Abstract
LINE-1s are active human DNA parasites that are agents of genome dynamics in evolution and disease. These streamlined elements require host factors to complete their lifecycles, whereas hosts have developed mechanisms to combat retrotransposition’s mutagenic effects. As such, endogenous L1 expression levels are extremely low, creating a roadblock for detailed interactomic analyses. Here we describe a system to express and purify highly active L1 RNP complexes from human suspension cell culture and characterize the co-purified proteome, identifying 37 high-confidence candidate interactors. These datasets include known interactors PABPC1 and MOV10 and, with in-cell imaging studies, suggest existence of at least three types of compositionally and functionally distinct L1 RNPs. Among the novel findings, UPF1, a key nonsense-mediated decay factor, and PCNA, the polymerase-delta-associated sliding DNA clamp, were identified and validated. PCNA interacts with ORF2p via a PIP box motif; mechanistic studies suggest this occurs during or immediately after target-primed reverse transcription.
Introduction
Long INterspersed Element-1s (L1s or LINE-1s) are autonomous retrotransposons that continuously re-enter host genomes (Ostertag and Kazazian, 2001). Although most L1 copies are functionally inactive, about 90 retrotransposition-competent L1s inhabit the human genome (Brouha et al., 2003). Somatic L1 insertions are rare in most tissues, but are found in brain, testis, and many cancer types and have been implicated in carcinogenesis (Shukla et al., 2013). Most L1 insertions are detrimental to the host; thus, retrotransposition is tightly controlled by a network of host factors (Arjan-Odedra et al., 2012; Dai et al., 2012; Goodier et al., 2012; Niewiadomska et al., 2007; Peddigari et al., 2012; Suzuki et al., 2009). L1s are extinct in certain lineages (Khan et al., 2006; Malik et al., 1999) and may contribute to neural plasticity in others (Muotri et al., 2005).
The full-length L1 transcript is ∼6 kb long, bicistronic and includes its promoter sequence within its 5’ untranslated region (UTR) (Ostertag and Kazazian, 2001). The L1 mRNA encodes two non-overlapping proteins, ORF1p and ORF2p, and includes a short 3’-UTR and a poly(A) tail (Belancio et al., 2007). L1 has a complex life cycle beginning with transcription by RNA polymerase II (pol II). The 40 kDa ORF1p forms a homotrimeric protein with nucleic acid chaperone activities (Martin and Bushman, 2001). The ∼ 150 kDa ORF2p is multifunctional, with endonuclease (EN) (Feng et al., 1996) and reverse transcriptase (RT) activities (Mathias et al., 1991). ORF1p is efficiently translated but ORF2p translation occurs at low levels through an unconventional mechanism (Alisch et al., 2006), making mechanistic research on ORF2p challenging. ORF2p exhibits preferential interaction with the L1 mRNA encoding it (cis-preference), forming a ribonucleoprotein (RNP) particle presumed to constitute a direct retrotransposition intermediate (Kulpa and Moran, 2006; Wei et al., 2001). L1 likely transposes via target primed reverse transcription (TPRT) (Cost et al., 2002; Luan and Eickbush, 1995).
Being parasitic, L1 requires host factors to complete its life cycle (Beauregard et al., 2008). For example, several tissue-specific transcription factors promote L1 transcription by host-encoded pol II (e.g. Athanikar et al., 2004). Poly(A) binding proteins are important in RNP formation and retrotransposition (Dai et al., 2012), DNA repair factors affect L1 retrotransposition, most likely in TPRT (Suzuki et al., 2009). Host genomes have evolved mechanisms to restrict potentially harmful L1 retrotransposition, employing DNA methylation (Hata and Sakaki, 1997), RNAi pathways (Soifer et al., 2005; Yang and Kazazian, 2006), APOBEC3 cytidine deaminases (Niewiadomska et al., 2007), and RNA helicase MOV10 (Arjan-Odedra et al., 2012; Goodier et al., 2012). Two recent studies identified protein and RNA interactors in L1 RNP complexes via an ORF1 tag (Goodier et al., 2013; Mandal et al., 2013). However, while host factors have been found, technical limitations have hindered isolation of pure, active RNP complexes in analytically tractable quantities.
Although L1 is highly active in germ line and brain, it is effectively repressed in most somatic cells (Muotri et al. 2005). To evaluate L1 activity in cell culture, it is often paired with a constitutive, high-level promoter (Moran et al., 1996). Fractions containing expressed L1 RNP complexes have been prepared by sucrose cushion velocity sedimentation of cell lysates (Kulpa and Moran, 2005, 2006). These provided valuable insights into mechanism, but suffer from low purity, limiting their usefulness for studying L1-host interactomes. Traditional epitope tagging of ORF polypeptides has been used to study proteins co-purifying with the L1 RNP (Doucet et al., 2010; Goodier et al., 2012, 2013), but these conferred insufficient yield and purity for comprehensive proteomics with high specific activity.
In contrast, new methods implementing cryomilling/rapid affinity capture provide outstanding purification that significantly improves interactomic surveys (Cristea et al., 2005; Domanski et al., 2012; Oeffinger et al., 2007). Intact cells are flash-frozen in liquid nitrogen and milled in solid phase (cryomilling). Native intermolecular interactions are preserved during milling, producing a fine powder well-suited to subsequent affinity isolation of protein complexes (“pullouts”). Micron-scale antibody-conjugated magnetic beads enable rapid purifications with binding times as short as 5 minutes and facilitate binding of large complexes excluded from pores of traditional matrices (Oeffinger et al., 2007). When coupled with “Isotopic Differentiation of Interactions as Random or Targeted” (I-DIRT), stable interactions formed in vivo are distinguished from post-extraction artifacts (Tackett et al., 2005). These techniques have been applied to the study of protein complexes from many systems (Di Virgilio et al., 2013; Domanski et al., 2012; Lee et al., 2008; Oeffinger et al., 2007).
In this study, we constructed a series of inducible L1 expression vectors containing either synthetic (ORFeus-Hs) (An et al., 2011) or native (L1RP) L1 (Kimberland et al., 1999) elements and screened epitope tags on both L1 ORFs for effects on retrotransposition. Active constructs were expressed in suspension culture at large scale and RNPs were purified using cryomilling and rapid capture, facilitating enzymatic, transcriptomic, and proteomic assays. Purified RNPs had ∼70-fold higher specific activity than those made by previous techniques. RNA sequencing revealed L1 RNA as an abundant component along with U6 snRNA, commonly observed as part of hybrid elements with L1 (Buzdin et al., 2002). ORF1p to ORF2p ratio was unexpectedly low in tandem purified L1 particles. Cell imaging revealed most cells fail to express ORF2p. Mass spectrometric (MS) characterization identified 37 high confidence interactors, including known interactors PABPC1 and MOV10, and revealed that ORF1 and ORF2 RNPs exhibit both overlapping and distinct interactomes. UPF1, a helicase critical for nonsense-mediated decay (NMD), and PCNA, the polymerase-associated sliding clamp, were further validated for roles in the L1 life cycle.
Results
Producing highly purified L1-RNPs
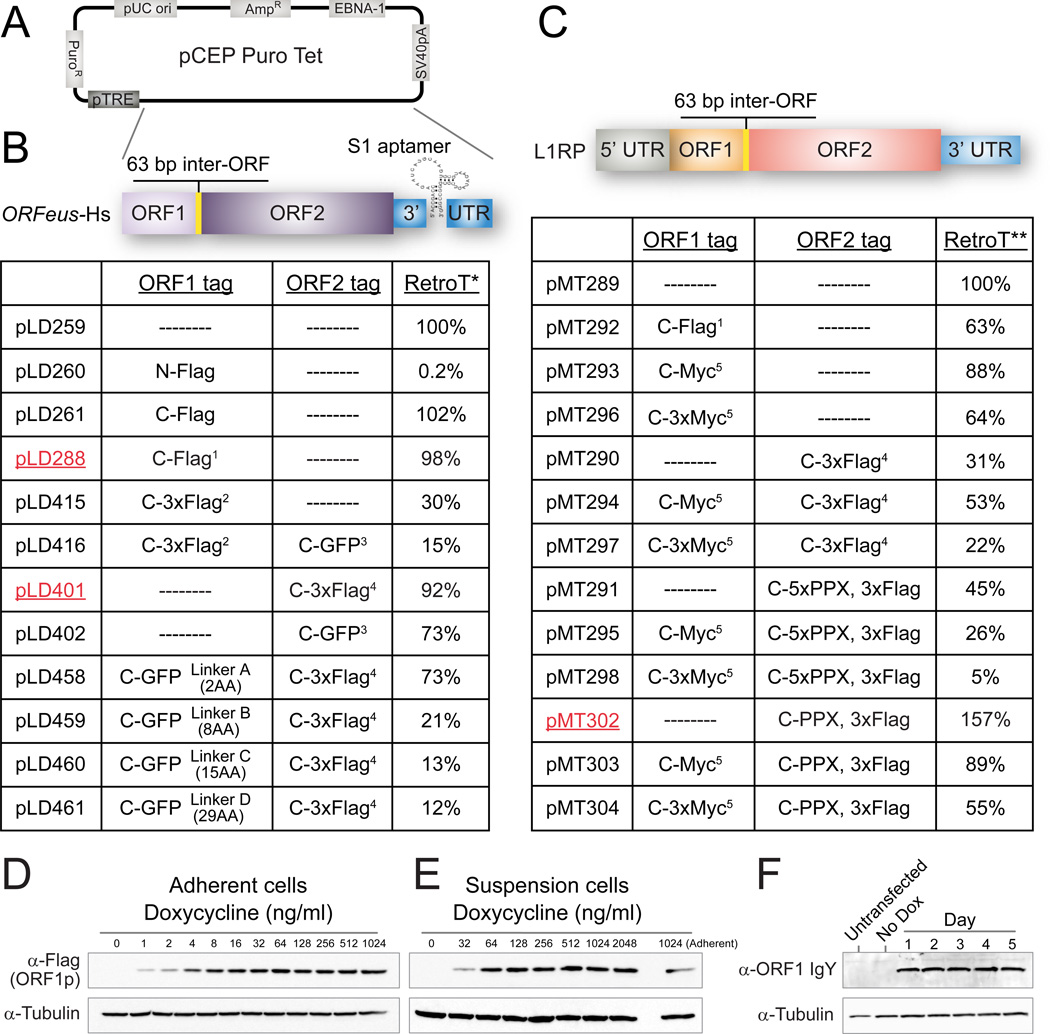
To isolate L1 RNP complexes at high yield and purity, we used synthetic human L1 ORFeus-Hs, which has high activity and produces >40 fold more RNA and 5–10 fold more ORF1p than a native counterpart, while encoding identical proteins (An et al., 2011). Uncontrolled L1 expression slows or eliminates growth; therefore, we leveraged expression from a tetracycline-regulated mini-CMV promoter (pTRE, Figure 1A) (O’Donnell et al., 2013). Due to a lack of appropriate antibodies, especially for ORF2p, we designed many epitope tag combinations and measured retrotransposition efficiency relative to untagged ORFeus-Hs (Figure 1B, supplemental text). Constructs pLD288 (ORF1p-Flag) and pLD401 (ORF2p-3xFlag) were best as they combined high L1 activity with short tag sequences.
