Abstract
Water plays essential structural and dynamical roles in protein-DNA recognition through contributing to enthalpic or entropic stabilization of binding complex and by mediating intermolecular interactions and fluctuations for biological function. These interfacial water molecules are confined by the binding partners in nanospace but in many cases they are highly mobile and exchange with outside bulk solution. Here, we report our studies of the interfacial water dynamics in the binary and ternary complexes of a polymerase (Dpo4) with DNA and an incoming nucleotide using a site-specific tryptophan probe with femtosecond resolution. By systematic comparison of the interfacial water motions and local sidechain fluctuations in the apo, binary and ternary states of Dpo4, we observed that the DNA binding interface and active site is dynamically solvent accessible and the interfacial water dynamics are similar to the surface hydration water fluctuations on picosecond time scales. Our molecular dynamics simulations also show the binding interface full of water molecules and nonspecific weak interactions. Such a fluid binding interface facilitates the polymerase sliding on DNA for fast translocation while the spacious and mobile hydrated active site contributes to the low fidelity of the lesion-bypass Y-family DNA polymerase.
Keywords: protein-DNA complex, interfacial water, nanospace confinement, ultrafast dynamics, fluid binding interface
Introduction
Water plays a critical role in protein-DNA recognition through entropic and enthalpic contributions in their binding energy for structural stability and through interfacial specificity and flexibility in their dynamic interactions for biological function.1–13 The binding interface is heavily solvated due to the high polarity of the negative phosphate groups on DNA and the abundance of charged groups on the protein. The extensive analyses of many protein-DNA complexes showed that water mediates interactions dominantly by screening unfavorable electrostatics and hydrogen bonding at the interface.2,5 Water can also behave as a “filler” to maintain packing densities at interface.3 For a large binding area, the specific recognition can occur at several positions to reach specificity by the entropic-driven water displacement and the enthalpic-enhanced direct interactions (hydrogen bonding, hydrophobic contact, van der Waals, electrostatic). The nonspecific interactions dominate the binding area with mobile water molecules acting various roles to maintain interfacial flexibility.6–13 In some cases, the interfacial water molecules can exchange with bulk solvent and the time scale was believed in nanoseconds,6,11–13 similar to the dynamics of trapped water molecules inside protein cavities.14 These dynamic water molecules actually maintain a partially disordered interface and behave as a molecule glue to increase structural adaptability and even to lubricate protein sliding on the DNA.1 Actually, these mobile interfacial water molecules may fluctuate even faster on a time scale shorter than nanoseconds,15 making experimental observation more difficult.
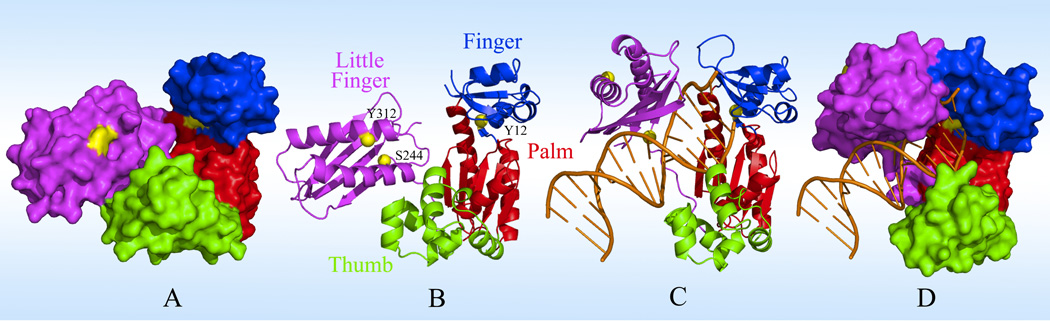
In this report, we studied the solvent dynamics at the binding interface of Sulfolobus solfataricus DNA polymerase IV (Dpo4) complex with DNA. Dpo4 is a model Y-family DNA polymerase that catalyzes DNA lesion bypass.16 It contains a typical polymerase core consisting of finger, thumb and palm domains which are structurally arranged in a right hand-like configuration, and a little finger domain which is only found in the Y-family members; see Figure 1. Comparison of the X-ray crystal structures of apo Dpo4 and its binary complex with DNA reveals a 131° rotation of the little finger domain relative to the polymerase core upon DNA binding.17 In the binary structure, the little finger and thumb domains hold the DNA duplex from the major and minor grooves, respectively.18 The active site of Dpo4 in the polymerase core is spacious and solvent-accessible due to the unusually small and stubby thumb and finger domains (Figure 1). During the binding of an incoming nucleotide to form the Dpo4-DNA-dNTP ternary complex, the active site residues undergo rearrangements but the polymerase core retains the same configuration.19 Since the ternary structure shows a flexible and solvent accessible active site,17,18,20 mobile water molecules must be involved in numerous nonspecific binding interactions. Interestingly, water molecules have been recently proposed to involve in the local active-site reorganization and the catalytic nucleotidyl-transfer reaction.21,22
Figure 1.
The X-ray structures of Dpo4 in both apo (PDB: 2RDI) and binary (PDB: 2RDJ) states. (A) and (B) show the surface-map and ribbon presentations of the apo structure with four domains of thumb (green), palm (red), finger (blue) and little finger (magenta) in the right hand-like architecture. Three mutation sites (Y12W, S244W and Y312W) are shown as yellow patches in (A) and balls in (B). (C) and (D) show the corresponding binary structure with the binding DNA (orange).
Here, we systematically characterized the solvent dynamics at residue Y12 in the finger domain, which is part of the active site of Dpo4, and at residue S244 in the little finger domain, which is within the DNA binding cleft, in the three states of Dpo4 (Dpo4 alone, the Dpo4-DNA binary complex and the Dpo4-DNA-dNTP ternary complex) (Figure 1).23 Since Dpo4 (352 amino acid residues, 40.2 kDa) does not possess a single tryptophan residue (W), we generated two single point mutants Y12W and S244W through site-directed mutagenesis. In addition, we prepared a single point mutant Y312W in order to monitor the solvent dynamics at residue Y312 in the little finger domain, which serves as a control site based on the fact that Y312 is on the surface of Dpo4 and is not involved in DNA binding and polymerase function (Figure 1). Using the methodology we developed before,23–26 we measured the femtosecond (fs)-resolved fluorescence dynamics of the probe tryptophan in the three states of Dpo4 and determined the water dynamics around the Dpo4 surface, at the complex binding interface, and on the active site with and without an incoming nucleotide.
