Abstract
Ribonuclease P (RNase P) catalyzes the maturation of the 5′ end of precursor-tRNAs (pre-tRNA) and is conserved in all domains of life. However, the composition of RNase P varies from bacteria to archaea and eukarya, making RNase P one of the most diverse enzymes characterized. Most known RNase P enzymes contain a large catalytic RNA subunit that associates with one to 10 proteins. Recently, a protein-only form of RNase P was discovered in mitochondria and chloroplasts of many higher eukaryotes. This proteinaceous RNase P (PRORP) represents a new class of metallonucleases. Here we discuss our recent crystal structure of PRORP1 from Arabidopsis thaliana and speculate on the reasons for the replacement of catalytic RNA by a protein catalyst. We conclude, based on an analysis of the catalytic efficiencies of ribonucleoprotein (RNP) and PRORP enzymes, that the need for greater catalytic efficiency is most likely not the driving force behind the replacement of the RNA with a protein catalyst. The emergence of a protein-based RNase P more likely reflects the increasing complexity of the biological system, including difficulties in importation into organelles and vulnerability of organellar RNAs to cleavage.
Keywords: RNase P, tRNA processing, RNA world, ribozyme, PRORP, mitochondrial tRNA, MRPP
The Varying Composition of RNase P Enzymes
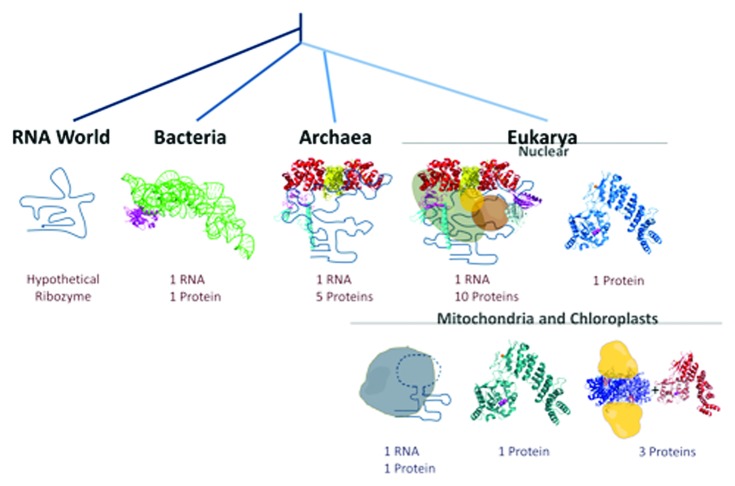
TRNA (tRNA) primary transcripts are extensively processed and modified before they participate in translation. One of the initial steps in precursor tRNA (pre-tRNA) processing is the removal of extra nucleotides flanking the 5′ and 3′ ends. Ribonuclease P (RNase P) is the endonuclease responsible for 5′ end cleavage (Fig. 1) and is a ribonucleoprotein (RNP) throughout all three domains of life. The RNA component is responsible for catalytic activity and associates with either one, five or 10 proteins in bacteria, archaea and eukaryotes, respectively (Fig. 2). To date, the archeon Nanoachaeum equitans is the only known organism without RNase P, presumably because the pre-tRNAs in this organism are transcribed without leader sequences.1 The RNA components in bacteria, archaea and eukaryotic nuclear RNase P are structurally related.2 However, deviations from the consensus structure exist in mitochondrial and chloroplast genomes. This was first observed in many fungi, where the consensus structure contains only two of the 11 conserved helices found within the minimal bacterial consensus structure.3 Furthermore, the Saccharomyes cerevisiae mitochondrial-encoded RNA associates with a single 105-kDa nuclear-encoded RPM2 protein that shares no homology to any other known RNase P proteins. The deviation in sequence and increased size of RPM2 suggests that the protein moiety could play a more substantial role in catalysis or molecular recognition.4
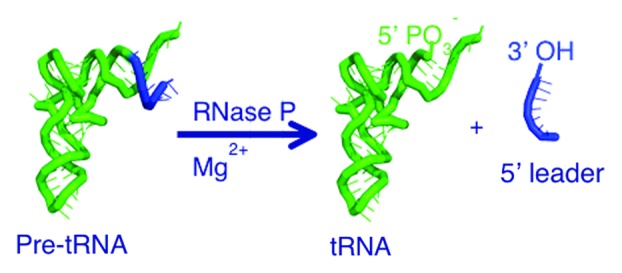
Figure 1. RNase P catalyzes the cleavage of 5′ leader sequences from precursor tRNAs. RNase P enzymes use divalent metal ions to catalyze hydrolysis of a specific phosphodiester bond in pre-tRNA, resulting in the formation of a mature 5′-end containing a phosphate and a leader with 3′ hydroxyl.