Figure 1. L1 expression and retrotransposition constructs for purification.
(A) Backbone plasmid pCEP-puro-Tet. See abbreviations for details. (B) Constructs in ORFeus-Hs. Complete tag and linker sequences (protein and DNA) are detailed in Table S4. 1, Linker-Tev-V5-HA-Flag; 2, Linker-Tev-V5-HA-3xFlag; 3, Linker-LAP tag (His8-2xPrescission-GFP); 4, Linker-His8-2xPrescission-2xMyc-3xFlag. RetroT*: retrotransposition efficiency of all ORFeus-Hs tagged constructs is relative to untagged L1 (pLD259), assigned as 100%. (C) Constructs in L1RP. 5, Linker-Tev-Myc or Linker-Tev-3xMyc. RetroT**: retrotransposition efficiency of all tagged L1RP constructs were relative to untagged L1 (pMT289), assigned as 100%. (D, E) L1 expression (pLD288) in adherent and suspension Tet-On HEK293LD cells at various Dox concentrations. (F) Untagged L1 expression (pLD259) in suspension cells over time with 1 µg/ml Dox.
We introduced these constructs into adherent Tet-On HEK293TLD (Dai et al., 2012) and selected transfected cells for one week or more with puromycin. Doxycycline (Dox) concentration and times had similar effects in monolayer and suspension cultures (Figure 1D–F); expression of both ORFs peaked 24 hours post-induction. Although ORF1p levels were relatively stable over the next 3 days, ORF2p levels decreased within 48 hours (Figures 1F, 2H, S2).
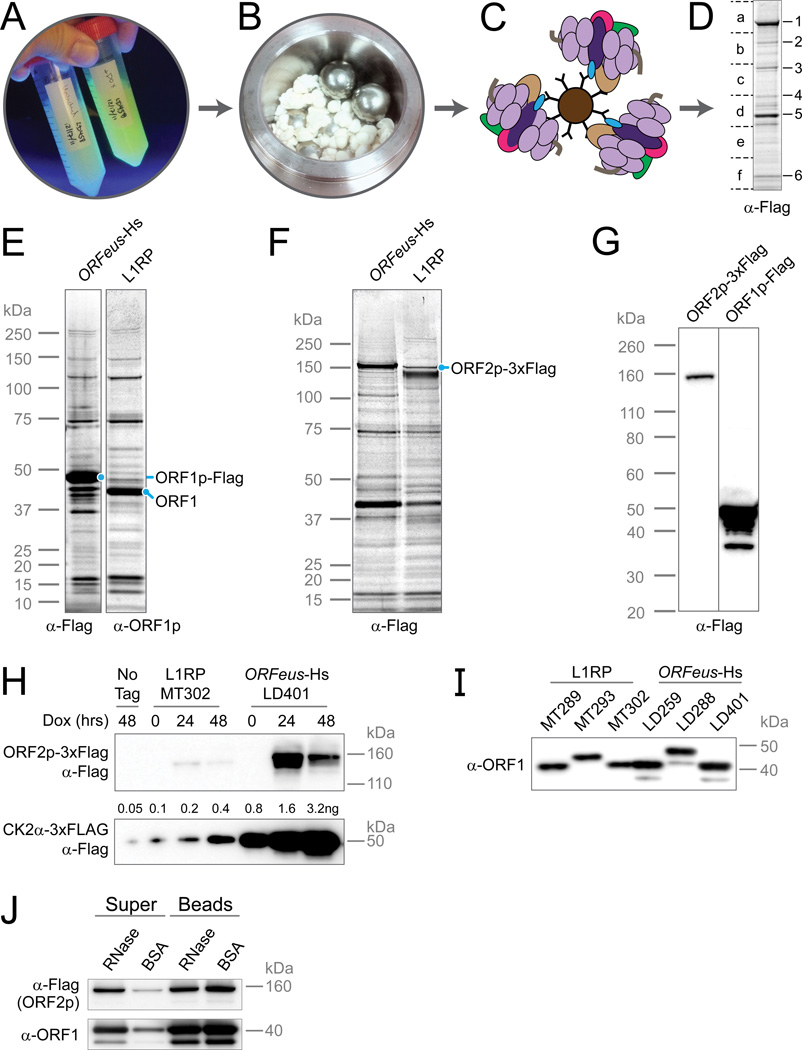
Figure 2. Interactomics workflow applied to L1 produces tractable quantities of both ORF proteins from both L1RP and ORFeus-Hs.
(A) Transposon-expressing human cell cultures are grown in suspension and induced for 24 hours. (B) Frozen cell “BBs” (∼3 mm diameter spherical globules of cells previously frozen in LN2) are cryomilled using stainless steel ball bearings under liquid nitrogen, resulting in a fine powder stored at −80°C. (C) Antibody-conjugated µm-scale magnetic medium is used for rapid pullout. (D) Gel-based proteomics: purified proteins are subjected to MALDI-TOF-PMF for identification of major species (1–6) or LC-MS/MS (a–f). (E) ORF1p and co-purified factors for ORFeus-Hs (pLD288) and L1RP (pMT289) by ORF1p purification. (F) ORF2p and co-purified factors for ORFeus-Hs (pLD401) and L1RP (pMT302) by ORF2p (anti-Flag) purification. (G) Comparison of ORF1p–Flag (pLD288) and ORF2p-3xFlag (pLD401) expression levels in puro-selected adherent cells; 20µg protein loaded in each lane. Please see Figures S2 for detailed calculations. (H) ORF2p expression levels in L1RP vs ORFeus-Hs. CK2α-3xFlag was loaded as control. 20µg protein loaded in each lane. Please see Figures S2 for detailed calculations. (I) ORF1p expression levels in L1RP vs ORFeus-Hs. 2µg protein loaded in each lane. (J) ORF1p–ORF2p interactions is RNase sensitive. L1 was affinity-purified with anti-ORF1 Dynabeads from pLD401 and then treated on-bead with RNases A/T1 or mock-treated with BSA. “Super”, proteins released into the supernatant; “Beads”, sample retained on beads after washing and eluted with LDS sample buffer.
Multi-liter suspension cultures were induced, harvested, cryomilled and pullouts were optimized (Figures 2A–D and S1). Both L1 proteins were readily observed by Coomassie brilliant blue (CBB) staining (Figures 2E–F). We also tagged native L1RP, which has lower protein expression levels (Figure 1C). Surprisingly, the C-terminal Flag and 3xFlag tag combinations on ORF1p and ORF2p that worked well in ORFeus-Hs were poorly tolerated in L1RP. We designed and screened new tags and linkers; a single Myc-tag at the C-terminus of ORF1p was tolerated (pMT293, pMT303), but did not provide high quality affinity isolations. By shortening the linker on ORF2p-3xFLAG we preserved L1RP retrotransposition efficiency (pMT302, pMT303). An improved αORF1p antibody obviated need for an epitope tagged L1RP ORF1p and thus pMT302 (L1RP w/ ORF2p-3xFlag) was selected (Figures 2E–F).
Both synthetic (pLD401) and native (pMT302) constructs gave robust ORF2p expression (Figures 2G–H). Taking into account the different detection sensitivities exhibited by the two tags (Hernan et al., 2000), ORF1p is apparently expressed 1,000–10,000 fold higher than ORF2p (Figures 2G and S2). Comparing ORFeus-Hs and L1RP constructs, ORF1p levels differ by ∼2.5-fold, but ORF2p levels differ by ∼20–40 fold compared to a purified standard (Figures 2H–I and S2).
The expression constructs, cell lines and approach collectively constitute an analytical suite unhindered by typical limitations in protein abundance, detection sensitivity, or capture affinity.
L1-RNP properties
Eluting native L1 RNPs by gentle displacement with excess 3xFlag peptide revealed both co-purifying species and associated enzymatic activity. ORF2p shows an RNase sensitive association with ORF1p pullouts as determined by immunoblotting (Figure 2J) and electrospray MS (Figure 4B). Initially, interactors were characterized by MALDI-TOF PMF (see abbreviations) of selected protein bands observed after CBB staining. Numerous protein bands were more abundant or uniquely present in ORF pullouts as compared to mock pullouts (Figure 3A). These included PABPC1 and MOV10, recently characterized components of an otherwise sparsely defined L1-host interactome (Arjan-Odedra et al., 2012; Dai et al., 2012; Goodier et al., 2012).
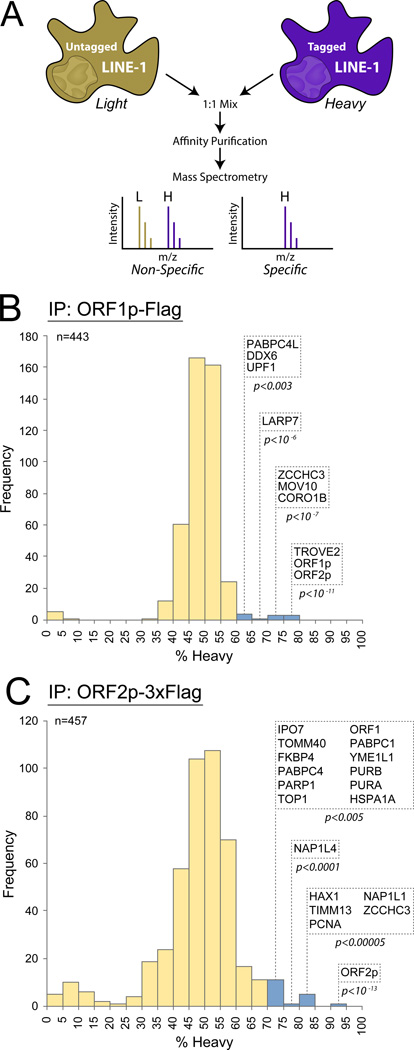
Figure 4. High confidence interactors of ORF1p and ORF2p from I-DIRT experiments.
