Abstract
Little consideration has been given to the effect of different segmentation methods on the variability of data derived from microarray images. Previous work has suggested that the significant source of variability from microarray image analysis is from estimation of local background. In this study, we used Analysis of Variance (ANOVA) models to investigate the effect of methods of segmentation on the precision of measurements obtained from replicate microarray experiments. We used four different methods of spot segmentation (adaptive, fixed circle, histogram and GenePix) to analyse a total number of 156 172 spots from 12 microarray experiments. Using a two-way ANOVA model and the coefficient of repeatability, we show that the method of segmentation significantly affects the precision of the microarray data. The histogram method gave the lowest variability across replicate spots compared to other methods, and had the lowest pixel-to-pixel variability within spots. This effect on precision was independent of background subtraction. We show that these findings have direct, practical implications as the variability in precision between the four methods resulted in different numbers of genes being identified as differentially expressed. Segmentation method is an important source of variability in microarray data that directly affects precision and the identification of differentially expressed genes.
INTRODUCTION
Expression profiling using microarrays offers a powerful technology to gain novel insights into different biological phenotypes through studying genome-wide differences. However, the technique suffers from an inherent lack of precision owing to the multiple sources of variability in processing a microarray experiment (1,2). The extent of this variability can preclude the production of interpretable results. It is therefore very important to understand and optimize the variables that may introduce noise into the data analysis.
Variability in microarray experiments can arise from pre-scanning and post-scanning steps. Pre-scanning steps include methods of RNA extraction (3,4), different types of probe preparation (3,5), probe labelling (6), hybridization and slide quality (7,8). The second category includes image acquisition and image/data analysis. Interestingly, relatively little attention has been given to the variability introduced by image analysis methods as a potential source of noise. A previous report suggested that variability introduced by image analysis is predominantly determined by the method of estimating signal background from a spot, and not the method of segmentation (which identifies the individual pixels that make up a feature) (9). However, a pixel represents the basic unit from which intensity values are derived. Brown et al. (10) have shown that small- and large-scale fluctuations in pixel intensities within a spot lead to uncertainty in microarray quantitation, and that pixel-to-pixel variability correlates with variability between replicate spots on duplicate slides. It therefore follows that methods of summarizing individual pixel data by segmentation could have major effects on the precision of the data.
The algorithms used by different segmentation methods have been previously described (9). Although the overall aim of each of the methods is to summarize data obtained from individual pixel intensities, there are striking differences in how this is achieved. For example, the histogram method samples only 15% of the pixel centiles for foreground and background estimation. In contrast, the adaptive method summarizes all the available pixels regardless of their centile values. The implications of these variations on repeatability have not been formally investigated. Most investigators are unaware of these issues and simply use the commercial software provided with their microarray scanner. Moreover, further variability may be introduced as software packages can offer a choice of segmentation algorithms or allow the centile range for sampling to be determined by the user.
In this report, we show that the choice of segmentation method significantly influences the precision of the data, and this effect is independent of background subtraction but dependent upon the fluor intensity of the hybridized probe. In order to analyse differences between methods, independently of other sources of possible noise, we initially performed significance analysis on the correlation data from adaptive, fixed circle and histogram segmentation using identical grid placement from a single analysis package (QuantArray). Having identified significant differences between the methods we then used a more generalizable method of comparison, the coefficient of repeatability, to confirm these findings and included an additional method (GenePix) in the analysis (11).