Figure 2. Evolutionary spread of RNase P. RNase P is conserved in all three domains of life. Bacterial RNase Ps consist of one RNA (green) and one protein (magenta) (pdb 3Q1R). Model of archaeal RNase P, which contains one RNA (secondary structure in blue) and at least four proteins [red: PH1877 (PDB 2CZV), yellow: PH1481 (PDB 2CZV), magenta: PH1771 (PDB: 2ZAE), cyan: PH1601 (PDB 2CZV)]. Proteins are arbitrary positioned. Most eukaryal nuclear RNase Ps are RNP based (left), which contain one RNA (blue) and at least nine proteins (four archaeal homologs, green: POP1, brown: POP3, purple: POP6 (PDB 3IAB), silver: POP7 (PDB 3IAB), orange: POP8). The proteins are arbitrary positioned. Some nuclear RNase Ps are proposed to be protein-only with homology to PRORP1 (i.e., T. brucei and A. thaliana; structure of PRORP1 shown). Mitochondrial and chloroplast RNase Ps from left to right: yeast; plants, some algae and some protists (A. thaliana and T. brucei); mammals (human). The mitochondrial yeast RNase P contain one RNA (blue) and one large protein, RPM2, (gray). Plant, some algae and protists mitochondrial/chloroplast RNase Ps are single proteins (teal) that have homology to A. thaliana PRORP1. Mammalian mitochondrial RNase Ps consist of three nuclear encoded proteins TRMT10C, (MRPP1), SDR5C1 (MRPP2) and PRORP (MRPP3) shown in yellow cartoon, blue tetramer (PDB 1U7T) and red (homology model based of A. thaliana PRORP1), respectively. The positioning of the TRMT10C/SDR5C1 subunits has not yet been demonstrated.
A striking deviation from the canonical RNA-dependent RNase P is found in human mitochondria, where RNase P is devoid of an RNA subunit and instead is composed of three proteins: tRNA m1G methyltransferase (MRPP1/TRMT10C), hydroxysteroid 17-β dehydrogenase 10 (MRPP2/SDR5C1) and a metallonuclease (MRPP3/PRORP).5 The methyltransferase and dehydrogenase form a complex with a proposed stoichiometry of 2:4 and the metallonuclease does not tightly associate with either subunit.5,6 The tRNA methyltransferase activity of TRMT10C is activated by the dehydrogenase (SDR5C1); however, the catalytic activity of either the methyltransferase or the dehydrogenase is not required for pre-tRNA cleavage catalyzed by PRORP.6 One possible explanation for the requirement of TRMT10C and SDR5C1 to activate PRORP activity is that the TRMT10C-SDR5C1 complex associates with a pre-tRNA substrate to induce conformational changes required for pre-tRNA cleavage.
In contrast to the multi-subunit human mitochondrial RNase P, the RNase P in the mitochondria/chloroplasts of most plants, algae and some protists is predicted to be a single protein enzyme (PRORP).7,8 This was first shown in A. thaliana, where three isoforms are found: PRORP1, 2 and 3. PRORP1 localizes to the mitochondria and chloroplasts, whereas PRORP2 and 3 localize to the nucleus.7 All three of these enzymes catalyze pre-tRNA processing in their respective localized organelles and in vitro.9 Attempts to identify RNase P RNA genes in plant genomes by sequence homology have not yielded any potential candidates for a canonical nuclear RNase P.10-12 This suggests that A. thaliana may be devoid of an RNA-based RNase P thereby catalyzing 5′ end processing using only PRORP enzymes.13 Alternatively, the plant RNase P RNA may be non-canonical, thus making it hard to identify by sequence homology searches. Evidence in support of this hypothesis comes from RNase P activity detected by immunoprecipitation of the protein POP1, which is a shared protein component of MRP and nuclear RNase P in other eukaryotes.14 The protozoan Trypanosoma brucei harbors 2 PRORP isoforms, both of which have 5′ pre-tRNA processing activity in vitro.8 One isoform (PRORP1) localizes to the nucleus and the second (PRORP2) to the mitochondrion. Strikingly, T. brucei PRORP1 can substitute for yeast nuclear RNase P in vivo, demonstrating that a single protein can complement a RNP complex composed of nine proteins and a ~400 nucleotide catalytic RNA.8 In addition, this result shows that T. brucei PRORP1 catalyzes all of the other non-canonical, yet vital functions of nuclear yeast RNase P, which may include processing of non-canonical RNAs (see below).
