Abstract
Organismal development is the most dynamic period of the life cycle, yet we have only a rough understanding of the dynamics of gene expression during adolescent transition. Here we show that adolescence in Caenorhabditis elegans is characterized by a spectacular expression shift of conserved and highly polymorphic genes. Using a high resolution time series we found that in adolescent worms over 10,000 genes changed their expression. These genes were clustered according to their expression patterns. One cluster involved in chromatin remodelling showed a brief up-regulation around 50 h post-hatch. At the same time a spectacular shift in expression was observed. Sequence comparisons for this cluster across many genotypes revealed diversifying selection. Strongly up-regulated genes showed signs of purifying selection in non-coding regions, indicating that adolescence-active genes are constrained on their regulatory properties. Our findings improve our understanding of adolescent transition and help to eliminate experimental artefacts due to incorrect developmental timing.
In the eukaryote life cycle the transformation from a sub-adult or larvae into an adult, ‘adolescence’, is a key transitional stage. Apart from an increase in size and shape of the body, its principal physical expression is the development of gonads, reproductive organs and secondary sexual characteristics, all of which provide a complete reproductive competency. Chronological age provides only a rough marker of adolescence and the variation between individuals and especially between genetic lines in the speed of transition is a key life history attribute1,2,3,4,5. In many animals the precise sequence of developmental events and the relation with gene regulation of ‘adolescence’ are not well known, but in principle would provide a clear staging post for comparison among individuals, populations and species. To investigate genome-wide gene expression dynamics during adolescence we sampled mRNA from Caenorhabditis elegans populations starting in late juvenile L3 stage, through the entire L4 stage and into adulthood at hourly intervals (44–58 h; Fig. 1a). Previous analyses reported a few hundred genes having different expression levels between stages2,3,6,7,8,9,10,11,12,13. Only recently, it was found that over 10,000 genes have different expression levels over the successive larval stages14. Using a high resolution time series we found that in adolescent worms alone over 10,000 genes displayed changes in expression. Specific patterns of changing gene expression were found which could be linked to patterns of natural and induced genetic polymorphisms showing different types of selection pressure. Lastly, we show how to use developmental time series for detecting small but significant developmental differences in gene expression experiments.
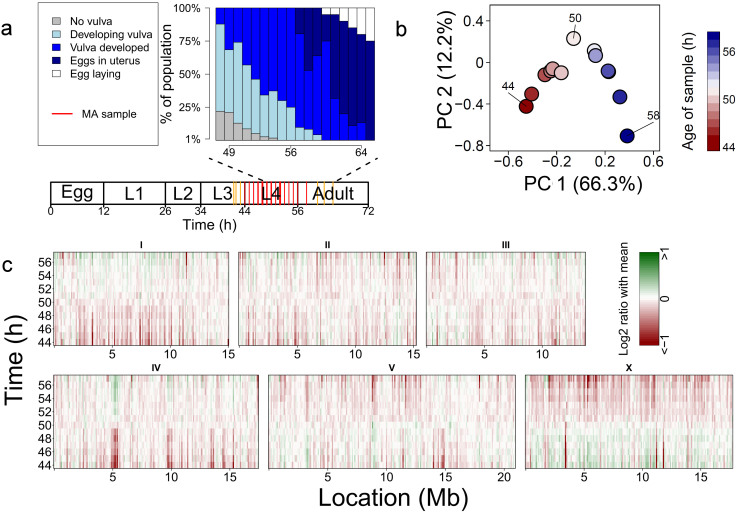
Figure 1. Experimental setup and an overview of the expression changes.
(a) Sampling setup. At 20°C, eggs hatch 10–12 hours after they were laid, leading to L1 stage worms. First, 2nd, 3rd and 4th molts occur 26, 34, 44 and 56 hours after egg laying resp., leading to L2, L3, L4 and adult worms. Microscopic observations of the developmental stage of the vulva at several time points are summarized in the blue graph. The red lines in the diagram below the graph represent the time points at which samples for the microarrays (MA) were drawn. The yellow lines show sampling points that were used later to confirm that the MA data could be used to predict the age of a sample (see Supplementary methods for more information). (b) A Principal component analysis of the relative gene expression shows that 66.3% of the variation sorts the time-points in a chronological order. Notably, the 50 h time-point is very distant from the others, indicating the sudden switch in expression. (c) The relative expression in log2 ratio of the mean, shown in 100 kb bins. There is a pronounced shift in the expression activity of the chromosomal regions, centring around 50 hrs.
