Abstract
In a cell, the chromatin state is controlled by the highly regulated interplay of epigenetic mechanisms ranging from DNA methylation and incorporation of different histone variants to posttranslational modification of histones and ATP-dependent chromatin remodeling. These changes alter the structure of the chromatin to either facilitate or restrict the access of transcription machinery to DNA. These epigenetic modifications function to exquisitely orchestrate the expression of different genes, and together constitute the epigenome of a cell. In the skin, different epigenetic regulators form a regulatory network that operates to guarantee skin stem cell maintenance while controlling differentiation to multiple skin structures. In this review, we will discuss recent findings on epigenetic mechanisms of skin control and their relationship to skin pathologies.
In the skin, various epigenetic regulators guarantee stem cell maintenance while controlling differentiation. For example, SWI/SNF activity is essential for proper differentiation in the epidermis.
The mammalian genome is organized into a highly compacted structure that allows a 6-µm nucleus to accommodate 3 billion base pairs of DNA (Redi and Capanna 2012; Van Bortle and Corces 2012). Strikingly, the nuclear architecture and the level of genomic compaction are highly dynamic and depend on the state of the cell with the chromatin structure changing to regulate gene expression (Hemberger et al. 2009; Bickmore and van Steensel 2013). These so-called epigenetic modifications change the accessibility of DNA to transcriptional machinery in such a way that chromatin state can be inherited.
Different epigenetic regulators have specific enzymatic actives that modify DNA or chromatin. One mechanism includes changing the chemical composition of DNA by the addition of a methyl group that is usually associated with transcriptional repression (Fig. 1) (Smith and Meissner 2013). DNA is wrapped around eight histone proteins to form nucleosomes (Fig. 2A), and a second mechanism involves modifying specific amino acid residues on the histone tails (Fig. 2B) (Andrews and Luger 2011). These posttranslational histone modifications are able to recruit additional proteins that either positively or negatively affect transcription (Fig. 2C) (Barski et al. 2007; Wang et al. 2008). Different epigenetic complexes can be classified by enzymatic activity, and together they interact to establish the epigenetic state of the cell (Berger et al. 2009; Ho and Crabtree 2010; Botchkarev et al. 2012).
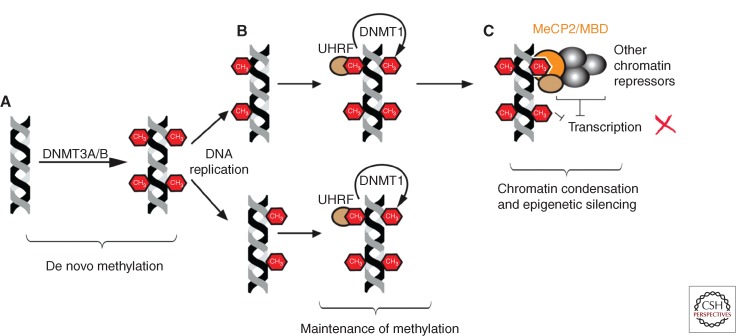
Figure 1.
DNA methylation. (A) DNA methyltransferases 3A and 3B (DNMT3A/B) catalyze the de novo addition of a methyl group to cytosine in the CpG context. (B) After cell division, DNMT1 is responsible for copying the DNA methylation pattern from the mother strand to the daughter strand to maintain the DNA methylation pattern. This process is aided by UHRF, which binds hemimethylated CpG sequences. (C) The presence of methyl groups in the DNA can block the binding of transcriptions factors or be recognized by other methyl-binding proteins as MeCP2 or MBD that recruit other chromatin repressors (e.g., histone deacetylases [HDACs]), resulting in chromatin condensation and silencing of gene expression. See main text for details.
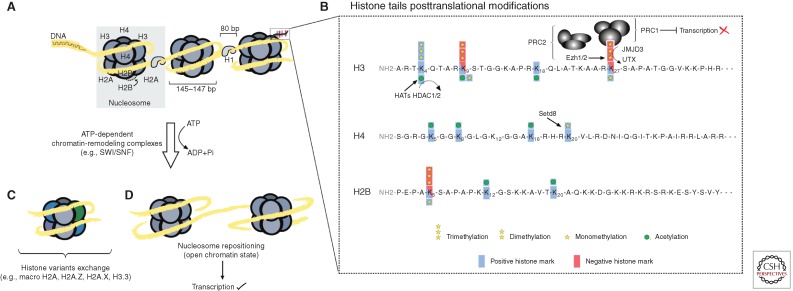
Figure 2.
Chromatin remodeling and histone code. (A) Inside the nucleolus, ∼146 bp of DNA are wrapped around the nucleosome core, an octamer protein complex composed of four histone heterodimers: two H2A-H2B and two H3-H4. Between protein octamers, histone H1 helps to stabilize the linker DNA. The amino terminus of each core histone protrudes outside the protein octamer. These “histone tails” are the target of posttranslational modifications. Histone tail modifications described in the text are presented in (B). The histone acetyltransferases (HATs) catalyze the addition of acetyl residues on specific lysine residues. Other conserved lysine residues are methylated. In particular, the activity of the polycomb repressive complex 2 (PRC2; through the enzymatic activity of Ezh1 and Ezh2) mediates the di- and trimethylation of the Lysine 27 on the Histone H3. This modification is then recognized by the PRC1 mediating the condensation and epigenetic silencing of a DNA region. See main text for details. The lysine residue modified by the methyltransferase activity of Setd8 is also represented. (C) Under some circumstances, histone variants are introduced into the nucleosome by histone chaperones. See main text for details. (D) Nucleosome positioning can be altered by ATP-dependent chromatin-remodeling complexes that results in a more relaxed chromatin permissive for transcription.
In stem cells, the epigenetic control of gene expression plays an essential role in regulating stem cell maintenance by repressing expression of differentiation genes while allowing cell-cycle progression and cell renewal (Goldberg et al. 2007; Spivakov and Fisher 2007). On differentiation, the epigenetic machinery represses stemness genes, but promotes expression of cell-commitment genes. Although it has been shown that the chromatin state of a cell can be reverted (as in iPS reprogramming) (Papp and Plath 2013), under physiological conditions the epigenome sets a genetic memory that is preserved through cell division. The epigenetic control of stem cells is present through all life stages, maintaining the equilibrium between self-renewal and differentiation to regulate tissue homeostasis. It is thus not surprising that genetic aberrations in components of epigenetic machinery are observed in many human diseases, including cancer (Widschwendter 2007; Chi et al. 2010).
Recent studies in skin have revealed a critical role for the chromatin regulators in control of development and homeostasis. In this review, we will describe in detail the mechanisms of DNA methylation, histone modifications, and nuclear topology in skin control. Most of our insight into the role the different chromatin remodelers play in the skin comes from genetic studies performed in vivo with murine models or in vitro with human and murine keratinocytes; these studies will be discussed throughout the review and are summarized in Table 1. We will also discuss the role of different activating and repressing epigenetic marks during epidermal stem cell differentiation, focusing on the orchestration of skin stem cell control and the diseases that can result from epigenetic abnormalities.
Table 1.