(A) I-DIRT methodology. Light and heavy represent stable isotope labeling of Arg and Lys residues. (B) Interactors identified by ORF1p (Flag) IP with 5 min binding. (C) Interactors identified by ORF2p (Flag) IP with 30 min binding. Histograms plot the number of recovered proteins as compared to their heavy isotope content. Yellow bars, non-significant groups; blue bars, statistically significant specificity after Benjamini–Hochberg correction for false discovery. P-values were determined by Perseus software. Please see Figures S6 for results of all six I-DIRT experiments.
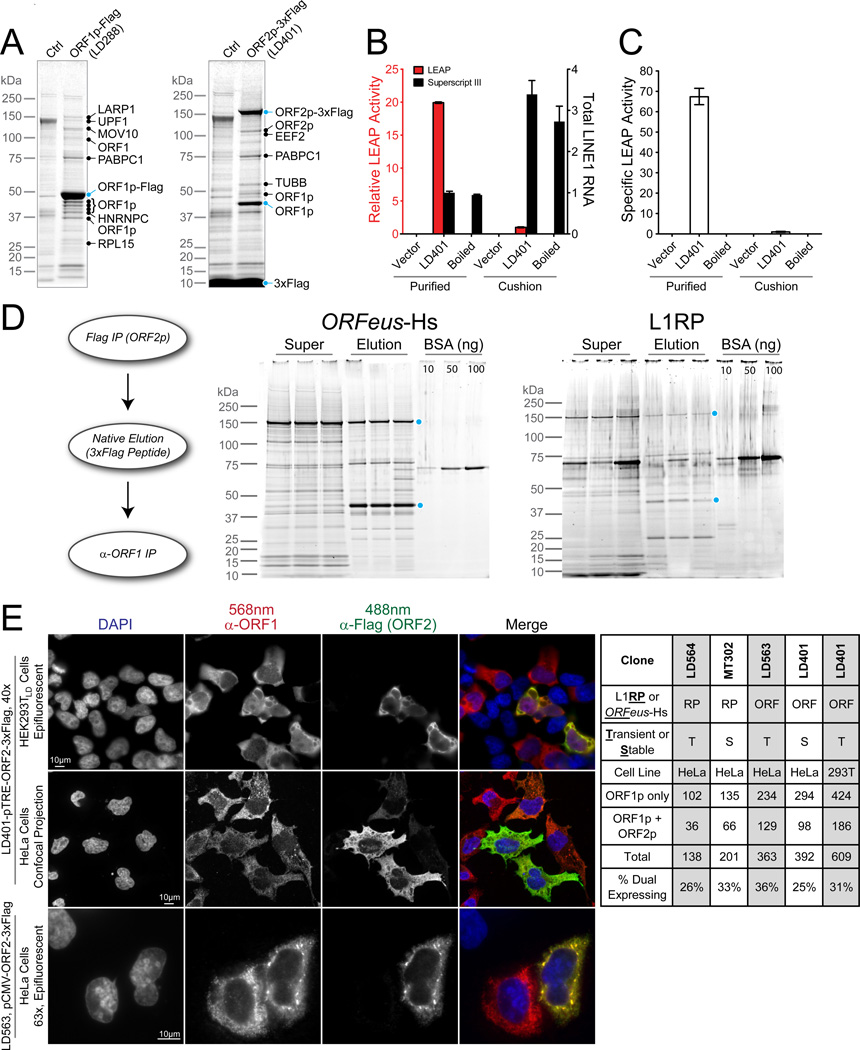
Figure 3. Characterization of highly purified L1 RNPs.
(A) L1 interactors characterized by MALDI-TOF-PMF MS. Ctrl – empty vector-containing cells. (B) Relative LEAP activity from affinity purified RNPs and RNPs made by sucrose cushion velocity sedimentation. Vector, RNP purified from cells transfected with empty pCEP puro vector; Boiled, RNP sample was boiled for 10 min at 100°C before the reaction. Data are represented as mean +/− SEM. (C) Specific LEAP activity normalized to total L1 mRNA in each sample. LEAP activity of RNP prepared from sucrose cushion was assigned as 1. Please see Figures S3 for calculations. Data are represented as mean +/− SEM. (D) Tandem affinity purification of L1 RNPs. L1 RNPs were first purified from ORF2p (3xFlag) and then ORF1p. Sypro Ruby stained supernatant (super) and elution from triplicate ORF1p purifications are shown for both ORFeus-Hs and L1RP; ORF1p and ORF2p levels were quantified against a BSA standard (Figure S4). (E) Indirect immunofluorescent imaging of ORF1p and ORF2p in HeLa and HEK293T cells. Ratio calculation for HeLa and HEK293T cells expressing ORF1p alone or both ORF1p and ORF2p is shown. Numbers in the table are counts of cells expressing ORF1p alone or both ORFs above background. Please see Figures S5 for more imaging and controls.
We assayed RT activity in our preparations with an exogenous homopolymeric template/primer. Pullouts from both tagged ORF1p (pLD288) and ORF2p (pLD401) expressing cells were active; ORF2p preparations exhibited ∼40-fold more activity (Figure S3), requiring dilution to prevent saturation of the assay. Signal in both types of purified RNPs, but not controls, was ∼doubled by addition of RNase A/T1, suggesting that additional RT activity may be masked by RNA-based interactions in RNPs.
We also used the LEAP (L1 Element Amplification Protocol) assay, a more stringent test measuring the ability of RNPs to reverse transcribe endogenous RNA upon addition of a specialized DNA primer. LEAP has demonstrated cis-preference of ORF2p in vitro; it is currently the best biochemical assay for functional coassembly of L1 RNA and proteins (Kulpa and Moran, 2006). Pullout RNPs exhibited 20-fold more LEAP activity than those isolated from traditional sucrose cushions (Figure 3B) but has 3.4-fold less L1 RNA, resulting in 67-fold higher specific activity (Figure 3C); product electrophoretic mobilities were similar. When signal was normalized to total RNA, pullout RNPs had >13,000 fold higher LEAP activity (Figure S3).
In RNASeq of pullout RNPs, L1 RNA represented 8.3–10.3% and 18.0–28.2% of mappable reads for ORF1p and ORF2p, respectively. U6 snRNA, but not other snRNAs, was enriched 2.5–5.9 fold in ORF2p pullouts as compared to total RNA, but not enriched in ORF1p pullouts, relative to housekeeping mRNA controls. This finding mechanistically supports U6/L1 hybrid elements commonly found in the human genome (Buzdin et al., 2002). Interestingly Alu RNA was not significantly enriched, possibly because Alu and ORF2 interactions are weaker or transient and may not have been retained in the stringent purification condition used. Alu RNA was identified in a similar study in which a crosslinking method was used (Mandal et al., 2013).
Relative amounts of L1 proteins
It is widely believed that L1 RNA is coated by ORF1p trimers to form an RNP containing one or two molecules of ORF2p, and this is supported by relative expression levels observed for ORF1p and ORF2p (Beck et al., 2011; Burns and Boeke, 2012; Doucet et al., 2010; Goodier and Kazazian, 2008). Each ORF1p trimer can protect ∼27 nucleotides and trimers can be spaced approximately every ∼45 nucleotides (Khazina et al., 2011). Thus a fully coated L1 RNA could contain as many as one ORF1p trimer every ∼75 nt, to a maximum of 80 trimers or 240 ORF1p molecules per 6kb L1 RNA, giving an ORF1p:ORF2p ratio of up to 240:1. In contrast, our initial observations, based on CBB staining of ORF2–3xFlag pullouts in both ORFeus-Hs and L1RP, suggested a dramatically lower ORF1p:ORF2p molar ratio of ∼3:1 (Figure S1).
Due to overexpression, some of the affinity captured ORF2–3xFlag protein may not be bound within a physiological L1 RNP, and this may skew the ORF1p:ORF2p ratio. To mitigate this possibility, we performed tandem pullout experiments: purifying first by ORF2p-3xFlag, natively eluting the complexes with 3xFlag peptide, and then re-purifying by ORF1p affinity. Hence, every ORF2p observed in the elution from a tandem pullout was physically linked to ORF1p throughout. We measured the relative ratio within these more homogenous L1 preparations containing both polypeptides (Figure 3D and Figure S4). Based on both Sypro and CBB staining intensity, we estimate that the ratio of ORF1p:ORF2p in tandem affinity isolated RNPs is approximately 6:1–9:1 for ORFeus and L1RP constructs (Figure S4). In an alternative approach, we utilized MS and implemented “Intensity-Based Absolute Quantification” (iBAQ) (Schwanhaeusser et al., 2011; Smits et al., 2013) to assess stoichiometry of ORF proteins. We found ratios of 47:1 ± 2 for particles derived from ORFeus, and 27:1 ± 7 for particles derived from L1RP (Table S5). Since the former method accounts only for full-length proteins and not partially degraded protein that survives the tandem pullout procedure, and the latter can detect peptides derived from either full length or partially degraded proteins, these results are not incompatible. Some degradation likely occurs during the tandem pullout despite the use of protease inhibitors. The higher ORF1:ORF2p ratios determined by iBAQ may better reflect actual ratios within RNPs, but is significantly lower than the ratio expected if L1 RNA were stably coated with ORF1p trimers.