MATERIALS AND METHODS
Microarrays
Expression microarrays containing 6528 pairs of duplicate cDNA spots were used (Cancer Research UK DNA Microarray Facility at the Institute of Cancer Research; CR-UK DMF Human 6.5k genome-wide array). All microarrays used were from the same printing batch. Total RNA was obtained from the cell line HCT116 and an isogenic daughter line with a targeted disruption of the EP300 gene derived by homologous recombination (N.G.Iyer, S.-F.Chin, H.Ozdag, Y.Daigo, D.-E.Hu, M.Cariati, K.Brindle, S.Aparicio and C.Caldas, submitted). Total RNA was used for reverse transcription and indirect labelling with Cy3 and Cy5 dyes (Amersham) using random hexamers as previously described (12). Measurements of the amount of purified cDNA and Cy3/Cy5 incorporation were made before hybridization using the Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies, Inc.). For all hybridizations, the fluor incorporation was highly correlated to the mass of cDNA (data not shown). Two sets of experiments (A and B) were carried out, using six slides in each with a balanced dye-swap design (three slides for each dye). Experiments A and B were identical but used 10 and 15 µg total RNA for labelling for each hybridization. Scanning was performed using the ScanArray 4000 (Perkin Elmer) at a resolution of 10 µm at maximum laser power and photomultiplier tube voltage of 50–60%. Segmentation was performed using QuantArray (Perkin Elmer) and GenePix Pro 4.1 (Axon Instruments, Inc.) software. All three methods of segmentation available within the QuantArray package were evaluated. The default settings for centile sampling were used for all the analyses. The histogram, fixed circle and adaptive methods sampled the foreground intensity from centiles 80–95, 45–95 and 1–99, respectively. The background was estimated by measuring centiles 5–20, 5–55 and 1–99, respectively. The GenePix method used all centiles within the defined foreground and background areas. All raw image and derived data files are available at the GEO repository (http://www.ncbi.nlm.nih.gov/geo/; accession numbers GSM16895–42; series entity GSE1054).
Statistical methods
All statistical analysis was conducted using the R environment (13) and the R package ‘Statistics for Microarray Analysis’ (14). Log intensity ratios for each spot were obtained with and without background subtraction. All spots from each microarray were included in the analysis. Data normalization was performed using scaled loess normalization and differential genes were identified using an empirical Bayes method for analysing replicated microarray data (15). Data precision was initially assessed by using correlation coefficients. Correlation data were divided into three groups according to the segmentation method used (adaptive, fixed circle or histogram). Analysis of variance (ANOVA) was used for testing the correlation differences between the groups. Homogeneity of the correlation values within the different groups was tested using the Levene test of Homogeneity and between-group comparisons of correlation values were performed using the Tukey HSD test. Coefficient of repeatability was used as an alternative to testing correlation values (11). An annotated script for the entire analysis is available as Supplementary Material.
RESULTS
Segmentation method significantly influences within-slide correlations
To investigate whether the segmentation method was an important determinant of precision, we first determined the correlation between data obtained from within-slide replicates as these data are relatively independent of variations in slide printing or sample preparation. We used log ratios for expression differences as these are more robust than single channel data for normalization purposes and are more biologically meaningful (15). We calculated the Pearson’s correlation coefficient (r) for the M ratio values obtained from 6528 pairs of replicate spots from each of six hybridizations (experiment A). The microarray images were analysed using fixed circle, histogram and adaptive segmentation methods resulting in 18 correlation coefficients. To further minimize noise, we used an identical grid placement for each method as minor differences in grid registration can significantly affect correlations (data not shown). To avoid multiple pairwise testing of differences between the means of categories (A<–>F, A<–>H and F<–>H), a one-way ANOVA analysis was conducted using the segmentation method as the independent variable and the r values as the outcome.
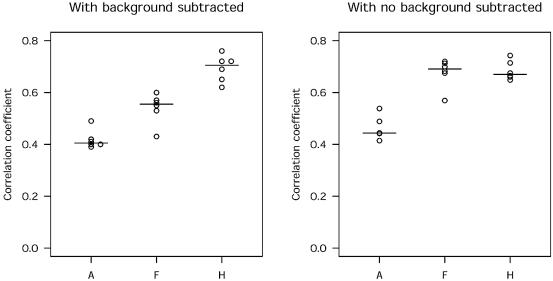
We found a significant difference (P < 0.001) between the three methods of segmentation at each level of comparison, with the histogram method giving the highest correlation and the adaptive method giving the lowest correlation (Fig. 1). As the ANOVA model assumes equal variance we confirmed this in our data by carrying out formal testing for homogeneity (Levine test; P = 0.6) and assessed the goodness of fit using the R-squared test (R2 = 0.84) as well as quantile–quantile plots (data not shown). To evaluate whether the observed differences were merely because of differences in estimating background values, the analysis was repeated without background subtraction. The overall difference between the methods remained (P < 0.001) although the difference between the histogram and fixed circle method was no longer significant (Fig. 1).