Materials and Methods
Sample Preparation
S. solfataricus Dpo4 gene was cloned into the NdeI and XhoI sites of pET22b and a His6 tag was attached to the C-terminal. We also prepared the plasmids encoding three point mutants of Dpo4 (Y12W, S244W and Y312W) by site-directed mutagenesis. The four plasmids were individually transformed into E. coli BL21(DE3). Then, Dpo4 and the mutants were expressed and purified by following the procedures reported previously.27 The purified proteins were stored in a buffer solution containing 50 mM Tris-HCl (pH 7.5), 200 mM NaCl, 5 mM MgCl2, 1 mM DTT, 0.5 mM EDTA, and 5% glycerol. The concentration of each Dpo4 mutant used in the laser experiments was around 800 µM. DNA was purchased from Integrated DNA Technologies (Coralville, IA) and then purified and annealed before mixing with the polymerase. The sequences of the duplex DNA substrate are 8-mer 5´-AATCGCCG-3´ and 6-mer 5´-CGGCGA-3´ as the template and primer, respectively. The mixing ratio of Dpo4 to duplex DNA concentrations is 1 to 1.1 (Kd=~10 nM). The incoming nucleotide used is ddTTP (Kd=~230 µM) and is recognized at the active site but without the catalytic reaction after one incorporation due to the removal of the hydroxyl group at the 3’ position. The kinetic competency of Y12W, S244W and Y312W was examined by pre-steady state kinetic analysis, and their structures were analyzed by circular dichroism (CD) spectroscopic analysis.28 These mutants were concluded to have the same structure and function as wild-type Dpo4.
Femtosecond Fluorescence Spectroscopy
All fs-resolved measurements were carried out using the fluorescence up-conversion method as described before.29 Briefly, the pump wavelength was centered at 295 nm from a series of nonlinear mixing and doubling in 0.2-mm-thick barium borate crystals (BBO, type I) with a repetition rate of 1 kHz. The pump pulse energy was typically attenuated to 140 nJ prior to being focused into the motor-controlled rotating sample cell. Using 295-nm wavelength to excite the sample minimized tyrosine absorption in Dpo4 and the observed fluorescence was essentially from the excited tryptophan emission. The fluorescence emission was collected by a pair of parabolic mirrors and then mixed with a gating pulse (800 nm) in a 0.2-mm-thick BBO crystal through a nonlinear configuration. The up-converted signal ranging from 218 to 292 nm was detected by a photomultiplier coupled with a double-grating monochromator. The instrument response time under the current noncollinear geometry is between 350 and 450 fs as determined from the water Raman signal around 327 nm. The pump-pulse polarization was set at the magic angle (54.7°) with respect to the acceptance axis (vertical) of the up-conversion crystal, and the gating-pulse polarization was set parallel to this axis. For all fluorescence anisotropy measurements, the pump-pulse polarization was rotated to either parallel or perpendicular to the acceptance axis to obtain the parallel (I//) and perpendicular (I⊥) signals, respectively. These transients were used to construct the fs-resolved anisotropy dynamics: r(t)=(I//− I⊥)/(I//+2I⊥).
Results and Discussion
Steady-State Emission, Ultrafast Fluorescence Transients and Solvent Accessibility
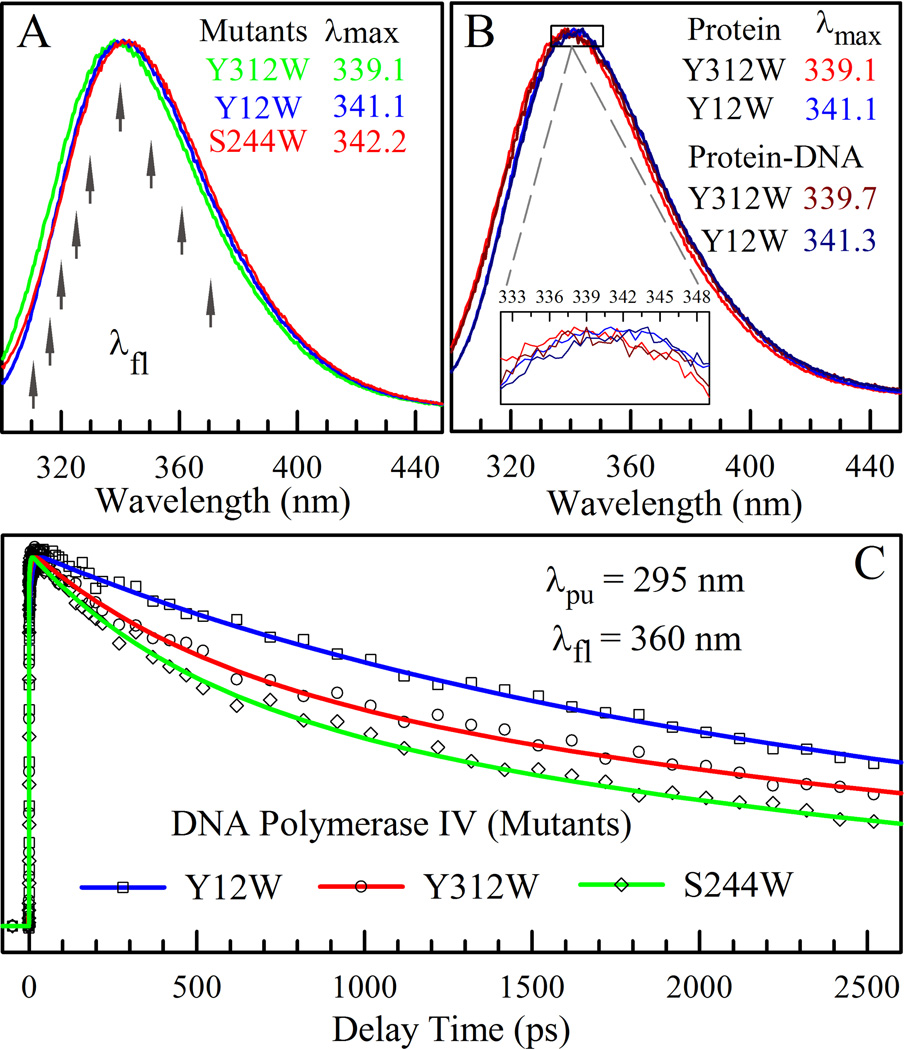
Figures 2A and 2B show the steady-state emission spectra of three mutants in different states. All three mutants have the emission peaks around 340 nm in the apo state, indicating that the three tryptophan residues are located at the protein surface and exposed to solvent molecules.30 Surprisingly, upon DNA binding, the emission peaks of the Y12W and S244W mutants show negligible changes and the recognition have little effect on the local polarity around the probe. The residue Y12W is located at the active site of Dpo4.18 After DNA binding, the emission peak changes from 341.1 to 341.3 nm and with an incoming nucleotide in the ternary state to 341.1 nm again. This observation clearly indicates that the active site in the complex is fully solvent accessible and the incoming nucleotide by displacement of a few water molecules has a little perturbation on the local polarity. The little finger residue S244W is located within the major groove of DNA in the Dpo4-DNA binary complex and the emission peak is 342 nm, slightly blue-shifted from 342.2 nm in the apo state, even with a large conformation swinging during the interactions with DNA.17 In the ternary state, the emission remains the same, indicating that the local structure and polar environment around S244W have negligible changes with an incoming nucleotide. The Y312 mutant was chosen as a control and is located on the surface of the little finger domain and far from the DNA binding pocket. Similarly, the local environment around Y312W shows a minor change from 339.1 nm in the apo state to 339.7 nm in the two complex states, still exposed to surface solvent. Figure 2C shows the different fluorescence transients of the lifetime emission of the three mutants at 360 nm in the apo state. These lifetimes remain similar upon complexation with DNA, further indicating the similar polar environments of the probe in the three states of Dpo4 and thus a lot of water molecules staying at the interface.
