Abstract
MicroRNAs (miRNAs) are present in both plant and animal kingdoms and represents a growing family of non-coding RNAs. These tiny RNAs act as small guides and direct negative regulations usually in the process of development through sequence complementarity to target mRNAs. Although a large number of miRNAs have been identified from various animals, so far plant miRNA studies have focused mainly on Arabidopsis. Here we describe the identification of 20 miRNAs from a rice cDNA library. All the miRNAs were presumably processed from precursors with stem–loop structures and were positively detected in rice cells from at least one tissue, some of which showed tissue-specific expression. Twenty-three unique rice genes were identified to be feasible targets for seven rice miRNAs, including four members of Scarecrow-like transcription factor, the targets of miR-39 that had been characterized in Arabidopsis. Lacking long complementarity, the regulatory targets of 13 miRNAs remain to be further investigated. A possible mechanism of translational repressor for plant miRNAs that lack perfect complementarity to target mRNAs is discussed.
INTRODUCTION
MicroRNAs (miRNAs) ranging in size from 20 to 25 nt represent a new family of non-coding RNAs that function in post-transcriptional regulation of gene expression (1–3). These tiny RNAs act as small guides and direct negative regulations through sequence complementarity to the 3′-untranslated regions (UTRs) or coding sequences of target mRNAs (4–6). The first characterized endogenous miRNAs were lin-4 and let-7, both of which were shown in the pathway controlling the timing of larval development in the nematode Caenorhabditis elegans (7,8). Since then, more than 300 miRNAs have been identified from different organisms including both animals and plants (9–14). In general, miRNAs are processed from longer precursors that form stem–loop hairpin structures by the RNase III-like enzyme, Dicer in animals (15) or Dicerlike1 (DCL1) in plants (16), and are then incorporated into a ribonucleoprotein complex (miRNP), probably identical to the RNA-induced silencing complex (RISC) which is involved in RNA interference (RNAi) (17). One of the intriguing aspects of the miRNAs is their similarity to small interfering RNAs (siRNAs) which direct mRNA cleavage during RNAi and related processes (18). siRNAs are similar in length to miRNAs but generally differ from miRNAs in their origin. They are derived from long, double-stranded RNAs that are transcribed endogenously or introduced into cells by viral infection or transfection. It has been shown that if an miRNA encounters a target with perfect sequence complementarity, it can enter the RNAi pathway and trigger target cleavage (19,20). Some miRNAs naturally contain sequence complementarity to mRNA and seem to function as endogenous siRNA by directing cleavage of their mRNA targets (21). Now miRNA genes are recognized as a pervasive and widespread feature of animal and plant genomes. The total number of miRNAs in each organism is unknown, but is estimated to represent ∼1% of all genes (22,23), implying that a myriad of miRNAs and their roles in cellular processes, such as development and size control of organisms, remain to be discovered.
A large number of miRNAs have been characterized from different animals such as the worm Caenorhabditis elegans, the fly Drosophila as well as mammals including both the human and the mouse. However, plant miRNA studies have focused mainly on Arabidopsis, and only a handful of miRNAs have been identified (14,24,25). Rice is an economically important food crop. Recent progress in the Rice Genome Project has revealed 50 000–60 000 genes in the rice genome (26), nearly twice the number of genes predicted for Arabidopis thaliana (27), which made rice a favorite plant for functional genomic research. It is therefore interesting to investigate miRNAs in rice that represents a monocotyledon. In this study, we constructed and screened a rice cDNA library of size-fractionated RNAs. Our results showed a very abundant and diverse population of small RNAs in rice, from which 20 miRNAs were further characterized.
MATERIALS AND METHODS
Nucleic acid isolation
Total cellular RNAs were isolated from different tissues of Oryza sativa L. ssp. indica, A.thaliana and maize by guanidine thiocyanate/phenol–chloroform extraction as described (28). A fraction enriched with small-sized RNAs was obtained according to a protocol described previously (24). Briefly, 400 µl (1–2 mg) of total cellular RNAs were combined with 50 µl each of 50% PEG8000 and 5 M NaCl, incubated on ice for 2 h, and centrifuged at 15 000 g for 10 min. After adding 1/10 vol of 3 M sodium acetate and 2 vols of 95% ethanol to the supernatant, small-sized RNA was spun down at 15 000 g following incubation at –20°C for 2 h, washed with 75% ethanol, dried briefly, and re-suspended in RNase-free water.