Figure 1.
Segmentation method significantly influences precision for within-slide correlations. The dot plots show correlations between 6528 replicate spots from six slides by segmentation method. The medians are indicated by horizontal lines. Left and right panels show the effect of background correction. The Tukey HSD test showed no difference between the fixed circle and the histogram method when background was not subtracted (A, adaptive; F, fixed circle and H, histogram method).
Histogram segmentation gives lower pixel-to-pixel variability
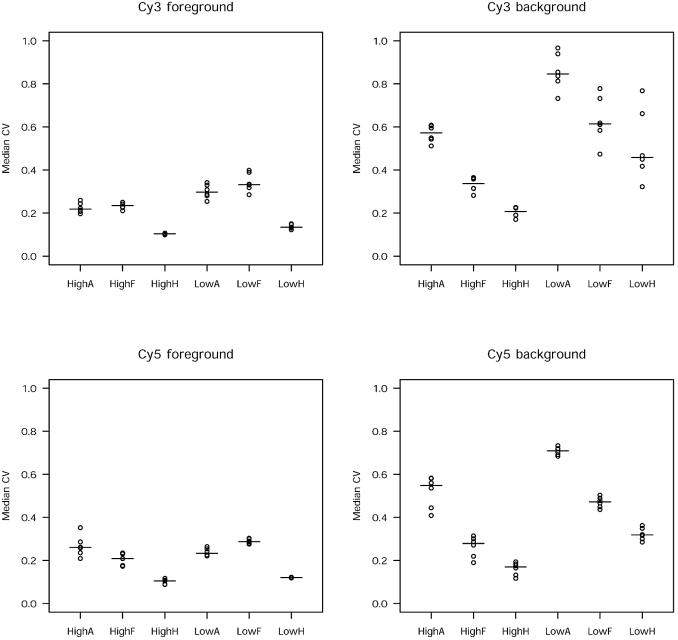
We hypothesized that the better precision for the histogram method was because of less variability in pixel intensity, as fluctuations in pixel values have been shown to increase noise (10). The histogram method summarizes centiles of pixel intensities obtained from a square centred around the true spot. From this, it follows that a narrow window of centiles will reduce the within-spot variability as compared to the other methods of segmentation used here. We therefore calculated the coefficient of variability (CV) for foreground and background pixels for each feature in experiments A and B in both Cy3 and Cy5 channels (Fig. 2). Experiments A and B differed only in the amount of labelled RNA sample hybridized to each array (see later). The histogram method had the lowest CV values in both foreground and background.
Figure 2.
The histogram method yields less pixel-to-pixel variability compared to other methods of segmentation. The dot plot shows the distribution of median within-spot CV values by segmentation method and low and high amounts of labelled probe. The medians are indicated by horizontal lines (A, adaptive; F, fixed circle; H, histogram; low, experiment A; high, experiment B).
Dye-swapping confounds the precision of between-slide comparisons
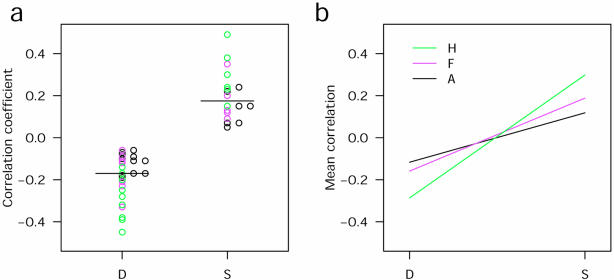
We next studied the effect of segmentation on between-slide variability by deriving a matrix of correlations for all possible pair-wise comparisons between the slides for each method (15 comparisons for each of three segmentation methods). A one-way ANOVA was conducted as above. In contrast to our results from within-slide comparisons, no significant differences in the correlations were found. The correlation coefficients between slides with dye-swapping were mostly negative, indicating low overall repeatability of the data (Fig. 3a). As dye-swapping would be expected to alter correlations between slides, we reanalysed the data by restricting the comparisons to those between replicates in which cDNA probes had been labelled with the same fluors. As expected, comparisons between replicates with the same dyes had higher correlations than between slides with swapped dyes (Fig. 3a). However, the beneficial effect of histogram segmentation on correlation was observed for slides with the same dye (Fig. 3b).