Figure 2.
(A) Normalized steady-state emission spectra of three Dpo4 mutants in the apo state. The arrows indicate the fluorescence wavelengths gated in fs-resolved measurements. (B) Normalized steady-state emission spectra of the mutant Y12W at the active site in the finger domain and Y312W as a control in the little finger domain with and without binding to DNA. The inset shows a close-up view around the emission peaks and the peak differences are minor. (C) Femtosecond-resolved fluorescence transients of the three Dpo4 mutants gated at 360 nm in apo state. The symbols are the experimental data and the solid lines are the best exponential fit.
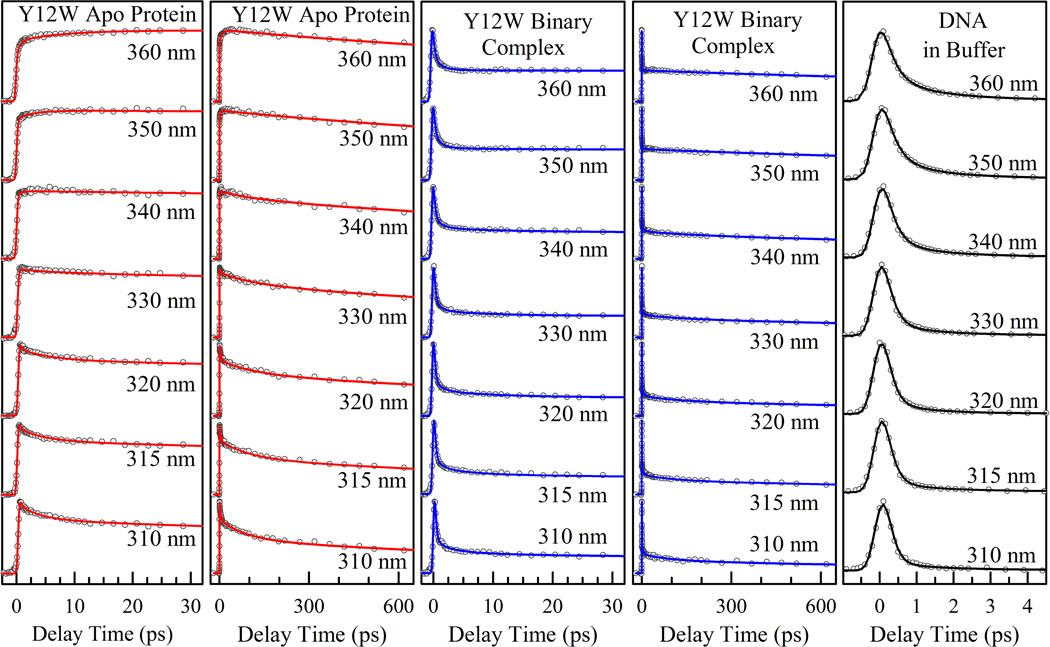
Figure 3 shows the fs-resolved fluorescence transients of Y12W at several typical wavelengths gated from the blue to red side of the emission spectra (Figure 2A) in the apo and binary states along with the fluorescence signals of DNA alone in buffer solution. The DNA signals decay in femtoseconds due to the ultrafast deactivation of excited state through conical intersections.31,32 The signals in the complex can be directly obtained from the wild-type (no tryptophan residue) or Y312 mutant (no interactions with DNA) binary state and are similar to those in buffer solution (Figure S1), indicating the DNA base-pair structure and hydration unchanged in the different states. The transients of the Y12W ternary state and other two mutants S244W and Y312W in the different states are given in the SI (Figures S2–S4). Similar to many proteins we studied,23–26, 33–35 the transients of Y12W in the apo state show the decay at the blue side and rise on the red end of the emission spectra, a typical signature of solvation dynamics mainly from relaxation of neighboring water molecules. The transients exhibit three decay components (330–500 fs, 3.6–7.5 ps and 85–110 ps) at the blue side and two rise components (0.4–1.1 ps and 8.9–15.4 ps) besides two long lifetime emissions. These time scales typically reflect surface hydration water relaxation within ~10–15 Å.36,37 Systematic studies have indicated that the time scale less than a few picoseconds represents the water-network local relaxation and the dynamics in tens of picoseconds results from the water-network rearrangements, coupled with local protein fluctuations, from a nonequilibrium configuration to an equilibrated state.23–26,33,38–41 In the binary and ternary states, the transients show a similar pattern and slightly slow down with three decay components around 0.65–1 ps, 4.5–10 ps and 85–137 ps. Interestingly, the dynamics of residue S244W are similar to those of Y12W with three distinct time scales and also slow down upon DNA binding. For the probe in residue Y312W, the relaxation is the same for all three states of Dpo4 with three distinct decay components because Y312W is not directly involved in the interactions with DNA.
Figure 3.
The left two columns show the normalized fs-resolved fluorescence transients of Y12W from seven selected wavelengths in the apo state on the short and long time ranges. The middle two columns are the corresponding normalized fs-resolved fluorescence transients of Y12W in the binary state with binding of DNA. The last column shows the normalized fs-resolved fluorescence transients of DNA alone in buffer solution. All the experimental data are shown in circles and the solid lines are the best exponential fit.
Both the steady-state emission and the fluorescence transients show the slight changes upon DNA binding, clearly showing a significant amount of water molecules in the DNA binding cleft and active site of Dpo4. Thus, the interface is highly solvated and solvent accessible. These water molecules are partially confined by two sides of the polymerase and DNA, respectively, but we did not observe any dramatic changes of the dynamics. Thus, those water molecules are not trapped at the interface and must exchange with bulk solvent. The interactions between Dpo4 and DNA are loose and the interface is dynamically solvent accessible.