Construction and screen of rice cDNA library from low molecular weight RNAs
cDNAs synthesis, cloning and sequencing were carried out as described previously (29) with some modifications. Briefly, small RNAs with a size from 16 to 28 nt were recovered from denaturing 15% polyacrylamide gel fractionation (8 M urea, 1× TBE buffer), and then polyadenylated by using poly(A) polymerase (Takara). Synthesis of the first strand of cDNA was performed with 1 µg of poly(A)-tailed RNA in a 20 µl reaction mix containing 0.1 µg of primer HindIII (dT)16 5′-CCCCAAAGCT16-3′ and 200 U of MMLV reverse transcriptase (Promega) for 45 min at 42°C. The cDNAs were poly(G) tailed at the 3′ end by using terminal deoxynucleotidyl transferase (Takara), and then amplified by PCR with primers HindIII (dT)16 and BamHI (dC)16, 5′-GGAATTCGGATC16-3′ and cloned into plasmid pTZ18 as described previously (30). The recombinant plasmid-carrying fragments were sequenced with the BigDye terminator cycle sequencing kit (PE Applied Biosystems) and were analyzed on an ABI377 DNA sequencer.
Northern blot analysis
A 30 µg aliquot of small-sized RNAs from different tissue of rice and other plants was separated on a denaturing 15% polyacryamide–8 mol/l urea gel at 300 V for 3 h, then were electrophoretically transferred to Zeta-probe GT membranes (Bio-Rad) by using a Trans-Blot Electrophoretic Transfer (Pharmacia LKB). After electroblotting, the RNAs were fixed to the membrane by UV cross-linking (1200 µJ, Stratalinker; Stratagene) and by baking in a vacuum oven at 80°C for 1 h. DNA oligonucleotides complementary to miRNA sequences were synthesized (Sangon, Shanghai). The 5′ ends of the DNA probes were labeled with [γ-32P]ATP (Yahui Co.) using T4 polynucleotide kinase (Promega) and submitted to purification according to standard laboratory protocols as described previously (31). The membrane was pre-hybridized in 7% SDS, 0.3 M NaCl, and 50 mM phosphate buffer (pH 7.2) at 42°C for at least 1 h. Membranes were hybridized with 32P-end-labeled oligonucleotide probes at a temperature of 10–15°C below the calculated dissociation temperature (Td) for 16 h. The blots were washed twice with 2× SSPE/0.5% SDS and once with 0.5× SSPE/0.1% SDS at 40°C. The northern blots were quantified by using a phosphorimager apparatus (Typhon 8600, Amersham Bioscience).
Computational analyses
RNA sequences were subjected to BLAST analyses against the rice genome (http://www.ncbi.nlm.nih.gov/blast, http://btn.genomics.org.cn/rice). Secondary structures of RNA precursor were predicted by using longer genomic sequences of cloned RNAs and the m-fold program [(32) http://www.bioinfo.rpi.edu/applications/mfold/old/rna/form1.cgi]. Potential targets for rice miRNAs are identified by BLAST analyses according to the search algorithm in which only three or fewer mismatches are allowed to be present in the complementarity between miRNAs and their targets, and gaps are not allowed (21).
RESULTS
Identification of 20 miRNAs from a rice cDNA library
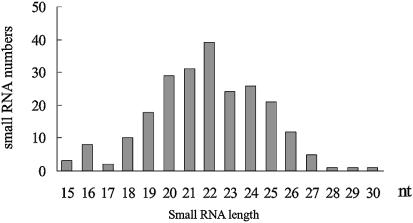
A cDNA library was generated from size-fractional RNAs between ∼16 and 28 nt. A total of 236 insert-containing clones randomly selected from the library were analyzed, and 203 unique sequences were obtained. Although the sequences ranged in length from 15 to 30 nt, they had a much tighter length distribution, centering on 20–25 nt (Fig. 1), which was coincident with the known specificity of Dicer processing. This profile was very similar to that of miRNA populations isolated from A.thaliana and C.elegans, and siRNA generated in a green fluorescent protein (GFP) and double-stranded GFP silencing system (12,25). Obviously, most of the cloned sequences had a composition preference that began with a uridine at the 5′ terminus, as had been observed in miRNAs from different organisms (10,11,14).
Figure 1.
Size and distribution of small RNAs isolated from the rice cDNA library.
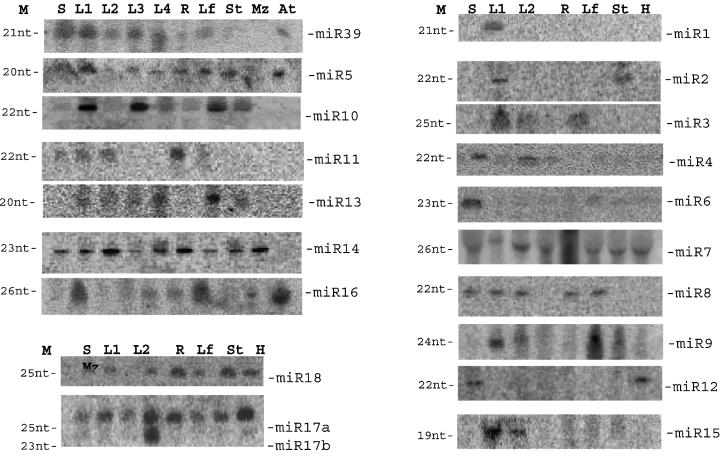
The 203 cloned RNA sequences were subjected to BLAST analysis against the rice genome. Only two sequences originated from fragments of tRNA or rRNA. A total of 122 sequences had at least one nucleotide mismatch with the rice genome and they were not further analyzed. The remaining 79 small RNA sequences matched perfectly with the rice genome, and their possible genomic locations were identified. In the analysis of secondary structures, 32 genomic sequences that contain putative miRNAs were capable of forming stem–loop structures characteristic of miRNA precursors. Subsequently, 20 of the 32 putative miRNAs were further confirmed by northern blot (Fig. 2). Thus the 20 miRNAs were identified from rice, and their sequences and precursor structures are shown in Table 1 and Figure 3, respectively.