Results and discussion
We generated a high-resolution temporal map of developmental gene expression during L4 development (Supplementary fig. S1 contains expression profiles of all genes). This is an increase in resolution compared to previous experiments where global gene expression profiles were analysed in separate stages2,3,6,7,8,9,10,11,12,13.
Principal component analysis (PCA) was used to explore the sources of variation of gene expression (Fig. 1b). The first principal component (capturing 66% of the variation) consistently placed the time points in chronological order. It shows that development during adolescence is causal for the majority of gene expression variation. The first two components (capturing 78% of the variation) place time point 50 h at a striking position, indicating that global gene expression patterns become remarkably different at this point.
The expression was plotted across the genome showing that gene expression shifts profoundly and very rapidly during adolescence (Fig. 1c). Moreover, genes with a similar expression pattern are clustered in the genome as shown by the enriched loci. Correlation analysis supports the PCA and shows a separation of adolescence in an early and a late phase, reversing around 50 h (Supplementary fig. S2).
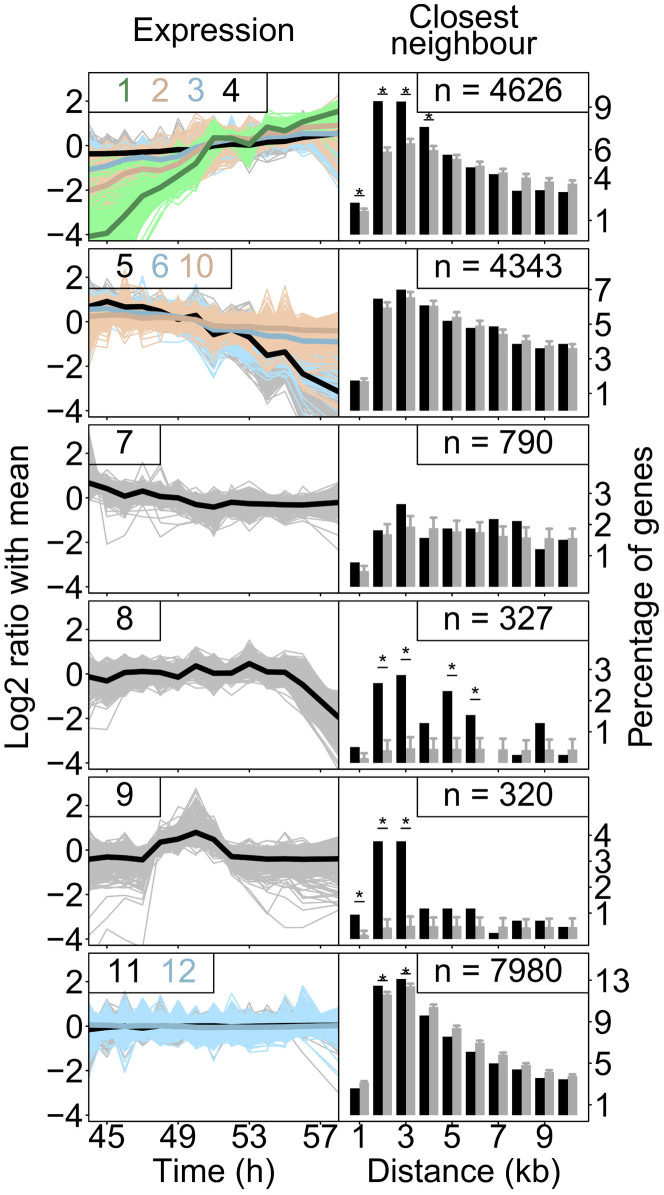
The dynamics of gene expression changes were explored via k-means clustering into 12 clusters (Fig. 2, Supplementary fig. S3 and Supplementary table S1). Fig. 2 shows the 12 resulting clusters, each having different dynamics. The largest group (43%) comprises genes that showed no change in expression over time (clusters 11 and 12). The rest, 57% of all genes, showed a dynamic pattern during development. This is substantially more than previously detected within and between different stages2,3,6,7,8,9,10,11,12,13, however, less than the recently detected 88% of all genes showing a dynamic expression pattern comparing all the larval stages14. The clusters can be grouped into six specific patterns. Most genes with a dynamic pattern show either a continuous increase (clusters 1, 2, 3 and 4; 4626 genes) or continuous decrease (clusters 5, 6 and 10; 4343 genes) at different intensities. A decrease in expression early L4 was found in cluster 7 (790 genes) and late L4 in cluster 8 (327 genes). In cluster 9 a striking, relatively short lived up-regulation was detected immediately followed by down-regulation, around 50 h.