Figure 3.
Dye-swapping decreases reproducibility at low probe concentration. (a) Dot plots show correlations for different dye (D) or same dye (S) comparisons for six slides from experiment A (black circles, adaptive; magenta circles, fixed; green circles, histogram). (b) Interaction plot shows the effects of dye-swapping on the mean correlation coefficients between replicate slides for each segmentation method. Note that for slides with the same dye, the histogram method performed best and this was confounded by the dye swap (A, adaptive; F, fixed circle; H, histogram).
Precision is determined by choice of segmentation method and amount of labelled probe
The impact of the quantity of RNA used on the overall precision of microarray data has been previously reported (16). In experiment A, we labelled 10 µg of total RNA for each slide which yielded a median of 2.1 µg [interquartile range (IQR) 1.1–3.1] of cDNA probe after purification, and incorporated a median of 151 pmol (IQR 104–206) of each fluor. In order to identify whether the low between-slide correlations were caused by inadequate specific activity of our samples, we repeated the experiment using starting material of 15 µg of total RNA for each sample (experiment B; median labelled cDNA 5.5 µg, IQR 4.6–6.6, median incorporation of each fluor 463 pmol, IQR 384–534). To examine both the effect of the amount of labelled sample and the segmentation method, we performed two-way ANOVA analyses by combining the full data sets from experiments A and B (36 within-slide correlations and 90 between-slide correlations).
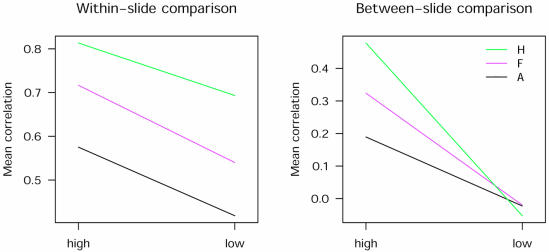
For within-slide correlations, the amount of labelled sample and method of segmentation independently and significantly (P < 0.001) influenced correlations (Fig. 4). There was no significant interaction between the two variables and this effect was still significant with or without background subtraction (P < 0.001) (data not shown).
Figure 4.
Histogram segmentation improves within- and between-slide correlations. Interaction plots show the effect of the amount of labelled probe on the mean correlations for all data from experiments A and B for each of the three segmentation methods. Left and right panels show within-slide (six data points per category) and between-slide correlations (15 data points per category). The amount of labelled probe has a significant effect (P < 0.001) as indicated by the downward slope of the lines from high to low amounts. In between-slide comparisons, the benefit of histogram segmentation is confounded by low amounts of labelled probe (A, adaptive; f, fixed circle; H, histogram).
For between-slide comparisons, using a larger amount of labelled sample significantly(P < 0.001) improved the correlations independently of the segmentation method used (Fig. 4). However, significant interaction was observed between the amount of labelled sample and the segmentation method. Therefore, while there was no advantage for any segmentation method when low amounts of labelled sample were used, there were marked differences for the methods when using higher amounts. These differences were independent of background subtraction (data not shown).
Coefficient of repeatability confirms higher precision
A low value for the correlation coefficient does not necessarily mean low repeatability as the correlation coefficient is not a measure of sameness (17). Previous reports have shown discrepancies between correlation coefficients and repeatability coefficients (11,17). In order to confirm our findings, we repeated the analysis using the coefficient of repeatability (CR) values to compare between the three different methods of segmentation and included a fourth proprietary method encoded within the GenePix software package. The box plots for the sigma factors obtained for each feature from the slides from experiment B (no background subtraction) showed that the histogram methods had the lowest median CR value (CR = 0.14) followed by the fixed circle method (CR = 0.15), the GenePix method (CR = 0.16), and the adaptive method (CR = 0.2) (Fig. 5).
Figure 5.