Figure 2.
Detection of the miRNAs by northern blot with oligonucleotide probes complementary to miRNA sequences. M, mark; S, sprout; L1–L4, 1–4 leaf stage of rice; H, heading stage of rice; R, root (heading stage); Lf, leaf (heading stage); St, stem (heading stage); At, Arabidopsis; Mz: maize.
Table 1. MicroRNAs identified from rice.
| miRNA genes | miRNA sequence | Length (nt) | Folback arm/ nt | Chromo some | Distance to nearest gene | Orientation |
|---|---|---|---|---|---|---|
| MIR 39 | UGAUUGAGCCGUGCCAAUAUC | 21 | 3′/117 | 10 | 4334 bp from start of OSJNBb0015I11.25 | Antisense |
| 3′/100 | 3 | 3432 bp from start of OJ1006F06.3 | Antisense | |||
| MIR 1 | UGAAGAGUUUACCUUUUUACU | 21 | 5′/102 | 1 | 0.274 kb from end of P0004A09.27 | Sense |
| MIR 2 | GAAGGUUUGGUGUGCUUUUGUC | 22 | 3′/82 | Contig 649 from 23 042 to 23 021 | ||
| MIR 3 | UUAUAUAAGAACAACAUUUUGUGUC | 25 | 5′/119 | 0.259 kb from end of OsNAS1 gene | Sense | |
| MIR 4 | UAAGCUAUUAAAUUGGCUAUAG | 22 | 5′/362 | 1 | 0.526 kb from start of unnamed protein gene | Antisense |
| 5′/365 | 3 | 0.283 kb from end of OSJNBa0032G08.11 | Sense | |||
| 5′/362 | 4 | 9.988 kb from start of P0076O17.11 | Sense | |||
| MIR 5 | UUCAUUAACAAGUUACACCA | 20 | 5′/160 | 7 | 0.114 kb from end of OJ1753_E03.12 | Antisense |
| MIR 6 | UCUAUGGAAAAUGGGUCAUUGCC | 23 | 3′/68 | 5 | Clone OJ1576_F01; 63 609–63 587 | |
| MIR 7 | GUGUGGUGUGGCGGCAUGGGGAUGUC | 26 | 3′/62 | 1 | 0.85 kp from start of P0557A01.12 | Antisense |
| 3′/64 | 4 | 5.799 kp from start of OSJNBa0042F21.24 | Sense | |||
| 3′/62 | 7 | 3.745 kp from start of OSJNBa0067K15.15 | Antisense | |||
| 3′/64 | 10 | 69 bp from start of OSJNBb0005J14.tRNA-Ile-1 | Sense | |||
| MIR 8 | UCUGUACAGUUUACAAUUCCCC | 22 | 3′/180 | 1 | 0.186 kb from end of P0434B04.13 | Antisense |
| MIR 9 | UGAGUAAAGAAUUAUACUUCCUAC | 24 | 5′/170 | 1 | 94 bp from end of the predicted gene, AU057948 | Sense |
| MIR 10 | UAUCUGAAGUUCUUGCAAGAAC | 22 | 3′/60 | 2 | In BAC clone:OSJNBa0016D04; 31 528–31 507 | |
| MIR 11 | UAUAGCCCUUGGUAGAGUUACU | 22 | 5′/150 | 1 | In CR of P0557A01.24; 106 115–106 136 | Sense |
| 5′/150 | 3 | In clone:OSJNBa0048D11; 54 622–601 | Sense | |||
| 5′/154 | 4 | 2.145 kb from end of OSJNBb0022F23.11 | Sense | |||