Summary of the effects of removal of the different epigenetic regulators from the epidermis
| Protein | Function | Mutant/type of inactivation | Phenotype | References |
|---|---|---|---|---|
| DNA methylation | ||||
| DNA methyltransferases (DNMTs) | DNA methylation | DNMT1 cKO in mouse epidermis | Loss of the methylation-dependent repression of epidermal differentiation genes leads to differentiation of basal cells, uneven thickened epidermis and sebaceous gland hyperplasia. Up-regulation of cell-cycle inhibitor p16INK4A results in loss of proliferation, progressive aging, and increased apoptosis in the hair follicle stem cells that leads to progressive alopecia. | Li et al. 2012 |
| KD of DNMT1 in human keratinocytes xeonografted in immune-deficient mice | Premature differentiation and epidermal hypoplasia. Decreased proliferation potential and cell-cycle arrest because of up-regulation of the Ink4a/Arf/Ink4b locus and abnormal expression of differentiation genes leads to tissue loss. | Sen et al. 2010 | ||
| Chemical inhibition of DNMTs with 5-azacytidine in human keratinocytes | Differentiation and inhibition of growth, in part by affecting expression of genes of the epidermal differentiation complex (EDC). | Okada et al. 1984; Rosl et al. 1988; Elder and Zhao 2002 | ||
| UHRF1 | DNMT1 targeting by binding to hemimethylated DNA | In vitro KD of UHRF1 in human keratinocytes | UHRF1 depletion results in derepression of differentiation genes and reduced proliferation. | Sen et al. 2010; Mulder et al. 2012 |
| Gadd45A/B | DNA demethylation by interaction with DNA excision repair complexes | In vitro KD or overexpression of Gadd45A/B in human keratinocytes | Lack of Gadd45A or B impairs epidermal differentiation while their forced expression induces premature differentiation. | Sen et al. 2010 |
| Posttranslational histone modification | ||||
| Polycomb repressive complex2 (PRC2) | Trimethylation of lysine 27 on histone H3 (H3K27me3) | Ezh2 cKO in mouse embryonic epidermis | Loss of H3K27me3 results in up-regulation of epidermal differentiation genes (including many of genes of the EDC cluster) and precocious epidermal development. | Ezhkova et al. 2009 |
| Ezh1/2 2KO in mouse embryonic epidermis | Loss of Ezh1/2 from epidermal progenitors results in an increase in the number of differentiated Merkel cells, as a result of loss H3K27me3 repression of Sox2, a master regulator of Merkel cell differentiation. Loss of Ezh1/2 also results in decreased proliferation and subsequent degeneration of the hair follicles caused by activation of the Ink4a/Arf/Ink4b locus. | Ezhkova et al. 2011; Bardot et al. 2013; Lesko et al. 2013 | ||
| JMJD3 | Lysine-specific demethylase | In vitro KD and o/e of JMJD3 in human keratinocytes | Loss of JMJD3 demethylation activity arrests differentiation of human epidermal keratinocytes while expression of an active JMJD3 results in premature differentiation. | Sen et al. 2008 |
| Setd8 | Monomethylation of lysine 20 on histone H4 (H4K20mel) | Setd8 cKO in mouse embryonic and adult epidermis | Loss of Setd8-dependent histone methylation resulted in loss of proliferation and impaired differentiation, followed by loss of the interfollicular epidermis and sebaceous glands. | Driskell et al. 2012 |
| Histone deacetylases (HDACs) | Histone deacetylation | HDAC1/2 cKO in mouse embryonic epidermis | Removal of HDAC1/2 from the embryonic epidermis results in loss of stem cell proliferation, loss of stratification of the epidermis, lack of hair follicles, and eventual apoptosis in the basal layer because of increased expression of the p63-repressed genes p16INK4a, p21, and p53. | LeBoeuf et al. 2010 |
| Treatment of adult mouse skin with HDAC inhibitor trichostatin A | HDAC inhibition in the adult skin induces the hair follicle stem cells to proliferate and exit the stem cell compartment. | Frye et al. 2007 | ||
| In vitro treatment of human keratinocytes or epidermal explants with HDAC inhibitors | HDAC inhibition in human keratinocytes results in arrest of proliferation and premature expression of differentiation markers. | Saunders et al. 1999; Elder and Zhao 2002; Frye et al. 2007; Markova et al. 2007 | ||
| Chromatin remodeling | ||||
| SWI/SNF | Chromatin remodeling | Brg1 and Brm 2KO in mouse embryonic epidermis | Loss of Brg1 and Brm in the developing epidermis leads to aberrant expression of differentiation genes and results in defective formation of the skin barrier. | Indra et al. 2005 |
| ACTL6a cKO in mouse adult epidermis and KD in human keratinocytes | Loss of ACTL6a results in loss of stem cell proliferation and exit from the cell cycle as well as premature differentiation of the epidermis, in part caused by activation of KLF4 expression. | Bao et al. 2013 | ||
| Brg1 cKO in hair follicle stem cells | Loss of Brg1 in the bulge results in loss of the hair follicle stem cells over time, partly because of ectopic expression of the cell-cycle inhibitor p27. | Xiong et al. 2013 | ||
| Nuclear architecture remodeling | ||||
| Satb1 | Large-scale chromatin remodeler | Satb1 KO in mouse embryonic and postnatal epidermis | Removal of Satb1 from epidermis increases the volume and spacing of the EDC locus within the nucleus, accompanied by down-regulation of the EDC and thinner epidermis. | Fessing et al. 2011 |
DNA METHYLATION
Conversion of cytosine bases in DNA to 5-methyl-cytosine (5mC) is one of the best-characterized epigenetic modifications in vertebrates and is mostly associated with transcriptional repression (Reik 2007; Smith and Meissner 2013). This postreplication modification occurs predominantly in the context of the CpG dinucleotide, although in embryonic stem (ES) cells, 5mC has also been detected in the CpA and CpT contexts (Ramsahoye et al. 2000).
The presence of 5mC is generally believed to repress transcriptional activity by blocking the binding of transcription factors to DNA or by recruitment of methyl-DNA-binding proteins, such as MeCP2 or MDB1, that then recruit chromatin repressor complexes (Fig. 1) (Klose and Bird 2006). In mammalian genomes, up to 80% of the CpG sequences are methylated, and most of the unmethylated CpG are found in regions of high CpG content, known as CpG islands (Reik 2007), close to the promoters and first exons of actively transcribed genes. Although DNA methylation is generally associated with transcriptional repression, there are some examples in which 5mC is thought to mediate gene expression by recruitment of the mCpG-binding protein C/EBPα (CRE enhancer-binding protein alpha-isoform), which functions as a transcriptional activator (Oh et al. 2007; Rishi et al. 2010).
The Role of DNA Methylation in Skin Differentiation
During skin stem cell differentiation, DNA methylation changes occur specifically in regulatory regions of developmental genes. These changes consistently happen in the CpG island shores and occur in two directions: (1) gain of methylation-dependent repression of nonskin genes on cell commitment, and (2) demethylation of epidermal lineage-specific genes (Khavari et al. 2010; Bock et al. 2012).
There are three DNA methyltransferase enzymes that are responsible for the DNA methylation state in mammalian tissues (Smith and Meissner 2013). DNA methylation is maintained by DNMT1, which copies the pattern of methyl marks from the parent strand to the daughter strand after cell division (Fig. 1), and loss of the ability to maintain DNA methylation results in early embryonic lethality (Li et al. 1992). The de novo methylation of unmodified DNA is catalyzed by the DNMT3A and 3B methyltransferases (Fig. 1) (Okano et al. 1999; Bird 2002). In the skin, DNMT3A and 3B are expressed in the basal layer of the epidermis (Sen et al. 2010; Nandakumar et al. 2011) where they are thought to play an important role in establishing DNA methylation in nonepidermal genes during skin stem cell differentiation. Consistent with this, about 20% of the repressed genes are methylated de novo during epidermal differentiation (Sen et al. 2010). However, the exact role of DNMT3A and 3B in skin homeostasis and differentiation is still not fully understood.
DNMT1 is expressed in the hair follicle and in the basal layer of the epidermis, and its expression rapidly diminishes on differentiation (Sen et al. 2010; Li et al. 2012). Conditional ablation of DNMT1 from mouse epidermis results in sebaceous hyperplasia, thickened epidermis, and up-regulation of some differentiation markers (Li et al. 2012). This phenotype probably results from the aberrant differentiation of basal cells caused by the loss of the methylation-dependent repression of epidermal differentiation genes. The animals lacking DNMT1 in the epidermis also show signs of premature and progressive alopecia during aging as a result of reduced proliferation and increased apoptosis in the hair follicle stem cells (HFSCs; Li et al. 2012). This effect is attributed to lack of methylation-dependent repression (and the consequent up-regulation) of the cell-cycle inhibitor p16INK4A (Table 1).
Knockdown of DNMT1 in human epidermis generated from xenografted keratinocytes implanted onto immune-deficient mice results in premature differentiation and epidermal hypoplasia (Sen et al. 2010). The tissue also showed decreased proliferation and loss of cell renewal capabilities, accompanied by cell-cycle arrest because of up-regulation of the Ink4a/Arf/Ink4b locus (Table 1) (Sen et al. 2010). Concordantly, depletion of UHRF1, a protein that aids to direct DNMT1 to hemimethylated DNA and is expressed in undifferentiated basal cells, also resulted in up-regulation of differentiation genes and decreased proliferation (Sen et al. 2010; Mulder et al. 2012). Thus, the activity of DNMT1/UHRF1 mammalian skin stem cells seems to be fundamental to maintain the equilibrium between preventing differentiation by repressing differentiation genes and allowing stem cell proliferation by repressing genes that block cell-cycle progression (Sen et al. 2010; Mulder et al. 2012).
This complex DNA methylation dynamic is consistent with initial observations showing that exposure of human keratinocytes to 5-aza-cytidine (a nucleoside analog that inhibits DNMTs) results in differentiation and inhibition of growth (Table 1) (Okada et al. 1984; Rosl et al. 1988). The effects of this agent were particularly interesting at the epidermal differentiation complex (EDC), a 1.5-Mb cluster of genes involved in late epidermal differentiation that undergo coordinated expression during keratinocyte differentiation (Bazzi et al. 2007). Treatment with 5-aza-cytidine induced expression of SPRR1/2 and involucrin, but repressed the expression of S100A2 (Elder and Zhao 2002).
In this regard, it has been shown that in keratinocytes, the transcription factor C/EBPα has a higher affinity for promoters that contain a methylated cAMP repressor element (TGACGTCA) (Rishi et al. 2010). Expression of C/EBPα increases on differentiation and seems to be specific to the suprabasal layers, which suggests it might play a role in epidermal differentiation (Rishi et al. 2010). Further, the overexpression of C/EBPα in the skin leads to hyperplasia of the basal layer of the epidermis whereas its down-regulation in vitro results in inhibition of differentiation (Oh et al. 2007; Rishi et al. 2010). These phenotypes are compatible with the role of C/EBPα in inducing expression of methylated genes involved in skin differentiation. Remarkably, the methylation status of the C/EBPα targeted epidermal genes does not change during differentiation, and thus it is the gain in C/EBPα expression on differentiation that induces expression of the methylated promoters (Rishi et al. 2010). Taken together, this data shows that during epidermal differentiation DNA methylation can act in opposing ways to affect gene expression: repression via DNMTs or activation via C/EBPα recruitment.
Alternatively, some epidermal genes undergo active demethylation during the differentiation process (Sen et al. 2010). Among them are S100P and EphA2, a tyrosine kinase receptor important for skin terminal differentiation (Sen et al. 2010; Bock et al. 2012). Although our understanding of the demethylation mechanism operating in mammals is incomplete, it seems that in the skin, loss of 5mC is in part mediated by the activity of Gadd45, a protein capable of promoting demethylation via DNA repair (Barreto et al. 2007; Zhang et al. 2011). Gadd45 is up-regulated on skin stem cell differentiation, and its knockdown in skin stem cells prevents induction of differentiation genes in vitro (Sen et al. 2010). Accordingly, overexpression of Gadd45 in skin stem cells induces premature differentiation, probably as result of demethylation and activation of epidermal differentiation genes (Sen et al. 2010). Recently, it has been proposed that hydroxylation of 5mC mediated by ten-eleven translocation (TET) dioxygenases can lead to active demethylation of DNA, and this modification plays a fundamental role in tuning the balance between pluripotency and differentiation in ES cells (Ito et al. 2010). However, the role of this novel DNA modification and the TET proteins in epidermal differentiation are yet to be elucidated.