To better understand the discrepancy between total and RNP ORF1p content, we examined intracellular distribution of the proteins by immunofluorescence in Tet-On HEK293TLD, Tet-On HeLa, and standard HeLa cells. The predominant localization of both proteins was cytoplasmic, although we noted fewer foci and greater staining uniformity than reported elsewhere (Doucet et al., 2010; Goodier et al., 2010; Goodier et al., 2007). Unexpectedly, we found only ∼30% of cells expressing ORF1p coexpress ORF2p (Figure 3E). This trend held in both Tet-On HEK293TLD and HeLa, when the inducible Tet promoter was replaced with a CMV promoter, and in both L1RP and ORFeus-Hs construct (Figures 3E and S5). In cells expressing both proteins, ORF1p and ORF2p colocalize. To verify that this ∼30% expression was due to L1 context and not an inherent property of ORF2p, we expressed ORF2p alone from a TRE promoter and found that >90% of cells express ORF2p (Figure S5, pLD561, diagrammed in Figure 5B).
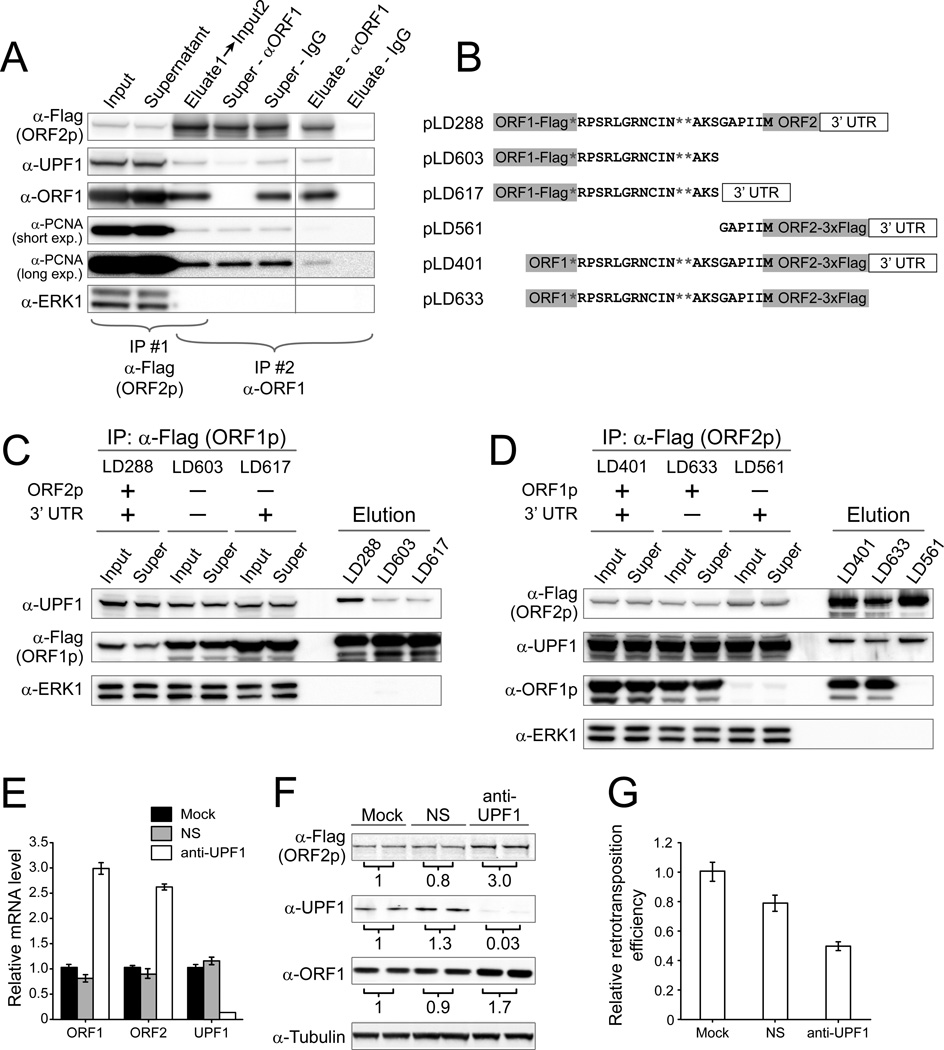
Figure 5. UPF1 has multiple effects on L1 expression and retrotransposition.
(A) UPF1 is co-purified in particles containing both ORF1p and ORF2p. L1 from pLD401 was affinity purified sequentially using anti-Flag first and, second, either anti-ORF1 or, as a control, IgG conjugated magnetic beads. Super, supernatant after purification. (B) Constructs used to test co-purification of UPF1. (C) UPF1 binding to L1 RNPs was significantly reduced without ORF2 and is not affected by the 3’ UTR. Please see Figures S7 for detailed calculations. (D) UPF1 and PCNA binding to the L1 RNP was unaffected by absence of ORF1 (LD561) or 3’ UTR (pLD633). (E–F) Knockdown of endogenous UPF1 increased L1 mRNA and protein levels. NS, non-silencing siRNA. Protein levels were normalized to a tubulin loading control, and numbers below each panel are the average fold change relative to mock transfection from two independent experiments and standard error is shown. (G) Knockdown of endogenous UPF1 reduced L1 retrotransposition efficiency. The retrotransposition efficiency from mock-treated cells was assigned as 1 and the values are averages of four independent experiments.
MS of L1-RNPs
Bolstered by CBB stain detection of PABPC1 and MOV10, we implemented I-DIRT (Tackett et al., 2005) in order to distinguish proteins that tightly associate with L1 RNPs from non-specific contaminants (Figure 4A). High-sensitivity LC-MS/MS (see abbreviations) analyses of six independent ORF1p and ORF2p pullouts from ORFeus-Hs- or L1RP-expressing cells identified 37 specific interactors from 755 sequenced candidates (Figure S6; Table S1). Results of a 5-min ORF1p-Flag (pLD288) and 30-min ORF2p-3xFlag (pLD401) pullout are shown in Figures 4B and C, respectively.
Although ORF1p and ORF2p reciprocally co-precipitate as expected, they also yield a number of differentially co-precipitated factors. For example, TROVE2, the Ro autoantigen known to bind mis-folded RNA (Fuchs et al., 2006), was found with high specificity in ORF1p pullouts only. PCNA, found with high specificity but low abundance in only ORF2p I-DIRT analyses, was followed up as a novel potentially nuclear factor. Other nuclear factors include DNA repair, transcription and chromatin factors (PARP1, PURA, PURB, RUVBL2, NAP1, TOP1 and ZCCHC3 proteins) an apoptosis regulator (HAX1), and a nuclear import factor (IPO7). These results are consistent with the possibility of ORF1p and ORF2p having both shared and distinct host interactomes. Notably, PABPC1, PABPC4, MOV10, UPF1, and ZCCHC3 were specific interactors in both ORF1p and ORF2p RNP preparations. While all these hits are candidates for mechanistic studies, we focused initial follow-up on UPF1 and PCNA. UPF1 was chosen because of its high abundance and obvious potential connections between bicistronic L1 mRNA and NMD.
UPF1 interaction requires ORF2 and downregulates L1 expression
We validated the L1/ UPF1 interaction by immunoblotting for UPF1 in ORF1p-Flag and ORF2p-3xFlag pullouts from cell lines expressing pLD288 and pLD401, respectively. UPF1 was detected in both ORF1p and ORF2p elutions and copurified through tandem pullouts from pLD401 expressing cells (Figure 5A). The L1/ UPF1 interaction in purified fractions was sensitive to RNase treatment (Figures 6G–H, see below).
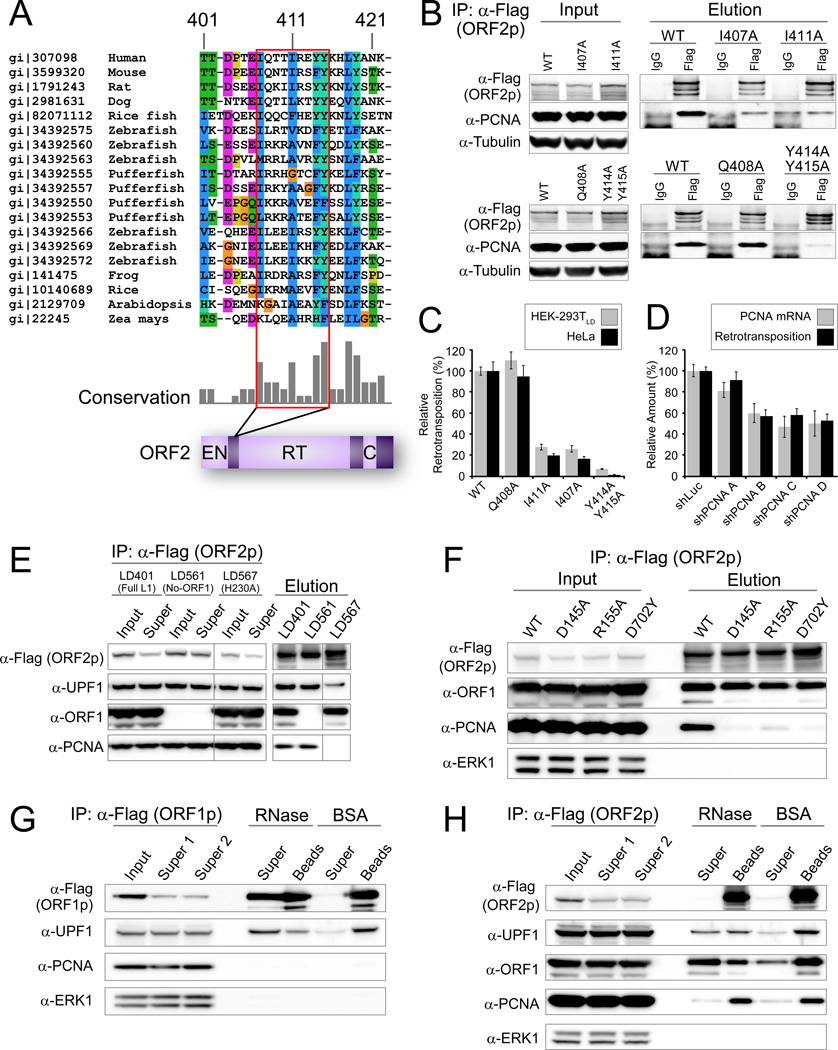
Figure 6. PCNA specifically interacts with conserved region within ORF2p in an EN and RT dependent way.