Hydration Dynamics, Local Fluctuations and Interfacial Water Mobility
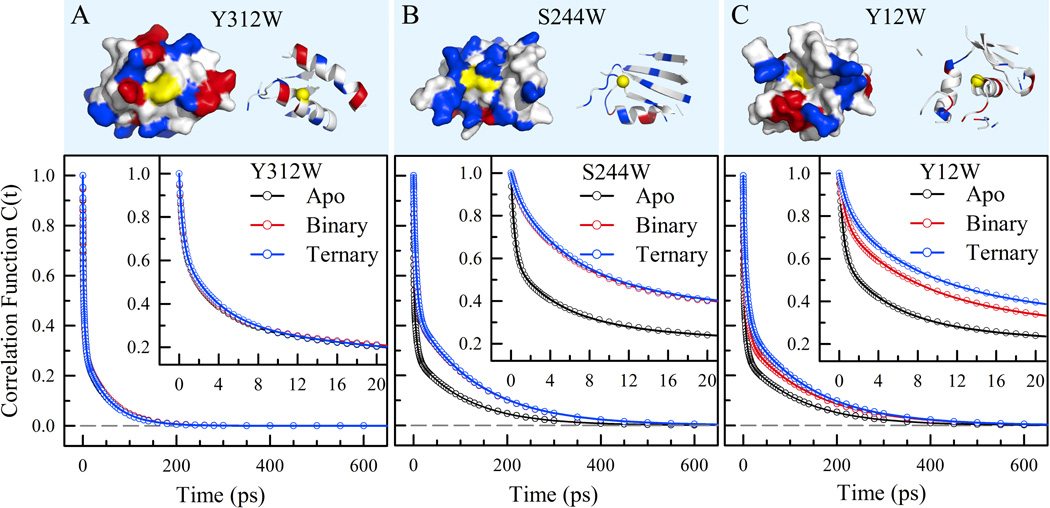
With the methodology we developed23,26,42 and careful analyses of the complex signals (see SI), we can quantitatively examine the dynamics and flexibility of those interfacial water molecules and determine how mobile they are at the interface. Figure 4 shows the final correlation functions of the local relaxation dynamics of three mutant positions with the local structural and chemical properties displayed at the top. Overall, the dynamics of Y312W are the fastest among the three mutants due to the convex structure and exposed site. The dynamics of S244 and Y12W are longer because of the densely positive-charged neighboring environment and the concave structure at the bottom of the binding pocket, respectively. Specifically, for Y312W, the correlation functions in the three states are nearly the same, consistent with their structural results that show no direct interactions with DNA.18 Thus, the dynamics represent the surface-water motions in the hydration layers. For S244W, the dynamics in the binary state slow down owing to the local confinement by the interactions with DNA, but keep similar in the ternary state due to its long distance from the active site. For Y12W, the dynamics in three states are different, gradually slowing down with complexation, indicating the changes by interactions in the binary state and further addition of an incoming nucleotide at the active site in the ternary state.
Figure 4.
Local protein properties and solvation correlation functions for the mutant Y312W (A), S244W (B) and Y12W (C) of Dpo4 in the apo, binary and ternary states. (Upper) Surface-map and ribbon representations of the local structures within 12Å from the tryptophan probe with positive (blue), negative (red) and neutral (white) residues as well as mutation sites (yellow). The yellow balls in the ribbon structures indicate the specific mutation sites. (Lower) Solvation correlation functions in three different states. The circles are the derived experimental data and the solid lines are the best exponential fit. Y312W as a control shows no changes in the three states, S244W in the binding site results in slowdown from the apo state to the complex states, and Y12W in the active site shows gradual changes from the apo, to binary and to ternary states.
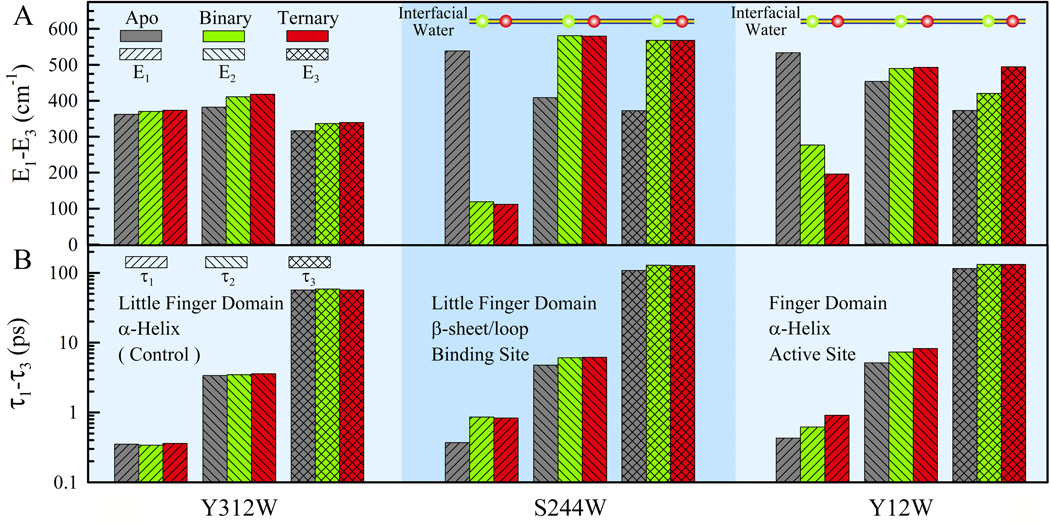
Figure 5 shows the detailed relaxation energies and time scales probed by tryptophan at the three positions in the three states (Table S1). For the control position of Y312W, the three time scales in the three states are all around 0.35, 3.5 and 52 ps, a typical hydration water relaxation around a rigid protein. The 0.35-ps dynamics (τ1) results from the outer-layers water relaxation in the hydration shell, close to bulk water. The 3.5-ps time scale (τ2) reflects inner-layers water relaxation near the protein surface. The longtime relaxation of 52 ps (τ3) dominantly represents the water-network rearrangements, mostly translational, coupled with local protein fluctuations.23–26,38,41,43,44 The total energy relaxation is about 1100 cm−1 and the corresponding energy distributions are around 33% (E1), 37% (E2) and 30% (E3). The slight increase (20–30 cm−1) of E2 and E3 in the binary and ternary states is from the slightly red-shifted emission (Figure 2B) due to a large conformation change (Figure 1). For S244W, the three time scales are 0.37, 4.8 and 111 ps in the apo state with the corresponding relaxation energies of 539, 407 and 372 cm−1, respectively. The three time scales are consistent with the surface-water relaxation around a dense-charge site observed in apomyoglobin.24,25 Upon DNA binding, the E1 energy drops significantly to 119 cm−1, but the corresponding time τ1 increases to 0.86 ps. The other energies of E2 and E3 increase to 579 and 564 cm−1 (Figure 5A) and their time scales of τ2 and τ3 lengthen to 6.1 and 136 ps (note the log scale in Figure 5B), respectively. These results are completely consistent with the DNA binding; the outer-layers water molecules around the binding site of S244W are partially confined and could be reduced, leading to the significant change of the first ultrafast component both in the relaxed energy and on the time scale. The total relaxation energy (~1300 cm−1) and local polarity (λmax~342 nm) do not change dramatically after the DNA binding and thus the partially confined water molecules take slightly longer times (τ2 and τ3) to finish complete relaxation with more solvation energy release at the interface. However, all these changes are not significant and again the water molecules are not trapped at the interface because the trapped water molecules relax in nanoseconds or longer.14,45,46 Thus, those water molecules around the binding site are still very mobile. With an incoming nucleotide, the dynamics and released solvation energies are similar, consistent with the minor structural changes at the distant binding site.
Figure 5.
(A) The derived solvation energies and (B) the corresponding relaxation times for the three mutants of Dpo4 in three different states of apo (gray), binary (green) and ternary (red). Y312W is a control and not directly involved in the interactions with DNA and thus shows no obvious changes in three different states for three energies and three time scales. S244W, located at and interacting with the major groove of DNA but far from the active site, shows changes from the apo state to the complex states both in energy and in time. Y12W, around the active site and directly probing the binding of DNA and an incoming nucleotide, shows gradual changes in energy and time from the apo state to the binary complex and then to the ternary state. See text.