| 5′/156 | 10 | 983 bp from start of OSJNBa0057L21.12, | Sense | |||
| MIR 12 | UGAAUAAACGGCAGUUCCGUUG | 22 | 3′/74 | 12 | In BAC OSJNBa0035N13; 26 598–26 577 | |
| MIR 13 | AACUCUUUGAUGAAAAGAUG | 20 | 5′/114 | 1 | 2.231 kb from end of P0557A01.32, | Antisense |
| 5′/114 | 3 | In BAC OSJNBb0106M04; 80 529–80 510; | ||||
| 5′/114 | 10 | 0.462kb from start of OSJNBa0022D10.3 | Sense | |||
| 5′/114 | 12 | In BAC OSJNBa0021D06; 20 182–201 | ||||
| MIR 14 | UGCCACGUUAGCAAAUAUUUUGC | 23 | 5′/164 | Contig 10140; 7163–7141 | ||
| MIR 15 | AUUUGAUUCGUCGAUCUUC | 19 | 3′/63 | 2 | BAC clone:P0476H10; 99 788–99 770 | |
| 3′/63 | 4 | 808 bp from end of OSJNBa0061C08.5 | Antisense | |||
| 3′/63 | 10 | In CR of OSJNBb0005J14.18, rps14 | Sense | |||
| MIR 16 | AAAAUGUAGUACAUACAUGUUUACUG | 26 | 5′/71 | 3 | 205 bp from end of OSJNBa0059G06.17 | Sense |
| MIR 17a | AAUGCCGAAUCGACGACCUAUGUAU | 25 | 5′/124 | 4 | 12.566 kb from end of OSJNBa0042F21.25 | Sense |
| 5′/124 | 10 | In intron of OSJNBa0034L04.43 | Sense | |||
| MIR 17b | AAUGCCGAAUCGACGACCUAUGU | 23 | 5′/124 | 4 | 12.566 kb from end of OSJNBa0042F21.25 | Sense |
| 5′/124 | 10 | In intron of OSJNBa0034L04.43 | Sense | |||
| MIR 18 | UUAAUCCAUUUAAAUCAAUUAAAUC | 25 | 3′/87 | 1 | 616 bp from end of P0432B10.17 | Sense |
| 3′/87 | 1 | In clone:P0011D01, in 3′LTR; 119 418–119 442; | ||||
| 3′/87 | 3 | 0.232 kb from start of OSJNBb0006O08.5 | Antisense | |||
| 3′/87 | 4 | 562 bp from end of OSJNBb0115I09.11 | Sense | |||
| 3′/76 | 7 | 219 bp from start of OJ1060_D03.123 | Sense | |||
| 3′/87 | 8 | In BAC clone:B1142B04; 65 524–65 570 | ||||
| 3′/87 | 8 | In BAC clone:OJ1449; 45 090–45 114 | ||||
| 3′/76 | 10 | In intron of OSJNBa0017E08.18 | Sense | |||
| 3′/76 | 10 | 655 bp from end of OSJNBb0015K05.1 | Sense | |||
| 3′/76 | 10 | 563 bp from end of OSJNBa0051D19.15 | Sense | |||
| 3′/87 | 12 | In BAC OSJNBa0096K13; 107 920–107 956; | ||||
| 3′/87 | In retrotransposon Osr1 DNA; 5447–5471 | Sense |
Genomic locations of microRNAs are presented as chromosome number and distance to nearest gene. The multiple copies of microRNAs whose precursors can form stem–loop structures are also included in the table.
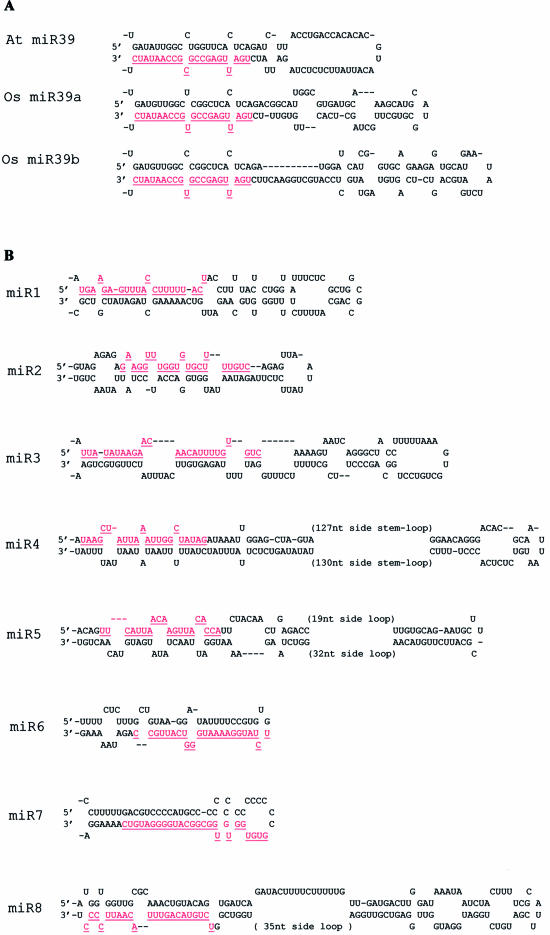
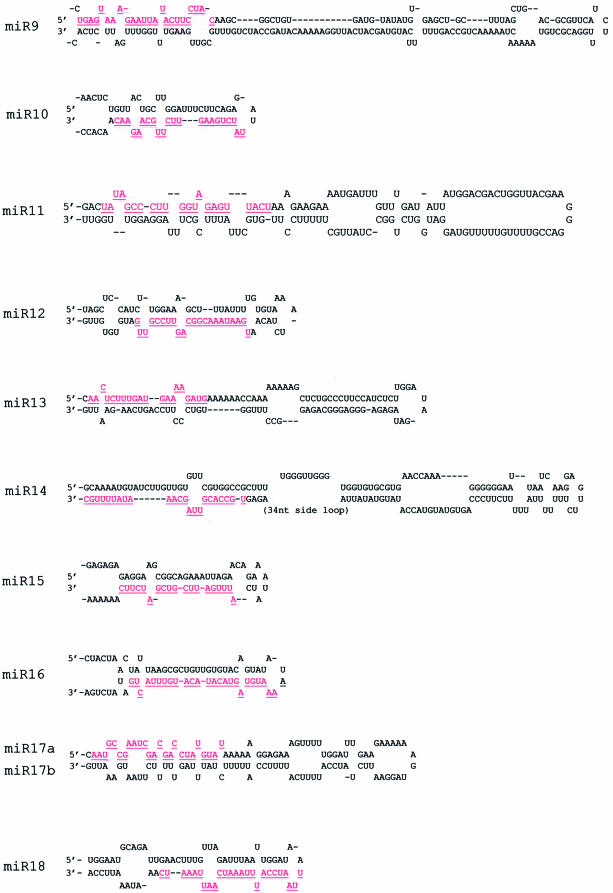
Figure 3.
