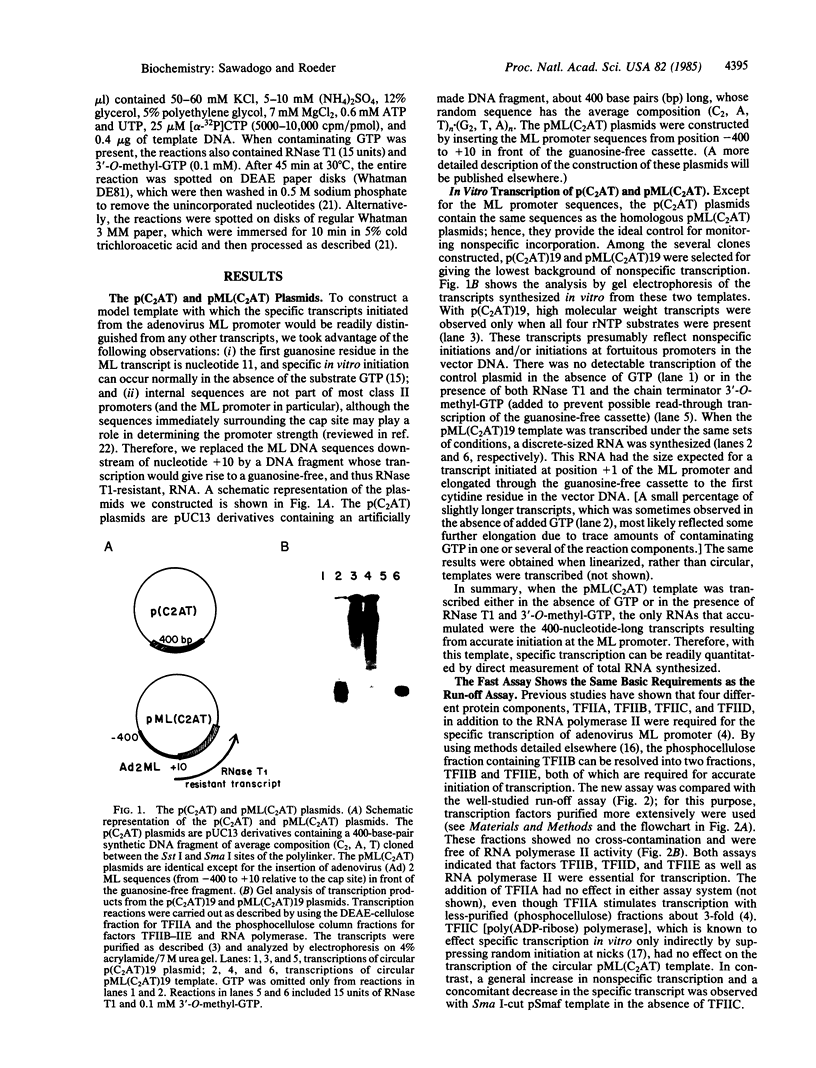
Abstract
We describe a new assay system that allows a rapid, direct, and quantitative detection of promoter-dependent in vitro transcription by RNA polymerase II. The template used is a hybrid plasmid containing the adenovirus major late promoter linked to a synthetic 400-base-pair DNA fragment that lacks cytidine residues on the transcribed strand--i.e., generates a transcript with no guanosine residues. In vitro transcriptions are carried out in the absence of GTP or, if the reactions contain GTP, in the presence of RNase T1 and the chain terminator 3'-0-methyl-GTP. Under these conditions the only RNAs that can accumulate, whether from a circular or linearized DNA template, are the 400-nucleotide RNase T1-resistant transcripts resulting from accurate initiation at the major late promoter. Thus, specific transcription can be directly monitored by conventional RNA quantitation methods. Using this fast assay, we show that three basic transcription factors, TFIIB, TFIID, and TFIIE, are absolutely required, in addition to the RNA polymerase II, for specific transcription initiation from the adenovirus major late promoter. Units of activity can be defined for each of these individual components. The applicability of this kind of assay to other systems is discussed.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bitter G. A. Purification of DNA-dependent RNA polymerase II from Saccharomyces cerevisiae. Anal Biochem. 1983 Feb 1;128(2):294–301. doi: 10.1016/0003-2697(83)90378-0. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Brent R., Ptashne M. A bacterial repressor protein or a yeast transcriptional terminator can block upstream activation of a yeast gene. Nature. 1984 Dec 13;312(5995):612–615. doi: 10.1038/312612a0. [DOI] [PubMed] [Google Scholar]
- Bunick D., Zandomeni R., Ackerman S., Weinmann R. Mechanism of RNA polymerase II--specific initiation of transcription in vitro: ATP requirement and uncapped runoff transcripts. Cell. 1982 Jul;29(3):877–886. doi: 10.1016/0092-8674(82)90449-4. [DOI] [PubMed] [Google Scholar]
- Davison B. L., Egly J. M., Mulvihill E. R., Chambon P. Formation of stable preinitiation complexes between eukaryotic class B transcription factors and promoter sequences. Nature. 1983 Feb 24;301(5902):680–686. doi: 10.1038/301680a0. [DOI] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J. D., Martin P. L., Shastry B. S., Roeder R. G. Eukaryotic gene transcription with purified components. Methods Enzymol. 1983;101:582–598. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. Isolation of transcription factors that discriminate between different promoters recognized by RNA polymerase II. Cell. 1983 Mar;32(3):669–680. doi: 10.1016/0092-8674(83)90053-3. [DOI] [PubMed] [Google Scholar]
- Egly J. M., Miyamoto N. G., Moncollin V., Chambon P. Is actin a transcription initiation factor for RNA polymerase B? EMBO J. 1984 Oct;3(10):2363–2371. doi: 10.1002/j.1460-2075.1984.tb02141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A., Samuels M., Sharp P. A. Interactions between RNA polymerase II, factors, and template leading to accurate transcription. J Biol Chem. 1984 Feb 25;259(4):2509–2516. [PubMed] [Google Scholar]