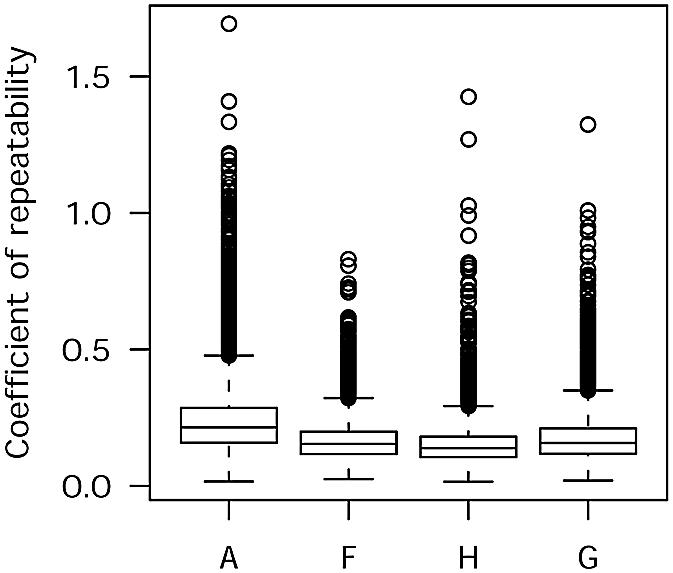
Histogram segmentation results in the lowest coefficient of repeatability (CR). Box plots show the distribution of the CR values of each of the 6528 spots in experiment B by four different methods of segmentation (A, adaptive; F, fixed circle; H, histogram; G, GenePix).
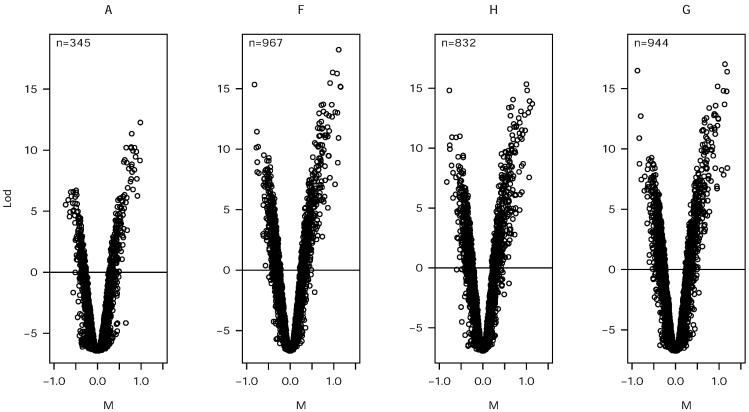
The probability of a gene to be differentially expressed is dependent on the variability of the data for that gene across the replicates of an experiment (1). It follows from our findings that the segmentation method could have a direct effect on the number of differentially expressed genes identified. In order to test this assumption, we used a Bayesian method to estimate the number of differentially expressed genes at a P value of 0.01 from the data set of experiment B (15). The number of genes identified varied considerably depending on the method of segmentation: 944, 967, 832 and 345 genes were identified for the GenePix, fixed circle, histogram and adaptive methods, respectively (Fig. 6).
Figure 6.
Volcano plots show likelihood of difference (LOD) for genes in experiment B by method of segmentation. Inset shows number of genes with LOD > 0 (M, log ratio; A, adaptive; F, fixed circle; H, histogram; G, GenePix).
DISCUSSION
A critical component of carrying out microarray experiments is the segmentation of images following scanning. We show here significant differences in precision and the number of differentially expressed genes using commonly used segmentation algorithms.
Differences between segmentation methods could result from differences in summarizing pixel-level data as pixel-to-pixel variability has been shown to have important effects on data quality (10). We were unable to obtain consistent findings to support this view. The histogram method yields the lowest within-spot variability as measured by the pixel CV values (Fig. 2) and performed best for within- and between-slide variability (Fig. 4). However, pixel CV values for the fixed circle and adaptive methods were very similar for foreground Cy3 and Cy5 measurements, but within- and between-slide correlations were very different. There were larger differences in pixel CVs for background which would be consistent with the finding that background estimation significantly affects precision (9). However, the loss of precision we observed for within-slide correlations was independent of background subtraction. These results could be explained by the sensitivity of CV calculation to outlying pixel values and in these cases using either the IQR or the repeatability coefficient could provide a more robust and therefore more reliable measure of variation. We were unable to conduct this form of analysis as none of the segmentation packages used report individual pixel data.