Figure 2. The temporal expression as identified by k-means clustering.
The 12 clusters (also see Supplementary fig. S3 and table S1) are shown, grouped by the direction of the gene expression changes. The six distinct expression changes found are: up-regulation (clusters 1, 2, 3 and 4), down-regulation (clusters 5, 6 and 10), down-regulation in early L4 (cluster 7), down regulation in late L4 (cluster 8), no change (cluster 11 and 12) and up-regulation mid-L4 (cluster 9). In the line-graphs on the left the expression (log2 ratio of the mean) of all the genes in a cluster (light colours) and the average expression of all these genes (dark colours) are shown over time (in hours). The numbers of the clusters depicted are shown in the graph. For example: the expression patterns of all genes falling in category 6 are shown in light-blue and their average is shown in blue. The bar-charts show the proximity of genes within the same group as a percentage of genes having their neighbour at a particular distance (black). The grey bars show what to expect based on a random set of genes with the same size. The error-bars indicate the standard deviation found over 1,000 random sets. The asterisks indicate significant increases of genes found at a specific distance (FDR < 0.01). Four out of these six categories consist of genes that are physically closer together than expected based on equally sized random sets.
Very recently, Kim et al. (2013) investigated postembryonic gene expression dynamics in C. elegans larval stages L1 to L4 using two hour intervals14. By k-means clustering their data could be split up into 12 different clusters, for which the clearest patterns were: oscillating, stable, and up-regulation after moulting to L4. Genes showing up-regulation specifically after moulting to L4 (Kim et al. cluster 8 and 9) are also showing up-regulation in our experiment (clusters 1, 2 and 3; Supplementary figure S4). Our results show that the up-regulation of the Kim clusters 8 and 9 continues after their last time point (48 hours). The oscillating Kim clusters (1 to 6) are linked to genes showing down-regulation during L4 in our experiment. Furthermore, a large group of genes show stable (non-changing) expression levels in both experiments; and therefore throughout the L1 to L4 stage. Furthermore gene lin-4 marking the period just after moulting14 shows a decreasing expression level (Supplementary figure S5) which confirms the start of our experiment at the late L3 to early L4 molt.
The genomic locations of genes with similar expression patterns were compared by measuring the physical distance between the nearest gene with the same pattern (Fig. 2; for individual k-mean clusters see Supplementary fig. S6). A significantly smaller distance between neighbouring genes was found for genes with a continuously increasing expression levels (cluster 1, 2, 3 and 4) and genes in cluster 8 and cluster 9 (FDR < 0.01) compared to the other clusters. Additionally, genomic hot-spots were found for the individual clusters (Supplementary fig. S7), showing that these genes also co-localize on a larger scale. On average, 19.3% of the genes belonging to an expression pattern cluster can be found in the genomic hotspots. Because of their higher recombination frequency15 it is interesting that the chromosomal arms were enriched for non-dynamic genes (Supplementary fig. S7). Especially, because the non-dynamic clusters 11 and 12 are not enrichment for genes with a “lethal” phenotype in RNAi experiments, whereas all the dynamic clusters are (Supplementary table S2). This is in agreement with the finding that lethal genes are mainly located on the chromosome centres16 and shows that they have a typical stable expression pattern during L4.
To gain insight in the biological processes underlying the dynamics we investigated the clusters for overrepresentation of several gene classifications (Supplementary table S2 and Supplementary fig. S8–S13 for details per classification). The genes in the clusters with the strongest up-regulation (1, 2 and 3) were involved in many processes related to reproduction, like oogenesis, spermatogenesis and formation of the egg. The genes in the down-regulated clusters (5, 6 and 10) were involved in the completion of the adult body as shown by the enrichment of for example molting-, cuticle-, neurogenesis- and metabolism-related genes. In cluster 7 many genes related to larval development and growth were over-represented. In cluster 8 also genes related to the cuticle were enriched. Cluster 9 was enriched for transcription-factor-, histone-, and nucleosome-genes which marks the expression change around the 50 h reversal. This, together with the identified pattern-specific loci, indicates the involvement of chromatin remodelling as a source of higher order regulation during the transition from adolescent into adult form.