Ostreococcus tauri, an alga and one of the smallest eukaryote species, may be a living transitional organism representing the possible switch from RNA- to protein-based RNase P activity. The chloroplast and mitochondrial genomes of O. tauri encode distinct individual RNase P RNA genes and the nucleus encodes both a bacterial-like RNase P protein component, and a PRORP enzyme.15 The organellar RNase P RNAs are expressed in vivo, however under in vitro conditions, catalysis of pre-tRNA cleavage is not observed even when associated with the nuclear encoded bacterial-like protein. O. tauri is the only organism studied thus far that encodes RNase P RNAs in both organellar genomes and contains a nuclear encoded PRORP. The localization of the nuclear-encoded PRORP is currently unknown. Thus, further studies are required to help understand why O. tauri has retained the organellar encoded RNase P RNA genes.
Transitioning RNase P from the Ancient (RNA) to the Modern (Protein) World
Our recent crystal structure of the A. thaliana PRORP1 revealed that nature pieced together three distinct domains to replace the ancient RNA enzyme: a pentatricopeptide repeat (PPR) domain tethered to a metallonuclease domain through a structural-zinc binding site.16 None of the domains have homology to any of the protein components of eukaryotic or bacterial RNase P enzymes, consistent with bioinformatic data suggesting their disparate evolution. The PPR domain enhances the affinity for pre-tRNA binding and is proposed to play an important role in orienting the pre-tRNA substrate for cleavage.17 PPR domains are helical repeat motifs present in a large family of RNA-binding proteins involved in mitochondrial and chloroplast gene expression. PPR domains in other proteins are proposed to interact with single-stranded RNAs in a sequence-specific manner17 leading to the speculation that the PPR domain of PRORP enzymes could interact with the single-stranded 5′ leader or 3′ trailer of pre-tRNA. A sequence-specific interaction seems unlikely since PRORP enzymes must recognize almost all of the pre-tRNAs transcribed within a genome (37 tRNAs are encoded in A. thaliana chloroplast, and 22 tRNAs are encoded in human mitochondria), and there is little sequence conservation in pre-tRNA leader and trailer sequences in the A. thaliana chloroplast and mitochondrial genomes. Alternatively, it is possible that the PPR motifs in PRORP could interact directly with the tRNA body (Fig. 3). Identification of the pre-tRNA-binding site in PRORP enzymes will not only reveal how the PPR motifs in PRORP interact with pre-tRNA, but may also yield insights into how PPR motifs can generally recognize their substrates.
