.1462-5822.2012.01792.x. [DOI] [PubMed] [Google Scholar]
- [90].Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- [91].von Muhlinen N, Thurston T, Ryzhakov G, Bloor S, Randow F. NDP52, a novel autophagy receptor for ubiquitin-decorated cytosolic bacteria. Autophagy. 2010;6:288–9. doi: 10.4161/auto.6.2.11118. [DOI] [PubMed] [Google Scholar]
- [92].Kirkin V, Lamark T, Sou YS, Bjorkoy G, Nunn JL, Bruun JA, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33:505–16. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- [93].Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–33. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Yoshikawa Y, Ogawa M, Hain T, Yoshida M, Fukumatsu M, Kim M, et al. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nat Cell Biol. 2009;11:1233–40. doi: 10.1038/ncb1967. [DOI] [PubMed] [Google Scholar]
- [95].Dupont N, Lacas-Gervais S, Bertout J, Paz I, Freche B, Van Nhieu GT, et al. Shigella phagocytic vacuolar membrane remnants participate in the cellular response to pathogen invasion and are regulated by autophagy. Cell Host Microbe. 2009;6:137–49. doi: 10.1016/j.chom.2009.07.005. [DOI] [PubMed] [Google Scholar]
- [96].Shahnazari S, Yen WL, Birmingham CL, Shiu J, Namolovan A, Zheng YT, et al. A diacylglycerol-dependent signaling pathway contributes to regulation of antibacterial autophagy. Cell Host Microbe. 2010;8:137–46. doi: 10.1016/j.chom.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Kumar D, Nath L, Kamal MA, Varshney A, Jain A, Singh S, et al. Genome-wide analysis of the host intracellular network that regulates survival of Mycobacterium tuberculosis. Cell. 2010;140:731–43. doi: 10.1016/j.cell.2010.02.012. [DOI] [PubMed] [Google Scholar]
- [98].Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–66. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- [99].Wong KW, Jacobs WR., Jr Critical role for NLRP3 in necrotic death triggered by Mycobacterium tuberculosis. Cell Microbiol. 2011;13:1371–84. doi: 10.1111/j.1462-5822.2011.01625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Watson RO, Manzanillo PS, Cox JS. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell. 2012;150:803–15. doi: 10.1016/j.cell.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Ponpuak M, Davis AS, Roberts EA, Delgado MA, Dinkins C, Zhao Z, et al. Delivery of cytosolic components by autophagic adaptor protein p62 endows autophagosomes with unique antimicrobial properties. Immunity. 2010;32:329–41. doi: 10.1016/j.immuni.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Alonso S, Pethe K, Russell DG, Purdy GE. Lysosomal killing of Mycobacterium mediated by ubiquitin-derived peptides is enhanced by autophagy. Proc Natl Acad Sci U S A. 2007;104:6031–6. doi: 10.1073/pnas.0700036104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Kim BH, Shenoy AR, Kumar P, Das R, Tiwari S, MacMicking JD. A family of IFN-gamma-inducible 65-kD GTPases protects against bacterial infection. Science. 2011;332:717–21. doi: 10.1126/science.1201711. [DOI] [PubMed] [Google Scholar]
- [104].Zhao Z, Fux B, Goodwin M, Dunay IR, Strong D, Miller BC, et al. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe. 2008;4:458–69. doi: 10.1016/j.chom.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Liesenfeld O, Parvanova I, Zerrahn J, Han SJ, Heinrich F, Munoz M, et al. The IFN-gamma-inducible GTPase, Irga6, protects mice against Toxoplasma gondii but not against Plasmodium berghei and some other intracellular pathogens. PLoS One. 2011;6:e20568. doi: 10.1371/journal.pone.0020568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Portillo JA, Okenka G, Reed E, Subauste A, Van Grol J, Gentil K, et al. The CD40-autophagy pathway is needed for host protection despite IFN-Gamma-dependent immunity and CD40 induces autophagy via control of P21 levels. PLoS One. 2010;5:e14472. doi: 10.1371/journal.pone.0014472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Orvedahl A, Alexander D, Talloczy Z, Sun Q, Wei Y, Zhang W, et al. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe. 2007;1:23–35. doi: 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