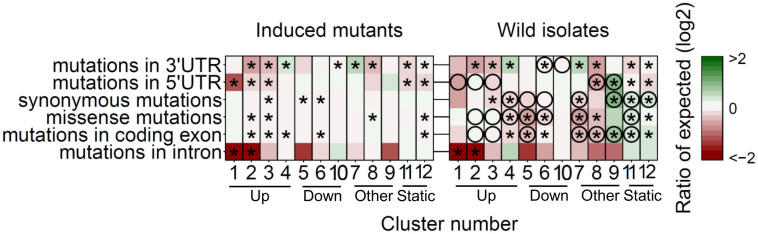
Organismal development is a very robust process regardless of the environment17. This means that the developmental program is buffered against environmental variation18. Hence, purifying selection is expected to act on core-developmental genes, as development is robust whereas diversifying selection can be expected for the genes which enable buffering against environmental variation as environments can be variable between sites. Therefore, we asked if the specific dynamic patterns show different mutation frequencies. To investigate this, we obtained data from the million mutation project19, which includes both mutagenized (2007 genotypes) and wild-type (40 genotypes) genome-wide variation. This dataset provides an insight in the occurrence of mutations per-se (induced mutations) and the occurrence of variation under natural selection (wild isolates). A substantial number of natural polymorphisms are locally linked even though between-strain variation is larger than the between-isolation site variation20,21,22. Moreover, given the recent selective sweeps in natural populations of C. elegans23 and natural selection in general, the distribution of mutations in wild isolates is vastly different from induced mutants15,19. We have focused on the mutations in both these datasets, which are providing 758,335 (induced mutants) and 597,777 (natural isolates) individual mutations (Supplementary table S3). The genes in the 12 expression pattern clusters were enriched for, or depleted for, mutations compared to a random set of genes (of the same size) from both the induced and natural set of polymorphisms (Fig. 3). Interestingly, specific functional structures within the gene (untranslated regions, exons, introns) showed different mutation frequencies.
Figure 3. Mutation frequencies coupled to the expression profile clusters.
Mutation frequencies19 are shown as the log2 ratio versus expected (based on random group of genes of the same size) and an asterisk indicates significant in- and decreases (FDR < 0.01). Circles in right heat-map indicate differences between frequencies found in the induced mutants and the wild isolates. The clusters are ordered based on their expression pattern.
In the induced mutants the non-coding regions of the three most highly up-regulated clusters are largely unaffected by mutations (the introns of clusters 1 and 2, 5'UTR of clusters 1, 2 and 3 and the 3'UTR of clusters 2 and 3; FDR < 0.01). Given the nature of the induced mutants, this indicates that these regulatory elements are of vital importance in the developmental progression to a reproductive adult. Also a slight, but significant, conservation was seen in the UTRs of the non-responsive genes (cluster 11 and 12). Interestingly, we also detected significantly increased mutation frequencies, but only in the 3'UTR of two clusters (4 and 7). This suggests that the post-transcriptional expression regulation of these genes is less likely to be of importance for reproduction.
Compared to the induced mutants we saw more extreme differences between the clusters in mutation frequencies for the wild isolates. Intriguingly, we also found that variants in the regulatory regions of the three most highly up-regulated clusters were selected against, especially in the introns (cluster 1 and 2, FDR < 0.01) and the 3'UTR (cluster 2 and 3, FDR < 0.01), but not significantly so in the 5'UTR. For many of the clusters we saw up to a 2-fold reduction in the mutation frequency within the coding regions (cluster 4, 5, 6, 7 and 8, FDR < 0.01). Increased mutation frequencies were observed more often, especially in the genes of cluster 9 suggesting diversifying selection and possibly adaptation. Genes with constant expression levels (clusters 11 and 12), were more likely to have genetic polymorphisms and the majority was previously found to be highly polymorphic between local populations20.
Combining the results from the induced mutants and the wild isolates shows potential adaptation, diversifying- and purifying-selection. The most pronounced difference is a decrease in mutation frequency in the exons of the wild isolates. Especially, the frequency of missense mutations is low, a feature reflected in the dataset as a whole19. This is most noticeable in the wild-isolates which is likely due to purifying-selection acting on mutations with only minor fitness penalties. Another profound difference is the increased variation found in wild isolates in the coding regions of the genes of clusters 9, 11 and 12. This implies that these genes are under strong diversifying selection pressure, which could indicate adaptation.