The introduction of dye-swapping has been proposed as a measure for accounting for preferential gene–dye interactions. The evidence to support this hypothesis is weak (18). Our findings suggest that dye-swapping introduces considerable variability in the experiment. The use of a common reference design could eliminate the need for dye-swapping (18). Indirect comparison to a common reference does introduce more variance as compared to a direct comparison between two samples, and therefore increases the number of replicates required in an experiment (19). In a two-sample comparison such as the one conducted in this paper, we feel that the decision to use dye-swapping or a common reference design should be based on the comparison of the noise introduced by each method in pilot experiments. Our results, however, demonstrate that at low probe concentration, dye-swapping yields uninterpretable data and mostly negative correlations. This has detrimental effects on gene discovery microarray experiments.
In this report, we used both the correlation coefficient and the coefficient of repeatability as measures of data quality. However, having a higher precision should not automatically be interpreted as better data. The investigator needs to understand the principles of segmentation analysis and to use the most appropriate methods for the experimental conditions. For example, the histogram method may give the best overall precision for slides that have low background, but it performs badly when local background is higher than the foreground spot intensity. These spots (‘ghost spots’) will be erroneously reported with inverted foreground and background values. The use of plots and multiple comparisons across the raw data set is of utmost importance in deciding on the appropriate segmentation method. This should then lead to careful consideration of whether the segmentation method should be included as a variable in the statistical model for identifying significantly differentially expressed genes. Such measures are easily automatable within a powerful statistical environment as provided by the R language.
These findings raise questions about how microarray data should be reported and compared, as results could be significantly different when obtained by different segmentation methods. The MIAME standard defines information about the type of scanning software used in microarray experiments and the Microarray and Gene Expression Object Model (MAGE-OM) includes the software version number (20). We believe that it is absolutely essential to also report the exact method used in segmentation. Ideally, raw image data should always be publicly available to allow the most robust comparison between different experiments.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
A.A.A. is a Medical Research Council Clinical Research Fellow and a Sackler Fellow. J.D.B. is a Cancer Research UK Senior Clinical Research Fellow. A.A.A. conceived the study, conducted the analysis and drafted the manuscript. M.V. conducted the microarray experiments. N.G.I. provided the cell lines and helped in conducting the microarray experiments. C.C. participated in the coordination of the study and in manuscript preparation. J.D.B. participated in designing the experiments, coordinating the study, data analysis and manuscript preparation. All authors read and approved the final manuscript. The authors declare that they have no competing financial interests.
REFERENCES
- 1.Lee M.L., Kuo,F.C., Whitmore,G.A. and Sklar,J. (2000) Importance of replication in microarray gene expression studies: statistical methods and evidence from repetitive cDNA hybridizations. Proc. Natl Acad. Sci. USA, 97, 9834–9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tseng G.C., Oh,M.K., Rohlin,L., Liao,J.C. and Wong,W.H. (2001) Issues in cDNA microarray analysis: quality filtering, channel normalization, models of variations and assessment of gene effects. Nucleic Acids Res., 29, 2549–2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang E., Miller,L.D., Ohnmacht,G.A., Liu,E.T. and Marincola,F.M. (2000) High-fidelity mRNA amplification for gene profiling. Nat. Biotechnol., 18, 457–459. [DOI] [PubMed] [Google Scholar]