For Y12W around the active site, the three time scales in the apo state are 0.43, 5.2 and 120 ps, similar to those of S244W, with the corresponding energies of 534, 453 and 372 cm−1, respectively. Although there are less charged residues around Y12W at the active site, the probe (W12) is at the bottom of the binding pocket with a concave local structure and thus water molecules are expected to move relatively slower.47,48 With complexation of DNA and a nucleotide, the time scales become gradually longer, 0.62, 7.4 and 140 ps for the binary state and 0.91, 8.31 and 140 ps for the ternary state. Similarly, the first energy components become smaller due to the confinement by the DNA binding (277 cm−1) and further incorporation of a nucleotide (196 cm−1). However, in the binary state both the second and third components of E2 and E3 keep slight increases to 488 and 419 cm−1, respectively. This observation indicates more water molecules confined, leading to energy increases of E2 and E3. With an incoming nucleotide, the E2 energy is nearly the same, and E3 increases to 494 cm−1, consistent with the local structural reorganization induced by the nucleotide binding. Overall, these changes are also not significant and even less than those observed in S244W, consistent with the large water-exposed open area around the active site (Figure 1).
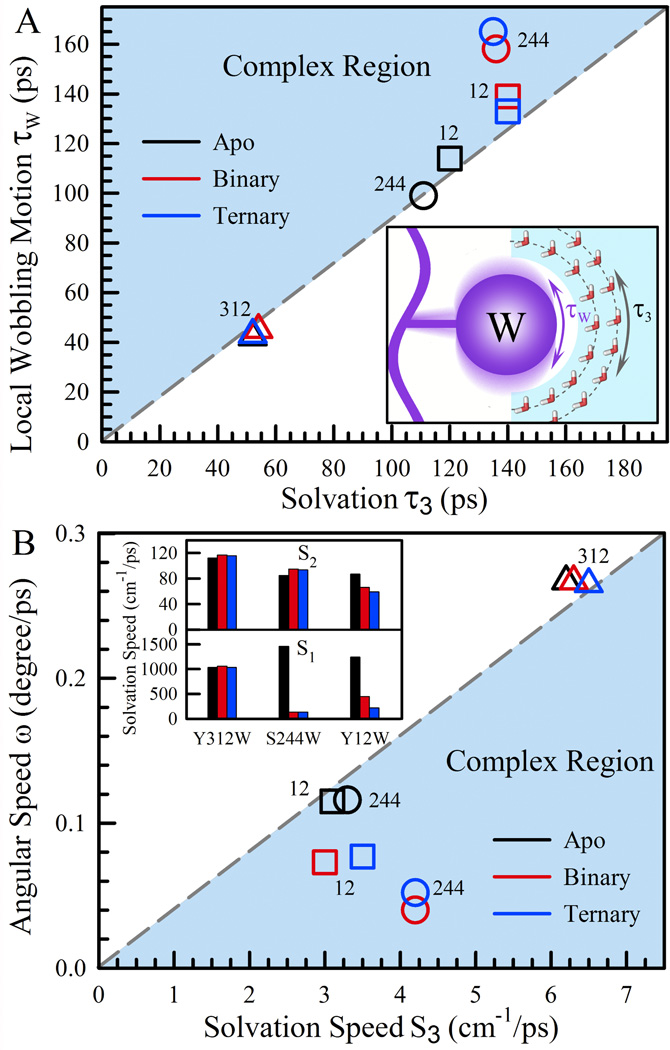
We also studied the local sidechain flexibility in the three states by measuring the fs-resolved anisotropy dynamics of the probe tryptophan (Fig. S5) and obtained the wobbling angles (θ) and related time scales (τw) besides that we observed a longer tumbling time for the binding complex than that for the apo protein alone.49 In our systematic studies of apomyoglobin,25 we observed that the solvation relaxation τ3 has an intrinsic relationship with the local wobbling time τw of tryptophan. Figure 6A shows the relationship we observed for all three states. Surprisingly, the observed wobbling and long solvation times at the three positions in the apo state are linearly correlated, indicating that the local wobbling motion is inherently related to the coupled water-protein relaxation (insert in Figure 6A). Note that the correlated line starts at the original point of (0, 0). Interestingly, the data of Y12W in the binary and ternary states also nearly follow the linear line, indicating that the active site in the two states of Dpo4 is highly solvated, similar to the apo-state property. But, for S244W, the times in the two complex states of Dpo4 do not follow the line any more, falling into the complex region where the increase of the local structural constraints by the binding of DNA and dNTP causes longer τw.
Figure 6.
(A) Correlation of the long solvation time τ3 and the local wobbling time τw. The blue area is the complex region that the binding interactions result in a longer τw. S244W in the complex states falls into this region. The other two mutants follow the correlated line indicating the intrinsic relationship between the coupled water-protein relaxation and local sidechain motion. The inset shows a cartoon representation of the two inherent motions. (B) Correlation between the solvation speed S3 and the angular speed ω. The blue area is the corresponding complex region where the sidechain relaxation speed is reduced due to the constraints from DNA and dNTP binding. The data of three mutants in the apo state follow a straight line. The inset shows the solvation speeds S1 and S2 of the three mutants in all three states of apo (black), binary (red) and ternary (blue). Note that the solvation speeds of S2 and S3 for S244W slightly increase in the complex states.
We also defined the solvation speed (Si=Ei/τi, i=1–3) and the angular speed (ω=θ/τw) as two parameters to quantify how fast the water network relaxes and how flexible the sidechain wobbles, respectively.25 In Figure 6B, we plotted the angular speed ω of tryptophan relative to the solvation speed S3. Again, the data in the apo state surprisingly exhibit a linear correlation starting at (0, 0). But, the data in the binary and ternary states fall into the complex region due to the certain constraints by the interactions at the interface, reducing the angular speed. Surprisingly, we observed the solvation speeds (S3) of S244W in the complex states are faster than that in the apo state, implying a more mobile environment around S244 at the interface. In insert of Figure 6B, it shows the significant decreases of S1 for both S244W and Y12W sites owing to the binding of DNA and dNTP. For S244W, similar to S3, S2 increases in the complex states, further indicating the more mobile water networks at the interface. For Y12W, S2 gradually decreases in the binary and ternary states due to the binding of DNA and dNTP. For Y312W as a control outside of the binding area, the data are similar for all three states in Figures 6A and 6B.
The systematic analyses of the fluorescence transient behaviors and local anisotropy dynamics clearly show a characteristic of hydration water dynamics around the protein surface in the apo state with two major water-network relaxations of a local orientational motion in femtoseconds (outer-layers water) to a few picoseconds (inner-layers water) and a water-protein coupled translational motion in tens to a hundred of picoseconds (inner-layers water). With the binding of DNA and an incoming dNTP, those dynamics show changes, reflecting the nature of interactions by imposing certain constraints on the interfacial water molecules. However, the observed changes in the DNA binding cleft (S244W) and the active site (Y12W) are small and insignificant, revealing the mobile interfacial water molecules at the DNA binding cleft and active site of Dpo4.