Stem–loop structures of putative miRNA precursors. The miRNA sequences are in red and underlined. The actual size of the precursors is not identified experimentally and may be slightly shorter or longer than represented. Multicopy miRNAs and their corresponding precursor structures are not shown. (A) Conserved precursor structures of miR-39 between rice and Arabidopsis. (B) Precursor structures predicted for rice miR-1 to miR-18.
Positive detection and tissue-specific expression of the rice miRNAs
The expression of the rice miRNAs was examined by northern blot with low molecular weight RNAs isolated from different tissues of rice. Northern blot confirmed that 20 miRNAs were stably expressed in rice cells from at least one tissue (Fig. 2). The accumulations of some miRNAs appear to be developmental or tissue specific. For example, miR-1, miR-6 and miR-10 were somewhat more strongly expressed as seedlings compared with the adult plant, or vice versa. miR-11 and miR-16 accumulate at a higher level in roots and leaves, respectively. A probe to miR-17 detected both miR-17a and miR-17b with a size of 25 and 23 nt, respectively. Interestingly, miR-17a accumulated in different tissues and developmental stages, but miR-17b with 23 nt is only most highly expressed in the rice roots. Other miRNAs were not uniformly expressed through all tissues, and large variations in the expression levels were observed. In some cases, miRNA precursors were detected as described in C.elegans (12).
Probes complementary to rice miRNAs were used to test total cellular RNAs from other plants in the same assays. The homologs of miR-39, miR-5 and miR-16 were found to be present in A.thaliana, and miR-7, miR-12, miR-14, miR-16, miR-17 and miR-18 were detected in maize (Fig. 2), suggesting the conservation of these miRNAs in flowering plants.
Prediction of the rice miRNA targets
To identify potential targets, we searched for rice mRNAs with complementarity to the newly identified miRNAs by an algorithm employed upon the discovery of Arabidopsis miRNA targets, in which most antisense hits with three or fewer mismatches appeared to be potential miRNA targets (21). Twenty-three unique rice genes were identified to be feasible targets for seven rice miRNAs, because there are high stringent complementarities with three or fewer mismatches between the miRNAs and their targets (Table 2).
Table 2. Prediction of rice miRNA targets.
| MicroRNA | Target protein family | Target gene names (no. of mismatches) |
|---|---|---|
| miR-39 | GRAS domain proteins (SCARECROW-like) | Os.2406 (1), Os.20826 (1), Os.21578 (1), Os.31716 (1), Os.46878 (2) |
| miR-1 | Unknown protein | Os.19366 (3) |
| miR-4 | Similarity to Arabidopsis putative MAP3K α protein kinase | Os.27038 (3) |
| Similarity to Arabidopsis mucin-like protein | Os.23008 (3) | |
| MRP-like ABC transporter | AJ535074 (3) | |
| Unknown or hypothetical proteins | Os.1798 (3), Os.35458 (3), Os.38137 (3), AK062268 (3), AK062967 (3), AK068031 (3), AK100013 (3), AK100599 (3) | |
| miR-5 | Similarity to Arabidopsis trehalose-6-phosphate phosphatase | Os.25244 (0) |
| miR-10 | Putative Cf2/Cf5 disease resistance protein | Os.24878 (3) |
| Unknown protein | Os.11923 (3) | |
| miR-13 | Similarity to Arabidopsis aluminum-induced-like protein | Os.9392 (3) |
| Putative protein | AK064680 (3) | |
| miR-15 | PDR-like ABC transporter | AJ535053 (0) |
Prediction was performed as described by Rhoades et al. (21). Rice genes are labeled by their UniGene numbers or accession numbers in GenBank. For each gene, the number of mismatches between the miRNA and the mRNA is tallied in parentheses.