- [108].Liang C, E X, Jung JU. Downregulation of autophagy by herpesvirus Bcl-2 homologs. Autophagy. 2008;4:268–72. doi: 10.4161/auto.5210. [DOI] [PubMed] [Google Scholar]
- [109].Li Y, Zhang C, Chen X, Yu J, Wang Y, Yang Y, et al. ICP34.5 protein of herpes simplex virus facilitates the initiation of protein translation by bridging eukaryotic initiation factor 2alpha (eIF2alpha) and protein phosphatase 1. J Biol Chem. 2011;286:24785–92. doi: 10.1074/jbc.M111.232439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].E X, Hwang S, Oh S, Lee JS, Jeong JH, Gwack Y, et al. Viral Bcl-2-mediated evasion of autophagy aids chronic infection of gammaherpesvirus 68. PLoS Pathog. 2009;5:e1000609. doi: 10.1371/journal.ppat.1000609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Kyei GB, Dinkins C, Davis AS, Roberts E, Singh SB, Dong C, et al. Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. J Cell Biol. 2009;186:255–68. doi: 10.1083/jcb.200903070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Eekels JJ, Sagnier S, Geerts D, Jeeninga RE, Biard-Piechaczyk M, Berkhout B. Inhibition of HIV-1 replication with stable RNAi-mediated knockdown of autophagy factors. Virol J. 2012;9:69. doi: 10.1186/1743-422X-9-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Blanchet FP, Moris A, Nikolic DS, Lehmann M, Cardinaud S, Stalder R, et al. Human immunodeficiency virus-1 inhibition of immunoamphisomes in dendritic cells impairs early innate and adaptive immune responses. Immunity. 2010;32:654–69. doi: 10.1016/j.immuni.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Harman AN, Kraus M, Bye CR, Byth K, Turville SG, Tang O, et al. HIV-1-infected dendritic cells show 2 phases of gene expression changes, with lysosomal enzyme activity decreased during the second phase. Blood. 2009;114:85–94. doi: 10.1182/blood-2008-12-194845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Zhou D, Spector SA. Human immunodeficiency virus type-1 infection inhibits autophagy. AIDS. 2008;22:695–9. doi: 10.1097/QAD.0b013e3282f4a836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Arroyo DS, Soria JA, Gaviglio EA, Rodriguez-Galan MC, Iribarren P. Toll-like receptors are key players in neurodegeneration. Int Immunopharmacol. 2011;11:1415–21. doi: 10.1016/j.intimp.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Galea I, Bechmann I, Perry VH. What is immune privilege (not)? Trends Immunol. 2007;28:12–8. doi: 10.1016/j.it.2006.11.004. [DOI] [PubMed] [Google Scholar]
- [118].Frank-Cannon TC, Alto LT, McAlpine FE, Tansey MG. Does neuroinflammation fan the flame in neurodegenerative diseases? Mol Neurodegener. 2009;4:47. doi: 10.1186/1750-1326-4-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Rivest S. Regulation of innate immune responses in the brain. Nat Rev Immunol. 2009;9:429–39. doi: 10.1038/nri2565. [DOI] [PubMed] [Google Scholar]
- [120].Popovich PG, Longbrake EE. Can the immune system be harnessed to repair the CNS? Nat Rev Neurosci. 2008;9:481–93. doi: 10.1038/nrn2398. [DOI] [PubMed] [Google Scholar]
- [121].Streit WJ. Microglia as neuroprotective, immunocompetent cells of the CNS. Glia. 2002;40:133–9. doi: 10.1002/glia.10154. [DOI] [PubMed] [Google Scholar]
- [122].Glass CK, Saijo K, Winner B, Marchetto MC, Gage FH. Mechanisms underlying inflammation in neurodegeneration. Cell. 2010;140:918–34. doi: 10.1016/j.cell.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Hanisch UK. Microglia as a source and target of cytokines. Glia. 2002;40:140–55. doi: 10.1002/glia.10161. [DOI] [PubMed] [Google Scholar]
- [124].Rosello A, Warnes G, Meier UC. Cell death pathways and autophagy in the central nervous system and its involvement in neurodegeneration, immunity and central nervous system infection: to die or not to die--that is the question. Clin Exp Immunol. 2012;168:52–7. doi: 10.1111/j.1365-2249.2011.04544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Winslow AR, Rubinsztein DC. Autophagy in neurodegeneration and development. Biochim Biophys Acta. 2008;1782:723–9. doi: 10.1016/j.bbadis.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Nixon RA, Yang DS, Lee JH. Neurodegenerative lysosomal disorders: a continuum from development to late age. Autophagy. 2008;4:590–9. doi: 10.4161/auto.6259. [DOI] [PubMed] [Google Scholar]