(A) Partial sequences alignment of LINE ORF2p from various species indicating the PIP box, a known PCNA interaction motif, outlined in red. PIP: QXX(V/L/M/I)XX(F/Y)(F/Y). (B) PCNA specifically co-ips with ORF2p and this interaction is dependent on the PIP box. (C) The PIP box is important for L1 retrotransposition activity in both Hela and HEK293T cells. The retrotransposition efficiency of the wild type L1 was assigned as 1; values are the average of three independent experiments standard error is shown. (D) Knockdown of endogenous PCNA decreased L1 retrotransposition and data are represented as mean +/− SEM. (E) PCNA-ORF2p co-ip is abolished in the ORF2p EN- mutant. UPF1 but not PCNA binding is retained in a mutant ORF2p lacking endonuclease activity (H230A). (F) PCNA-ORF2p co-ip is abolished in an RT- mutant (D702Y) and an additional ORF2p EN- mutant D145A; partially active EN mutant R155A has reduced PCNA recovery. (G) The interaction between UPF1 and ORF1p is RNase sensitive. pLD288 purified with anti-Flag Dynabeads and treated on-bead with RNases A/T1 or mock-treated with BSA. “Super”, supernatants; “Beads” samples retained on beads after washing and eluted with LDS sample buffer. (H) ORF2p–PCNA and ORF2p–ORF2p interactions are RNase resistant, and ORF2p–ORF1p and ORF2p–UPF1 interactions are RNase sensitive. pLD401 material was purified with anti-Flag Dynabeads and treated as in 6G.
In addition to recognizing mRNAs bearing premature nonsense codons, UPF1 affects expression of intronless and bicistronic mRNAs (e.g. MAP3K14, ARHGEF18) (Mendell et al., 2004). Within the L1 mRNA, a 63 nt inter-ORF region contains two additional stop codons in frame with ORF1 (Figure 5B). We hypothesized that the three inter-ORF stop codons in conjunction with a long “3’UTR-mimic” (∼4 kb ORF2 plus the native 3’ UTR) might be recognized by UPF1 (Hogg and Goff, 2010). It is known that UPF1 also binds to and scans 3’ UTRs of many cellular mRNAs (Shigeoka et al., 2012). We constructed several plasmids with different combinations of ORF, interORF and 3’ UTR sequences to map interacting sequences (Figure 5B).
ORF1p affinity isolations demonstrate that removal of ORF2p results in higher yields of both ORF1p protein and L1 RNA but dramatically reduced yield of UPF1; removal of the 3’ UTR had no effect (Figure S7 and Figure 5C). Similarly, in ORF2p affinity isolations, removal either the 3’ UTR or ORF1p had no effect on the yield of UPF1 (Supplemental text, Figures 5D and S7). Although the yield of ORF2p in the three purifications differed slightly, the UPF1 and ORF2p signal intensities precisely correlate (r2=0.995) (Figure S7G). In summary, the ORF2 coding sequence or ORF2p expression, rather than the interORF sequence, ORF1p sequence, bicistronic context, or 3’UTR, enable the L1-UPF1 interaction.
Next we investigated the impact of UPF1 on L1 expression by knocking down endogenous UPF1 by RNA interference. L1 mRNA and polypeptides were all up-regulated in the knockdown cells. Surprisingly, L1 retrotransposition efficiency decreased by ∼50% (Figures 5E–G and Figures S7C–F). To minimize the possibility of off-target RNAi effects, we co-expressed L1 with 5 different anti-UPF1 shRNAs. Four of these reduced retrotransposition significantly by as much as 90%, suggesting UPF1 depletion decreases retrotransposition. Knockdown of two other proteins that act in concert with UPF1 in NMD, UPF2 and UPF3a, also reduced retrotransposition and increased L1 mRNA levels (Table S6).
PCNA binding to an ORF2 PIP-box is critical for retrotransposition
PCNA was found as a low-abundance but high specificity interactor of ORF2p but not ORF1p. We identified a canonical PCNA Interacting Protein (PIP) box (Qxx[V/L/M/I]xx[F/Y][F/Y]) in residues 407–415 of ORF2p, located between EN and RT domains (Figure 6A). Within the PIP sequence, four amino acid residues (I407, I411, Y414 and Y415) are highly conserved across L1 ORF2s from diverse species, suggesting a crucial role for a PIP box in ORF2 binding to PCNA. Mutation of these residues to alanine (A) disrupted ORF2p/PCNA copurification, while mutation of Q408, less conserved in ORF2p, does not affect it (Figure 6B). Mutations that disrupt the ORF2p /PCNA interaction correspondingly decrease retrotransposition efficiency of L1, whereas the Q408A mutation shows no effect on retrotransposition (Figure 6C). Overall, these data support an ORF2p/PCNA interaction through a PIP box and that this interaction is critical for retrotransposition. We also knocked down endogenous PCNA using four shRNAs and observed down-regulation of L1 retrotransposition well-correlated with decreased PCNA levels (r2 = 0.93, Figure 6D). To investigate the L1 lifecycle stage at which L1/PCNA interaction functions, we assayed for maintenance of this interaction in cells expressing an ORF2p EN domain catalytic mutant (H230A) (Figure 6E), previously shown to abolish DNA nicking and L1 retrotransposition (Feng et al., 1996). Co-purification of PCNA was greatly reduced in RNPs isolated from the endonuclease domain mutant (Figure 6E). To rule out the possibility that mutations in the EN domain disrupt overall ORF2p folding, we repeated the experiment with 6 additional well-studied EN Mutants (E43A, D145A, R155A, T192Y, I204Y D205G; Figures 6F and S7) (Repanas et al., 2009; Repanas et al., 2007; Weichenrieder et al., 2004). We measured RT activity of these EN mutants and all were highly active suggesting proper folding (Figure S7). Similarly, a catalytic RT mutant (D702Y) also abolished ORF2p-PCNA interaction (Figure 6F). These observations suggest that PCNA may be recruited to ORF2 after completion of the target DNA nicking and during or after L1 cDNA synthesis steps.
Since PCNA is involved in DNA repair pathways, we also studied other L1 RNP factors with possible roles in DNA repair in the L1 retrotransposition assay. We found that knocking down PARP1, RUVBL1, and RUVBL2 reduced L1 activity (Table S6), consistent with possible roles as retrotransposition host factors.
Connectivity and forms of L1 RNPs
To investigate the RNA-dependent connectivity within these L1 RNP particles, affinity captured ORF1p-Flag and ORF2–3xFlag fractions were incubated with a mixture of RNases A and T1 or subjected to mock treatments while immobilized on magnetic beads. When ORF1p was immobilized, ORF1p itself and UPF1 and ORF2p were all RNase sensitive, suggesting interaction via L1 RNA (Figures 2J and 6G). PCNA was undetectable following ORF1p purification. When ORF2p was immobilized, ORF1p and UPF1 were RNase sensitive, but PCNA and ORF2p itself were insensitive (Figure 6H). PCNA was dramatically reduced following ORF1p purification (Figure 5A).
The combination of the RNase sensitivity data, the immunofluorescence data, and the differential purification leads us to propose (at least) these three distinct RNP forms (Figure 7A): (I) cytoplasmic and (II) nuclear RNPs in ORF1p+ORF2p expressing cells and (III) cytoplasmic RNPs in cells expressing ORF1p alone. However, it is likely that this simplistic classification hides additional complexity in the form of additional RNP types.
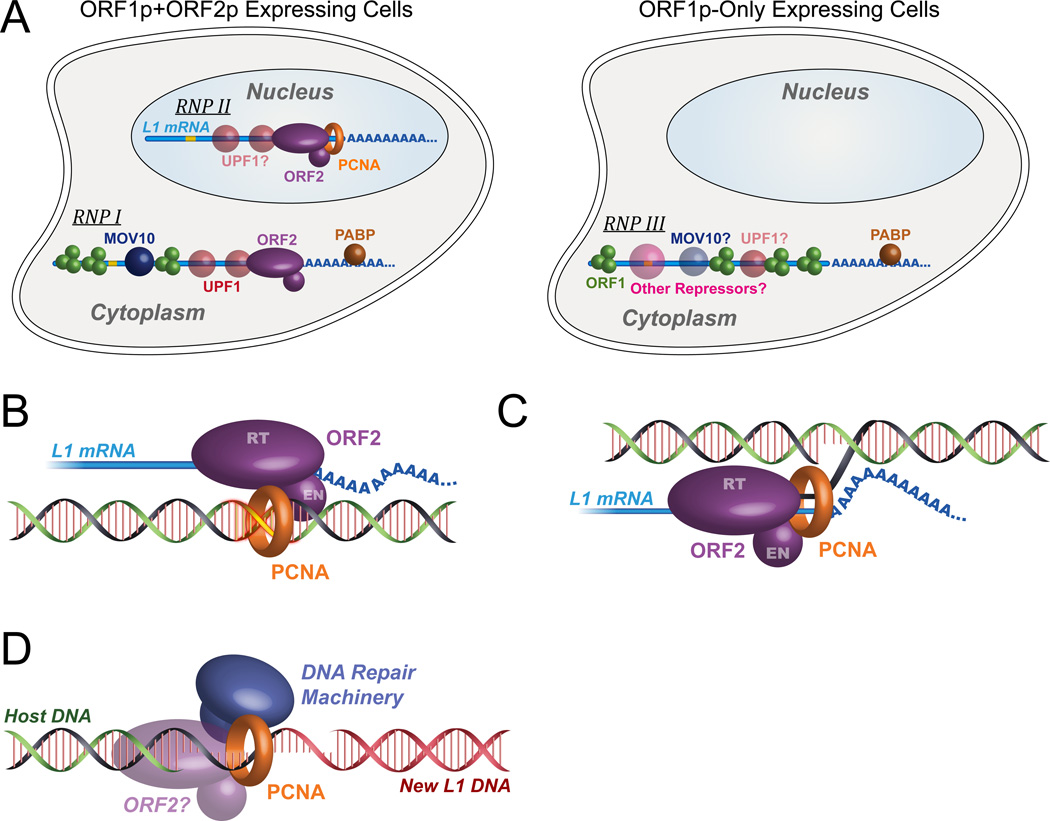
Figure 7. Proposed models for L1 RNP complex and its interaction with UPF1 and PCNA.