Among the seven miRNAs, three miRNAs are perfect antisenses matched to the mRNAs of protein-coding genes. miR-39 targets the coding region of mRNAs from four members of Scarecrow-like transcription factor that controls a wide range of developmental processes. This miRNA appears homologous to Arabidopsis miR-39 (or miR-171) that had been predicted to be present in rice by RNA hybridization (14,19). In fact, miR-39 varies slightly in sequence (one nucleotide change) between the two plants, but a U/G pair still maintains a perfect duplex of miR-39–target mRNA in rice. Interestingly, two rice miR-39 precursors display secondary structures very similar to that of Arabidopsis despite considerably divergent sequences outside the miRNAs (Fig. 3A), lending support to the evolutionary conservation of this miRNA in flowering plants. A novel target predicted for miR-5 is the mRNA of a protein-coding gene whose function remains annotated. Being different from the two precedents, miR-15 has a perfect antisense match to the fourth intron of PDR-like ABC transporter, implying a possible role for miRNA in the stage of RNA splicing. Lacking longer complementarity, the targets of 13 miRNAs remain to be further investigated.
Genomic organization of the miRNAs
Among the 20 miRNAs, 11 were each encoded by a single copy in the rice genome, whereas another 10 miRNAs had multiple loci in the genome (Table 1), probably because of duplications that were still active in the rice genome (26). Comparative analyses showed that the multiple genomic sequences containing the miRNAs are often completely or partially conserved in the rice genome. For example, miR-18 corresponds to 12 genomic sequences in long terminus repeat sequences or in the Osr1 retrotransposon. Twenty-eight genomic loci representing 15 rice miRNAs were mapped to intergenic regions. Three copies of miRNAs were found in the sense orientation within introns of coding transcripts. Two copies of miRNAs were located in the exons of potential protein-coding genes. Another 13 copies from nine miRNAs were found in genomic regions that have no annotation in the genome. Therefore, miRNAs are either transcribed from their own promoters or derived from a pre-mRNA that frequently codes for an additional gene product.
DISCUSSION
miRNAs are widespread in eukaryotes and usually isolated from cDNA libraries of size-fractionated RNAs. To identify the miRNAs from rice, we used an improved cDNA cloning procedure derived from experimental RNomics (33), which has been successfully applied to plant snmRNA isolation (29,34). Through screening of the cDNA library of rice, numerous small endogenous RNAs were isolated. The profile of size distribution of small RNAs is very similar to that of the miRNA population from Arabidopsis and animals (12,25), proving the effectiveness in cloning of miRNAs from rice by this method as an alternative to the RNA ligation strategy (35). Meanwhile, a non-redundant set of more than 200 sequences from the cDNA library showed the high diversity of small RNAs in rice. In this study, we obtained many small RNAs that had at least one mismatch to the rice genome, indicating the incomplete database of the rice genome, probably in some highly repeated intergenic regions. It is believed that lots of small RNA genes may be hidden in these regions, as has been shown in Arabidopsis and Drosophila (25,36).
Although numerous small RNAs have been isolated from Arabidopsis, and from rice in this study, not all small endogenous RNAs are miRNAs. Conversely, only a small fraction of the plant small RNA population previously described meets the strict definition of miRNAs (1). An important criterion required for novel miRNAs is the secondary structure of precursors, which are stem–loop structures recognized and processed subsequently by the endonuclease Dicer. In animals, the precursors are usually ∼60–80 nt, but the sizes of plant precursors appear more variable (14,24), which may make plant miRNA identification more complicated than that of an animal. We noted that most of the rice miRNA precursors were 60–200 nt and the miRNAs could be processed from either the 5′ or 3′ arm of the foldback precursor. However, each miRNA with multiple matches to the genome is always present on the same arm of its potential precursors, suggesting that these loci share a common origin through local or distant duplications in the rice genome.
miRNAs are usually involved in developmental controls of animals and plants, but the mechanism(s) of miRNAs in regulating gene expressions appears different as if there were two functional classes of miRNA in different organisms (2). In animals, miRNAs, such as lin-4 and mir-23, partially complementary to the 3′-UTR or part of the coding region of target mRNAs, act as repressors of translation (37), but do not reduce the abundance of target mRNAs. However, in plants, some miRNAs with full or near complementarity to the coding region of target mRNAs, such as miR-39 (miR-171) (19,21) and miRNA-JAW (miR-159) (38), direct the cleavage of target mRNAs at precise positions, showing a degraded mRNA pathway that resembles RNAi guided by siRNAs. Recently, Zeng et al. (20) demonstrated that miRNAs and siRNAs may be functionally interchangeable in human, and the alternative mechanisms may depend largely or entirely on the degree of complementarity between the small guide RNAs and the mRNA target. The principle revealed from animals would be applicable to plants. It is possible that plant miRNAs with partial complementarities to target mRNAs are able to inhibit the mRNA expression by the same mechanism of translational repressor as those of animals. This speculation will be useful to the functional research of plant miRNAs because there are growing numbers of newly identified miRNAs that lack perfect complementarity to target mRNAs, as has been shown previously (24) and in this study. Interestingly, evidence that an miRNA, miR-172, in Arabidopsis downregulates APETALA2-like target genes by a translational mechanism rather than by RNA cleavage supports this hypothesis (39). Nevertheless, in a manner similar to translational repression, the minimum number of base pairs needed for a functional duplex of miRNA–target mRNA remains to be defined. The phenomenon of RNA-mediated post-transcriptional gene silencing was first discovered in plants (40,41); how many miRNAs exist and their functions in plants remain to be deciphered.