- [127].Wang QJ, Ding Y, Kohtz DS, Mizushima N, Cristea IM, Rout MP, et al. Induction of autophagy in axonal dystrophy and degeneration. J Neurosci. 2006;26:8057–68. doi: 10.1523/JNEUROSCI.2261-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Wang Y, Martinez-Vicente M, Kruger U, Kaushik S, Wong E, Mandelkow EM, et al. Tau fragmentation, aggregation and clearance: the dual role of lysosomal processing. Hum Mol Genet. 2009;18:4153–70. doi: 10.1093/hmg/ddp367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Dello Russo C, Lisi L, Feinstein DL, Navarra P. mTOR kinase, a key player in the regulation of glial functions: relevance for the therapy of multiple sclerosis. Glia. 2013;61:301–11. doi: 10.1002/glia.22433. [DOI] [PubMed] [Google Scholar]
- [130].Lucin KM, Wyss-Coray T. Immune activation in brain aging and neurodegeneration: too much or too little? Neuron. 2009;64:110–22. doi: 10.1016/j.neuron.2009.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Lucin KM, O'Brien CE, Bieri G, Czirr E, Mosher KI, Abbey RJ, et al. Microglial beclin 1 regulates retromer trafficking and phagocytosis and is impaired in Alzheimer's disease. Neuron. 2013;79:873–86. doi: 10.1016/j.neuron.2013.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Wong E, Cuervo AM. Autophagy gone awry in neurodegenerative diseases. Nat Neurosci. 2010;13:805–11. doi: 10.1038/nn.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Bove J, Martinez-Vicente M, Vila M. Fighting neurodegeneration with rapamycin: mechanistic insights. Nat Rev Neurosci. 2011;12:437–52. doi: 10.1038/nrn3068. [DOI] [PubMed] [Google Scholar]
- [134].Shibata M, Lu T, Furuya T, Degterev A, Mizushima N, Yoshimori T, et al. Regulation of intracellular accumulation of mutant Huntingtin by Beclin 1. J Biol Chem. 2006;281:14474–85. doi: 10.1074/jbc.M600364200. [DOI] [PubMed] [Google Scholar]
- [135].Nixon RA, Wegiel J, Kumar A, Yu WH, Peterhoff C, Cataldo A, et al. Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J Neuropathol Exp Neurol. 2005;64:113–22. doi: 10.1093/jnen/64.2.113. [DOI] [PubMed] [Google Scholar]
- [136].Lee JH, Yu WH, Kumar A, Lee S, Mohan PS, Peterhoff CM, et al. Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell. 2010;141:1146–58. doi: 10.1016/j.cell.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Pickford F, Masliah E, Britschgi M, Lucin K, Narasimhan R, Jaeger PA, et al. The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J Clin Invest. 2008;118:2190–9. doi: 10.1172/JCI33585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Price DL, Tanzi RE, Borchelt DR, Sisodia SS. Alzheimer's disease: genetic studies and transgenic models. Annu Rev Genet. 1998;32:461–93. doi: 10.1146/annurev.genet.32.1.461. [DOI] [PubMed] [Google Scholar]
- [139].Sherrington R, Rogaev EI, Liang Y, Rogaeva EA, Levesque G, Ikeda M, et al. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer's disease. Nature. 1995;375:754–60. doi: 10.1038/375754a0. [DOI] [PubMed] [Google Scholar]
- [140].Berger Z, Ravikumar B, Menzies FM, Oroz LG, Underwood BR, Pangalos MN, et al. Rapamycin alleviates toxicity of different aggregate-prone proteins. Hum Mol Genet. 2006;15:433–42. doi: 10.1093/hmg/ddi458. [DOI] [PubMed] [Google Scholar]
- [141].Wang H, Ma J, Tan Y, Wang Z, Sheng C, Chen S, et al. Amyloid-beta1-42 induces reactive oxygen species-mediated autophagic cell death in U87 and SH-SY5Y cells. J Alzheimers Dis. 2010;21:597–610. doi: 10.3233/JAD-2010-091207. [DOI] [PubMed] [Google Scholar]
- [142].Cheung ZH, Ip NY. Autophagy deregulation in neurodegenerative diseases - recent advances and future perspectives. J Neurochem. 2011;118:317–25. doi: 10.1111/j.1471-4159.2011.07314.x. [DOI] [PubMed] [Google Scholar]
- [143].Lesage S, Brice A. Parkinson's disease: from monogenic forms to genetic susceptibility factors. Hum Mol Genet. 2009;18:R48–59. doi: 10.1093/hmg/ddp012. [DOI] [PubMed] [Google Scholar]