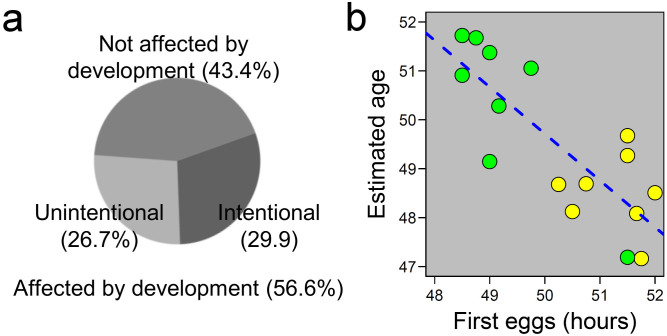
Our findings on the rapid gene expression shift in adolescent worms prompted us to determine the precise developmental age of samples in other published experiments for which gene expression levels had been determined (Supplementary fig. S14, Supplementary methods, Supplementary discussion & Supplementary fig. S15). We found that ~57% of the 143 different gene expression studies in the co-expression database SPELL24 were affected by a difference in development between the samples (Fig. 4a & Supplementary fig. S15). While many experiments were conducted with the purpose to study aging and development where these differences could be expected, other experiments set out to study the effect of genetic or environmental factors, like wild isolates (natural genetic variation), temperature, or different bacterial foods or pathogens2,3,20,25,26,27,28,29 (Supplementary discussion). Apparently, these factors can have a small, yet profound, effect on the developmental speed as most of the samples were taken at one time point. For example bacterial diet can affect the transcript profiles of many genes26. The differences in expression between the different bacterial food sources were enriched with genes strongly up-regulated during L4 development. This suggests a difference in developmental speed between worms grown on different bacteria. To confirm this we used different bacterial foods (E. coli OP50 and Erwinia rhapontici20) to grow different genotypes and measured two things, gene expression at 48 hours and the time until reproduction (first egg) (Fig. 4b & Supplementary fig. S14). We found a strong negative correlation between the developmental age determined by the transcription profiles and the time until the first eggs were laid. This negative correlation shows that both genotype and bacterial foods can affect developmental speed and transcription profiles. Combining classical phenotyping and transcription profiling can show a relatively small but significant and highly influential developmental difference.
Figure 4. Consequences for previous and future studies.
(a) The transcriptomic ruler was applied to all studies deposited in the SPELL database24, which showed that >50% of the studies has a developmental component, which is unintentional in 26% of all cases. (b) In a follow-up experiment the influence of food-source (Escherichia coli in yellow and Erwinia rhapontici in green) was shown to have a small but significant impact on the speed of development, C. elegans develops faster on E. rhapontici (R2 = 0.63, p < 0.001; also see Supplementary discussion).
In conclusion, we present here the first intensive time-series analysis of gene regulatory dynamics during adolescence in C. elegans, the attainment of a reproductively viable adult from larval tissues. We show that transcription levels can change surprisingly fast (in ~1 hour) and that transcription-factor-, histone-, and nucleosome-genes are particularly affected. The identified genes in the dynamic patterns are prone to either purifying- or diversifying-selection leading to variation in the arrangements of polymorphisms.
We show that the results generated by L4 gene expression studies in C. elegans are sensitive to variation in development, and illustrate that interpretation of data without knowledge of the expression dynamics can influence the outcome of studies. Given the increasing application of large-scale gene expression studies, using expression dynamics enables an even better interpretation of the results.