(A) Proposed model of distinct L1 RNPs in the cytoplasm and nucleus. Green sphere, ORF1p; purple oval, ORF2p; red sphere, UPF1; blue sphere, MOV10; brown sphere, PABPC1, orange ring, PCNA; blue line, L1 mRNA, yellow line, inter-ORF region of the L1 RNA, containing three in-frame stop codons. (B) PCNA Model 1: ORF2p uses PCNA to find insertion sites, at pre-existing nicks or sites of DNA damage (yellow). (C) PCNA Model 2: PCNA is a processivity factor, and may coordinate RNase activities on the newly-synthesized DNA-RNA Hybrid. (D) PCNA Model 3: PCNA is recruited by ORF2p to coordinate post-insertion DNA repair, perhaps including second-strand synthesis.
Discussion
As a result of its high copy number and ability to mobilize itself and non-autonomous retro-elements like Alu, SVA, and processed pseudogenes, L1 is considered the most successful parasite in the human genome. Viewed in this light, physical interactions between L1 proteins and host factors likely reflects a long-standing evolutionary battle between the parasite, which exploits opportunities to proliferate, and the host, which fights to suppress L1 accumulation. Dissecting the dynamics of this L1-host “arms race” will help better decipher our genome dynamics in relation to cancer, brain development, reproduction, and potentially to other human diseases.
The construction of synthetic LINE-1 elements has facilitated L1 expression in mammalian cells for in vitro and in vivo studies (An et al., 2011; Han and Boeke, 2004). Another facilitator of this study was the adaptation of adherent cells to suspension culture, dramatically reducing space, cost, and labor needed to maintain 109–1010 cells. Exploration of epitope tags unexpectedly revealed that some tags compatible with ORFeus-Hs were not necessarily compatible with L1RP at the same position. This may reflect differences in local RNA structure, overall translational context, or complex effects on protein/RNP structure. Cryogenic milling of frozen cells allowed us to prepare large batches of frozen solid powder usable over long periods of experimentation. Experiments within and between batches were easily calibrated to use the same mass of cell powder in each experiment. These advantages allowed us to minimize experimental variation, perform extensive optimization, and develop efficient pullout procedures including only five minutes of binding.
Comparing native and synthetic elements, ORF1p levels differed by ∼2.5-fold but we found that ORF2p levels differ by ∼20–40-fold, mirroring the ∼40-fold difference seen previously in the RNA levels (An et al., 2011). Given the modest increase in retrotransposition observed in ORFeus-Hs expressing cells, bulk L1 expression may not limit retrotransposition in these models. With absolute efficiencies under 20% in even the most permissive cell lines, host defenses may result in seemingly stochastic expression of ORF2p in just 30% of cells, which may in part explain the limitation. Alternatively, lack of balance in expression of the RNA and the two ORFs may explain observed nonlinear increases in retrotransposition. Conversely, the sensitivity of the L1RP to tag sequences that are well-tolerated in ORFeus-Hs may suggest that higher expression can compensate for impaired L1 protein function.
Unlike other non-LTR transposons, such as the R2 element, L1 ORF2p is translated sparingly. ORF2p detection has been challenging even by immunoblotting with the help of epitope tags. This is now routine, and moreover our method provides L1 RNPs at previously unobtainable yield and purity. Most importantly, affinity isolated RNPs were highly active in vitro in LEAP and RT assays. The high activity of the RNPs in vivo and in vitro should facilitate further biochemical dissection of the L1 retrotransposition mechanism.
Pullout experiments with ORF1 or ORF2 shed light on relative stoichiometry in RNPs. A low ORF1p:ORF2p ratio challenges widely held assumptions about RNP composition. RNase insensitivity of immobilized ORF2p in the context of RNase sensitivity of other RNP components is consistent with a single ORF2p copy per mRNA; however, we cannot rule out ORF2p dimerization, dual tethering to the affinity matrix, or RNase-insensitive interactions. Instead of a constant L1 protein ratio in each cell, imaging studies revealed that only a minority of cells express both proteins. These data suggest that the host may combat L1 by regulating ORF2p translation or stability to produce deleterious ORF1p:ORF2p ratios. This dual nature of cell types may be cell-cycle regulated, epigenetically controlled, or a response to other signals. In light of this, some interactors found bound only to ORF1p may represent interactions formed in cells not expressing ORF2p. Surprisingly, we do not observe nuclear localization of ORF2p by immunofluorescence. At least a fraction of it must be nuclear, given that the target DNA is located there and that ORF2p/PCNA interaction requires ORF2p EN and RT activities.
In this study, we identify 37 high confidence interactors among 755 candidates. Many known interactors were recovered with confidence; some were not recovered, presumably because our methods enrich stable interactors. A recent study reported 96 L1 ORF1 interactors (Goodier et al., 2013) of which we found 69; seven (10%) were significant using the I-DIRT threshold employed (Table S7).
Models for UPF1’s role and L1 expression effects
Nonsense-mediated decay (NMD) is an important pathway for maintenance of high fidelity gene expression. In NMD, UPF1 is a central player, recruiting other factors to initiate targeted degradation of mRNAs containing premature termination codons. UPF1 also targets bicistronic and intronless mRNAs, and RNA sequencing of UPF1-depleted HeLa cells revealed hundreds of down-regulated endogenous transcripts (Mendell et al., 2004; Tani et al., 2012). L1 bicistronic mRNA seemed reasonable as a target of UPF1. Consistent with this model, UPF1 co-purification with ORF1p-Flag was greatly reduced when ORF2p coding sequence was removed. However, UPF1 co-purification with ORF2p-3xFLAG was retained on deletion of ORF1 and the inter-ORF region (Figures 5B–D). Results from additional constructs ruled out the 3’UTR as critical for UPF1 binding (Figure 5C). Thus ORF1, the stop codons, inter-ORF region, bicistronic transcript structure, and 3’ UTR are all dispensable for UPF1 recruitment. Therefore, UPF1 recognition of L1 RNA does not resemble canonical NMD. Additionally, RNA is critical to the interaction, as the ORF2p/UPF1 interaction exhibits sensitivity to RNase treatment (Figure 6G).
Taken together, these data suggest that L1-UPF1 interaction occurs through one of three modes: i) through ORF2 RNA; ii) through the ORF2p-L1 RNA complex; or iii) through the ORF2 protein, but stabilized when ORF2p is also bound to RNA. The first model seems unlikely because both ORFeus and L1RP recruit UPF1 even though their ORF2 RNA sequences are quite different. Indeed, UPF1 presents as a positive regulator of retrotransposition activity, despite negatively regulating the levels of L1 constituents (Figures 5E–G), leaving open the possibility of UPF1 affecting other RNP-dependent processes. Furthermore, like the UPF1, UPF2 and UPF3a knockdowns paradoxically increased L1 RNA levels while decreasing retrotransposition. In conclusion, the data support a positive role for UPF1, UPF2, and UPF3a in L1 RNP function while at the same time demonstrating repression of protein and RNA levels through a mechanism distinct from the canonical NMD pathway.
Models for PCNA’s role in the L1 lifecycle
We showed that the ORF2 protein binds to the sliding DNA clamp protein PCNA through a canonical PIP domain. Disruption of the conserved amino acid residues within the ORF2 PIP sequence abrogated ORF2p binding to PCNA and inhibited L1 retrotransposition. The well-established role of PCNA in DNA replication and DNA damage repair led to three hypotheses for how PCNA supports L1 retrotransposition: 1) ORF2p may be recruited by PCNA onto genomic DNA (or led to it) and thereby enable efficient scanning of gDNA until a preferred endonuclease site or a preformed nick is found in target DNA (Figure 7B); 2) PCNA may act as a processivity factor for L1 RT (Figure 7C); or 3) PCNA may be recruited by ORF2p to repair the “damage” in the form of nicks, gaps, or RNA flaps at junctions between retrotransposon and host DNAs (Figure 7D). These hypotheses, especially #2 and #3, are not mutually exclusive.
According to the first model, PCNA may function as a scaffolding protein that “scans” genomic DNA either during DNA replication or during DNA damage repair, to find permissive sites for L1 retrotransposition. It has been previously speculated (Cost et al., 2002; Farkash et al., 2006) that L1 may use DNA damage sites as targets for retrotransposition (Coufal et al., 2011). Also, in cell lines deficient for non-homologous end-joining repair (NHEJ), L1 can use DNA damage sites for retrotransposition and this event is independent of the ORF2p endonuclease activity (Morrish et al., 2002). All of these findings make such a scanning model of some interest. However, in contrast with this model, our co-immunoprecipitation experiments using EN-mutant or RT-mutant elements clearly show that PCNA binding to ORF2p depends on EN and RT activities, and are thus more consistent with its recruitment during or subsequent to TPRT. These observations are most consistent with a model in which PCNA is involved in a step after target DNA nicking and L1 cDNA synthesis. All EN mutants maintained high RT activity, and several of them have crystal structures closely resembling the wild-type enzyme, minimizing the possibility that EN mutations disrupt overall folding of ORF2p and thereby the physical interaction between ORF2p and PCNA.
Further, PCNA is known to be capable of recruiting repair enzymes like lesion bypass polymerases and ligases (Beattie and Bell, 2011; Ulrich, 2011), and most interestingly, has been specifically implicated in recruiting RNase H2 to RNA/DNA hybrids in genomic DNA (Bubeck et al., 2011). Mammalian L1 elements do not encode a recognizable RNase H activity, unlike most other retrotransposons (including non-LTR elements like L1 in other organisms such as the Drosophila I factor), even though the activity is presumably required to degrade the original RNA strand, allowing second DNA strand synthesis. Thus RNase H binding activity of PCNA would be highly consistent with a role in DNA elongation during reverse transcription, and provide a convenient link to the RNase H domain of reverse transcriptase that is “missing” from mammalian L1 elements. While this model may be appealing, the lack of interaction with the RT mutant presents a challenge to it; however, it is formally possible that only a few base pairs of synthesis are required to enable PCNA engagement.