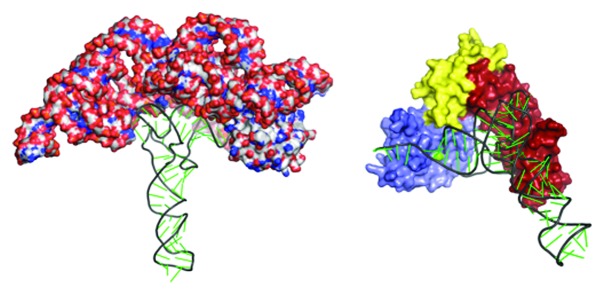
Figure 3. Comparison of a proposed model of PRORP-tRNA interaction and RNP-based RNase P. Left panel shows the crystal structure of bacterial RNase P (shown in spheres) in complex with tRNA (surface representation) (pdb 3Q1R). The right panel shows a hypothetical model of PRORP1 bound to tRNA. PRORP1 is shown in a surface representation with the PPR domain in red, central domain in yellow and the metallonuclease domain in blue. An active site metal is colored green and is in close proximity to the 5′ end of the tRNA. The tRNA was manually docked onto PRORP1. Pre-tRNAs substrates lacking an anticodon arm are cleaved by both enzymes, suggesting some similarity in recognition mechanisms.7,47 The D-TΨC loops in tRNA, a region that contacts bacterial RNase P RNA, are also recognized by PRORP1.47
The PRORP metallonuclease domain represents a novel class of nucleases called Nedd4-BP1, YacP nucleases (NYN), which share structural homology to the FLAP nuclease family.18 Our structure of A. thaliana PRORP1 revealed an active site that can bind two manganese atoms via conserved aspartate residues.16 This observation led us to propose a two-metal ion catalytic mechanism, similar to those previously proposed for RNA-based RNase P and members of the FLAP nuclease family. Despite the two-metal ion similarity with RNA-based RNase P, we propose that active site amino acid chains act as general acid/base catalysts in PRORP enzymes. This differs from RNA-based enzymes, which are proposed to use metal-bound waters for acid/base chemistry.19 The enhanced range in functionality of protein side chains is proposed to increase the catalytic efficiency of protein catalysts and this enhanced efficiency is one of the reasons most often proposed for the shift from a RNA-world to a protein-world.20,21 However, RNA-dependent RNase P is an extremely efficient enzyme. The catalytic efficiency (kcat/KM) for pre-tRNA cleavage of the yeast nuclear RNase P holoenzyme is diffusion-controlled (~1 × 108 M−1s−1) at 37þC,22 leaving minimal room for improvement. In contrast, the activities so far measured for the protein-only RNase P enzymes in A. thaliana (PRORP1 and 3) are ~one to two orders of magnitude slower (~1 × 105 M−1s−1)9,16,23 than even the simplest form of RNA-based RNase P (one RNA with one protein) from bacteria (~4 × 106 M−1s−1)24 (Table 1). To date, no other macromolecular cofactors have been identified for plant PRORPs that enhance the cleavage efficiency to a level comparable to the RNA-based enzymes. Thus, when compared with the extremely efficient RNA-based RNase P enzyme, the driving force for the development of a protein-only RNase P is unlikely to be mainly an enhanced catalytic efficiency.
Table 1. PRORP enzymes are less catalytically efficient than RNP-based RNase Ps in vitro.
| Species | Composition | kcat (sec-1) | Km (nM) | kcat/Km (M-1 sec-1) | Fold (kcat/Km) |
|---|---|---|---|---|---|
| B. subtilisa | RNP | 1 | 230 | 4 × 106 | 40 |
| Nuclear S. cerevisiaeb |
RNP | 2 | 20 | 1 × 108 | 1,000 |
| Mito/Chloro A. thaliana PRORP1c |
Protein | 0.1 | 850 | 1 × 105 | 1 |
A comparison of the catalytic efficiencies (kcat/Km) for RNase P enzymes that have been kinetically characterized reveal that PRORP enzymes may be up to several orders of magnitude less efficient. Values reported are from reactions performed at 37°C in the pH range of 7.8–8.0 with respective species specific pre-tRNAs containing 35 nt:CCA, 12 nt:0 nt and 5 nt:1 nt leader to trailer ratios for B. subtilis, S. cerevisiae and A. thaliana, respectively. Reactions performed with RNP-based RNase P contain higher concentrations of magnesium in order to correctly fold and stabilize the large catalytic RNA. a(24) b(22) c [Unpublished results which are comparable to PRORP3 kinetic parameters in (9)] Numbers reported are rounded.