Acknowledgments
ACKNOWLEDGEMENTS
We gratefully acknowledge the technical assistance of Xiao-Hong Chen and Dai-Ying Xu. Special thanks to Robin Pachman for revising the manuscript. This research was supported by the National Natural Science Foundation of China (key project 30230200 and 30170216) and by the Fund for Distinguished Young Scholars from the Ministry of Education of China.
REFERENCES
- 1.Ambros V., Bartel,B., Bartel,D.P., Burge,C.B., Carrington,J.C., Chen,X., Dreyfuss,G., Eddy,S.R., Griffiths-Jones,S., Marshall,M., Matzke,M., Ruvkun,G. and Tuschl,T. (2003) A uniform system for microRNA annotation. RNA, 9, 277–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carrington J.C. and Ambros,V. (2003) Role of microRNAs in plant and animal development. Science, 301, 336–338. [DOI] [PubMed] [Google Scholar]
- 3.Benfey P.N. (2003) Molecular biology: microRNA is here to stay. Nature, 425, 244–245. [DOI] [PubMed] [Google Scholar]
- 4.Grishok A., Pasquinelli,A.E., Conte,D., Li,N., Parrish,S., Ha,I., Baillie,D.L., Fire,A., Ruvkun,G. and Mello,C.C. (2001) Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C.elegans developmental timing. Cell, 106, 23–34. [DOI] [PubMed] [Google Scholar]
- 5.Lai E.C. (2002) Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nature Genet., 30, 363–364. [DOI] [PubMed] [Google Scholar]
- 6.Bartel B. and Bartel,D.P. (2003) MicroRNAs: at the root of plant development. Plant Physiol., 132, 709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee R.C., Feinbaum,R.L. and Ambros,V. (1993) The C.elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell, 75, 843–854. [DOI] [PubMed] [Google Scholar]
- 8.Reinhart B.J., Slack,F.J., Basson,M., Pasquinelli,A.E., Bettinger,J.C., Rougvie,A.E., Horvitz,H.R. and Ruvkun,G. (2000) The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature, 403, 901–906. [DOI] [PubMed] [Google Scholar]
- 9.Lagos-Quintana M., Rauhut,R., Lendeckel,W. and Tuscl,T. (2001) Identification of novel genes coding for small expressed RNAs. Science, 294, 853–858. [DOI] [PubMed] [Google Scholar]
- 10.Lee R.C. and Ambros,V. (2001) An extensive class of small RNAs in Caenorhabditis elegans. Science, 294, 863–864. [DOI] [PubMed] [Google Scholar]
- 11.Lagos-Quintana M., Rauhut,R., Yalcin,A., Meyer,J., Lendeckel,W. and Tuschl,T. (2002) Identification of tissue-specific microRNAs from mouse. Curr. Biol., 12, 735–739. [DOI] [PubMed] [Google Scholar]
- 12.Lau N.C., Lim,L.P., Weinstein,E.G. and Bartel,D.P. (2001) An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science, 294, 858–862. [DOI] [PubMed] [Google Scholar]
- 13.Mourelatos Z., Dostie,J., Paushkin,S., Sharma,A., Charroux,B., Abel,L., Rappsilber,J., Mann,M. and Dreyfuss,G. (2002) miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev., 16, 720–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reinhart B.J., Weinstein,E.G., Rhoades,M.W., Bartel,B. and Bartel,D.P. (2002) MicroRNAs in plants. Genes Dev., 16, 1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Provost P., Dishart,D., Doucet,J., Frendewey,D., Samuelsson,B. and Radmark,O. (2002) Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J., 21, 5864–5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie Z., Kasschau,K.D. and Carrington,J.C. (2003) Negative feedback regulation of dicer-like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr. Biol., 13, 784–789. [DOI] [PubMed] [Google Scholar]
- 17.Hutvagner G. and Zamo,P.D. (2002) A microRNA in a multiple-turnover RNAi enzyme complex. Science, 297, 2056–2060. [DOI] [PubMed] [Google Scholar]
- 18.Cullen B.R. (2002) RNA interference: antiviral defense and genetic tool. Nature Immunol., 3, 597–599. [DOI] [PubMed] [Google Scholar]
- 19.Llave C., Xie,Z., Kasschau,K.D. and Carrington,J.C. (2002) Cleavage of scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science, 297, 2053–2056. [DOI] [PubMed] [Google Scholar]
- 20.Zeng Y., Yi,R. and Cullen,B.R. (2003) MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc. Natl Acad. Sci. USA, 100, 9779–9784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhoades M.W., Reinhart,B.J., Lim,L.P., Burge,C.B., Bartel,B. and Bartel,D.P. (2002) Prediction of plant microRNA targets. Cell, 110, 513–520. [DOI] [PubMed] [Google Scholar]