DNA Methylation Alterations in Skin Diseases
Alterations in DNA methylation have been reported in different human diseases including cancer (Bergman and Cedar 2013). In skin, DNA methylation alterations are common hallmarks of skin cancer. For example, hypermethylation of the tumor suppressor genes (TSGs) T-cadherin and E-cadherin is commonly observed in basal cell carcinoma (BCC) and squamous cell carcinoma (SCC). Generally, reduced expression of cadherin adhesion molecules aids in the invasiveness of the cancer cells, and thus reduced cadherin expression is directly associated with the aggressiveness of these kinds of skin cancers. Another group of TSGs that are commonly found hypermethylated in BCC are the cell-cycle inhibitors 14-3-3δ (Lodygin et al. 2003), p16INK4A, and p14ARF, whose down-regulation promotes the rapid cell growth and genomic instability typical of cancer cells.
Alterations in DNA methylation have also been reported in other skin diseases (Millington 2008). For example, in psoriasis, a chronic inflammatory skin disease, the hypermethylation and resulting down-regulation of p16INK4A seems to be more prevalent in severe lesions than in nonaffected skin areas (Zhang et al. 2007, 2010). In atopic dermatitis, another complex skin disease characterized by chronic and severe itching, the levels of DNMT1 seem to be lower when measured in blood of affected patients (Sehra et al. 2008) suggesting that DNA methylation levels may contribute to the pathogenesis of this disease.
The skin is the primary body barrier against environmental insults, and thus skin cells are especially susceptible to mutations induced by ultraviolet (UV) radiation. Consistent with this, exposure to UV light is the main etiological factor in skin cancer progression (Marks 1995). Interestingly, UV exposure increases the expression of DNMT1, 3a, and 3b in the epidermis (Nandakumar et al. 2011), and this up-regulation is even more dramatic in UV-light-induced mouse tumors or human BCC samples, in which it contributes to hypermethylation and silencing of p16INK4A and RASSF1A (Nandakumar et al. 2011; Brinkhuizen et al. 2012). This hypermethylation is accompanied by increased occupancy by MBD1, MeCP2, and histone deacetylases (see the Chromatin Modifications section), exemplifying the interplay between DNA methylation and histone modification enzymes in skin tumorigenesis. What role UV-light-induced silencing of TSG via DNA methylation plays physiologically is still an open question, but one likely hypothesis is that this mechanism is an effort by the skin to reconstitute the UV-damaged tissue by inducing epidermal proliferation.
Importantly, epigenetic misregulation could be treated by pharmacological approaches (e.g., the demethylating agent 5-aza-cytidine [Vidasa] could be used to treat BCC and SCC), with the expectation that demethylation of TSGs such as p16INK4A or RASSF1A could induce senescence of the cancer cells (Li et al. 2009; Brinkhuizen et al. 2012).
CHROMATIN MODIFICATIONS
Chromatin structure can be modified by the interplay of different molecular mechanisms including posttranscriptional modification of histones, ATP-dependent chromatin remodeling, or incorporation of histone variants (Fig. 2). These alterations in chromatin structure can act to activate or repress transcription by promoting an open chromatin conformation and recruitment of transcriptional components, or compacting the chromatin through recruitment of repressor complexes, respectively. In this section, we will discus the interaction of the different chromatin modifications during skin development and homeostasis.
Histone Posttranslational Modifications
Histones are subjected to a variety of posttranslational covalent modifications, such as acetylation, methylation, phosphorylation, ADP-ribosylation, ubiquitination, sumoylation, arginine deamination, and proline isomerization. These modifications occur mostly at the amino-terminal parts called histone tails, and both the type of modification and its location determine the effect on gene transcription (Fig. 2B). Posttranslational modifications alter histone–DNA and histone–histone interactions and, thus, regulate the accessibility of transcription factors and components of transcriptional machinery to the DNA (Allis et al. 2007; Clapier and Cairns 2009; Meissner 2010; Talbert and Henikoff 2010). The different histone modifications often act cooperatively and synergistically to either repress or activate transcription, and the specific combination of histone modifications will constitute the epigenetic state of the cell. This so-called “histone code” is akin to an epigenetic readout that determines the transcriptional state of a DNA region (Nessa et al. 2012).
In the epidermis, two well-characterized modifications are histone methylation and acetylation, which are dynamically regulated by methyltransferase/acetyltransferase enzymes that establish these modifications and demethylase/deacetylase enzymes that remove them (Wang et al. 2009). Histone methylation marks can have both activating and repressive roles depending on which residue of the histone tail is modified. Acetylation occurs on multiple lysine residues and normally positively affects gene expression (Fig. 2B). Histone acetylation is regulated by two families of enzymes: histone acetyl transferases (HATs), many of which are transcriptional coactivators, and histone deacetylases (HDACs), which are generally associated with gene repression (Allis et al. 2007; Wang et al. 2009; Suganuma and Workman 2011). As will be discussed in more detail below, the presence of distinct methylation and acetylation marks on histones are not entirely independent events, as the complexes that regulate the different enzymatic activities have been shown to interact with each other (van der Vlag and Otte 1999).
Regulation of the Repressive H3K27me3 Histone Mark
One of the best-studied repressive histone modifications is the trimethylation of lysine 27 on histone H3 (H3K27me3). Two chromatin modifiers with opposing activities regulate this mark and their mechanisms have been well described in skin (Botchkarev et al. 2012; Zhang et al. 2012). The polycomb repressive complex (PRC) acts to repress genes by methylating H3K27 through a mechanism involving two subcomplexes (PRC1/2), with the methyltransferase activity being catalyzed by the interchangeable Ezh1 or Ezh2 subunits (Fig. 2B) (Cao et al. 2002; Min et al. 2003; Margueron et al. 2008; Shen 2008). The counterparts of the PRC are JMJD3 and UTX, Jumanji family proteins, which are capable of removing methyl groups from H3K27 to release genes from polycomb repression (Agger et al. 2007; De Santa et al. 2007; Lan et al. 2007; Shaw and Martin 2009).
In the epidermis, the H3K27me3 histone mark is associated with numerous gene promoters, including those for late epidermal differentiation genes and multiple nonskin lineages (Ezhkova et al. 2009; Lien et al. 2011). Several polycomb subunit proteins (Ezh2, Eed, Bmi1, Cbx2, and Pcgf2) are highly expressed in mouse epidermal progenitors, and are down-regulated in differentiated suprabasal cells (Reinisch et al. 2007; Ezhkova et al. 2009). Functional studies have shown that Ezh2-mediated repression prevents binding of the transcription factor AP1 to its target genes, many of each encode for critical proteins required for epidermal barrier formation. Conditional ablation of Ezh2 from embryonic epidermal progenitors results in the selective up-regulation of epidermal differentiation genes, including many genes of the EDC cluster, and a resulting acceleration in epidermal barrier formation (Table 1) (Ezhkova et al. 2009). Interestingly, in vitro studies showed that human keratinocytes expressing a histone H3 K27-demethylase JMJD3 differentiate prematurely, whereras loss of JMJD3 leads to arrested differentiation (Table 1) (Sen et al. 2008). Further work will be needed to determine the mechanism of H3K27 demethylation in vivo in control of epidermal development and maintenance of epidermal stem cell state.
It has been shown that Ezh1, an Ezh2 paralog, is expressed in the epidermis and can compensate for loss of Ezh2 (Ezhkova et al. 2009). A new study of the embryonic epidermal progenitor cells shows that Ezh1 and Ezh2 also play a critical role in the differentiation program of the Merkel cells (Bardot et al. 2013), a specialized cell type of epithelial origin responsible for mechanotransduction of sensory stimuli (Haeberle and Lumpkin 2008; Maricich et al. 2009; Bardot et al. 2013). Recent work has proposed Sox2 as a master regulator of Merkel cell differentiation (Driskell et al. 2009; Bardot et al. 2013; Lesko et al. 2013), and loss of both Ezh1 and Ezh2 from epidermal progenitors leads to a dramatic increase in the number of fully differentiated Merkel cells, which is a direct result of loss H3K27me3 repression at Sox2 (Driskell et al. 2009; Bardot et al. 2013; Lesko et al. 2013). Together these studies show evidence for the role of polycomb-mediated repression in maintenance of epidermal progenitor cells by repressing both suprabasal and Merkel cell lineages. However, control of these two lineages differs mechanistically. During suprabasal layer differentiation, transcription factors including AP1 are present in the epidermal progenitors, but polycomb-mediated repression prevents their recruitment to target genes. On the other hand, for Merkel cell differentiation, the polycomb complex represses Sox2, the key transcription factor required for Merkel cell specification.
The H3K27me3 histone mark also plays important roles during skin homeostasis. In the absence of Ezh1 and Ezh2, the hair follicles suffer a decrease in proliferation and eventual degeneration (Table 1) (Ezhkova et al. 2011). The dissection of molecular mechanisms has shown that loss of Ezh1/2-triggered activation of the Ink4a/Arf/Ink4b locus, resulting in inability of HFSCs to proliferate and leading to loss of the hair follicles.