The difficulty of importing a large structured RNA into the mitochondrion might have been a driving force for the switch of RNA to protein-based RNase P. There are mechanisms for import of nuclear-encoded proteins into the mitochondria, with an estimated 900 imported proteins in human mitochondria.25 In contrast, the import of RNA into the mitochondria is less prevalent. Only a few nuclear-encoded tRNAs seem to be significantly present within human mitochondria. The tRNAsGlnUAA and LeuUAA are enriched in mitoplast preps and tRNAGlnUAA is imported via a mechanism that is distinct from protein import.26,27 Although the biological significance of importation of this tRNA is unknown, it demonstrates that RNA can be imported into the mitochondria. The import of larger structured RNAs such as the RNA components of MRP and RNase P into the mitochondria is controversial and remains to be fully resolved.28-31 However, the observation that the human mitochondrion encodes both rRNA components within its genome supports the hypothesis that large structured RNAs are not readily imported.
Aside from the complexity inherent in importing the catalytic RNA and eight or nine protein subunits into organelles, it is possible that the nuclear RNase P and RNase MRP have evolved into nucleases that are too dangerous to be allowed access to the organellar RNA. The simple bacterial enzyme primarily recognizes substrates via the tertiary structure of tRNA; the identified non-tRNA substrates resemble a tRNA near the cleavage site.32 In addition to cleaving tRNA-like structures, both nuclear RNase P and RNase MRP have much broader substrate recognition. Unlike the bacterial counterparts, both enzymes efficiently cleave single-stranded RNA in a manner that is not particularly sequence- or structure-specific.33,34 This ability presumably contributes to the participation of MRP in cytoplasmic mRNA turnover35,36 and of RNase P in other nuclear RNA processing and turnover pathways.37-41 For these functions, the increased complexity of the protein subunit content allows the ancient catalytic site in the RNase P RNA to efficiently recognize and cleave a much broader range of substrates. Substrate specificity for non-tRNAs must therefore depend on the ability of the protein subunits to guide the enzymes to the correct nuclear and cytoplasmic RNPs, either by subcellular localization or protein-protein contacts with protein-bound RNA substrates. Conversely, nuclear and cytoplasmic RNAs that are not appropriate substrates for RNase P or MRP cleavage are presumably protected in vivo by some combination of RNP structure and mutually exclusive localization. The contribution of RNP structure to encouraging appropriate and discouraging inappropriate cleavage is axiomatic,42 though this level of control has not been considered extensively for the highly structured pre-tRNA substrates. If these holoenzymes were imported into organelles, it is possible that they would be deleterious, since whatever the structures of organellar RNAs, they are unlikely to mimic the pathways adopted in the nucleus and cytoplasm. Thus, a possible driving force for evolving a new, protein-based enzyme is that the broad, promiscuous substrate recognition of the nuclear/cytoplasmic enzymes evolved and diverged from the more narrow recognition requirements of organellar pre-tRNA cleavage. Consistent with this hypothesis, an open reading frame RNA shown to be cleaved in multiple locations by nuclear RNase P33 is not cleaved by excess A. thaliana PRORP1 at any detectable rate (unpublished results). We hypothesize that the relatively simple protein-based nucleases with tRNA specificity might have evolved to circumvent the need to protect organelle RNA through RNP structures or compartmentalization that would limit access to the correct RNA substrates.
While the differences between the structure of nuclear and organellar pre-tRNA substrates could also have contributed to the driving force for evolution of the protein-based enzyme, there is little evidence for this proposal. A mitochondrial RNase P that specifically processes the non-canonical mammalian mitochondrial tRNAs could be beneficial.43 However, PRORP enzymes likely evolved in an early eukaryote, such as algae, where organellar tRNAs are canonical, resembling eubacterial tRNAs in both primary sequence and secondary structure.44 In plants, the localization of PRORP1 (chloroplast and mitochondria) and PRORP2 and 3 (nucleus) might suggest differential substrate requirements for these isozymes. Additionally, PRORP enzymes catalyze cleavage of T. thermophilus pre-tRNAGly despite alterations in catalytic mechanism.23 These observations suggest that protein-only RNase P enzymes in plants are not evolved to specifically recognize plant tRNAs, but rather catalyze cleavage of a wide range of canonical pre-tRNA substrates. Thus, it seems unlikely that pre-tRNA substrate specificity was a strong driving force for PRORP evolution. However, the need for recognizing non-canonical pre-tRNA substrates in human mitochondria could help explain why this organelle requires a multi-subunit proteinaceous RNase P.