Interface Fluidity, Active-Site Flexibility, and Low Fidelity
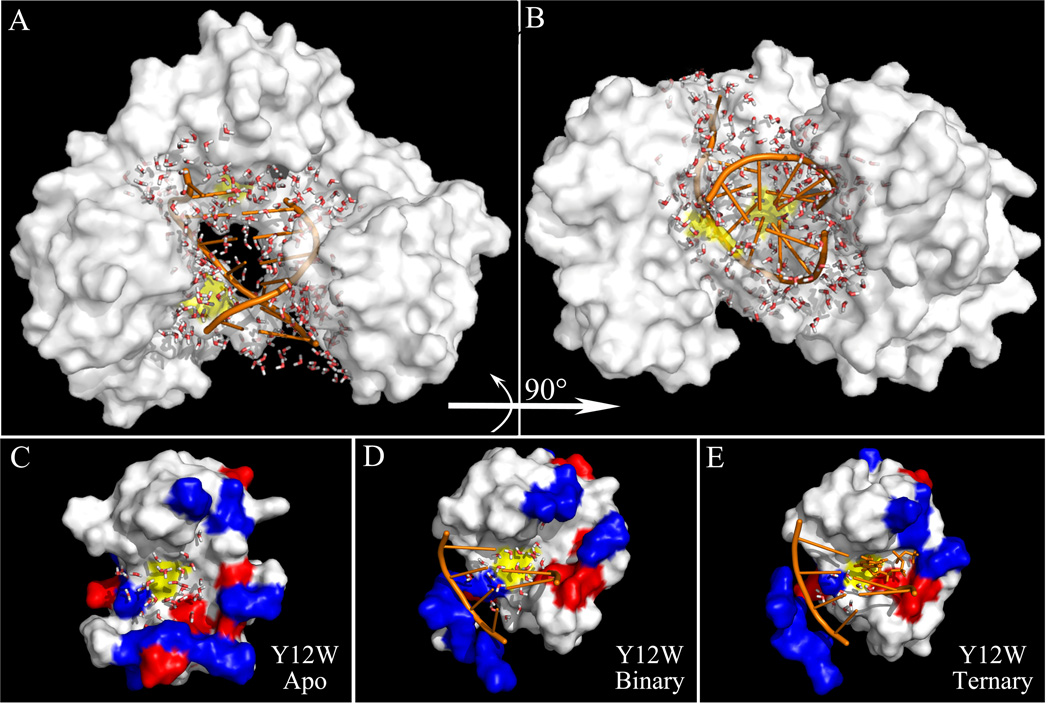
The X-ray binary structure shows that the little finger and thumb domains grip the dsDNA about eight base pairs across the major groove and the minor groove, respectively (Figure 1C). The interactions are mainly with phosphodiester moieties and a few deoxyriboses through hydrogen bonds and van der Walls contacts. No direct interactions with the DNA bases were observed.18 The S244 site is at a loop near the end of the β-11 strand in the little finger domain and interacts with the major groove of DNA (Figure 1C). Near the active site, a large crevice is formed between the little finger and finger domains and such a hole is solvent accessible (Figure 7A).18 The Y12W site is in the α-helix A in the finger domain close to the position of the incoming nucleotide and adjacent to the active site. Although the X-ray complex structures do not show many water molecules at the interface, our experimental observation clearly indicates a significant amount of mobile interfacial water molecules mediating DNA interaction and the X-ray scattering cannot capture the mobile water in structure.
Figure 7.
A snapshot from a molecular dynamics simulation in 4 ns. (A) and (B) show 274 interfacial water molecules between Dpo4 (white) and DNA (orange) from two different views. The mutation sites are shown by the yellow patches. (C), (D) and (E) show the local structures of Y12W within 12 Å and interfacial water molecules within 7.5 Å from the probe tryptophan in the three states. The positive and negative charged residues are in blue and red and the neutral residues in white. The mutation site is patched in yellow. The dsDNA is shown in orange while the nucleotide dCTP in orange sticks (E). Within 7.5 Å, 30, 21 and 10 water molecules were observed in the apo, binary and ternary states, respectively.
We performed molecular dynamics simulations with a time range of 4 ns with fixing the DNA and incoming nucleotide structures. Figure 7 shows a snapshot of our simulation results with a full of water at the interface. The interfacial water molecules in Figures 7A and 7B were calculated with a total distance from the polymerase and DNA within 10 Å and a total number of 274 interfacial water molecules were observed. Figures 7C–E show the local structure of Y12W within 12 Å in the three states with the interfacial water molecules within 7.5 Å from the indole ring of the tryptophan probe. Within 7.5 Å, there are 30, 21 and 11 water molecules around Y12W in the apo, binary and ternary states, respectively, but within 10 Å increasing to 67, 50 and 37 ones. For S244W, there are 61 and 33 water molecules within 7.5 Å and 136 and 70 ones within 10 Å in the apo and complex states, respectively. For Y312W, a total of 43 water molecules within 7.5 Å and 108 ones within 10 Å in all the three states. The binding clearly reduces the number of water molecules but a significant number of water molecules still remains at the interface.
Based on the experimental observations of static binding interactions from the X-ray structures, a significant amount of water molecules at the interface from our MD simulations, and dynamic water accessibility and ultrafast water mobility at the binding and active sites from our fs-resolved site-specific studies, a cohesive picture on the interaction interface emerges. The interface is full of dynamic water molecules and behaves fluid. Such fluidity maintains weak and loose interactions between the polymerase and DNA, facilitates the polymerase sliding on the DNA. The active site is spacious and highly solvent accessible. Such a mobile solvated active site in the flexible complex leads to an error-prone and lesion-bypass DNA synthesis and gives rise to the low fidelity of Dpo4. The mobile interfacial water molecules play a significant role in Dpo4 function.
Conclusions
We reported here our characterization of the interfacial water dynamics in the binary complex of the polymerase Dpo4 with DNA and in the ternary state with a nucleotide substrate. Using tryptophan as a local optical probe through site-direct mutagenesis, we studied two specific positions at the interface, a binding site located in DNA major groove in the little finger domain (S244W) and the active site (Y12W) in the finger domain, as well as one control site (Y312W) in the little finger domain far from the interaction interface. By the systematic comparison of the hydration water dynamics and local sidechain motions in the apo, binary and ternary states, we observed that the complex interface is dynamically solvent accessible and the water molecules are not trapped at the interfacial nanospace and are extremely mobile on the picosecond timescales, similar to the surface-water motions in the apo state without DNA interaction. The observed slight changes in dynamics in the complex states reflect that the confinement by the substrate binding is not severe and tight. The complex interactions are weak and loose, resulting in a fluid interface with a significant amount of water molecules to lubricate sliding between the polymerase and DNA with fast translocation. The spacious and water-flooding active site can accommodate various substrates and leads to an error-prone and lesion-bypass DNA synthesis with the low fidelity. The mobile interfacial water is critical to the flexible complex and active site of Dpo4 and plays a significant role in Dpo4 function. The method developed here is general and can be used to study other important protein-DNA complexes, especially for the replicative polymerases, and such work is currently under way.
Supplementary Material
ACKNOWLEDGEMENTS
This paper is dedicated to the “Terry Miller Festschrift” special issue and we thank him for his strong support and constant encouragement through the years at The Ohio State University. We also like to thank Sean A. Newmister and John Pryor for the help of purifying DNA substrate. The work is supported in part by the National Institute of Health (Grant GM095997), the National Science Foundation (Grant CHE-0748358), the Camille Dreyfus Teacher-Scholar and the Guggenheim fellowship to DZ and by the National Science Foundation (Grant MCB-0960961) to ZS.