Although the role of the polycomb complex in skin development and homeostasis is well studied, its role in the skin during stresses such as wound healing and age-related decline are poorly understood. The activation of the Ink4a/Arf/Ink4b locus is a hallmark of aging (Chen et al. 2009), and indeed expression of PRC subunits is often reduced in aged skin (Ressler et al. 2006; Cordisco et al. 2010). However, the specifics of how this activation leads to the familiar phenotypes associated with aging are unclear. On wound healing, it is known that PRC subunits are down-regulated as JMJD3 is simultaneously up-regulated during mouse skin repair (Shaw and Martin 2009). This results in a drastic loss of H3K27me3 at the wound site as the genes needed for repair are rapidly induced.
Many PRC subunits have been tied to the onset and progression of cancers from tissues including lung, prostate, breast, and skin (Kleer et al. 2003; Cha et al. 2005; Sudo et al. 2005; Matsukawa et al. 2006; Simon and Lange 2008; Karanikolas et al. 2009; Suva et al. 2009; Balasubramanian et al. 2010; Cao et al. 2011; Eckert et al. 2011). Although skin cancers with elevated levels of PRC subunits are well known, the specific mechanisms for how H3K27me3 controls skin tumorigenesis are unclear. The two main theories are related to the repressive action of H3K27me3. The first theory is that overexpression of PRC subunits could keep tumor-initiating cells in a highly proliferative state causing increased growth potential. The second hypothesizes that PRC expression can cause aberrant repression of tumor suppressors, which would normally act to prevent cancer formation. Knowing which genes are specifically and incorrectly targeted by H3K27me3 in skin cancers will help to determine what targets may be amenable for clinical treatments.
Dynamics in Histone Modifications on Stem Cell Activation and Differentiation
Recent studies have found that histone modifications are very dynamic and that changes in stem cell state correlate with changes in epigenetic modifications (Cui et al. 2009; Wang et al. 2009; Fisher and Fisher 2011). Actively transcribed genes are characterized by multiple histone modifications, the most clearly identified being trimethylation of histone 3 at lysine 4 (Fig. 2B, H3K4me3), which marks the transcription initiation site and is associated with transcriptional initiation. Di- or trimethylation of lysine 79 (H3K79me2/3) is also present at active genes, but is located at the 3′ region of the transcribed gene and is associated with transcriptional elongation (Wang et al. 2009). Analysis of the global distribution of histone modifications has shown that some gene promoters are simultaneously enriched in both active (H3K4me3) and repressive (H3K27me3) marks, which is thought to put genes in a “poised-to-go” state (Cui et al. 2009; Wang et al. 2009; Fisher and Fisher 2011). Bivalent promoters are very abundant in ES cells and have also been identified in other stem cell lineages, including a small number of genes in the HFSCs (Lien et al. 2011).
The hair follicle has a population of quiescent HFSCs that become active during the growth phase of the hair cycle and proliferate, giving rise to transit amplifying (TA) cells in the hair follicle (Blanpain and Fuchs 2009). Histone methylation profiling of these distinct populations of cells found that in the quiescent HFSCs, genes associated with the stem cell fate contain the active H3K4me3 and H3K79me2 marks, whereas genes associated with the TA fate only have the H3K27me3 repressive mark (Lien et al. 2011). In TA cells, the opposite pattern was observed, with repressive H3K27me3 marks present on the stem cell genes and activating H3K4me3 and H3K79me2 marks present on genes required for the TA fate (Lien et al. 2011). When the quiescent HFSCs become active, they start acquiring repressive H3K27me3 repressive marks on genes required for the stem cell fate, placing these genes in a bivalent state (Lien et al. 2011). The fact that only a small number of genes were bivalently modified in the HFSCs suggests that this state is less important in adult somatic stem cells than in ES cells, where it is critical to maintain a wider differentiation potential.
Transcription Activation by Setd8-Mediated Histone Methylation
Histone H4 monomethylation on lysine 20 (H4K20me1) is present at transcriptionally active genes and catalyzed by the histone methyltransferase Setd8 (Fig. 2B). Loss of Setd8 in the epidermis both during development and in adult skin resulted in loss of proliferation and impaired differentiation, accompanied by loss of the interfollicular epidermis and sebaceous glands (Table 1) (Driskell et al. 2012). Setd8 was shown to be a target of c-Myc and required for c-Myc-induced epidermal proliferation (Driskell et al. 2012). The loss of differentiation in Setd8-null skin is a result of the lack of H4K20me1-mediated activation of p63, a gene expressed in basal cells that is thought to function as a master regulator of the stratification of the developing epidermis (Blanpain and Fuchs 2007; Driskell et al. 2012). However, the failure of Setd8-null skin to proliferate is mostly because of increased expression of p53, which results in increased apoptosis in the basal layer of the epidermis (Driskell et al. 2012). These phenotypes illustrate the fine epigenetic control on both positive and negative regulators of differentiation and cell-cycle progression such as p63 and p53 by H4K20me1, and thus it is not surprising that global changes in H4K20me1 are a frequent hallmark of cancer (Fraga et al. 2005). It is therefore important to better understand the role that Setd8 plays in both tissue homeostasis and transformation in the skin.
The Role of Histone Acetylation in Epidermal Differentiation
Another well-characterized posttranslational modification on histones is acetylation (Fig. 2B). It has been shown that actively transcribed regions of DNA typically contain acetylated histones within the nucleosomes. By removing these acetyl marks from histones, HDACs promote chromatin compaction and repress transcription. Two members of the HDAC family, HDAC1 and HDAC2, have been shown to be associated with other transcriptional corepressors and can act with PRC2 to mediate gene repression (van der Vlag and Otte 1999). During development, removal of HDAC1 and HDAC2 from the basal cells of the epidermis results in loss of proliferation of the skin stem cells together with loss of stratification of the epidermis, lack of hair follicle formation, and eventual apoptosis in the basal layer (Table 1) (LeBoeuf et al. 2010).
This phenotype is very similar to the phenotype of the removal of p63 from the skin (Mills et al. 1999; Yang et al. 1999). Indeed, HDAC1/2 bind to the same promoter regions of the genes repressed by p63, and loss of HDAC1/2 results in increased expression of p63-repressed genes while not affecting the p63-activated genes (LeBoeuf et al. 2010). One of the truncated forms of p63, ΔN-p63α, functions as a survival factor in epidermal cells. Not surprisingly, ΔN-p63α has been found to have enhanced expression in 80% of SCCs, and was shown to function synergistically with HDAC1/2 to inhibit apoptosis (Rodrigues et al. 2012). Importantly, pharmaceutical inhibitors of HDACs also induced growth arrest in SCC cell lines, suggesting that HDACs are potential pharmaceutical targets for the treatment of skin cancers (Saunders et al. 1999).
Of the genes with increased expression in the HDAC1/2-null skin, the cell-cycle inhibitors p16INK4a and p21 are among the most dramatically up-regulated, consistent with other phenotypes showing epidermal proliferation and hair follicle abnormalities (LeBoeuf et al. 2010). HDAC1/2 are also required to prevent hyperacetylation of p53 in the developing epidermis. This mechanism is important to allow normal amplification of skin stem cells and, in fact, the increase in apoptosis observed in HDAC1/2-null epidermis is a result of increased p53 expression (LeBoeuf et al. 2010).
As mentioned previously, loss of either HDAC1/2 or Ezh2 in the developing epidermis results in increased p16INK4A expression (Ezhkova et al. 2009), consistent with the idea that HDAC1/2 interacts with PRC2 (van der Vlag and Otte 1999). However, Ezh2-null epidermis does not show a dramatic phenotype, possibly because of compensation by Ezh1 or the persistence of acetylation marks in the genome. As interaction between HDAC1/2 and PRC2 has not yet been characterized in the epidermis, it will be important to further understand whether these critical epigenetic regulators interact in keratinocytes and how.
In the adult skin, quiescent stem cells present in both the interfollicular epidermis (IFE) and the hair follicle bulge were found to have high levels of hypoacetylated histone H4, whereas Myc-induced exit from quiescence and proliferation resulted in increase acetylation of histone H4 (Frye et al. 2007), suggesting that histone acetylation might promote proliferation of quiescent epidermal stem cells. Consistent with this observation, treatment of adult skin with the HDAC inhibitor trichostatin A (TSA) induces the HFSCs to proliferate and exit the stem cell compartment (Table 1) (Frye et al. 2007). Other studies showed that, similarly to the described role for histone acetylation during embryonic development, in vitro treatment of human keratinocytes or epidermal explants with HDAC inhibitors, such as butyrate and TSA, results in arrest in proliferation by promoting Cdk1 expression, as well as premature expression of differentiation markers (Table 1) (Saunders et al. 1999; Elder and Zhao 2002; Frye et al. 2007; Markova et al. 2007).
Histone Variants
Exciting new fields of epigenetic regulation are also emerging. Important work from the last decade has found that the substitution of canonical histones by variants can change the local or global structure of chromatin, regulating the accessibility of DNA (Fig. 2D) (Talbert and Henikoff 2010). Recent studies have shown that a variant of the histone H2A, H2A.Z, promotes nucleosome dissociation, transcription factor binding, and efficient RNA polymerase II recruitment to DNA, and is required for differentiation of ES cells (Creyghton et al. 2008; Talbert and Henikoff 2010; Li et al. 2012). Although it is not known whether H2A.Z plays a role in the skin stem cells and differentiation, it will be important to investigate whether its functions are similar to those observed in other systems.