In conclusion, most mitochondrial or chloroplast RNase P enzymes identified thus far do not share significant homology with bacterial, archaeal or nuclear RNA-based RNase P enzymes. This suggests the presence of an early evolutionary driving force behind the replacement of RNA with a protein catalyst in organelles. The identity of this driving force(s) remains unknown, but it is not likely to be enhanced catalytic efficiency. We speculate that it could be a number of other factors which reflect an increase in biological complexity of the system and which are not mutually exclusive: substrate specificity, difficulties in importation and vulnerable organellar RNAs. Other selective pressures may include macromolecular stability and regulation. The higher pH and concentration of free radicals within organelles would be more deleterious toward RNA structure.45,46 These proposed driving forces are also coupled with the selective pressure to down-size or compact organellar genomes. Understanding the evolutionary driving forces and mechanisms behind the replacement of RNA for protein catalysts in RNase P enzymes may shed light on the possible transitions that may have occurred on early earth during the presumed RNA-to protein world transition.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Randau L, Schröder I, Söll D. Life without RNase P. Nature. 2008;453:120–3. doi: 10.1038/nature06833. [DOI] [PubMed] [Google Scholar]
- 2.Ellis JC, Brown JW. The RNase P family. RNA Biol. 2009;6:362–9. doi: 10.4161/rna.6.4.9241. [DOI] [PubMed] [Google Scholar]
- 3.Seif ER, Forget L, Martin NC, Lang BF. Mitochondrial RNase P RNAs in ascomycete fungi: lineage-specific variations in RNA secondary structure. RNA. 2003;9:1073–83. doi: 10.1261/rna.5880403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hollingsworth MJ, Martin NC. RNase P activity in the mitochondria of Saccharomyces cerevisiae depends on both mitochondrion and nucleus-encoded components. Mol Cell Biol. 1986;6:1058–64. doi: 10.1128/mcb.6.4.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holzmann J, Frank P, Löffler E, Bennett KL, Gerner C, Rossmanith W. RNase P without RNA: identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell. 2008;135:462–74. doi: 10.1016/j.cell.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 6.Vilardo E, Nachbagauer C, Buzet A, Taschner A, Holzmann J, Rossmanith W. A subcomplex of human mitochondrial RNase P is a bifunctional methyltransferase--extensive moonlighting in mitochondrial tRNA biogenesis. Nucleic Acids Res. 2012;40:11583–93. doi: 10.1093/nar/gks910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gobert A, Gutmann B, Taschner A, Gössringer M, Holzmann J, Hartmann RK, et al. A single Arabidopsis organellar protein has RNase P activity. Nat Struct Mol Biol. 2010;17:740–4. doi: 10.1038/nsmb.1812. [DOI] [PubMed] [Google Scholar]
- 8.Taschner A, Weber C, Buzet A, Hartmann RK, Hartig A, Rossmanith W. Nuclear RNase P of Trypanosoma brucei: a single protein in place of the multicomponent RNA-protein complex. Cell Rep. 2012;2:19–25. doi: 10.1016/j.celrep.2012.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gutmann B, Gobert A, Giegé P. PRORP proteins support RNase P activity in both organelles and the nucleus in Arabidopsis. Genes Dev. 2012;26:1022–7. doi: 10.1101/gad.189514.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartmann E, Hartmann RK. The enigma of ribonuclease P evolution. Trends Genet. 2003;19:561–9. doi: 10.1016/j.tig.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 11.Piccinelli P, Rosenblad MA, Samuelsson T. Identification and analysis of ribonuclease P and MRP RNA in a broad range of eukaryotes. Nucleic Acids Res. 2005;33:4485–95. doi: 10.1093/nar/gki756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenblad MA, López MD, Piccinelli P, Samuelsson T. Inventory and analysis of the protein subunits of the ribonucleases P and MRP provides further evidence of homology between the yeast and human enzymes. Nucleic Acids Res. 2006;34:5145–56. doi: 10.1093/nar/gkl626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldfarb KC, Borah S, Cech TR. RNase P branches out from RNP to protein: organelle-triggered diversification? Genes Dev. 2012;26:1005–9. doi: 10.1101/gad.193581.