- 4.Perou C.M., Jeffrey,S.S., van de Rijn,M., Rees,C.A., Eisen,M.B., Ross,D.T., Pergamenschikov,A., Williams,C.F., Zhu,S.X., Lee,J.C., Lashkari,D., Shalon,D., Brown,P.O. and Botstein,D. (1999) Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proc. Natl Acad. Sci. USA, 96, 9212–9217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu L., Wang,J., Baggerly,K., Wang,H., Fuller,G.N., Hamilton,S.R., Coombes,K.R. and Zhang,W. (2002) Obtaining reliable information from minute amounts of RNA using cDNA microarrays. BMC Genomics, 3, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.’tHoen P.A., de Kort,F., van Ommen,G.J. and den Dunnen,J.T. (2003) Fluorescent labelling of cRNA for microarray applications. Nucleic Acids Res., 31, e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hughes T.R., Mao,M., Jones,A.R., Burchard,J., Marton,M.J., Shannon,K.W., Lefkowitz,S.M., Ziman,M., Schelter,J.M., Meyer,M.R., Kobayashi,S., Davis,C., Dai,H., He,Y.D., Stephaniants,S.B., Cavet,G., Walker,W.L., West,A., Coffey,E., Shoemaker,D.D., Stoughton,R., Blanchard,A.P., Friend,S.H. and Linsley,P.S. (2001) Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat. Biotechnol., 19, 342–347. [DOI] [PubMed] [Google Scholar]
- 8.Taylor S., Smith,S., Windle,B. and Guiseppi-Elie,A. (2003) Impact of surface chemistry and blocking strategies on DNA microarrays. Nucleic Acids Res., 31, e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Y.H., Buckley,M.J., Dudoit,S. and Speed,T.P. (2002) Comparison of methods for image analysis on cDNA microarray data. J. Comp. Graph. Stat., 11, 108–136. [Google Scholar]
- 10.Brown C.S., Goodwin,P.C. and Sorger,P.K. (2001) Image metrics in the statistical analysis of DNA microarray data. Proc. Natl Acad. Sci. USA, 98, 8944–8949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jenssen T.K., Langaas,M., Kuo,W.P., Smith-Sorensen,B., Myklebost,O. and Hovig,E. (2002) Analysis of repeatability in spotted cDNA microarrays. Nucleic Acids Res., 30, 3235–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richter A., Schwager,C., Hentze,S., Ansorge,W., Hentze,M.W. and Muckenthaler,M. (2002) Comparison of fluorescent tag DNA labeling methods used for expression analysis by DNA microarrays. Biotechniques, 33, 620–628. [DOI] [PubMed] [Google Scholar]
- 13.Ihaka R. and Gentleman,R. (1996) R: A language for data analysis and graphics. J. Comput. Graph. Stat., 5, 299–314. [Google Scholar]
- 14.Dudoit S., Yang,Y.H., Callow,M.J. and Speed,T.P. (2002) Statistical methods for identifying differentially expressed genes in replicated cDNA microarray experiments. Stat. Sin., 12, 111–139. [Google Scholar]
- 15.Lönnstedt I. and Speed,T. (2002) Replicated microarray data. Stat. Sin., 12, 31–46. [Google Scholar]
- 16.Yue H., Eastman,P.S., Wang,B.B., Minor,J., Doctolero,M.H., Nuttall,R.L., Stack,R., Becker,J.W., Montgomery,J.R., Vainer,M. and Johnston,R. (2001) An evaluation of the performance of cDNA microarrays for detecting changes in global mRNA expression. Nucleic Acids Res., 29, e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bland J.M. and Altman,D.G. (1986) Statistical methods for assessing agreement between two methods of clinical measurement. Lancet, 1, 307–310. [PubMed] [Google Scholar]
- 18.Dobbin K., Shih,J. and Simon,R. (2003) Questions and answers on design of dual-label microarrays for identifying differentially expressed genes. J. Natl Cancer Inst., 95, 1362–1369. [DOI] [PubMed] [Google Scholar]
- 19.Yang Y. and Speed,T. (2002) Design issues for cDNA microarray experiments. Nat. Rev. Genet., 3, 579–588. [DOI] [PubMed] [Google Scholar]
- 20.Brazma A., Hingamp,P., Quackenbush,J., Sherlock,G., Spellman,P., Stoeckert,C., Aach,J., Ansorge,W., Ball,C.A., Causton,H.C., Gaasterland,T., Glenisson,P., Holstege,F.C., Kim,I.F., Markowitz,V., Matese,J.C., Parkinson,H., Robinson,A., Sarkans,U., Schulze-Kremer,S., Stewart,J., Taylor,R., Vilo,J. and Vingron,M. (2001) Minimum information about a microarray experiment (MIAME)—toward standards for microarray data. Nature Genet., 29, 365–371. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.