CHROMATIN REMODELING BY SWI/SNF
In addition to the covalent modifications mentioned above, several chromatin remodeling complexes have been found to play a role in regulating DNA accessibility through rearrangement of nucleosomes (Fig. 2C). Chromatin remodeling complexes are multisubunit complexes containing SWI2/SNF2-like ATPases that hydrolize ATP to alter the interaction between histones and DNA within nucleosomes, leading to nucleosome sliding, histone eviction, or the exchange of histone variants (Clapier and Cairns 2009). These alterations to nucleosome structure function to destabilize histone–DNA interactions, resulting in an open chromatin state and transcriptional activation. The SWI/SNF family, also known as the BRG1/BRM-associated factor (BAF) complex, is one of the best-characterized chromatin remodeling complexes. It consists of the catalytic ATPase subunits Brg1 or Brm as well as eight to 14 regulatory subunits (Clapier and Cairns 2009). These subunits can be encoded by at least 20 different genes, resulting in 288 possible combinatorial assemblies, with these different complexes having potentially different DNA target affinities and functions (Clapier and Cairns 2009; Wu et al. 2009). Interaction between the components of SWI/SNF and a plethora of regulators has been characterized, including interaction with master regulators of differentiation (AP1), cell-cycle progression (c-MYC, Rb), nuclear hormone receptors (β-catenin), and malignant transformation (BRCA1, p53) (de la Serna et al. 2006; Clapier and Cairns 2009; Wu et al. 2009).
In the epidermis, the activity of SWI/SNF complexes is essential for proper execution of the differentiation program. Loss of expression of Brg1 and Brm in the epidermis does not considerably affect proliferation, but does lead to aberrant expression of differentiation genes and results in defective formation of the skin barrier (Table 1) (Indra et al. 2005). This is consistent with the known role of SWI/SNF in mediating exit from cell cycle in combination with Rb and HDAC (Zhang et al. 2000). In spite of Brg1 being expressed in all layers of the epidermis, SWI/SNF activity is restricted to suprabasal differentiation layers. This is because of differential expression of the SWI/SNF subunit actin-like 6a (ACTL6a/BAF53a) that binds the SWI/SNF complex and prevents it from binding to its target genes (Bao et al. 2013). Loss of ACTL6a results in loss of stem cell proliferation and exit from the cell cycle as well as premature differentiation of the epidermis, in part owing to activation of KLF4 expression (Table 1) (Bao et al. 2013), a critical regulator of epidermal differentiation (Segre et al. 1999). Studies in other systems have shown that ACTL6a is also required in neuronal stem cells and hematopoietic stem cells to maintain stem cell proliferation (Lessard et al. 2007; Krasteva et al. 2012), suggesting that ACTL6a might be a general regulator of stem cell proliferation and commitment (Perdigoto et al. 2013). It is not yet known how ACTL6a is down-regulated in the epidermis, however, microRNAs are required for the repression of ACTL6a in neuronal stem cells (Yoo et al. 2009) and a similar mechanism might be taking place in the skin.
A recent study has identified a novel role for Brg1 in regulating proliferation of the HFSCs during the hair cycle (Xiong et al. 2013). Brg1 expression increases in activated HFSCs as they start to proliferate, and its activity is required in the bulge for HFSC maintenance and proliferation by repressing the expression of the cell-cycle inhibitor p27 (Table 1) (Xiong et al. 2013). Interestingly, in TF cells located in the matrix region of the hair follicles, Brg1 binds to the promoter of the Sonic hedgehog (Shh), a regulator of the hair cycle (Blanpain and Fuchs 2009) that will in turn lead to increased expression of Brg1 in the proliferating cells of the bulge and hair germ/hair follicle (Xiong et al. 2013). It will be valuable to investigate the subunit composition of the SWI/SNF active in the hair follicle and whether the difference in subunits can explain why SWI/SNF has played such different roles in the hair follicle and the interfollicular epidermis.
Understanding how SWI/SNF activity is being regulated in the skin will be critical because SWI/SNF components are frequently mutated or down-regulated in many human cancers, and mutations in different SWI/SNF subunits confer proliferative advantages in several tumor cell lines (Wu et al. 2009). In human skin cancers, both Brg1 and Brm are shown to be down-regulated in SCCs and BCCs (Moloney et al. 2009; Bock et al. 2011). The regulatory subunits of the SWI/SNF complex are thus potential therapeutic targets because targeting them is likely to be more tissue-specific and less likely to affect the global function of SWI/SNF.
There are three other SWI2/SNF2-like subfamilies that use different ATPases in chromatin remodeling (ISWI, CHD, and INO80), and they all use ATP hydrolysis to change the packaging state of chromatin by moving, ejecting, or restructuring nucleosomes. Studies in human keratinocytes have implicated the ISWI-containing NURF complex and the CHD-containing NuRD complex in maintaining stem cells in an undifferentiated state (Mulder et al. 2012). Although there are no other described functions for these other complexes in differentiation of the skin, they are essential, along with SWI/SNF, for DNA repair. Specifically, they are required to expose DNA bases to the repair machinery as well as to repair double-stranded breaks through recombination; this process is especially critical in the skin, which is particularly susceptible to environmental DNA damaging agents (Clapier and Cairns 2009; Bao 2011). The future function of SWI/SNF complex in control of DNA repair and skin carcinogenesis will require further investigation.
NUCLEAR DYNAMICS DURING EPIDERMAL DIFFERENTIATION
During the process of epidermal differentiation, the basal cells of the epidermis asymmetrically divide to generate the spinous layer that then terminally differentiates to the granular and cornified layers that provide the impermeable barrier of the skin. During this process, the nucleolus transitions from a state of active transcription in the basal cells to a fully inactive state in the stratum corneum (Botchkarev et al. 2012). Remarkable work performed in vivo has shown that during this process, the size of the nucleus decreases and changes in orientation from perpendicular to parallel with respect to the basal layer (Gdula et al. 2013). These alterations in nuclear shape are accompanied by an increase in pericentromeric heterochromatin clusters (regions of highly compacted, silent chromatin) and a reduction in nucleoli that are actively repositioned to the center of the nucleus. The nucleoli are sites of ribosomal RNA maturation and ribosome production (Sirri et al. 2008), and thus the decrease in the number of nucleoli reflects the reduced transcriptional levels observed on differentiation.
Although the majority of gene transcription is down-regulated during epidermal differentiation, up-regulation of the EDC cluster is required to undergo terminal differentiation. During this process, the EDC remains associated with active nucleoli and is localized to the nuclear periphery where active genes are located (Gdula et al. 2013). Accordingly, in cells in which the EDC in not expressed, such as hematopoietic stem cells, the cluster is localized to internal, silenced nuclear domains (Williams et al. 2002). Strikingly, the proper expression of the EDC depends on Satb1, a large-scale chromatin remodeler (Fessing et al. 2011) expressed exclusively in undifferentiated basal cells that directly binds the EDC, keratin type I/II loci, and keratin-associated protein locus to allow the establishment of large domains of active chromatin (Gdula et al. 2013). Surprisingly, absence of Satb1 in the epidermis increases the volume and spacing of the EDC within the nucleus, but was accompanied by down-regulation of the EDC and thinner epidermis (Table 1) (Fessing et al. 2011). This seemingly conflicting data indicates that Satb1 is required for higher-order DNA organization within the nucleus, and that this organization is important for proper EDC expression. This phenotype resembles p63-null skin, and, in fact, Satb1 is a downstream target of p63, proving that this genome organizer is part of the p63 regulatory network.
Thus, during epidermal differentiation, changes to both epigenetic modifications and nuclear organization at the DNA and chromatin levels work to silence nonepidermal genes and maintain accessibility of the EDC and other epidermal differentiation genes. Further understanding the precise mechanisms controlling this process, as well as characterizing the nuclear dynamics at play, will be fundamental to approaching skin diseases.
Chromatin architecture and nuclear dynamics should be further explored, as it is becoming increasingly clear that these processes play a critical role in tissue regulation. The nuclear localization of stemness and differentiation genes needs to be correlated with stem cell state. Additionally, inter- and intrachromatin interactions of key skin regulatory genes need to be determined as they might highlight an additional level of control of gene expression and skin regulation.
CONCLUSIONS
In this review, we summarized the most relevant data for understanding how epigenetic modulators regulate stem cell biology and differentiation in the skin (Fig. 3), including implications for human skin diseases. Yet, although there is evidence to suggest a link between multiple skin pathologies and epigenetic aberrations, these correlations are mostly understudied and many questions remain. In this final section, we will address some of the most promising areas for future research.