Other co-purified factors may be associated with nuclear L1 RNPs through PCNA or in relation to PCNA. PURA, PURB, MEPCE, and PCNA have been shown to be associated with PCNA clamp-loaders RFC1-5 (Havugimana et al., 2012; Kubota et al., 2013). Similarly, RUVBL1 and RUVBL2 are co-purified ring-shaped proteins implicated in DNA damage response (Jha and Dutta, 2009; Rosenbaum et al., 2013). PARP1 may provide a link between nuclear and mitochondrial-derived interactors of ORF2p via DNA damage repair and apoptosis (Hong et al., 2004), and interestingly, PARP1 modifies both PCNA and TOP1, both high-confidence interactors of ORF2p (Simbulan-Rosenthal et al., 1999). We did not recover any APOBEC3 family cytidine deaminases, known inhibitors of LINE1 activity with mixed nuclear and cytoplasmic localizations (Bogerd et al., 2006); however, these proteins may not be expressed highly (or at all) in HEK293T cells and this represents a general limitation of this study. Knockdown of endogenous PARP1, RUVBL1 and RUVBL2 resulted in reduced retrotransposition, suggesting additional roles for other enzymes involved in generic DNA repair in the retrotransposition pathway.
Distinct RNPs at Different Lifecycle Stages?
Our data are consistent with isolation of L1 RNPs of at least three distinct types, (Figure 7A). First, some RNPs purified using ORF1p are likely isolated from cells not expressing ORF2p, perhaps a state in which the host has defeated L1 by preventing ORF2p expression (RNP III). Focusing on the tandem pullout (Figure 5A), we identify two additional distinct populations: 1) the elution, containing all detectable ORF1p, the majority of UPF1, and only a trace amount of PCNA (RNP I), and 2) the supernatant depleted for ORF1p but containing the vast majority of the PCNA and a minority, but nevertheless substantial amount of UPF1 (RNP II).
Taken together with the idea that PCNA may be tightly and specifically bound to ORF2p only after initiation of TPRT, this suggests that these two populations of L1 RNPs, while both retaining ORF2p, interact with ORF1p, PCNA, and UPF1 in distinct ways: RNP I is the tandem-purified RNP, present in the cytoplasm, primarily containing ORF1p/ORF2p/UPF1 in which UPF1 is active in a negative regulatory capacity, and RNP II is a distinct complex in nuclei, primarily containing ORF2p/PCNA with lesser amounts of UPF1, and seemingly lacking ORF1p. This suggests ORF1p may be absent in steps subsequent to EN activity. Interestingly, UPF1 has been linked to Pol δ in DNA replication/repair (Azzalin and Lingner, 2006), and may play a second role here as a permissive host factor; this could explain the unexpectedly reduced retrotransposition in the context of UPF1 knockdown.
Experimental Procedures
Materials and experimental procedures are detailed in Extended Experimental Procedures.
Supplementary Material
Research Highlights.
Isolated highly active LINE-1 ribonucleoprotein particle complexes from human cells
37 identified interactors comprise known and novel factors, notably UPF1 and PCNA
L1 RNP linked by a both protein-protein (PCNA) and protein-RNA (UPF1) interactions
PCNA binds to ORF2p via a PIP box and is critical for retrotransposition
Acknowledgements
We thank Jeffrey Han for gifts of antibodies and helpful discussions. We thank Phil Cole and Carolyn Machamer for helpful discussion and critical reading of the manuscript. We thank Jennifer Wang, Chih-Yung Lee and Geraldine Seydoux for assistance with confocal microscopy and helpful discussions. We thank Yana Li, Dan Leahy, Jennifer Kavran, and Jacqueline McCabe for help with suspension cell culture. DNA template for artwork in Figure 7 was adapted from www.dragonartz.net, with permission. Supported in part by NIH grants U54 GM103511 to M.P.R. and B.T.C. R01 GM36481 and U54 GM103520 to J.D.B.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alisch RS, Garcia-Perez JL, Muotri AR, Gage FH, Moran JV. Unconventional translation of mammalian LINE-1 retrotransposons. Genes & development. 2006;20:210–224. doi: 10.1101/gad.1380406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An W, Dai L, Niewiadomska AM, Yetil A, O'Donnell KA, Han JS, Boeke JD. Characterization of a synthetic human LINE-1 retrotransposon ORFeus-Hs. Mobile DNA. 2011;2:2. doi: 10.1186/1759-8753-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arjan-Odedra S, Swanson CM, Sherer NM, Wolinsky SM, Malim MH. Endogenous MOV10 inhibits the retrotransposition of endogenous retroelements but not the replication of exogenous retroviruses. Retrovirology. 2012;9 doi: 10.1186/1742-4690-9-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanikar JN, Badge RM, Moran JV. A YY1-binding site is required for accurate human LINE-1 transcription initiation. Nucleic Acids Res. 2004;32:3846–3855. doi: 10.1093/nar/gkh698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzalin CM, Lingner J. The human RNA surveillance factor UPF1 is required for S phase progression and genome stability. Current Biology. 2006;16:433–439. doi: 10.1016/j.cub.2006.01.018. [DOI] [PubMed] [Google Scholar]
- Beattie TR, Bell SD. The role of the DNA sliding clamp in Okazaki fragment maturation in archaea and eukaryotes. Biochem Soc Trans. 2011;39:70–76. doi: 10.1042/BST0390070. [DOI] [PubMed] [Google Scholar]
- Beauregard A, Curcio MJ, Belfort M. The take and give between retrotransposable elements and their hosts. Annu Rev Genet. 2008;42:587–617. doi: 10.1146/annurev.genet.42.110807.091549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck CR, Garcia-Perez JL, Badge RM, Moran JV. LINE-1 elements in structural variation and disease. Annu Rev Genomics Hum Genet. 2011;12:187–215. doi: 10.1146/annurev-genom-082509-141802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belancio VP, Whelton M, Deininger P. Requirements for polyadenylation at the 3' end of LINE-1 elements. Gene. 2007;390:98–107. doi: 10.1016/j.gene.2006.07.029. [DOI] [PubMed] [Google Scholar]
- Bogerd HP, Wiegand HL, Hulme AE, Garcia-Perez JL, O'Shea KS, Moran JV, Cullen BR. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:8780–8785. doi: 10.1073/pnas.0603313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, Kazazian HH., Jr Hot L1s account for the bulk of retrotransposition in the human population. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:5280. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubeck D, Reijns MA, Graham SC, Astell KR, Jones EY, Jackson AP. PCNA directs type 2 RNase H activity on DNA replication and repair substrates. Nucleic Acids Res. 2011;39:3652–3666. doi: 10.1093/nar/gkq980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns KH, Boeke JD. Human transposon tectonics. Cell. 2012;149:740–752. doi: 10.1016/j.cell.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzdin A, Ustyugova S, Gogvadze E, Vinogradova T, Lebedev Y, Sverdlov E. A new family of chimeric retrotranscripts formed by a full copy of U6 small nuclear RNA fused to the 3' terminus of l1. Genomics. 2002;80:402. doi: 10.1006/geno.2002.6843. [DOI] [PubMed] [Google Scholar]
- Cost GJ, Feng Q, Jacquier A, Boeke JD. Human L1 element target-primed reverse transcription in vitro. Embo J. 2002;21:5899. doi: 10.1093/emboj/cdf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coufal NG, Garcia-Perez JL, Peng GE, Marchetto MC, Muotri AR, Mu Y, Carson CT, Macia A, Moran JV, Gage FH. Ataxia telangiectasia mutated (ATM) modulates long interspersed element-1 (L1) retrotransposition in human neural stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:20382–20387. doi: 10.1073/pnas.1100273108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature biotechnology. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Cristea I, Williams R, Chait B, Rout M. Fluorescent proteins as proteomic probes. Molecular & cellular proteomics : MCP. 2005;4:1933–1941. doi: 10.1074/mcp.M500227-MCP200. [DOI] [PubMed] [Google Scholar]
- Dai L, Taylor MS, O'Donnell KA, Boeke JD. Poly(A) binding protein C1 is essential for efficient L1 retrotransposition and affects L1 RNP formation. Molecular and cellular biology. 2012 doi: 10.1128/MCB.06785-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Virgilio M, Callen E, Yamane A, Zhang W, Jankovic M, Gitlin AD, Feldhahn N, Resch W, Oliveira TY, Chait BT, et al. Rif1 prevents resection of DNA breaks and promotes immunoglobulin class switching. Science. 2013;339:711–715. doi: 10.1126/science.1230624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domanski M, Molloy K, Jiang H, Chait BT, Rout MP, Jensen TH, LaCava J. Improved methodology for the affinity isolation of human protein complexes expressed at near endogenous levels. BioTechniques. 2012;0:1–6. doi: 10.2144/000113864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet AJ, Hulme AE, Sahinovic E, Kulpa DA, Moldovan JB, Kopera HC, Athanikar JN, Hasnaoui M, Bucheton A, Moran JV, et al. Characterization of LINE-1 ribonucleoprotein particles. PLoS genetics. 2010;6 doi: 10.1371/journal.pgen.1001150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkash EA, Kao GD, Horman SR, Prak ET. Gamma radiation increases endonuclease-dependent L1 retrotransposition in a cultured cell assay. Nucleic Acids Res. 2006;34:1196–1204. doi: 10.1093/nar/gkj522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- Fuchs G, Stein AJ, Fu C, Reinisch KM, Wolin SL. Structural and biochemical basis for misfolded RNA recognition by the Ro autoantigen. Nature structural & molecular biology. 2006;13:1002–1009. doi: 10.1038/nsmb1156. [DOI] [PubMed] [Google Scholar]
- Goodier JL, Cheung LE, Kazazian HH., Jr MOV10 RNA helicase is a potent inhibitor of retrotransposition in cells. PLoS Genet. 2012;8:e1002941. doi: 10.1371/journal.pgen.1002941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier JL, Cheung LE, Kazazian HH., Jr Mapping the LINE1 ORF1 protein interactome reveals associated inhibitors of human retrotransposition. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier JL, Kazazian HH., Jr Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell. 2008;135:23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- Goodier JL, Mandal PK, Zhang L, Kazazian HH., Jr Discrete subcellular partitioning of human retrotransposon RNAs despite a common mechanism of genome insertion. Human molecular genetics. 2010;19:1712–1725. doi: 10.1093/hmg/ddq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier JL, Zhang L, Vetter MR, Kazazian HH., Jr LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex. Mol Cell Biol. 2007;27:6469–6483. doi: 10.1128/MCB.00332-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JS, Boeke JD. A highly active synthetic mammalian retrotransposon. Nature. 2004;429:314–318. doi: 10.1038/nature02535. [DOI] [PubMed] [Google Scholar]