Footnotes
Supporting Information
The description of data analyses of fluorescence transients in the complex states, the fluorescence transients of DNA and the three mutants in the three states, and the related fitting results. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declared no competing financial interest.
REFERENCES
- 1.Levy Y, Onuchic JN. Water Mediation in Protein Folding and Molecular Recognition. Annu. Rev. Biophys. Biomol. Struct. 2006;35:389–415. doi: 10.1146/annurev.biophys.35.040405.102134. [DOI] [PubMed] [Google Scholar]
- 2.Jayaram B, Jain T. The Role of Water in Protein-DNA Recognition. Annu. Rev. Biophys. Biomol. Struct. 2004;33:343–361. doi: 10.1146/annurev.biophys.33.110502.140414. [DOI] [PubMed] [Google Scholar]
- 3.Reddy CK, Das A, Jayaram B. Do Water Molecules Mediate Protein-DNA Recognition? J. Mol. Biol. 2001;314(3):619–632. doi: 10.1006/jmbi.2001.5154. [DOI] [PubMed] [Google Scholar]
- 4.Janin J. Wet and Dry Interfaces: The Role of Solvent in Protein-Protein and Protein-DNA Recognition. Struct. Fold. Des. 1999;7(12):R277–R279. doi: 10.1016/s0969-2126(00)88333-1. [DOI] [PubMed] [Google Scholar]
- 5.Schwabe JWR. The Role of Water in Protein DNA Interactions. Curr. Opin. Struct. Biol. 1997;7(1):126–134. doi: 10.1016/s0959-440x(97)80016-4. [DOI] [PubMed] [Google Scholar]
- 6.Billeter M, Guntert P, Luginbuhl P, Wuthrich K. Hydration and DNA Recognition by Homeodomains. Cell. 1996;85(7):1057–1065. doi: 10.1016/s0092-8674(00)81306-9. [DOI] [PubMed] [Google Scholar]
- 7.Fraenkel E, Pabo CO. Comparison of X-Ray and NMR Structures for the Antennapedia Homeodomain-DNA Complex. Nat. Struct. Biol. 1998;5(8):692–697. doi: 10.1038/1382. [DOI] [PubMed] [Google Scholar]
- 8.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent Mediated Interactions in the Structure of the Nucleosome Core Particle at 1.9 Angstrom Resolution. J. Mol. Biol. 2002;319(5):1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 9.Ferreiro DU, Sanchez IE, Gay GD. Transition State for Protein-DNA Recognition. Proc. Natl. Acad. Sci. U. S. A. 2008;105(31):10797–10802. doi: 10.1073/pnas.0802383105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsui V, Radhakrishnan I, Wright PE, Case DA. NMR and Molecular Dynamics Studies of the Hydration of a Zinc Finger-DNA Complex. J. Mol. Biol. 2000;302(5):1101–1117. doi: 10.1006/jmbi.2000.4108. [DOI] [PubMed] [Google Scholar]
- 11.Sunnerhagen M, Denisov VP, Venu K, Bonvin AMJJ, Carey J, Halle B, Otting G. Water Molecules in DNA Recognition I: Hydration Lifetimes of Trp Operator DNA in Solution Measured by NMR Spectroscopy. J. Mol. Biol. 1998;282(4):847–858. doi: 10.1006/jmbi.1998.2033. [DOI] [PubMed] [Google Scholar]
- 12.Kosztin D, Bishop TC, Schulten K. Binding of the Estrogen Receptor to DNA. The Role of Waters. Biophys. J. 1997;73(2):557–570. doi: 10.1016/S0006-3495(97)78093-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorfe AA, Caflisch A, Jelesarov I. The Role of Flexibility and Hydration on the Sequence-Specific DNA Recognition by the Tn916 Integrase Protein: A Molecular Dynamics Analysis. J. Mol. Recognit. 2004;17(2):120–131. doi: 10.1002/jmr.658. [DOI] [PubMed] [Google Scholar]
- 14.Otting G, Liepinsh E, Wuthrich K. Protein Hydration in Aqueous-Solution. Science. 1991;254(5034):974–980. doi: 10.1126/science.1948083. [DOI] [PubMed] [Google Scholar]
- 15.Zhong D, Pal SK, Zewail AH. Femtosecond Studies of Protein-DNA Binding and Dynamics: Histone I. ChemPhysChem. 2001;2(4):219–227. doi: 10.1002/1439-7641(20010417)2:4<219::AID-CPHC219>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 16.Yang W. Damage Repair DNA Polymerases Y. Curr. Opin. Struct. Biol. 2003;13(1):23–30. doi: 10.1016/s0959-440x(02)00003-9. [DOI] [PubMed] [Google Scholar]
- 17.Wong JH, Fiala KA, Suo Z, Ling H. Snapshots of a Y-family DNA Polymerase in Replication: Substrate-Induced Conformational Transitions and Implications for Fidelity of Dpo4. J. Mol. Biol. 2008;379(2):317–330. doi: 10.1016/j.jmb.2008.03.038. [DOI] [PubMed] [Google Scholar]
- 18.Ling H, Boudsocq F, Woodgate R, Yang W. Crystal Structure of a Y-family DNA Polymerase in Action: A Mechanism for Error-Prone and Lesion-Bypass Replication. Cell. 2001;107(1):91–102. doi: 10.1016/s0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 19.Fiala KA, Suo Z. Mechanism of DNA Polymerization Catalyzed by Sulfolobus Solfataricus P2 DNA Polymerase IV. Biochemistry. 2004;43(7):2116–2125. doi: 10.1021/bi035746z. [DOI] [PubMed] [Google Scholar]
- 20.Rechkoblit O, Malinina L, Cheng Y, Kuryavyi V, Broyde S, Geacintov NE, Patel DJ. Stepwise Translocation of Dpo4 Polymerase During Error-Free Bypass of an OxoG lesion. PLoS Biol. 2006;4(1):25–42. doi: 10.1371/journal.pbio.0040011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang LH, Yu XY, Hu P, Broyde S, Zhang YK. A Water-Mediated and Substrate-Assisted Catalytic Mechanism for Sulfolobus Solfataricus DNA Polymerase IV. J. Am. Chem. Soc. 2007;129(15):4731–4737. doi: 10.1021/ja068821c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Schlick T. Quantum Mechanics/Molecular Mechanics Investigation of the Chemical Reaction in Dpo4 Reveals Water-Dependent Pathways and Requirements for Active Site Reorganization. J. Am. Chem. Soc. 2008;130(40):13240–13250. doi: 10.1021/ja802215c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhong D. Hydration Dynamics and Coupled Water-Protein Fluctuations Probed by Intrinsic Tryptophan. Adv. Chem. Phys. 2009;143:83–149. [Google Scholar]
- 24.Zhang L, Wang L, Kao YT, Qiu W, Yang Y, Okobiah O, Zhong D. Mapping Hydration Dynamics Around a Protein Surface. Proc. Natl. Acad. Sci. U. S. A. 2007;104(47):18461–18466. doi: 10.1073/pnas.0707647104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang L, Yang Y, Kao YT, Wang L, Zhong D. Protein Hydration Dynamics and Molecular Mechanism of Coupled Water-Protein Fluctuations. J. Am. Chem. Soc. 2009;131(30):10677–10691. doi: 10.1021/ja902918p. [DOI] [PubMed] [Google Scholar]