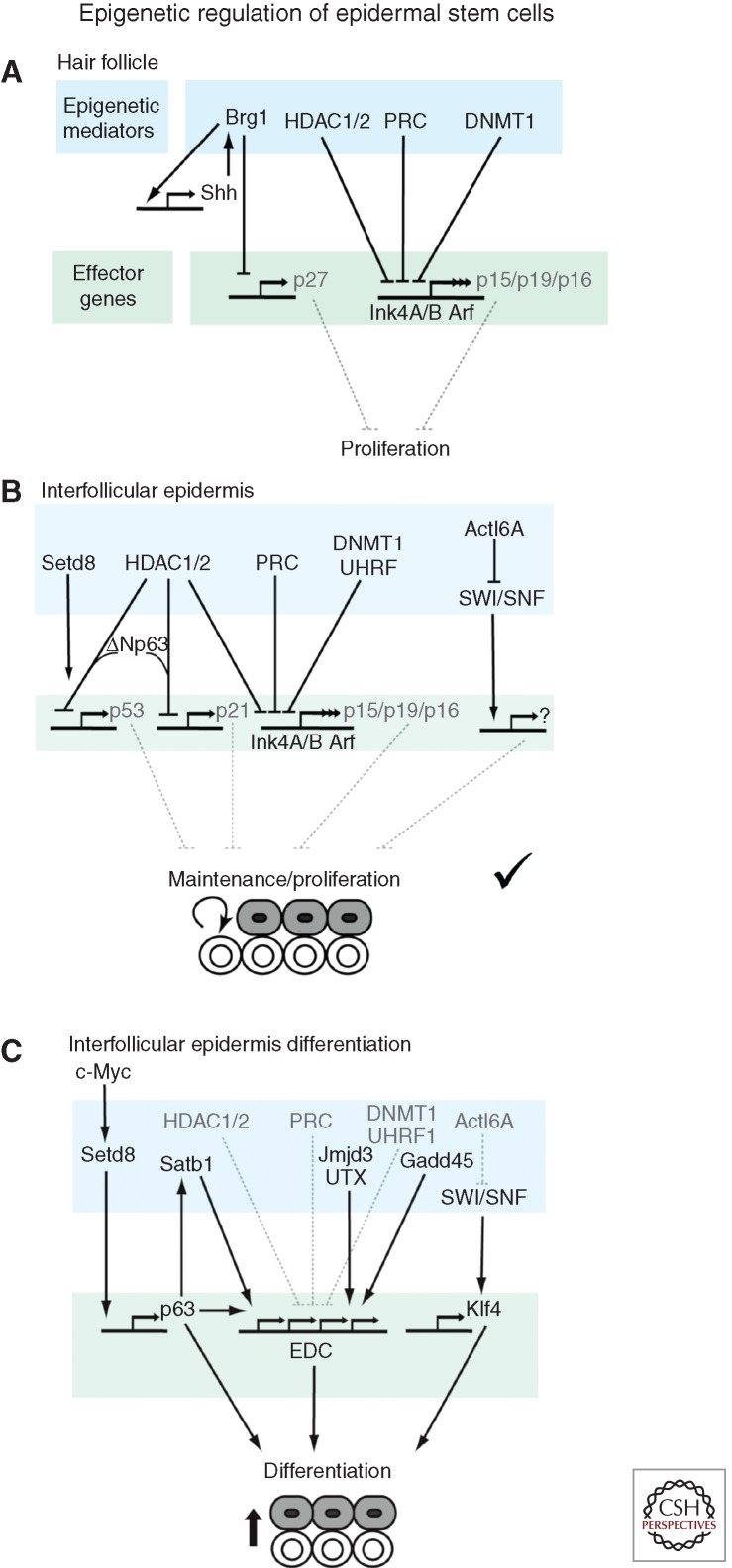
Figure 3.
Effects of epigenetic regulation on skin stem cells. Different epigenetic mediators operate to repress multiple effector genes to maintain stem cell proliferation and differentiation in (A) the hair follicle, and (B,C) the interfollicular epidermis. See main text for details.
The majority of chromatin regulatory complexes consist of multiple proteins with a few subunits possessing catalytic activity and others performing structural roles. Although most functional studies are focused on the analysis of the catalytic proteins in tissue control, the roles of other subunits of the complexes are unclear. More importantly, as some catalytic subunits are shared between different complexes and also have been shown to perform nontranscription or complex-independent functions, the conclusions from these types of studies have to be drawn very carefully (de Ruijter et al. 2003; Kouzarides 2007; Clapier and Cairns 2009; Sauvageau and Sauvageau 2010). It is possible that the catalytic subunits act in a context-dependent manner with different subunit combinations influencing functionality or conferring DNA-binding specificity. Thus, functional studies need to be expanded to the analysis of multiple subunits of chromatin complexes before their value can be fully realized.
Regulation of gene expression is a complex process and is controlled by the orchestrated action of multiple chromatin regulatory complexes. In vitro studies in human keratinocytes have indicated that stem cell maintenance and differentiation is regulated cooperatively by multiple epigenetic regulators that modify DNA methylation, histone acetylation and methylation states, and chromatin remodelers that work together in regulatory networks (Mulder et al. 2012). More work needs to be performed to understand how chromatin regulators interact with each other to regulate tissue homeostasis and determine the key regulatory genes altered in skin diseases. It will also be important to determine whether the function of chromatin regulators in skin control changes during stress conditions, such as wound healing and UV irradiation, as the skin is subjected to these assaults daily.
From data discussed herein, it appears that epigenetic regulators control either proliferation or differentiation in skin cells (Fig. 3). Although it is shown that these complexes target genes of multiple developmental lineages (Ezhkova et al. 2009; Lien et al. 2011; Bock et al. 2012), functional studies revealed that only genes of skin lineages become affected on ablation of chromatin regulator function (Ezhkova et al. 2009; Li et al. 2012). This paradox raises questions into why other lineages are not affected, what other players are active in the process, and how multiple factors interact to maintain the skin lineage. It will be important to uncover how genes of other developmental lineages are regulated and how memory of tissue state is maintained.
Finally, mutations in genes encoding for chromatin regulators were identified in different types of human malignancies. Importantly, in some cases, a direct role for chromatin regulators in promoting tumorigenesis has been determined. As altered expression of chromatin regulator complex components have been reported in human skin cancers, it will be crucial to determine the functional significance of these alterations for tumorigenesis and uncover whether the function of the whole complex is affected when one subunit is affected. Further, determination of downstream target genes that might affect cancer might help to devise better strategies for cancer treatment. This is especially important as single molecule inhibitors of HDAC and DNA methylation enzymes are currently in clinical trials, and thus can be easily transitioned to treat skin malignancies (Li et al. 2009; Brinkhuizen et al. 2012; Robert and Rassool 2012). We expect that in the near future further mechanistic insights will fuel both basic and clinically relevant advances in understanding the role of chromatin regulators in skin control.
Footnotes
Editors: Anthony E. Oro and Fiona M. Watt
Additional Perspectives on The Skin and Its Diseases available at www.perspectivesinmedicine.org
REFERENCES
- Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K 2007. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449: 731–734 [DOI] [PubMed] [Google Scholar]
- Allis C, Berger S, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, et al. 2007. New nomenclature for chromatin-modifying enzymes. Cell 131: 633–636 [DOI] [PubMed] [Google Scholar]
- Andrews AJ, Luger K 2011. Nucleosome structure(s) and stability: Variations on a theme. Annu Rev Biophys 40: 99–117 [DOI] [PubMed] [Google Scholar]
- Balasubramanian S, Adhikary G, Eckert RL 2010. The Bmi-1 polycomb protein antagonizes the (–)-epigallocatechin-3-gallate-dependent suppression of skin cancer cell survival. Carcinogenesis 31: 496–503 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Bao Y 2011. Chromatin response to DNA double-strand break damage. Epigenomics 3: 307–321 [DOI] [PubMed] [Google Scholar]
- Bao X, Tang J, Lopez-Pajares V, Tao S, Qu K, Crabtree G, Khavari P 2013. ACTL6a enforces the epidermal progenitor state by suppressing SWI/SNF-dependent induction of KLF4. Cell Stem Cell 12: 193–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardot ES, Valdes VJ, Zhang J, Perdigoto CN, Nicolis S, Hearn SA, Silva JM, Ezhkova E 2013. Polycomb subunits Ezh1 and Ezh2 regulate the Merkel cell differentiation program in skin stem cells. EMBO J 32: 1990–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreto G, Schafer A, Marhold J, Stach D, Swaminathan SK, Handa V, Doderlein G, Maltry N, Wu W, Lyko F, et al. 2007. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature 445: 671–675 [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh T-Y, Schones D, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Bazzi H, Fantauzzo KA, Richardson GD, Jahoda CA, Christiano AM 2007. Transcriptional profiling of developing mouse epidermis reveals novel patterns of coordinated gene expression. Dev Dyn 236: 961–970 [DOI] [PubMed] [Google Scholar]
- Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A 2009. An operational definition of epigenetics. Genes Dev 23: 781–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman Y, Cedar H 2013. DNA methylation dynamics in health and disease. Nat Struct Mol Biol 20: 274–281 [DOI] [PubMed] [Google Scholar]
- Bickmore WA, van Steensel B 2013. Genome architecture: Domain organization of interphase chromosomes. Cell 152: 1270–1284 [DOI] [PubMed] [Google Scholar]
- Bird A 2002. DNA methylation patterns and epigenetic memory. Genes Dev 16: 6–21 [DOI] [PubMed] [Google Scholar]
- Blanpain C, Fuchs E 2007. p63: Revving up epithelial stem-cell potential. Nat Cell Biol 9: 731–733 [DOI] [PubMed] [Google Scholar]
- Blanpain C, Fuchs E 2009. Epidermal homeostasis: A balancing act of stem cells in the skin. Nat Rev Mol Cell Biol 10: 207–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock V, Lyons J, Huang X, Jones A, McDonald L, Scolyer R, Moloney F, Barnetson R, Halliday G 2011. BRM and BRG1 subunits of the SWI/SNF chromatin remodelling complex are downregulated upon progression of benign skin lesions into invasive tumours. Br J Dermatol 164: 1221–1227 [DOI] [PubMed] [Google Scholar]
- Bock C, Beerman I, Lien WH, Smith ZD, Gu H, Boyle P, Gnirke A, Fuchs E, Rossi DJ, Meissner A 2012. DNA methylation dynamics during in vivo differentiation of blood and skin stem cells. Mol Cell 47: 633–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchkarev V, Gdula M, Mardaryev A, Sharov A, Fessing M 2012. Epigenetic regulation of gene expression in keratinocytes. J Investigat Dermatol 132: 2505–2521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkhuizen T, van den Hurk K, Winnepenninckx VJ, de Hoon JP, van Marion AM, Veeck J, van Engeland M, van Steensel MA 2012. Epigenetic changes in basal cell carcinoma affect SHH and WNT signaling components. PLoS ONE 7: e51710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y 2002. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298: 1039–1043 [DOI] [PubMed] [Google Scholar]
- Cao Q, Mani RS, Ateeq B, Dhanasekaran SM, Asangani IA, Prensner JR, Kim JH, Brenner JC, Jing X, Cao X, et al. 2011. Coordinated regulation of polycomb group complexes through microRNAs in cancer. Cancer Cell 20: 187–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha TL, Zhou BP, Xia W, Wu Y, Yang CC, Chen CT, Ping B, Otte AP, Hung MC 2005. Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science 310: 306–310 [DOI] [PubMed] [Google Scholar]
- Chen H, Gu X, Su IH, Bottino R, Contreras JL, Tarakhovsky A, Kim SK 2009. Polycomb protein Ezh2 regulates pancreatic β-cell Ink4a/Arf expression and regeneration in diabetes mellitus. Genes Dev 23: 975–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi P, Allis CD, Wang GG 2010. Covalent histone modifications—Miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer 10: 457–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier C, Cairns B 2009. The biology of chromatin remodeling complexes. Ann Rev Biochem 78: 273–304 [DOI] [PubMed] [Google Scholar]