Methods
Detailed information available in the Supplementary Methods and Materials. Caenorhabditis elegans strain Bristol N2 was grown on NGM at 20°C with E. coli OP50 as food source30. Morphological changes in worms between the age of 48 and 67 hours after synchronisation were detected by analysing pictures of individual worms that were taken at a magnification of 100×. Samples for microarray analysis were drawn every hour between 44 and 58 hours after synchronisation by bleaching. RNA was isolated with either a RNEasy Micro Kit from Qiagen or with use of a Maxwell® 16 AS2000 instrument with a Maxwell® 16 LEV simplyRNA Tissue Kit (both Promega). After RNA isolation, one, two or three independent replicates per time point were analysed with microarrays (C. elegans (V2) Gene Expression Micorarray 4X44K slides from Agilent) following the ‘Two-Color Microarray-Based Gene Expression Analysis’-protocol from Agilent. Data were extracted with the Agilent Feature Extraction Software. All statistical analyses were performed in the ‘R’ statistical programming software (version 2.13.1 × 64). For normalization the Limma package was used with for Agilent array recommended settings31. For k-means clustering, 20 iterations on 10 different starting situations were performed. Twelve k-means clusters were formed to be able to visualise gene expression changes properly. Enrichment studies were done using a hyper-geometric test. The physical distance from a gene to its closest neighbour within its k-means cluster was measured as the distance from start-site to start-site. The mutation frequency19 of the genes from the k-means clusters was calculated using a permutation analysis. Heat-maps were constructed with the ‘heatmap’ function. Data of published gene expression studies were obtained from SPELL24. Bacteria and genotypes for the bacterial food experiment are from20. Data was stored in WormQTL32,33 (www.WormQTL.org).
Author Contributions
L.B.S., M.G.S. and R.J.M.V. designed the experiments. M.K., K.J.B., R.P.J.B., J.A.G., R.J.M.V. and M.G.S. conducted the experiments. L.B.S. and M.G.S. analysed the data. L.B.S., M.G.S. and R.J.M.V. wrote the manuscript together with G.S., A.R.C. and J.E.K.
Supplementary Material
Supplementary file
Table S1
Table S2
Table S5
Acknowledgments
We thank Jaap Bakker for helpful suggestions to the mss., K. Joeri van der Velde and Morris A. Swertz (Groningen Bioinformatics Centre, Groningen, The Netherlands) for assistance with WormQTL. We also would like to thank WormBase for being a versatile and important resource for C. elegans research. LBS and ARC were funded by the ERASysbio-plus ZonMW project GRAPPLE (project nr. 90201066). MGS was supported by Graduate School Production Ecology & Resource Conservation. RJMV was funded by the NWO-ALW (project 855.01.151), JAGR and JEK were funded by PANACEA EU FP7 project contractnr. 222936.
References
- Gutteling E. W., Riksen J. A., Bakker J. & Kammenga J. E. Mapping phenotypic plasticity and genotype-environment interactions affecting life-history traits in Caenorhabditis elegans. Heredity 98, 28–37, 10.1038/sj.hdy.6800894 (2007). [DOI] [PubMed] [Google Scholar]
- Viñuela A., Snoek L. B., Riksen J. A. & Kammenga J. E. Genome-wide gene expression regulation as a function of genotype and age in C. elegans. Genome research 20, 929–937, 10.1101/gr.102160.109 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viñuela A., Snoek L. B., Riksen J. A. & Kammenga J. E. Aging Uncouples Heritability and Expression-QTL in Caenorhabditis elegans. G3 2, 597–605, 10.1534/g3.112.002212 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francesconi M. & Lehner B. The effects of genetic variation on gene expression dynamics during development. Nature, 10.1038/nature12772 (2013). [DOI] [PubMed] [Google Scholar]
- Doroszuk A., Snoek L. B., Fradin E., Riksen J. & Kammenga J. A genome-wide library of CB4856/N2 introgression lines of Caenorhabditis elegans. Nucleic Acids Res 37, e110, 10.1093/nar/gkp528 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein M. B. et al. Integrative Analysis of the Caenorhabditis elegans Genome by the modENCODE Project. Science 330, 1775–1787, 10.1126/science.1196914 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capra E. J., Skrovanek S. M. & Kruglyak L. Comparative developmental expression profiling of two C. elegans isolates. PloS one 3, e4055, 10.1371/journal.pone.0004055 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier L. W. et al. Massively parallel sequencing of the polyadenylated transcriptome of C. elegans. Genome research 19, 657–666, 10.1101/gr.088112.108 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke V., Gil I. S., Ward S. & Kazmer K. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development 131, 311–323, 10.1242/dev.00914 (2004). [DOI] [PubMed] [Google Scholar]
- Reinke V. et al. A global profile of germline gene expression in C. elegans. Molecular cell 6, 605–616 (2000). [DOI] [PubMed] [Google Scholar]