- 26.Qin Y, Chang CW, Wang L, Zhong D. Validation of Response Function Construction and Probing Heterogeneous Protein Hydration by Intrinsic Tryptophan. J. Phys. Chem. B. 2012;116(45):13320–13330. doi: 10.1021/jp305118n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fiala KA, Suo Z. Pre-Steady-State Kinetic Studies of the Fidelity of Sulfolobus Solfataricus P2 DNA Polymerase IV. Biochemistry. 2004;43(7):2106–2115. doi: 10.1021/bi0357457. [DOI] [PubMed] [Google Scholar]
- 28.Fiala KA, Sherrer SM, Brown JA, Suo Z. Mechanistic Consequences of Temperature on DNA Polymerization Catalyzed by a Y-family DNA Polymerase. Nucleic Acids Res. 2008;36(6):1990–2001. doi: 10.1093/nar/gkn004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang L, Kao YT, Qiu W, Wang L, Zhong D. Femtosecond Studies of Tryptophan Fluorescence Dynamics in Proteins: Local Solvation and Electronic Quenching. J. Phys. Chem. B. 2006;110(37):18097–18103. doi: 10.1021/jp063025e. [DOI] [PubMed] [Google Scholar]
- 30.Lakowicz JR. Principles of Fluorescence Spectroscopy. Springer; 2007. [Google Scholar]
- 31.Matsika S. Three-state conical intersections in nucleic acid bases. J. Phys. Chem. A. 2005;109(33):7538–7545. doi: 10.1021/jp0513622. [DOI] [PubMed] [Google Scholar]
- 32.Hudock HR, Martinez TJ. Excited-State Dynamics of Cytosine Reveal Multiple Intrinsic Subpicosecond Pathways. ChemPhysChem. 2008;9(17):2486–2490. doi: 10.1002/cphc.200800649. [DOI] [PubMed] [Google Scholar]
- 33.Qiu W, Zhang L, Kao YT, Lu W, Li T, Kim J, Sollenberger GM, Wang L, Zhong D. Ultrafast Hydration Dynamics in Melittin Folding and Aggregation: Helix Formation and Tetramer Self-Assembly. J. Phys. Chem. B. 2005;109(35):16901–16910. doi: 10.1021/jp0511754. [DOI] [PubMed] [Google Scholar]
- 34.Qiu W, Zhang L, Okobiah O, Yang Y, Wang L, Zhong D, Zewail AH. Ultrafast Solvation Dynamics of Human Serum Albumin: Correlations with Conformational Transitions and Site-Selected Recognition. J. Phys. Chem. B. 2006;110(21):10540–10549. doi: 10.1021/jp055989w. [DOI] [PubMed] [Google Scholar]
- 35.Qiu W, Kao YT, Zhang L, Yang Y, Wang L, Stites WE, Zhong D, Zewail AH. Protein Surface Hydration Mapped by Site-Specific Mutations. Proc. Natl. Acad. Sci. U. S. A. 2006;103(38):13979–13984. doi: 10.1073/pnas.0606235103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li T, Hassanali AA, Kao YT, Zhong D, Singer SJ. Hydration Dynamics and Time Scales of Coupled Water-Protein Fluctuations. J. Am. Chem. Soc. 2007;129(11):3376–3382. doi: 10.1021/ja0685957. [DOI] [PubMed] [Google Scholar]
- 37.Li T, Hassanali AA, Singer SJ. Origin of Slow Relaxation Following Photoexcitation of W7 in Myoglobin and the Dynamics of Its Hydration Layer. J. Phys. Chem. B. 2008;112(50):16121–16134. doi: 10.1021/jp803042u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang J, Zhang L, Wang L, Zhong D. Femtosecond Conical Intersection Dynamics of Tryptophan in Proteins and Validation of Slowdown of Hydration Layer Dynamics. J. Am. Chem. Soc. 2012;134(40):16460–16463. doi: 10.1021/ja305283j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pal SK, Peon J, Bagchi B, Zewail AH. Biological Water: Femtosecond Dynamics of Macromolecular Hydration. J. Phys. Chem. B. 2002;106(48):12376–12395. [Google Scholar]
- 40.Pal SK, Zewail AH. Dynamics of Water in Biological Recognition. Chem. Rev. 2004;104(4):2099–2123. doi: 10.1021/cr020689l. [DOI] [PubMed] [Google Scholar]
- 41.Zhong D, Pal SK, Zewail AH. Biological Water: A Critique. Chem. Phys. Lett. 2011;503(1–3):1–11. [Google Scholar]
- 42.Lu W, Qiu W, Kim J, Okobiah O, Hu H, Gokel GW, Zhong D. Femtosecond Studies of Crown Ethers: Supramolecular Solvation, Local Solvent Structure and Cation-Pi Interaction. Chem. Phys. Lett. 2004;394(4–6):415–422. [Google Scholar]
- 43.Grebenkov DS, Goddard YA, Diakova G, Korb JP, Bryant RG. Dimensionality of Diffusive Exploration at the Protein Interface in Solution. J. Phys. Chem. B. 2009;113(40):13347–13356. doi: 10.1021/jp9048082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Armstrong BD, Choi J, Lopez C, Wesener DA, Hubbell W, Cavagnero S, Han S. Site-Specific Hydration Dynamics in the Nonpolar Core of a Molten Globule by Dynamic Nuclear Polarization of Water. J. Am. Chem. Soc. 2011;133(15):5987–5995. doi: 10.1021/ja111515s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jesenska A, Sykora J, Olzynska A, Brezovsky J, Zdrahal Z, Damborsky J, Hof M. Nanosecond Time-Dependent Stokes Shift at the Tunnel Mouth of Haloalkane Dehalogenases. J. Am. Chem. Soc. 2009;131(2):494–501. doi: 10.1021/ja804020q. [DOI] [PubMed] [Google Scholar]
- 46.Nucci NV, Pometun MS, Wand AJ. Site-Resolved Measurement of Water-Protein Interactions by Solution NMR. Nat. Struct. Mol. Biol. 2011;18(2):245–249. doi: 10.1038/nsmb.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang CW, Guo L, Kao YT, Li J, Tan C, Li T, Saxena C, Liu Z, Wang L, Sancar A, Zhong D. Ultrafast Solvation Dynamics at Binding and Active Sites of Photolyases. Proc. Natl. Acad. Sci. U. S. A. 2010;107(7):2914–2919. doi: 10.1073/pnas.1000001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chang CW, He TF, Guo L, Stevens JA, Li T, Wang L, Zhong D. Mapping Solvation Dynamics at the Function Site of Flavodoxin in Three Redox States. J. Am. Chem. Soc. 2010;132(36):12741–12747. doi: 10.1021/ja1050154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steiner RF. In: In Topics in Fluorescence Spectroscopy. Lakowicz JR, editor. Vol.2. New York: Plenum Press; 1991. pp. 1–51. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.