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krehan M, Heubeck C, Menzel N, Seibel P, Schön A. RNase MRP RNA and RNase P activity in plants are associated with a Pop1p containing complex. Nucleic Acids Res. 2012;40:7956–66. doi: 10.1093/nar/gks476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai LB, Bernal-Bayard P, Mohannath G, Lai SM, Gopalan V, Vioque A. A functional RNase P protein subunit of bacterial origin in some eukaryotes. Mol Genet Genomics. 2011;286:359–69. doi: 10.1007/s00438-011-0651-y. [DOI] [PubMed] [Google Scholar]
- 16.Howard MJ, Lim WH, Fierke CA, Koutmos M. Mitochondrial ribonuclease P structure provides insight into the evolution of catalytic strategies for precursor-tRNA 5′ processing. Proc Natl Acad Sci USA. 2012;109:16149–54. doi: 10.1073/pnas.1209062109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barkan A, Rojas M, Fujii S, Yap A, Chong YS, Bond CS, et al. A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 2012;8:e1002910. doi: 10.1371/journal.pgen.1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anantharaman V, Aravind L. The NYN domains: novel predicted RNAses with a PIN domain-like fold. RNA Biol. 2006;3:18–27. doi: 10.4161/rna.3.1.2548. [DOI] [PubMed] [Google Scholar]
- 19.Cassano AG, Anderson VE, Harris ME. Analysis of solvent nucleophile isotope effects: evidence for concerted mechanisms and nucleophilic activation by metal coordination in nonenzymatic and ribozyme-catalyzed phosphodiester hydrolysis. Biochemistry. 2004;43:10547–59. doi: 10.1021/bi049188f. [DOI] [PubMed] [Google Scholar]
- 20.Narlikar GJ, Herschlag D. Mechanistic aspects of enzymatic catalysis: lessons from comparison of RNA and protein enzymes. Annu Rev Biochem. 1997;66:19–59. doi: 10.1146/annurev.biochem.66.1.19. [DOI] [PubMed] [Google Scholar]
- 21.Doudna JA, Lorsch JR. Ribozyme catalysis: not different, just worse. Nat Struct Mol Biol. 2005;12:395–402. doi: 10.1038/nsmb932. [DOI] [PubMed] [Google Scholar]
- 22.Hsieh J, Walker SC, Fierke CA, Engelke DR. Pre-tRNA turnover catalyzed by the yeast nuclear RNase P holoenzyme is limited by product release. RNA. 2009;15:224–34. doi: 10.1261/rna.1309409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pavlova LV, Gössringer M, Weber C, Buzet A, Rossmanith W, Hartmann RK. tRNA processing by protein-only versus RNA-based RNase P: kinetic analysis reveals mechanistic differences. Chembiochem. 2012;13:2270–6. doi: 10.1002/cbic.201200434. [DOI] [PubMed] [Google Scholar]
- 24.Kurz JC, Niranjanakumari S, Fierke CA. Protein component of Bacillus subtilis RNase P specifically enhances the affinity for precursor-tRNAAsp. Biochemistry. 1998;37:2393–400. doi: 10.1021/bi972530m. [DOI] [PubMed] [Google Scholar]
- 25.Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76:723–49. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- 26.Mercer TR, Neph S, Dinger ME, Crawford J, Smith MA, Shearwood AM, et al. The human mitochondrial transcriptome. Cell. 2011;146:645–58. doi: 10.1016/j.cell.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rubio MA, Rinehart JJ, Krett B, Duvezin-Caubet S, Reichert AS, Söll D, et al. Mammalian mitochondria have the innate ability to import tRNAs by a mechanism distinct from protein import. Proc Natl Acad Sci USA. 2008;105:9186–91. doi: 10.1073/pnas.0804283105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiss T, Filipowicz W. Evidence against a mitochondrial location of the 7-2/MRP RNA in mammalian cells. Cell. 1992;70:11–6. doi: 10.1016/0092-8674(92)90528-K. [DOI] [PubMed] [Google Scholar]