- Heintz N., Roeder R. G. Transcription of human histone genes in extracts from synchronized HeLa cells. Proc Natl Acad Sci U S A. 1984 May;81(9):2713–2717. doi: 10.1073/pnas.81.9.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hen R., Sassone-Corsi P., Corden J., Gaub M. P., Chambon P. Sequences upstream from the T-A-T-A box are required in vivo and in vitro for efficient transcription from the adenovirus serotype 2 major late promoter. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7132–7136. doi: 10.1073/pnas.79.23.7132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley J. L., Fire A., Cano A., Sharp P. A., Gefter M. L. DNA-dependent transcription of adenovirus genes in a soluble whole-cell extract. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3855–3859. doi: 10.1073/pnas.77.7.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T., Segall J., Weil P. A., Roeder R. G. Multiple factors required for accurate initiation of transcription by purified RNA polymerase II. J Biol Chem. 1980 Dec 25;255(24):11992–11996. [PubMed] [Google Scholar]
- Parker C. S., Topol J. A Drosophila RNA polymerase II transcription factor binds to the regulatory site of an hsp 70 gene. Cell. 1984 May;37(1):273–283. doi: 10.1016/0092-8674(84)90323-4. [DOI] [PubMed] [Google Scholar]
- Parker C. S., Topol J. A Drosophila RNA polymerase II transcription factor contains a promoter-region-specific DNA-binding activity. Cell. 1984 Feb;36(2):357–369. doi: 10.1016/0092-8674(84)90229-0. [DOI] [PubMed] [Google Scholar]
- Roeder R. G. Multiple forms of deoxyribonucleic acid-dependent ribonucleic acid polymerase in Xenopus laevis. Isolation and partial characterization. J Biol Chem. 1974 Jan 10;249(1):241–248. [PubMed] [Google Scholar]
- Samuels M., Fire A., Sharp P. A. Separation and characterization of factors mediating accurate transcription by RNA polymerase II. J Biol Chem. 1982 Dec 10;257(23):14419–14427. [PubMed] [Google Scholar]
- Sawadogo M., Roeder R. G. Energy requirement for specific transcription initiation by the human RNA polymerase II system. J Biol Chem. 1984 Apr 25;259(8):5321–5326. [PubMed] [Google Scholar]
- Scheer U., Hinssen H., Franke W. W., Jockusch B. M. Microinjection of actin-binding proteins and actin antibodies demonstrates involvement of nuclear actin in transcription of lampbrush chromosomes. Cell. 1984 Nov;39(1):111–122. doi: 10.1016/0092-8674(84)90196-x. [DOI] [PubMed] [Google Scholar]
- Schwartz L. B., Roeder R. G. Purification and subunit structure of deoxyribonucleic acid-dependent ribonucleic acid polymerase II from the mouse plasmacytoma, MOPC 315. J Biol Chem. 1975 May 10;250(9):3221–3228. [PubMed] [Google Scholar]
- Slattery E., Dignam J. D., Matsui T., Roeder R. G. Purification and analysis of a factor which suppresses nick-induced transcription by RNA polymerase II and its identity with poly(ADP-ribose) polymerase. J Biol Chem. 1983 May 10;258(9):5955–5959. [PubMed] [Google Scholar]
- Tolunay H. E., Yang L., Anderson W. F., Safer B. Isolation of an active transcription initiation complex from HeLa cell-free extract. Proc Natl Acad Sci U S A. 1984 Oct;81(19):5916–5920. doi: 10.1073/pnas.81.19.5916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai S. Y., Tsai M. J., Kops L. E., Minghetti P. P., O'Malley B. W. Transcription factors from oviduct and HeLa cells are similar. J Biol Chem. 1981 Dec 25;256(24):13055–13059. [PubMed] [Google Scholar]
- Weil P. A., Luse D. S., Segall J., Roeder R. G. Selective and accurate initiation of transcription at the Ad2 major late promotor in a soluble system dependent on purified RNA polymerase II and DNA. Cell. 1979 Oct;18(2):469–484. doi: 10.1016/0092-8674(79)90065-5. [DOI] [PubMed] [Google Scholar]