- 22.Lim L.P., Glasner,M.E., Yekta,S., Burge,C.B. and Bartel,D.P. (2003) Vertebrate microRNA genes. Science, 299, 1540. [DOI] [PubMed] [Google Scholar]
- 23.Grad Y., Aach,J., Hayes,G.D., Reinhart,B.J., Church,G.M., Ruvkun,G. and Kim,J. (2003) Computational and experimental identification of C.elegans microRNAs. Mol. Cell, 11, 1253–1263. [DOI] [PubMed] [Google Scholar]
- 24.Park W., Li,J., Song,R., Messing,J. and Chen,X. (2002) CARPEL FACTORY, a Dicer homolog and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol., 12, 1484–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Llave C., Kasschau,K.D., Rector,M.A. and Carrington,J. (2002) Endogenous and silencing-associated small RNAs in plants. Plant Cell, 14, 1605–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu J., Hu,S., Wang J., Wong,G.K., Li,S., Liu,B., Deng,Y., Dai,L., Zhou,Y., Zhang,X. et al. (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science, 296, 79–92. [DOI] [PubMed] [Google Scholar]
- 27.The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature, 408, 796–815. [DOI] [PubMed] [Google Scholar]
- 28.Chomczynski P. and Sacchi,N. (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal. Biochem., 162, 732–735. [DOI] [PubMed] [Google Scholar]
- 29.Chen C.L., Liang,D., Zhou,H., Zhuo,M., Chen,Y.Q. and Qu,L.H. (2003) The high diversity of snoRNAs in plants: identifiation and comparative study of 120 snoRNA genes from Oryza sativa. Nucleic Acids Res., 31, 2601–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou H., Chen,Y.-Q., Du,Y.-P. and Qu,L.-H. (2002) The Schizosaccharomyces pombe mgU6-47 snoRNA is required for the methylation of U6 snRNA at 41. Nucleic Acids Res., 30, 894–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 32.Zuker M. (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res., 31, 3406–3415,. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huttenhofer A., Kiefmann,M., Meier-Ewert,S., O’Brien,J., Lehrach,H., Bachellerie,J.P. and Brosius,J. (2001) RNomics: an experimental approach that identifies 201 candidates for novel, small, non-messenger RNAs in mouse. EMBO J., 20, 2943–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marker C., Zemann,A., Terhorst,T., Kiefmann,M., Kastenmayer,J.P., Green,P., Bachellerie,J.P., Brosius,J. and Huttenhofer,A. (2002) Experimental RNomics: identification of 140 candidates for small non-messenger RNAs in the plant Arabidopsis thaliana. Curr. Biol., 12, 2002–2013. [DOI] [PubMed] [Google Scholar]
- 35.Elbashir S.M., Lendeckel,W. and Tuschl,T. (2001) RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev., 15, 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aravin A.A., Lagos-Quintana,M., Yalcin,A., Zavolan,M., Marks,D., Snyder,B., Gaasterland,T., Meyer,J. and Tuschl,T. (2003) The small RNA profile during Drosophila melanogaster development. Dev. Cell, 5, 337–350. [DOI] [PubMed] [Google Scholar]
- 37.Kawasaki H. and Taira,K. (2003) Hes1 is a target of microRNA-23 during retinoic-acid-induced neuronal differentiation of NT2. Nature, 423, 838–842. [DOI] [PubMed] [Google Scholar]
- 38.Palatnik J.F., Allen,E., Wu,X., Schommer,C., Schwab,R., Carrington,J.C. and Weigel,D. (2003) Control of leaf morphogenesis by microRNAs. Nature, 425, 257–263. [DOI] [PubMed] [Google Scholar]
- 39.Aukerman M.J. and Sakai,H. (2003) Regulation of flowering and floral organ identity by a microRNA and its APETALA-like target genes. Plant Cell, 15, 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Napoli C., Lemieux,C. and Jorgensen,R. (1990) Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell, 2, 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jorgensen R.A., Cluster,P.D., English,J., Que,Q. and Napoli,C.A. (1996) Chalcone synthase cosuppression phenotypes in petunia flowers: comparison of sense vs. antisense constructs and single-copy vs. complex T-DNA sequences. Plant Mol. Biol., 31, 957–973. [DOI] [PubMed] [Google Scholar]