- [144].Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Choubey V, Safiulina D, Vaarmann A, Cagalinec M, Wareski P, Kuum M, et al. Mutant A53T alpha-synuclein induces neuronal death by increasing mitochondrial autophagy. J Biol Chem. 2011;286:10814–24. doi: 10.1074/jbc.M110.132514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J, et al. alpha-Synuclein locus triplication causes Parkinson's disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- [147].Winslow AR, Chen CW, Corrochano S, Acevedo-Arozena A, Gordon DE, Peden AA, et al. alpha-Synuclein impairs macroautophagy: implications for Parkinson's disease. J Cell Biol. 2010;190:1023–37. doi: 10.1083/jcb.201003122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–20. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142–51. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, et al. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8:688–99. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- [151].Marino G, Salvador-Montoliu N, Fueyo A, Knecht E, Mizushima N, Lopez-Otin C. Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in Atg4C/autophagin-3. J Biol Chem. 2007;282:18573–83. doi: 10.1074/jbc.M701194200. [DOI] [PubMed] [Google Scholar]
- [152].Kimmelman AC. The dynamic nature of autophagy in cancer. Genes Dev. 2011;25:1999–2010. doi: 10.1101/gad.17558811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Cully M, You H, Levine AJ, Mak TW. Beyond PTEN mutations: the PI3K pathway as an integrator of multiple inputs during tumorigenesis. Nat Rev Cancer. 2006;6:184–92. doi: 10.1038/nrc1819. [DOI] [PubMed] [Google Scholar]
- [155].Schwartz RA, Fernandez G, Kotulska K, Jozwiak S. Tuberous sclerosis complex: advances in diagnosis, genetics, and management. J Am Acad Dermatol. 2007;57:189–202. doi: 10.1016/j.jaad.2007.05.004. [DOI] [PubMed] [Google Scholar]
- [156].Wang RC, Wei Y, An Z, Zou Z, Xiao G, Bhagat G, et al. Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science. 2012;338:956–9. doi: 10.1126/science.1225967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Feng Z, Zhang H, Levine AJ, Jin S. The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci U S A. 2005;102:8204–9. doi: 10.1073/pnas.0502857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Crighton D, Wilkinson S, O'Prey J, Syed N, Smith P, Harrison PR, et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–34. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- [159].Janku F, McConkey DJ, Hong DS, Kurzrock R. Autophagy as a target for anticancer therapy. Nat Rev Clin Oncol. 2011;8:528–39. doi: 10.1038/nrclinonc.2011.71. [DOI] [PubMed] [Google Scholar]
- [160].Amaravadi RK, Yu D, Lum JJ, Bui T, Christophorou MA, Evan GI, et al. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007;117:326–36. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Voss V, Senft C, Lang V, Ronellenfitsch MW, Steinbach JP, Seifert V, et al. The pan-Bcl-2 inhibitor (−)-gossypol triggers autophagic cell death in malignant glioma. Mol Cancer Res. 2010;8:1002–16. doi: 10.1158/1541-7786.MCR-09-0562. [DOI] [PubMed] [Google Scholar]
- [162].Amaravadi RK, Lippincott-Schwartz J, Yin XM, Weiss WA, Takebe N, Timmer W, et al. Principles and current strategies for targeting autophagy for cancer treatment. Clin Cancer Res. 2011;17:654–66. doi: 10.1158/1078-0432.CCR-10-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Mancias JD, Kimmelman AC. Targeting autophagy addiction in cancer. Oncotarget. 2011;2:1302–6. doi: 10.18632/oncotarget.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Yang ZJ, Chee CE, Huang S, Sinicrope FA. The role of autophagy in cancer: therapeutic implications. Mol Cancer Ther. 2011;10:1533–41. doi: 10.1158/1535-7163.MCT-11-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Roccaro AM, Hideshima T, Richardson PG, Russo D, Ribatti D, Vacca A, et al. Bortezomib as an antitumor agent. Curr Pharm Biotechnol. 2006;7:441–8. doi: 10.2174/138920106779116865. [DOI] [PubMed] [Google Scholar]
- [166].Obeng EA, Carlson LM, Gutman DM, Harrington WJ, Jr, Lee KP, Boise LH. Proteasome inhibitors induce a terminal unfolded protein response in multiple myeloma cells. Blood. 2006;107:4907–16. doi: 10.1182/blood-2005-08-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Zhu K, Dunner K, Jr, McConkey DJ. Proteasome inhibitors activate autophagy as a cytoprotective response in human prostate cancer cells. Oncogene. 2010;29:451–62. doi: 10.1038/onc.2009.343. [DOI] [PMC free article] [PubMed] [Google Scholar]