- Spencer W. C. et al. A spatial and temporal map of C. elegans gene expression. Genome research 21, 325–341, 10.1101/gr.114595.110 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoemke K. et al. Genome-wide analysis of sex-enriched gene expression during C. elegans larval development. Developmental biology 284, 500–508, 10.1016/j.ydbio.2005.05.017 (2005). [DOI] [PubMed] [Google Scholar]
- Golden T. R., Hubbard A., Dando C., Herren M. A. & Melov S. Age-related behaviors have distinct transcriptional profiles in Caenorhabditis elegans. Aging cell 7, 850–865, 10.1111/j.1474-9726.2008.00433.x (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. H., Grun D. & van Oudenaarden A. Dampening of expression oscillations by synchronous regulation of a microRNA and its target. Nature genetics 45, 1337–1344, 10.1038/ng.2763 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman M. V. & Kruglyak L. Recombinational landscape and population genomics of Caenorhabditis elegans. PLoS genetics 5, e1000419, 10.1371/journal.pgen.1000419 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R. S. et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, 231–237, 10.1038/nature01278 (2003). [DOI] [PubMed] [Google Scholar]
- Kitano H. Biological robustness. Nature reviews. Genetics 5, 826–837, 10.1038/nrg1471 (2004). [DOI] [PubMed] [Google Scholar]
- Milloz J., Duveau F., Nuez I. & Felix M. A. Intraspecific evolution of the intercellular signaling network underlying a robust developmental system. Genes & development 22, 3064–3075, 10.1101/gad.495308 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson O. et al. The million mutation project: a new approach to genetics in Caenorhabditis elegans. Genome research 23, 1749–1762, 10.1101/gr.157651.113 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkers R. J. et al. Gene-environment and protein-degradation signatures characterize genomic and phenotypic diversity in wild Caenorhabditis elegans populations. BMC biology 11, 93, 10.1186/1741-7007-11-93 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter A. D. Nucleotide polymorphism and linkage disequilibrium in wild populations of the partial selfer Caenorhabditis elegans. Genetics 172, 171–184, 10.1534/genetics.105.048207 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrière A. & Félix M. A. High local genetic diversity and low outcrossing rate in Caenorhabditis elegans natural populations. Current biology: CB 15, 1176–1184, S0960-9822(05)00655-X [pii] 10.1016/j.cub.2005.06.022 (2005). [DOI] [PubMed] [Google Scholar]
- Andersen E. C. et al. Chromosome-scale selective sweeps shape Caenorhabditis elegans genomic diversity. Nature genetics, 10.1038/ng.1050 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibbs M. A. et al. Exploring the functional landscape of gene expression: directed search of large microarray compendia. Bioinformatics 23, 2692–2699, 10.1093/bioinformatics/btm403 (2007). [DOI] [PubMed] [Google Scholar]
- Li Y. et al. Mapping determinants of gene expression plasticity by genetical genomics in C. elegans. PLoS genetics 2, e222, 10.1371/journal.pgen.0020222 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coolon J. D., Jones K. L., Todd T. C., Carr B. C. & Herman M. A. Caenorhabditis elegans genomic response to soil bacteria predicts environment-specific genetic effects on life history traits. PLoS genetics 5, e1000503, 10.1371/journal.pgen.1000503 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman M. V., Skrovanek S. S. & Kruglyak L. Selection at linked sites shapes heritable phenotypic variation in C. elegans. Science 330, 372–376, 10.1126/science.1194208 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. et al. Global genetic robustness of the alternative splicing machinery in Caenorhabditis elegans. Genetics 186, 405–410, 10.1534/genetics.110.119677 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammenga J. E., Phillips P. C., De Bono M. & Doroszuk A. Beyond induced mutants: using worms to study natural variation in genetic pathways. Trends in genetics: TIG 24, 178–185, 10.1016/j.tig.2008.01.001 (2008). [DOI] [PubMed] [Google Scholar]
- Lewis J. A. & Fleming J. T. Basic culture methods. Methods in cell biology 48, 3–29 (1995). [PubMed] [Google Scholar]
- Zahurak M. et al. Pre-processing Agilent microarray data. BMC bioinformatics 8, 142, 10.1186/1471-2105-8-142 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snoek L. B. et al. WormQTL--public archive and analysis web portal for natural variation data in Caenorhabditis spp. Nucleic Acids Res 41, D738–743, 10.1093/nar/gks1124 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Velde K. J. et al. WormQTLHD--a web database for linking human disease to natural variation data in C. elegans. Nucleic Acids Res, 10.1093/nar/gkt1044 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file
Table S1
Table S2
Table S5