- Cordisco S, Maurelli R, Bondanza S, Stefanini M, Zambruno G, Guerra L, Dellambra E 2010. Bmi-1 reduction plays a key role in physiological and premature aging of primary human keratinocytes. J Invest Dermatol 130: 1048–1062 [DOI] [PubMed] [Google Scholar]
- Creyghton M, Markoulaki S, Levine S, Hanna J, Lodato M, Sha K, Young R, Jaenisch R, Boyer L 2008. H2AZ is enriched at polycomb complex target genes in ES cells and is necessary for lineage commitment. Cell 135: 649–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui K, Zang C, Roh T-Y, Schones D, Childs R, Peng W, Zhao K 2009. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell 4: 80–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Serna IL, Ohkawa Y, Imbalzano AN 2006. Chromatin remodelling in mammalian differentiation: Lessons from ATP-dependent remodellers. Nature Rev Genet 7: 461–473 [DOI] [PubMed] [Google Scholar]
- de Ruijter A, van Gennip A, Caron H, Kemp S, van Kuilenburg A 2003. Histone deacetylases (HDACs): Characterization of the classical HDAC family. Biochem J 370: 737–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G 2007. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell 130: 1083–1094 [DOI] [PubMed] [Google Scholar]
- Driskell RR, Giangreco A, Jensen KB, Mulder KW, Watt FM 2009. Sox2-positive dermal papilla cells specify hair follicle type in mammalian epidermis. Development 136: 2815–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driskell I, Oda H, Blanco S, Nascimento E, Humphreys P, Frye M 2012. The histone methyltransferase Setd8 acts in concert with c-Myc and is required to maintain skin. Embo J 31: 616–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert RL, Adhikary G, Rorke EA, Chew YC, Balasubramanian S 2011. Polycomb group proteins are key regulators of keratinocyte function. J Invest Dermatol 131: 295–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elder J, Zhao X 2002. Evidence for local control of gene expression in the epidermal differentiation complex. Exp Dermatol 11: 406–412 [DOI] [PubMed] [Google Scholar]
- Ezhkova E, Pasolli HA, Parker JS, Stokes N, Su IH, Hannon G, Tarakhovsky A, Fuchs E 2009. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell 136: 1122–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E, Lien WH, Stokes N, Pasolli HA, Silva JM, Fuchs E 2011. EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair. Genes Dev 25: 485–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fessing MY, Mardaryev AN, Gdula MR, Sharov AA, Sharova TY, Rapisarda V, Gordon KB, Smorodchenko AD, Poterlowicz K, Ferone G, et al. 2011. p63 regulates Satb1 to control tissue-specific chromatin remodeling during development of the epidermis. J Cell Biol 194: 825–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher C, Fisher A 2011. Chromatin states in pluripotent, differentiated, and reprogrammed cells. Curr Opin Genet Dev 21: 140–146 [DOI] [PubMed] [Google Scholar]
- Fraga M, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, Bonaldi T, Haydon C, Ropero S, Petrie K, et al. 2005. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet 37: 391–400 [DOI] [PubMed] [Google Scholar]
- Frye M, Fisher AG, Watt FM 2007. Epidermal stem cells are defined by global histone modifications that are altered by Myc-induced differentiation. PLoS ONE 2: e763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gdula MR, Poterlowicz K, Mardaryev AN, Sharov AA, Peng Y, Fessing MY, Botchkarev VA 2013. Remodeling of three-dimensional organization of the nucleus during terminal keratinocyte differentiation in the epidermis. J Invest Dermatol 133: 2191–2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AD, Allis CD, Bernstein E 2007. Epigenetics: A landscape takes shape. Cell 128: 635–638 [DOI] [PubMed] [Google Scholar]
- Haeberle H, Lumpkin EA 2008. Merkel cells in somatosensation. Chemosens Percept 1: 110–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemberger M, Dean W, Reik W 2009. Epigenetic dynamics of stem cells and cell lineage commitment: Digging Waddington’s canal. Nat Rev Mol Cell Biol 10: 526–537 [DOI] [PubMed] [Google Scholar]
- Ho L, Crabtree GR 2010. Chromatin remodelling during development. Nature 463: 474–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indra AK, Dupé V, Bornert J-M, Messaddeq N, Yaniv M, Mark M, Chambon P, Metzger D 2005. Temporally controlled targeted somatic mutagenesis in embryonic surface ectoderm and fetal epidermal keratinocytes unveils two distinct developmental functions of BRG1 in limb morphogenesis and skin barrier formation. Development 132: 4533–4544 [DOI] [PubMed] [Google Scholar]
- Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y 2010. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466: 1129–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanikolas BD, Figueiredo ML, Wu L 2009. Polycomb group protein enhancer of zeste 2 is an oncogene that promotes the neoplastic transformation of a benign prostatic epithelial cell line. Mol Cancer Res 7: 1456–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khavari D, Sen G, Rinn J 2010. DNA methylation and epigenetic control of cellular differentiation. Cell Cycle 9: 3880–3883 [DOI] [PubMed] [Google Scholar]
- Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, et al. 2003. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci 100: 11606–11611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Bird AP 2006. Genomic DNA methylation: The mark and its mediators. Trends Biochem Sci 31: 89–97 [DOI] [PubMed] [Google Scholar]
- Kouzarides T 2007. Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Krasteva V, Buscarlet M, Diaz-Tellez A, Bernard MA, Crabtree GR, Lessard JA 2012. The BAF53a subunit of SWI/SNF-like BAF complexes is essential for hemopoietic stem cell function. Blood 120: 4720–4732 [DOI] [PubMed] [Google Scholar]
- Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S, Iwase S, Alpatov R, Issaeva I, Canaani E, et al. 2007. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 449: 689–694 [DOI] [PubMed] [Google Scholar]
- LeBoeuf M, Terrell A, Trivedi S, Sinha S, Epstein J, Olson E, Morrisey E, Millar S 2010. Hdac1 and Hdac2 act redundantly to control p63 and p53 functions in epidermal progenitor cells. Dev Cell 19: 807–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesko MH, Driskell RR, Kretzschmar K, Goldie SJ, Watt FM 2013. Sox2 modulates the function of two distinct cell lineages in mouse skin. Dev Biol 382: 15–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, Wu H, Aebersold R, Graef IA, Crabtree GR 2007. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron 55: 201–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69: 915–926 [DOI] [PubMed] [Google Scholar]
- Li Y, Sawalha AH, Lu Q 2009. Aberrant DNA methylation in skin diseases. J Dermatol Sci 54: 143–149 [DOI] [PubMed] [Google Scholar]
- Li Z, Gadue P, Chen K, Jiao Y, Tuteja G, Schug J, Li W, Kaestner K 2012. Foxa2 and H2A.Z mediate nucleosome depletion during embryonic stem cell differentiation. Cell 151: 1608–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien W-H, Guo X, Polak L, Lawton L, Young R, Zheng D, Fuchs E 2011. Genome-wide maps of histone modifications unwind in vivo chromatin states of the hair follicle lineage. Cell Stem Cell 9: 219–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodygin D, Yazdi AS, Sander CA, Herzinger T, Hermeking H 2003. Analysis of 14-3-3σ expression in hyperproliferative skin diseases reveals selective loss associated with CpG-methylation in basal cell carcinoma. Oncogene 22: 5519–5524 [DOI] [PubMed] [Google Scholar]
- Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL, Dynlacht BD, Reinberg D 2008. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell 32: 503–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maricich SM, Wellnitz SA, Nelson AM, Lesniak DR, Gerling GJ, Lumpkin EA, Zoghbi HY 2009. Merkel cells are essential for light-touch responses. Science 324: 1580–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markova N, Karaman-Jurukovska N, Pinkas-Sarafova A, Marekov L, Simon M 2007. Inhibition of histone deacetylation promotes abnormal epidermal differentiation and specifically suppresses the expression of the late differentiation marker profilaggrin. J Invest Dermatol 127: 1126–1139 [DOI] [PubMed] [Google Scholar]
- Marks R 1995. An overview of skin cancers. Incidence and causation. Cancer 75: 607–612 [DOI] [PubMed] [Google Scholar]
- Matsukawa Y, Semba S, Kato H, Ito A, Yanagihara K, Yokozaki H 2006. Expression of the enhancer of zeste homolog 2 is correlated with poor prognosis in human gastric cancer. Cancer Sci 97: 484–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A 2010. Epigenetic modifications in pluripotent and differentiated cells. Nat Biotechnol 28: 1079–1088 [DOI] [PubMed] [Google Scholar]
- Millington GW 2008. Epigenetics and dermatological disease. Pharmacogenomics 9: 1835–1850 [DOI] [PubMed] [Google Scholar]
- Mills A, Zheng B, Wang X, Vogel H, Roop D, Bradley A 1999. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 398: 708–713 [DOI] [PubMed] [Google Scholar]