- 29.Li K, Smagula CS, Parsons WJ, Richardson JA, Gonzalez M, Hagler HK, et al. Subcellular partitioning of MRP RNA assessed by ultrastructural and biochemical analysis. J Cell Biol. 1994;124:871–82. doi: 10.1083/jcb.124.6.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang G, Chen HW, Oktay Y, Zhang J, Allen EL, Smith GM, et al. PNPASE regulates RNA import into mitochondria. Cell. 2010;142:456–67. doi: 10.1016/j.cell.2010.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rossmanith W. Of P and Z: mitochondrial tRNA processing enzymes. Biochim Biophys Acta. 2012;1819:1017–26. doi: 10.1016/j.bbagrm.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walker SC, Engelke DR. Ribonuclease P: the evolution of an ancient RNA enzyme. Crit Rev Biochem Mol Biol. 2006;41:77–102. doi: 10.1080/10409230600602634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marvin MC, Walker SC, Fierke CA, Engelke DR. Binding and cleavage of unstructured RNA by nuclear RNase P. RNA. 2011;17:1429–40. doi: 10.1261/rna.2633611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Esakova O, Perederina A, Quan C, Berezin I, Krasilnikov AS. Substrate recognition by ribonucleoprotein ribonuclease MRP. RNA. 2011;17:356–64. doi: 10.1261/rna.2393711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aulds J, Wierzbicki S, McNairn A, Schmitt ME. Global identification of new substrates for the yeast endoribonuclease, RNase mitochondrial RNA processing (MRP) J Biol Chem. 2012;287:37089–97. doi: 10.1074/jbc.M112.389023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gill T, Cai T, Aulds J, Wierzbicki S, Schmitt ME. RNase MRP cleaves the CLB2 mRNA to promote cell cycle progression: novel method of mRNA degradation. Mol Cell Biol. 2004;24:945–53. doi: 10.1128/MCB.24.3.945-953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chamberlain JR, Pagán-Ramos, Kindelberger DW, Engelke DR. An RNase P RNA subunit mutation affects ribosomal RNA processing. Nucleic Acids Res. 1996;24:3158–66. doi: 10.1093/nar/24.16.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Samanta MP, Tongprasit W, Sethi H, Chin C-S, Stolc V. Global identification of noncoding RNAs in Saccharomyces cerevisiae by modulating an essential RNA processing pathway. Proc Natl Acad Sci USA. 2006;103:4192–7. doi: 10.1073/pnas.0507669103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang L, Altman S. A noncoding RNA in Saccharomyces cerevisiae is an RNase P substrate. RNA. 2007;13:682–90. doi: 10.1261/rna.460607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilusz JE, Freier SM, Spector DL. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–32. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marvin MC, Clauder-Münster S, Walker SC, Sarkeshik A, Yates JR, 3rd, Steinmetz LM, et al. Accumulation of noncoding RNA due to an RNase P defect in Saccharomyces cerevisiae. RNA. 2011;17:1441–50. doi: 10.1261/rna.2737511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Han SP, Tang YH, Smith R. Functional diversity of the hnRNPs: past, present and perspectives. Biochem J. 2010;430:379–92. doi: 10.1042/BJ20100396. [DOI] [PubMed] [Google Scholar]
- 43.Helm M, Brulé H, Friede D, Giegé R, Pütz D, Florentz C. Search for characteristic structural features of mammalian mitochondrial tRNAs. RNA. 2000;6:1356–79. doi: 10.1017/S1355838200001047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marechal-Drouard L, Weil JH, Dietrich A. Transfer RNAs and Transfer RNA Genes in Plants. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:13–32. doi: 10.1146/annurev.pp.44.060193.000305. [DOI] [Google Scholar]
- 45.Llopis J, McCaffery JM, Miyawaki A, Farquhar MG, Tsien RY. Measurement of cytosolic, mitochondrial, and Golgi pH in single living cells with green fluorescent proteins. Proc Natl Acad Sci USA. 1998;95:6803–8. doi: 10.1073/pnas.95.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turrens JF. Mitochondrial formation of reactive oxygen species. J Physiol. 2003;552:335–44. doi: 10.1113/jphysiol.2003.049478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gobert A, Pinker F, Fuchsbauer O, Gutmann B, Boutin R, Roblin P, et al. Structural insights into protein-only RNase P complexed with tRNA. Nat Commun. 2013;4:1353. doi: 10.1038/ncomms2358. [DOI] [PMC free article] [PubMed] [Google Scholar]