- Hata K, Sakaki Y. Identification of critical CpG sites for repression of L1 transcription by DNA methylation. Gene. 1997;189:227–234. doi: 10.1016/s0378-1119(96)00856-6. [DOI] [PubMed] [Google Scholar]
- Havugimana PC, Hart GT, Nepusz T, Yang H, Turinsky AL, Li Z, Wang PI, Boutz DR, Fong V, Phanse S, et al. A census of human soluble protein complexes. Cell. 2012;150:1068–1081. doi: 10.1016/j.cell.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernan R, Heuermann K, Brizzard B. Multiple epitope tagging of expressed proteins for enhanced detection. BioTechniques. 2000;28:789–793. doi: 10.2144/00284pf01. [DOI] [PubMed] [Google Scholar]
- Hogg JR, Goff SP. Upf1 senses 3'UTR length to potentiate mRNA decay. Cell. 2010;143:379–389. doi: 10.1016/j.cell.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohjoh H, Singer MF. Sequence-specific single-strand RNA binding protein encoded by the human LINE-1 retrotransposon. EMBO J. 1997;16:6034–6043. doi: 10.1093/emboj/16.19.6034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SJ, Dawson TM, Dawson VL. Nuclear and mitochondrial conversations in cell death: PARP-1 and AIF signaling. Trends Pharmacol Sci. 2004;25:259–264. doi: 10.1016/j.tips.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Jha S, Dutta A. RVB1/RVB2: running rings around molecular biology. Mol Cell. 2009;34:521–533. doi: 10.1016/j.molcel.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan H, Smit A, Boissinot S. Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. Genome research. 2006;16:78–87. doi: 10.1101/gr.4001406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khazina E, Truffault V, Buttner R, Schmidt S, Coles M, Weichenrieder O. Trimeric structure and flexibility of the L1ORF1 protein in human L1 retrotransposition. Nature structural & molecular biology. 2011;18:1006–1014. doi: 10.1038/nsmb.2097. [DOI] [PubMed] [Google Scholar]
- Kimberland ML, Divoky V, Prchal J, Schwahn U, Berger W, Kazazian HH., Jr Full-length human L1 insertions retain the capacity for high frequency retrotransposition in cultured cells. Human molecular genetics. 1999;8:1557–1560. doi: 10.1093/hmg/8.8.1557. [DOI] [PubMed] [Google Scholar]
- Kubota T, Nishimura K, Kanemaki MT, Donaldson AD. The Elg1 Replication Factor C-like Complex Functions in PCNA Unloading during DNA Replication. Mol Cell. 2013;50:273–280. doi: 10.1016/j.molcel.2013.02.012. [DOI] [PubMed] [Google Scholar]
- Kulpa DA, Moran JV. Ribonucleoprotein particle formation is necessary but not sufficient for LINE-1 retrotransposition. Human molecular genetics. 2005;14:3237. doi: 10.1093/hmg/ddi354. [DOI] [PubMed] [Google Scholar]
- Kulpa DA, Moran JV. Cis-preferential LINE-1 reverse transcriptase activity in ribonucleoprotein particles. Nature structural & molecular biology. 2006;13:655–660. doi: 10.1038/nsmb1107. [DOI] [PubMed] [Google Scholar]
- Lee DJ, Busby SJW, Westblade LF, Chait BT. Affinity isolation and I-DIRT mass spectrometric analysis of the Escherichia coli O157:H7 Sakai RNA polymerase complex. Journal of bacteriology. 2008;190:1284–1289. doi: 10.1128/JB.01599-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan DD, Eickbush TH. RNA template requirements for target DNA-primed reverse transcription by the R2 retrotransposable element. Molecular and cellular biology. 1995;15:3882. doi: 10.1128/mcb.15.7.3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik HS, Burke WD, Eickbush TH. The age and evolution of non-LTR retrotransposable elements. Mol Biol Evol. 1999;16:793–805. doi: 10.1093/oxfordjournals.molbev.a026164. [DOI] [PubMed] [Google Scholar]
- Mandal PK, Ewing AD, Hancks DC, Kazazian HH., Jr Enrichment of processed pseudogene transcripts in L1-ribonucleoprotein particles. Human molecular genetics. 2013 doi: 10.1093/hmg/ddt225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SL, Bushman FD. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Molecular and cellular biology. 2001;21:467. doi: 10.1128/MCB.21.2.467-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathias SL, Scott AF, Kazazian HH, Jr, Boeke JD, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991;254:1808–1810. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nature genetics. 2004;36:1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- Morrish TA, Gilbert N, Myers JS, Vincent BJ, Stamato TD, Taccioli GE, Batzer MA, Moran JV. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nature genetics. 2002;31:159–165. doi: 10.1038/ng898. [DOI] [PubMed] [Google Scholar]
- Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- Niewiadomska AM, Tian C, Tan L, Wang T, Sarkis PT, Yu XF. Differential inhibition of long interspersed element 1 by APOBEC3 does not correlate with high-molecular-mass-complex formation or P-body association. JVirol. 2007;81:9577. doi: 10.1128/JVI.02800-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell KA, An W, Schrum CT, Wheelan SJ, Boeke JD. Controlled insertional mutagenesis using a LINE-1 (ORFeus) gene-trap mouse model. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:2706–2713. doi: 10.1073/pnas.1302504110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeffinger M, Wei KE, Rogers R, DeGrasse JA, Chait BT, Aitchison JD, Rout MP. Comprehensive analysis of diverse ribonucleoprotein complexes. Nature methods. 2007;4:951–956. doi: 10.1038/nmeth1101. [DOI] [PubMed] [Google Scholar]
- Ostertag EM, Kazazian HH., Jr Biology of mammalian L1 retrotransposons. AnnuRevGenet. 2001;35:501. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- Peddigari S, Li PW, Rabe JL, Martin SL. hnRNPL and nucleolin bind LINE-1 RNA and function as host factors to modulate retrotransposition. Nucleic Acids Res. 2013;41:575–585. doi: 10.1093/nar/gks1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repanas K, Fuentes G, Cohen SX, Bonvin AM, Perrakis A. Insights into the DNA cleavage mechanism of human LINE-1 retrotransposon endonuclease. Proteins. 2009;74:917–928. doi: 10.1002/prot.22201. [DOI] [PubMed] [Google Scholar]
- Repanas K, Zingler N, Layer LE, Schumann GG, Perrakis A, Weichenrieder O. Determinants for DNA target structure selectivity of the human LINE-1 retrotransposon endonuclease. Nucleic Acids Res. 2007;35:4914–4926. doi: 10.1093/nar/gkm516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum J, Baek SH, Dutta A, Houry WA, Huber O, Hupp TR, Matias PM. The emergence of the conserved AAA+ ATPases Pontin and Reptin on the signaling landscape. Sci Signal. 2013;6 doi: 10.1126/scisignal.2003906. pmr1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhaeusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Shigeoka T, Kato S, Kawaichi M, Ishida Y. Evidence that the Upf1-related molecular motor scans the 3'-UTR to ensure mRNA integrity. Nucleic Acids Res. 2012;40:6887–6897. doi: 10.1093/nar/gks344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla R, Upton KR, Munoz-Lopez M, Gerhardt DJ, Fisher ME, Nguyen T, Brennan PM, Baillie JK, Collino A, Ghisletti S, et al. Endogenous retrotransposition activates oncogenic pathways in hepatocellular carcinoma. Cell. 2013;153:101–111. doi: 10.1016/j.cell.2013.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simbulan-Rosenthal CM, Rosenthal DS, Iyer S, Boulares H, Smulson ME. Involvement of PARP and poly(ADP-ribosyl)ation in the early stages of apoptosis and DNA replication. Mol Cell Biochem. 1999;193:137–148. [PubMed] [Google Scholar]
- Smits AH, Jansen PWTC, Poser I, Hyman AA, Vermeulen M. Stoichiometry of chromatin-associated protein complexes revealed by label-free quantitative mass spectrometry-based proteomics. Nucleic Acids Research. 2013;41:e28. doi: 10.1093/nar/gks941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soifer HS, Zaragoza A, Peyvan M, Behlke MA, Rossi JJ. A potential role for RNA interference in controlling the activity of the human LINE-1 retrotransposon. Nucleic Acids Res. 2005;33:846–856. doi: 10.1093/nar/gki223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki J, Yamaguchi K, Kajikawa M, Ichiyanagi K, Adachi N, Koyama H, Takeda S, Okada N. Genetic evidence that the non-homologous end-joining repair pathway is involved in LINE retrotransposition. PLoS genetics. 2009;5:e1000461. doi: 10.1371/journal.pgen.1000461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tackett AJ, DeGrasse JA, Sekedat MD, Oeffinger M, Rout MP, Chait BT. I-DIRT, a general method for distinguishing between specific and nonspecific protein interactions. Journal of proteome research. 2005;4:1752–1756. doi: 10.1021/pr050225e. [DOI] [PubMed] [Google Scholar]
- Tani H, Imamachi N, Salam KA, Mizutani R, Ijiri K, Irie T, Yada T, Suzuki Y, Akimitsu N. Identification of hundreds of novel UPF1 target transcripts by direct determination of whole transcriptome stability. RNA biology. 2012;9:1370. doi: 10.4161/rna.22360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich HD. Timing and spacing of ubiquitin-dependent DNA damage bypass. FEBS Lett. 2011;585:2861–2867. doi: 10.1016/j.febslet.2011.05.028. [DOI] [PubMed] [Google Scholar]
- Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH, Boeke JD, Moran JV. Human L1 retrotransposition: cis preference versus trans complementation. Mol Cell Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichenrieder O, Repanas K, Perrakis A. Crystal structure of the targeting endonuclease of the human LINE-1 retrotransposon. Structure. 2004;12:975–986. doi: 10.1016/j.str.2004.04.011. [DOI] [PubMed] [Google Scholar]
- Yang N, Kazazian HH., Jr L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nature structural & molecular biology. 2006;13:763–771. doi: 10.1038/nsmb1141. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.