- Min J, Zhang Y, Xu RM 2003. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev 17: 1823–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moloney F, Lyons J, Bock V, Huang X, Bugeja M, Halliday G 2009. Hotspot mutation of Brahma in non-melanoma skin cancer. J Investigat Dermat 129: 1012–1015 [DOI] [PubMed] [Google Scholar]
- Mulder K, Wang X, Escriu C, Ito Y, Schwarz R, Gillis J, Sirokmány G, Donati G, Uribe-Lewis S, Pavlidis P, et al. 2012. Diverse epigenetic strategies interact to control epidermal differentiation. Nat Cell Biol 14: 753–763 [DOI] [PubMed] [Google Scholar]
- Nandakumar V, Vaid M, Tollefsbol TO, Katiyar SK 2011. Aberrant DNA hypermethylation patterns lead to transcriptional silencing of tumor suppressor genes in UVB-exposed skin and UVB-induced skin tumors of mice. Carcinogenesis 32: 597–604 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Nessa A, Rashid MH, E-Ferdous N, Chowdhury A 2012. Screening for and management of high-grade cervical intraepithelial neoplasia in Bangladesh: A cross-sectional study comparing two protocols. J Obstet Gynaecol Res 39: 564–571 [DOI] [PubMed] [Google Scholar]
- Oh WJ, Rishi V, Orosz A, Gerdes MJ, Vinson C 2007. Inhibition of CCAAT/enhancer binding protein family DNA binding in mouse epidermis prevents and regresses papillomas. Cancer Res 67: 1867–1876 [DOI] [PubMed] [Google Scholar]
- Okada N, Steinberg ML, Defendi V 1984. Re-expression of differentiated properties in SV40-infected human epidermal keratinocytes induced by 5-azacytidine. Exp Cell Res 153: 198–207 [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99: 247–257 [DOI] [PubMed] [Google Scholar]
- Papp B, Plath K 2013. Epigenetics of reprogramming to induced pluripotency. Cell 152: 1324–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdigoto C, Bardot E, Ezhkova E 2013. SWItching on epidermal cell fate. Cell Stem Cell 12: 141–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsahoye BH, Biniszkiewicz D, Lyko F, Clark V, Bird AP, Jaenisch R 2000. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc Natl Acad Sci 97: 5237–5242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redi CA, Capanna E 2012. Genome size evolution: Sizing mammalian genomes. Cytogenet Genome Res 137: 97–112 [DOI] [PubMed] [Google Scholar]
- Reik W 2007. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 447: 425–432 [DOI] [PubMed] [Google Scholar]
- Reinisch CM, Uthman A, Erovic BM, Pammer J 2007. Expression of BMI-1 in normal skin and inflammatory and neoplastic skin lesions. J Cutan Pathol 34: 174–180 [DOI] [PubMed] [Google Scholar]
- Ressler S, Bartkova J, Niederegger H, Bartek J, Scharffetter-Kochanek K, Jansen-Durr P, Wlaschek M 2006. p16INK4A is a robust in vivo biomarker of cellular aging in human skin. Aging Cell 5: 379–389 [DOI] [PubMed] [Google Scholar]
- Rishi V, Bhattacharya P, Chatterjee R, Rozenberg J, Zhao J, Glass K, Fitzgerald P, Vinson C 2010. CpG methylation of half-CRE sequences creates C/EBPα binding sites that activate some tissue-specific genes. Proc Natl Acad Sci 107: 20311–20316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert C, Rassool F 2012. HDAC inhibitors: Roles of DNA damage and repair. Adv Cancer Res 116: 87–129 [DOI] [PubMed] [Google Scholar]
- Rodrigues SM, Oliveira AC, Vargas AM, Moreira AN, E Ferreira EF 2012. Implications of edentulism on quality of life among elderly. Int J Environ Res Public Health 9: 100–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosl F, Durst M, zur Hausen H 1988. Selective suppression of human papillomavirus transcription in non-tumorigenic cells by 5-azacytidine. EMBO J 7: 1321–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders N, Dicker A, Popa C, Jones S, Dahler A 1999. Histone deacetylase inhibitors as potential anti-skin cancer agents. Cancer Res 59: 399–404 [PubMed] [Google Scholar]
- Sauvageau M, Sauvageau G 2010. Polycomb group proteins: Multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell 7: 299–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segre JA, Bauer C, Fuchs E 1999. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat Genet 22: 356–360 [DOI] [PubMed] [Google Scholar]
- Sehra S, Tuana FM, Holbreich M, Mousdicas N, Tepper RS, Chang CH, Travers JB, Kaplan MH 2008. Scratching the surface: Towards understanding the pathogenesis of atopic dermatitis. Crit Rev Immunol 28: 15–43 [DOI] [PubMed] [Google Scholar]
- Sen GL, Webster DE, Barragan DI, Chang HY, Khavari PA 2008. Control of differentiation in a self-renewing mammalian tissue by the histone demethylase JMJD3. Genes Dev 22: 1865–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen GL, Reuter JA, Webster DE, Zhu L, Khavari PA 2010. DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature 463: 563–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw T, Martin P 2009. Epigenetic reprogramming during wound healing: Loss of polycomb-mediated silencing may enable upregulation of repair genes. EMBO Rep 10: 881–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X 2008. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell 32: 491–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JA, Lange CA 2008. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res 647: 21–29 [DOI] [PubMed] [Google Scholar]
- Sirri V, Urcuqui-Inchima S, Roussel P, Hernandez-Verdun D 2008. Nucleolus: The fascinating nuclear body. Histochem Cell Biol 129: 13–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Meissner A 2013. DNA methylation: Roles in mammalian development. Nat Rev Genet 14: 204–220 [DOI] [PubMed] [Google Scholar]
- Spivakov M, Fisher AG 2007. Epigenetic signatures of stem-cell identity. Nat Rev Genet 8: 263–271 [DOI] [PubMed] [Google Scholar]
- Sudo T, Utsunomiya T, Mimori K, Nagahara H, Ogawa K, Inoue H, Wakiyama S, Fujita H, Shirouzu K, Mori M 2005. Clinicopathological significance of EZH2 mRNA expression in patients with hepatocellular carcinoma. Br J Cancer 92: 1754–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suganuma T, Workman J 2011. Signals and combinatorial functions of histone modifications. Ann Rev Biochem 80: 473–499 [DOI] [PubMed] [Google Scholar]
- Suva ML, Riggi N, Janiszewska M, Radovanovic I, Provero P, Stehle JC, Baumer K, Le Bitoux MA, Marino D, Cironi L, et al. 2009. EZH2 is essential for glioblastoma cancer stem cell maintenance. Cancer Res 69: 9211–9218 [DOI] [PubMed] [Google Scholar]
- Talbert P, Henikoff S 2010. Histone variants—Ancient wrap artists of the epigenome. Nat Rev Mol Cell Biol 11: 264–275 [DOI] [PubMed] [Google Scholar]
- Van Bortle K, Corces VG 2012. Nuclear organization and genome function. Annu Rev Cell Dev Biol 28: 163–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vlag J, Otte A 1999. Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Nat Genet 23: 474–478 [DOI] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. 2008. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 40: 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Schones D, Zhao K 2009. Characterization of human epigenomes. Curr Opin Genet Dev 19: 127–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widschwendter M 2007. Epigenetic stem cell signature in cancer. Nature Genet 39: 157–158 [DOI] [PubMed] [Google Scholar]
- Williams RR, Broad S, Sheer D, Ragoussis J 2002. Subchromosomal positioning of the epidermal differentiation complex (EDC) in keratinocyte and lymphoblast interphase nuclei. Exp Cell Res 272: 163–175 [DOI] [PubMed] [Google Scholar]
- Wu JI, Lessard J, Crabtree GR 2009. Understanding the words of chromatin regulation. Cell 136: 200–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Li W, Shang C, Chen RM, Han P, Yang J, Stankunas K, Wu B, Pan M, Zhou B, et al. 2013. Brg1 governs a positive feedback circuit in the hair follicle for tissue regeneration and repair. Dev Cell 25: 169–181 [DOI] [PubMed] [Google Scholar]
- Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson R, Tabin C, Sharpe A, Caput D, Crum C, et al. 1999. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 398: 714–718 [DOI] [PubMed] [Google Scholar]
- Yoo AS, Staahl BT, Chen L, Crabtree GR 2009. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature 460: 642–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Gavin M, Dahiya A, Postigo A, Ma D, Luo R, Harbour J, Dean D 2000. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell 101: 79–89 [DOI] [PubMed] [Google Scholar]
- Zhang K, Zhang R, Li X, Yin G, Niu X, Hou R 2007. The mRNA expression and promoter methylation status of the p16 gene in colony-forming cells with high proliferative potential in patients with psoriasis. Clin Exp Dermatol 32: 702–708 [DOI] [PubMed] [Google Scholar]
- Zhang P, Su Y, Chen H, Zhao M, Lu Q 2010. Abnormal DNA methylation in skin lesions and PBMCs of patients with psoriasis vulgaris. J Dermatol Sci 60: 40–42 [DOI] [PubMed] [Google Scholar]
- Zhang RP, Shao JZ, Xiang LX 2011. GADD45A protein plays an essential role in active DNA demethylation during terminal osteogenic differentiation of adipose-derived mesenchymal stem cells. J Biol Chem 286: 41083–41094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Bardot E, Ezhkova E 2012. Epigenetic regulation of skin: Focus on the Polycomb complex. Cell Mol Life Sci 69: 2161–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]